Abstract
In contrast to recessive conditions with biallelic inheritance, identification of dominant (monoallelic) mutations for Mendelian disorders is more difficult, because of the abundance of benign heterozygous variants that act as massive background noise (typically, in a 400:1 excess ratio). To reduce this overflow of false positives in next-generation sequencing (NGS) screens, we developed DOMINO, a tool assessing the likelihood for a gene to harbor dominant changes. Unlike commonly-used predictors of pathogenicity, DOMINO takes into consideration features that are the properties of genes, rather than of variants. It uses a machine-learning approach to extract discriminant information from a broad array of features (N = 432), including: genomic data, intra-, and interspecies conservation, gene expression, protein-protein interactions, protein structure, etc. DOMINO’s iterative architecture includes a training process on 985 genes with well-established inheritance patterns for Mendelian conditions, and repeated cross-validation that optimizes its discriminant power. When validated on 99 newly-discovered genes with pathogenic mutations, the algorithm displays an excellent final performance, with an area under the curve (AUC) of 0.92. Furthermore, unsupervised analysis by DOMINO of real sets of NGS data from individuals with intellectual disability or epilepsy correctly recognizes known genes and predicts 9 new candidates, with very high confidence. In summary, DOMINO is a robust and reliable tool that can infer dominance of candidate genes with high sensitivity and specificity, making it a useful complement to any NGS pipeline dealing with the analysis of the morbid human genome.
Main Text
By allowing the simultaneous identification of thousands of DNA variants at once, next-generation sequencing (NGS) has revolutionized the way human genetic diseases are investigated and diagnosed. Thanks to NGS and dedicated bioinformatics pipelines, both research and molecular diagnosis can be performed in a truly unsupervised way, by assessing thousands of DNA variants over entire genomes. However, this wealth of information is also a confounding factor when single events determining monogenic conditions are sought. Specifically, in Mendelian diseases, only one or two pathogenic mutations must be precisely identified among the myriad of innocuous variants that are naturally present in the human genome, roughly reducing NGS-based analyses to the recognition of one true positive (the actual mutation) from many false positives (benign DNA changes). The genome of a single individual typically carries 20,000 exonic variants, including ∼400 good-quality, nonsynonymous, and rare DNA changes.1, 2 In recessive conditions, two of such variants have forcibly to be present in the same gene to cause disease, lowering the number of candidate genes associated with the pathology to only 5–10, genome-wide.1, 3, 4 In contrast, any gene harboring one of these 400 variants in a heterozygous state represents potentially a gene associated with a dominant disorder, making it difficult to identify the cause of this class of genetic conditions (Figure 1A). As a consequence, NGS-based studies appear to be almost 10-fold more efficient in detecting genes associated to recessive disorders as compared to dominant ones.5 Prioritization of rare alleles as a function of their pathogenic potential at the heterozygous state represents therefore a crucial problem in solving dominant cases.
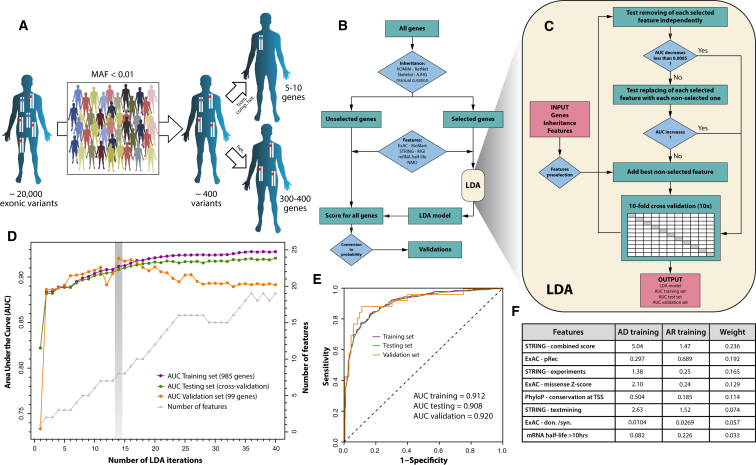
Figure 1.
Rationale and General Design of DOMINO
(A) A typical exome analysis identifies 20,000 variants, when compared to the human reference genome. After filtering by rarity in the general population (minor allele frequency, or MAF, < 1%) and by functional impact of each variant, approximately 400 DNA changes remain. These impact 300–400 genes, heterozygously (red dots), and 5–10 genes when they are present as homozygous or compound heterozygous variants (blue dots).
