Abstract
The protocol described here efficiently directs human pluripotent stem cells (hPSCs) to self-renewing epicardial cells in a completely defined, xeno-free system by temporal modulation of regulators of canonical Wnt signaling. Appropriate differentiation stage-specific application of Gsk3 inhibitor, Wnt inhibitor, then Gsk3 inhibitor is sufficient to produce cells expressing epicardial markers and exhibiting epicardial phenotypes with a high yield and purity from multiple hPSC lines in 16 days. Characterization of differentiated cells is performed via flow cytometry and immunostaining to assess quantitative expression and localization of epicardial cell-specific proteins. In vitro differentiation to fibroblasts and smooth muscle cells is also described. In addition, culture in the presence of TGFβ inhibitors allows long-term expansion of hPSC-derived epicardial cells for at least 25 population doublings. Functional human epicardial cells differentiated via this protocol may constitute a potential cell source for heart disease modeling, drug screening, and cell-based therapeutic applications.
INTRODUCTION
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), possess enormous potential for the study and treatment of cardiovascular diseases due to their capacity for unlimited self-renewal and ability to form any somatic cell type1,2. Functional epicardial cells and their progeny differentiated from hPSCs could be beneficial for many applications, including cardiac disease modeling, drug discovery and cellular therapies3. Realization of this potential will require protocols to differentiate hPSCs to cardiovascular cell lineages with high efficiency and reproducibility in a scalable and cost-effective manner. Moreover, therapeutic applications necessitate defined, xeno-free cell manufacturing processes.
Over the past decade, there has been significant progress in the generation of cardiomyocytes4–8, endothelial cells9–13, and smooth muscle cells (SMCs)14–16 from hPSCs. However, there have only been a few reports describing the differentiation of hPSCs to epicardial cells. Epicardial cells have been shown to contribute to fibroblast, smooth muscle, and vascular endothelial cell compartments in the developing heart, and also secrete trophic and regulatory factors involved in heart development and maintenance17,18. Initial efforts to differentiate hPSCs into epicardial cells implemented stage-specific application of BMP and Wnt ligands to embryoid bodies (EBs)19. In this approach19, treatment of EBs with BMP4 for 1 day and then BMP4, Activin A, and bFGF for 3 days induced mesoderm differentiation. The EBs were plated and treated with DKK1, VEGF, and SB431542 for 2 days to stimulate cardiovascular specification. Addition of BMP4 during this stage resulted in epicardial differentiation. Iyer et al.20 demonstrated differentiation of hPSCs to epicardial cells with a purity of at least 60% in multiple hPSC lines via a monolayer-based differentiation method. Treatment of hPSCs with bFGF, Ly294002, and BMP4 for 36 hr formed early mesoderm. An additional 3.5 days of bFGF and BMP4 treatment resulted in lateral plate mesoderm, which was then directed to epicardium by 10 days of treatment with BMP4, Wnt3a, and retinoic acid. These seminal studies pioneered efforts to differentiate hPSCs to epicardial cells, but the inclusion of expensive growth factors and/or xenogenic components in these protocols limits large-scale production and therapeutic applications. Furthermore, these hPSC-derived epicardial cells dedifferentiated in culture, similar to primary human epicardial cells21, complicating manufacturing and further applications.
To address these issues, we used a WT1-2A-eGFP knock-in hPSC line to show that temporal modulation of canonical Wnt signaling via small molecules is sufficient for epicardial induction from hPSCs under chemically-defined, xeno-free conditions, and that TGF-β inhibitor treatment permits long-term expansion of hPSC-derived epicardial cells22. After 48 days of expansion, the hPSC-derived epicardial cells exhibited uniform expression of epicardial markers and retained the capacity to differentiate to fibroblasts and smooth muscle cells. The majority of the hPSC-derived WT1+/TBX18+ cells also expressed TCF21, suggesting that this differentiation protocol might generate a subset of WT1+ cells identified in human epicardial cultures20 as well as chick and mouse epicardium23. In contrast, Iyer et al.20 reported populations of WT1+/TCF21+, WT1+/TCF21−, WT1−/TCF21+, and WT1−/TCF21− cells in their epicardial differentiation cultures. Differences between our study and the Iyer et al. report, including the starting cardiac progenitor cells and exposure to different developmental pathway modulators, may account for the generation of a more homogenous subpopulation of epicardium in our protocol. These findings improve our understanding of epicardial cell specification and self-renewal, and have implications for generating human epicardial cells for therapeutic applications.
In this protocol, we provide a detailed step-by-step process for 2D monolayer-based direct differentiation of hPSCs to epicardial cells. This protocol uses a completely defined, growth factor- and xeno-free system and applies temporal modulation of Wnt/β-catenin signaling via small molecules. This protocol is based on our earlier reports of cardiac progenitor and epicardial differentiation5,22 and is composed of four major stages: (steps 1–8) induction of cardiac progenitors from hPSCs by temporal modulation of canonical Wnt signaling under defined, albumin-free conditions, (steps 9–14) directed differentiation of cardiac progenitors to pro-epicardial then epicardial cells by Gsk3 inhibitor treatment, (steps 15 A-C) long-term maintenance of hPSC-derived epicardial cells under chemically defined conditions in the presence of a TGFβ inhibitor, and (steps 15 D) in vitro differentiation of epicardial cells to fibroblasts and SMCs. This protocol will enable efficient production of human epicardial cells for development and disease research, drug screening and testing, and advancing cardiac cellular therapies.