(B) Workflow of DOMINO methodology, showing the different steps of gene selection, annotation, and scoring.
(C) Details of the LDA algorithm. Relevant features are first preselected and then removed, replaced or added iteratively to the model, with specific acceptance criteria. 10 × 10-fold cross-validation is performed at each iteration.
(D) Performance of the model as a function of the iterations performed. AUCs of the training, testing and validation sets, as well as the number of features at each iteration are shown. The cut-off value retained corresponded to the 14th iteration and a set of 8 features. The model converges starting from the 36th iteration.
(E) ROC curves for the complete training, testing and validation sets, displaying AUC values of 0.912, 0.908, and 0.920, respectively.
(F) Features composing the selected model. Average values for AD and AR genes of the training set are shown, along with their relative weight. Units are as follows: for STRING entries, number of interactions;17 for ExAC-pRec, probability of being intolerant to homozygous but not heterozygous loss-of-function variants;18 for ExAC-missense Z score, value with respect to a distribution of expected number of missenses;18 PhyloP, average PhyloP score with respect to a 1,000-bp window centered on the TSS;19 ExAC-don./syn., number of variants at the donor splicing site, normalized to the number of synonymous variants in the coding sequence;20 mRNA half-life, 0 if ≤ 10 hr or 1 if > 10 hr.21
Several in silico tools have been developed to predict the damaging effect of DNA changes.6, 7 Yet, most of these methods focus on the deleteriousness of such variants on protein structure and/or function, rather than on making a distinction between mutations that are dominant or recessive. Other approaches predict haploinsufficiency of genes in the human genome.8, 9, 10, 11 These methods provide a partial solution to this problem, because dominant variants can produce a phenotype not only by haploinsufficiency, but also by gain-of-function or dominant-negative behavior.12
Here we propose an alternative approach, based on the scoring of features that distinguish genes associated with autosomal dominant (hereafter referred to as AD genes) versus autosomal recessive (referred to as AR genes) disorders, rather than on properties that are specific to a given DNA variant. To this end, we developed a predictive tool, called DOMINO, based on linear discriminant analysis (LDA), trained on a set of genes with known inheritance mode on a series of specific features, and finally validated with an independent group of genes.
We first collected a list of genes from different sources: hOMIM, a manually curated subset of OMIM13 (275 entries); RetNet, containing all genes involved in retinal degenerations and characterized by a high degree of genetic heterogeneity (99 entries); the Nosology of genetic skeletal diseases,14 listing genes linked to skeletal disorders (193 entries); and finally the full list of newly-discovered genes associated with Mendelian disorders published from 2009 to 2015 in the American Journal of Human Genetics (418 entries). To ensure quality, we manually curated these sources by discarding (1) all genes having both AD and AR inheritance, (2) genes directly linked to cancer, (3) genes carrying mutations that were not reported in the literature in more than one pedigree, and (4) genes associated with non-clinical phenotypes (Supplemental Methods and Table S1). We also removed all non-autosomal loci, as molecular evolution acts differently on autosomal versus X chromosome genes.15 This process resulted in the selection of 985 genes: 291 associated with AD phenotypes, and 694 with AR phenotypes, which were used as the “training set.”
To provide the highest a priori discrimination power to our tool, we used a wide range of features obtained from various databases and covering most of the attributes that genes can have, including general genetic, evolutionary, interactional, and functional information (Supplemental Methods and Table S2). Of the 700 different gene-specific features that could be extracted initially, 432 resulted to be available for protein-coding genes and allowed reliable scoring. These features were then filtered based on their significant differences between AD and AR genes of the training set, producing in the end 308 usable features.
An LDA-based algorithm was then chosen to allow machine-learning from the training set of genes, not only because of its recognized performance as a statistical method, but also to ensure the precise identification of the relevant features selected by the final model, allowing potentially to gain information on their biological relevance in the context of AD versus AR genes. To build a robust scoring system and to prevent over-fitting the training data, we devised an iterative process, able to identify the most discriminant features (Figure 1B, Figure 1, Figure 2, Figure 3). We first chose the one feature individually producing the highest area under the curve (AUC) from the receiver operating characteristic (ROC) function. Then, we iteratively tried to remove, replace or add features with specific criteria of acceptance (increase or decrease of the AUC, Figure 1C). Each time a change was accepted, 10 × 10-fold cross-validation16 was applied to the training set, to generate a “testing set” (Figure 1C). We let the algorithm run for 40 iterations and selected as best model the one for which there was an optimal AUC for the training and testing sets (Figure 1D). In other words, we selected the least complex model among those displaying similar AUC values. In our case, the best model was the one tested at the 14th iteration, composed of 8 features (Figure 1D) and displaying AUCs of 0.912 and 0.908 for the training and testing sets, respectively (Figure 1E). Starting from the 15th iteration, we observed a limited improvement of the testing set and a decreased performance for the validation set, clearly indicating over-fitting of the model on the training set, in support of the initial threshold selection. For each gene, in decreasing order of importance, the selected features were: (1) the number of interactions with AD genes of the training set from the combined score of STRING (a database regrouping functional protein association networks from various sources), with a confidence > 500 and a maximum of 8 interactions,17 (2) pRec (probability to be intolerant to homozygous but not heterozygous loss-of-function variants) as extracted from ExAC,18 (3) the number of interactions with AD genes of the training set from the experimental score of STRING, with a confidence > 400 and a maximum of 3 interactions,17 (4) the missense Z score from ExAC (intolerance to missenses),18 (5) the average PhyloP score for mammals across the transcriptional start site (TSS) (+/− 500 bp from the actual site),19 (6) the number of interactions with AD genes of the training set using the text-mining score of STRING, with a confidence > 300 and a maximum of 3 interactors,17 (7) the ratio between the number of donor site variants and synonymous variants present in ExAC,20 (8) a high mRNA half-life (> 10 hr) in mouse embryonic stem cells21 (Figure 1F, Figures S1).
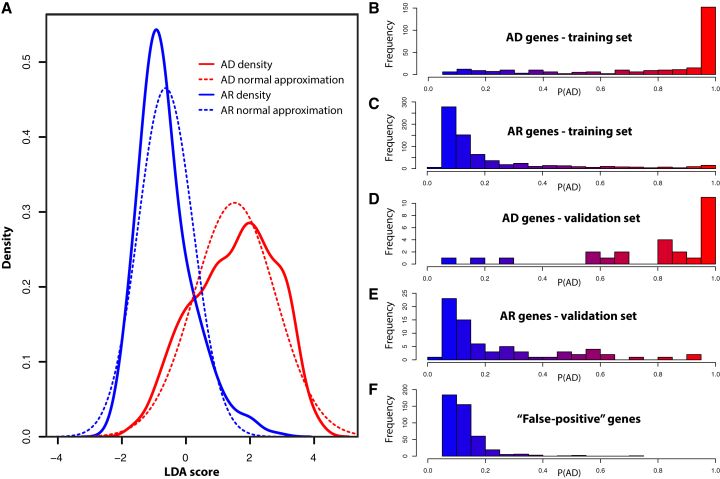
Figure 2.
Distributions of LDA Scores and Probabilities of Being Dominant, P(AD), for Genes in the Training and Validation Sets
(A) Density plots of LDA score for AD (red) and AR (blue) genes of the training set. Continuous lines refer to raw values, whereas dashed lines to their normal approximations.
(B–F) Histograms of P(AD) for: (B) AD genes of the training set, (C) AR genes of the training set, (D) AD genes of the validation set, (E) AR genes of the validation set, (F) Genes known to behave as false positives in NGS experiments, containing rare, non-pathogenic variants.
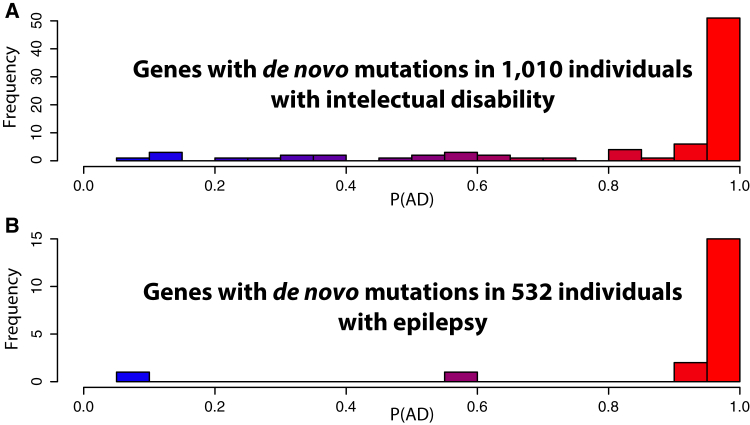
Figure 3.
Distributions of P(AD) for Genes with at Least Two De Novo Mutations in Different Individuals with Intellectual Disability or Epilepsy
Histograms of P(AD) for (A) 82 genes carrying de novo mutations in 1,010 individuals with intellectual disability or (B) 19 genes carrying de novo mutations in 532 individuals with epilepsy, as extracted from denovo-db.
At the end of this process, a score was computed for each gene, based on the LDA model. To facilitate the interpretation of the results by the end user, we transformed this score in a probability value, P(AD), measuring the probability for a gene to carry dominant mutations (Figure 2A, and Supplemental Methods), and developed a web-based interface (see Web Resources), enabling the interactive query of candidate genes and the scoring of their AD potential. As expected from the ROC curve (Figure 1E), most AD genes from the training set had a high P(AD), displaying the opposite trend when compared to AR genes (Figures 2B and 2C). At the maximal informedness point (LDA score = 0.225), computed by the Youden’s J equation (Jmax), the model had a specificity of 84.7% and a sensitivity of 80.4%. Interestingly, genes known to cause deleterious phenotypes by both dominant and recessive mechanisms, which we recovered from the pool of discarded genes from the training set and tested as new candidates, were scored either as AD or AR genes (Table S3). Specifically, out of 78 of such loci, 43 (55.1%) had a LDA score > 0.225, whereas the rest had P(AD)s comparable to those of genes associated with recessive disorders (Figure S2A), indicating the absence of an artifactual bias created by the model.
As a “validation set,” we used 99 genes with Mendelian mutations (26 AD genes and 73 AR genes) that we extracted from papers published from January 2016 to March 2017 in The American Journal of Human Genetics and in Nature Genetics, to mimic the discovery of newly-reported genes and confirm the absence of a potential bias toward well-studied and annotated genes, composing the bulk of the training set (Table S4). For the validation set, DOMINO predicted AD association with an AUC of 0.920 (Figures 1D and 1E) and specificity and sensitivity of 88.5% and 78.1% at Jmax, respectively (Table S4, Figures 2D and 2E). Specifically, 23 out of the 26 AD genes were correctly identified, confirming the reproducibility of the data obtained with the training set. For the remaining three dominant genes that were not recognized as such, namely: OVOL2 [MIM: 616441], KLHL24 [MIM: 611295], and SAMD9L [MIM: 611170], we noted unconventional mechanisms of pathogenicity. OVOL2 contains variants in the non-coding promoter region that results in a hyperactive promoter,22 while KLHL24 has a start-loss DNA change resulting in the use of a downstream alternative initiation site.23 The mechanisms of pathogenesis for SAMD9L are also rather unusual for a Mendelian condition and are characterized by particular chromosomal rearrangements.24
AD mutations can cause pathological phenotypes via different mechanisms, such as gain-of-function or haploinsufficiency. To examine the effectiveness of DOMINO in these two different cases, we evaluated AD genes from the training set as a function of the type of causative mutations they harbor. We reasoned that genes carrying exclusively pathogenic missenses (N = 107) would mainly cause disease by gain-of-function mechanisms, whereas those containing only truncating variants (N = 40) would be compatible with a haploinsufficient model of pathogenesis (genes carrying both types of variants were excluded, Table S5). Scores for the two groups were not statistically different (Figures S2B and S2C), with average P(AD) values of 0.66 and 0.74, respectively (p = 0.42, by Wilcoxon rank sum test with continuity correction). Therefore, in contrast to current tools, DOMINO’s effectiveness is not affected by the presence of specific mutations that a given gene might harbor, being a true predictor of AD features regardless of their mode of pathogenesis.
The performance of our model was also assessed by scoring the probability of being dominant for well-known false-positives for rare conditions in genome-wide screens,25 such as genes encoding mucins, taste and olfactory receptors, etc. Out of 436 genes from this set, only 4 had LDA scores higher than Jmax (Table S6, Figure 2F).
To assess the behavior of DOMINO on real sets of exome / genome data, we tested it on genotypes from denovo-db, a database of de novo variants identified by NGS,26 from which we extracted data from individuals with intellectual disability (ID) (N = 1,010) or with epilepsy (N = 532). Following a stringent filtering on allelic frequency (not see in ExAC or ESP),20 predicted effect on protein (nonsense, frameshift, missense) or on splicing (disruption of splicing sites), we selected all genes with at least two variants in different individuals (N = 82 for intellectual disabilities and N = 19 for epilepsy, Tables S7 and S8). By virtue of their heterozygous de novo inheritance (i.e., dominant in following generations), their presence in the same gene in more than one person, and of strict filtering procedures, all these DNA changes likely represent pathogenic mutations, and therefore all genes harboring them represent true AD genes detected by real NGS experiments. We then ranked all autosomal genes from the human genome according to their P(AD) and retained those for which P(AD) was ≥ 0.95, i.e., all genes that were predicted to be associated to dominant conditions with high confidence. Subsequently, we assessed the enrichment of genes with P(AD) ≥ 0.95 in these two groups of diseases within all human autosomal genes with P(AD) ≥ 0.95, by a hypergeometric test. We found that genes with at least two de novo variants from both the ID and epilepsy cohorts were significantly enriched for high P(AD) genes, with associated p-values of 1.8 × 10−35 (enrichment score = 18.9) and 9.6 × 10−14 (enrichment score = 43.1), respectively (Figure 3).
Remarkably, for cases with epilepsy, all 15 genes with at least two variants in different individuals and with high P(AD) were already known to be associated with dominant forms of the disease (4 were present in the training set). For ID, 39 out of 51 bona fide genes with high P(AD) were also already associated with AD forms of the diseases and allied conditions in OMIM (11 were present in the training set). Among the 12 remaining genes, three were previously predicted to be linked to this disorder by in silico analyses,27 whereas the other 9 represent excellent intellectual disability candidate genes that we propose for validation by forthcoming studies (Table 1). In more general terms, genes with high P(AD) genome-wide represent therefore either genes that were already identified to be associated with dominant conditions, or excellent new candidate genes for known or novel AD conditions. For instance, among the top 20 genes with highest P(AD), 10 were previously found to carry mutations for dominant disorders, while the remainder were not associated with any condition and might be considered in the future for disease association with very high confidence (Table 2).
Table 1.
Candidate ID-Associated Genes, as Predicted by DOMINO and Recurrent De Novo Mutations
| Gene Name | Protein Name | P(AD) | Function |
|---|---|---|---|
| AGO2 [MIM:606229] | Argonaute 2 | 0.999989 | Catalytic component of the RNA-induced silencing complex (RISC) |
| CACNA1E [MIM:601013] | Calcium Voltage-Gated Channel, Subunit Alpha1 E | 0.995065 | Calcium channels containing alpha-1E subunit. It could be involved in the modulation of firing patterns of neurons |
| CHD3 [MIM:602120] | Chromodomain Helicase DNA Binding Protein 3 | 0.999901 | Component of the histone deacetylase NuRD complex, participating in the remodelling of chromatin |
| FBXO11 [MIM:607871] | F-Box Protein 11 | 0.973952 | Part of a the SCF E3 ubiquitin-protein ligase complex, mediating protein ubiquitination and degradation |
| GRIA1 [MIM:138248] | Glutamate Ionotropic Receptor, AMPA Type, Subunit 1 | 0.980767 | Receptor for glutamate, mediating fast excitatory synaptic transmission in the central nervous system |
| KDM2B [MIM:609078] | Lysine Demethylase 2B | 0.989312 | Histone demethylase that demethylates Lys-4 and Lys-36 of histone H3 |
| LRP1 [MIM:107770] | LDL Receptor Related Protein 1 | 0.999963 | Endocytic receptor involved in endocytosis and in phagocytosis of apoptotic cells |
| PPP2CA [MIM:176915] | Protein Phosphatase 2, Catalytic Subunit Alpha | 0.999621 | Protein phosphatase 2A is one of the four major Ser/Thr phosphatases, implicated in the negative control of cell growth and division. |
| TCF7L2 [MIM:602228] | Transcription Factor 7 Like 2 | 0.999903 | Participates in the Wnt signaling pathway and modulates MYC expression |
Table 2.
Top 20 AD Genes, as Predicted by DOMINO
| Gene | P(AD) | In training set | Main OMIM description |
|---|---|---|---|
| SF3B1 [MIM:605590] | 0.999999 | No | Myelodysplastic syndrome, somatic/dominant [MIM:614286] |
| CSNK2A1 [MIM:115440] | 0.999998 | No | Okur-Chung syndrome, autosomal dominant [MIM:617062] |
| LHX2 [MIM:603759] | 0.999998 | No | Unassigned |
| DACH1 [MIM:603803] | 0.999998 | No | Unassigned |
| PAX6 [MIM:607108] | 0.999998 | Yes, AD | Aniridia, autosomal dominant [MIM:106210] |
| PRPF8 [MIM:607300] | 0.999996 | No | Retinitis pigmentosa, autosomal dominant [MIM:600059] |
| ATP2B1 [MIM:108731] | 0.999996 | No | Unassigned |
| DYNC1H1 [MIM:600112] | 0.999996 | Yes, AD | Charcot-Marie-Tooth disease, axonal, autosomal dominant [MIM:614228] |
| PIK3CA [MIM:171834] | 0.999995 | Yes, AD | Cowden syndrome 5, autosomal dominant [MIM:615108] |
| PTEN [MIM:601728] | 0.999995 | No | Bannayan-Riley-Ruvalcaba syndrome, autosomal dominant [MIM:153480] |
| TBL1XR1 [MIM:608628] | 0.999995 | No | Intellectual disability, autosomal dominant [MIM:616944] |
| HNRNPR [MIM:607201] | 0.999994 | No | Unassigned |
| TOP2B [MIM:126431] | 0.999994 | No | Unassigned |
| GSK3B [MIM:605004] | 0.999993 | No | Unassigned |
| CDK8 [MIM:603184] | 0.999992 | No | Unassigned |
| XPO1 [MIM:602559] | 0.999992 | No | Unassigned |
| SREBF1 [MIM:184756] | 0.999992 | No | Unassigned |
| PIAS1 [MIM:603566] | 0.999991 | No | Unassigned |
| NR2F2 [MIM:107773] | 0.999991 | Yes, AD | Congenital heart defects, autosomal dominant [MIM:615779] |
| BCL11B [MIM:606558] | 0.999990 | No | Immunodeficiency 49, autosomal dominant [MIM:617237] |
Finally, we took advantage of the LDA approach, allowing a transparent assessment of the features selected by the model, to gain possible insights on the general properties of AD versus AR genes. Interestingly, STRING components, accounting globally for the 47.5% of the weight of the model, are strong determinants of dominance, implying that organization in networks is seemingly rather important for AD genes/proteins. Moreover, among the many parameters measuring evolutionary pressure and conservation across species, only the PhyloP score at the TSS was retained (11.4% of the weight), while more classical scores, such as for instance the dN/dS ratio,28 appeared to be less relevant and were not included in the final model. Sequence-based features were nonetheless significant and have been retained in DOMINO, accounting for 37.8% of the weight. Their significance seems to be related to the global variation landscape in the human population, as identified in the ExAC project.20 Another intriguing result emerging from the selection of features is the fact that few AD genes have a long mRNA half-life. This finding could possibly be related to the observation that stable transcripts are enriched for mRNA encoding enzymes,21 which are usually associated with AR conditions. Also, our analysis of NGS data from individuals with intellectual disability or epilepsy showed that DOMINO has relevant predictive power for identifying genes that have not yet been studied or not yet found to carry pathogenic mutations.
In conclusion, DOMINO allows for an efficient prioritization of candidate genes associated with autosomal dominant Mendelian conditions, independently from the mutational events that a given gene might carry. Therefore, it can be used in combination with other predictors focusing on deleteriousness of DNA variants to reduce the number of false positives in mutational screens. In addition, the flexibility and modularity of the machine learning system enables the incorporation, at every update, of new informative features as they might emerge from future studies, making DOMINO a constantly evolving tool with progressively improving performances.
Acknowledgments
This work was supported by the Swiss National Science Foundation (grant # 156260, to C.R.) and by the PhD Fellowships in Life Science of the University of Lausanne (to M.Q.).
Published: October 5, 2017
Footnotes
Supplemental Information includes two figures and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.09.001.
Web Resources
DOMINO, https://wwwfbm.unil.ch/domino/
ExAC Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
RetNet – Retinal Information Network, https://sph.uth.edu/retnet/home.htm
STRING 9.0, http://www.string-db.org/
Supplemental Methods, https://wwwfbm.unil.ch/domino/supplementary.html
Supplemental Data
References
- 1.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., Broad GO. Seattle GO. NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamphans T., Sabri P., Zhu N., Heinrich V., Mundlos S., Robinson P.N., Parkhomchuk D., Krawitz P.M. Filtering for compound heterozygous sequence variants in non-consanguineous pedigrees. PLoS ONE. 2013;8:e70151. doi: 10.1371/journal.pone.0070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warr A., Robert C., Hume D., Archibald A., Deeb N., Watson M. Exome Sequencing: Current and Future Perspectives. G3 (Bethesda) 2015;5:1543–1550. doi: 10.1534/g3.115.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters-Sen L.C., Hashimoto S., Thrush D.L., Reshmi S., Gastier-Foster J.M., Astbury C., Pyatt R.E. Variability in pathogenicity prediction programs: impact on clinical diagnostics. Mol. Genet. Genomic Med. 2015;3:99–110. doi: 10.1002/mgg3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., 1000 Genomes Project Consortium A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norris M., Lovell S., Delneri D. Characterization and prediction of haploinsufficiency using systems-level gene properties in yeast. G3 (Bethesda) 2013;3:1965–1977. doi: 10.1534/g3.113.008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinberg J., Honti F., Meader S., Webber C. Haploinsufficiency predictions without study bias. Nucleic Acids Res. 2015;43:e101. doi: 10.1093/nar/gkv474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkie A.O. The molecular basis of genetic dominance. J. Med. Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonafe L., Cormier-Daire V., Hall C., Lachman R., Mortier G., Mundlos S., Nishimura G., Sangiorgi L., Savarirayan R., Sillence D. Nosology and classification of genetic skeletal disorders: 2015 revision. Am. J. Med. Genet. A. 2015;167A:2869–2892. doi: 10.1002/ajmg.a.37365. [DOI] [PubMed] [Google Scholar]
- 15.Wright A.E., Mank J.E. The scope and strength of sex-specific selection in genome evolution. J. Evol. Biol. 2013;26:1841–1853. doi: 10.1111/jeb.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastie T., Tibshirani R., Friedman J.H. Springer; New York, NY: 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- 17.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharova L.V., Sharov A.A., Nedorezov T., Piao Y., Shaik N., Ko M.S. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson A.E., Liskova P., Evans C.J., Dudakova L., Nosková L., Pontikos N., Hartmannová H., Hodaňová K., Stránecký V., Kozmík Z. Autosomal-Dominant Corneal Endothelial Dystrophies CHED1 and PPCD1 Are Allelic Disorders Caused by Non-coding Mutations in the Promoter of OVOL2. Am. J. Hum. Genet. 2016;98:75–89. doi: 10.1016/j.ajhg.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Z., Li S., Feng C., Yang S., Wang H., Ma D., Zhang J., Gou M., Bu D., Zhang T. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat. Genet. 2016;48:1508–1516. doi: 10.1038/ng.3701. [DOI] [PubMed] [Google Scholar]
- 24.Chen D.H., Below J.E., Shimamura A., Keel S.B., Matsushita M., Wolff J., Sul Y., Bonkowski E., Castella M., Taniguchi T. Ataxia-Pancytopenia Syndrome Is Caused by Missense Mutations in SAMD9L. Am. J. Hum. Genet. 2016;98:1146–1158. doi: 10.1016/j.ajhg.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shyr C., Tarailo-Graovac M., Gottlieb M., Lee J.J., van Karnebeek C., Wasserman W.W. FLAGS, frequently mutated genes in public exomes. BMC Med. Genomics. 2014;7:64. doi: 10.1186/s12920-014-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner T.N., Yi Q., Krumm N., Huddleston J., Hoekzema K., HA F.S., Doebley A.L., Bernier R.A., Nickerson D.A., Eichler E.E. denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2017;45:D804–D811. doi: 10.1093/nar/gkw865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lelieveld S.H., Reijnders M.R., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 28.Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.