Experimental design
Induction of cardiac progenitors from hPSCs (Steps 1–8)
A summary of cardiac progenitor generation (GiWi2 protocol5) is shown in Fig. 1. The hPSCs are initially cultured on Matrigel-coated plates or Synthemax-coated plates in mTeSR1 or E8 medium until fully confluent. For translational applications where fully-defined differentiation is important, a combination of Synthemax and E8 is recommended. The starting hPSC population should contain at least 95% Oct4+ cells with no detectable karyotypic abnormalities. Differentiation is initiated by removing the maintenance medium and adding RPMI basal medium containing a Gsk3 inhibitor, such as CHIR99021. 24 hr of culture in this medium generates a high percentage of brachyury-expressing cells (>95% by flow cytometry) (Fig. 1). In order to direct these brachyury-expressing mesendoderm progenitor cells to a cardiac progenitor fate, inhibition of canonical Wnt signaling by Wnt signaling inhibitors, such as Porcupine inhibitors IWP2 or IWP4, is performed on day 3. At days 5–6 this approach generates Nkx2.5+Isl1+ cardiac progenitor cells under chemically-defined, albumin-free differentiation conditions. The cardiac progenitor cells can also be efficiently generated in albumin-containing RPMI/B27-insulin medium with 12 μM CHIR99021 using our previous GiWi protocol6.
Figure 1.
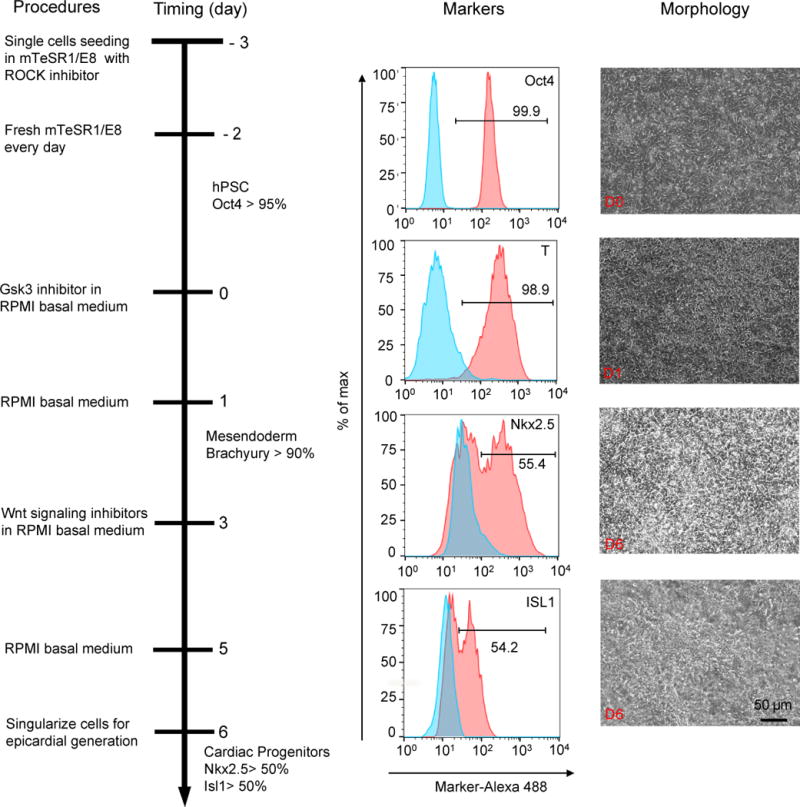
Schematic of protocol to differentiate cardiac progenitor cells from hPSCs with small molecule modulators of canonical Wnt signaling (Steps 1–8). Bright field images of the typical morphology of day 0, day 1 and day 6 cells differentiated from H13 hESCs are shown along with representative flow cytometry images of indicated markers analyzed as shown in Fig. S1. 10–50k cells were analyzed with percentages of Oct4: 99.5 ± 0.3, brachyury (T): 96.5 ± 2.5, Nkx2.5: 52.6 ± 2.6 and Isl1: 52.1 ± 1.9. Data are presented as mean ± S.D. from three independent experiments: Oct4 (99.8%, 99.5%, 99.3%); T (96.5%, 98.9%, 94%), Nkx2-2 (55.4%, 50.2%, 52.2%) and Isl1 (54.2%, 51.1%, 50.9%). Scale bar, 50 μm.
Directed differentiation of cardiac progenitors into pro-epicardial and epicardial cells (Steps 9–14)
A summary of this directed differentiation protocol is shown in Fig. 2. On day 6, hPSC-derived cardiac progenitor cells are singularized and seeded onto gelatin- or Synthemax-coated plates in albumin-containing LaSR13 basal medium (Advanced DMEM/F12 with 2.5 mM GlutaMAX and 100μg/mL ascorbic acid) or albumin-free RPMI/Vc/Ins medium (100 μg/mL ascorbic acid and1 μg/mL insulin) as needed. When cultured in these media without any treatment, the cardiac progenitor cells will spontaneously differentiate into functional contracting cardiomyocytes22. In order to direct these Nkx2.5+Isl1+ cardiac progenitor cells to an epicardial cell fate, activation of canonical Wnt signaling by Gsk3 inhibitors, such as CHIR99021, is performed for 48 hr from day 7 to day 9. WT1+TBX18+ epicardial cells are obtained on day 12 when cultured in LaSR basal or RPMI/Vc/Ins medium for another 3 days. Epicardial cell differentiation proceeds rapidly, and WT1 can be readily detected at day 10 of differentiation. The purity of WT1+ cells on day 12 should be greater than 90% (Fig. 2). At day 12 the population consists of WT1+/ALDH1A2- pro-epicardial cells but by day 16 these cells express ALDH1A2 and upregulate tight junction protein ZO-1 to become epicardial cells.
Figure 2.
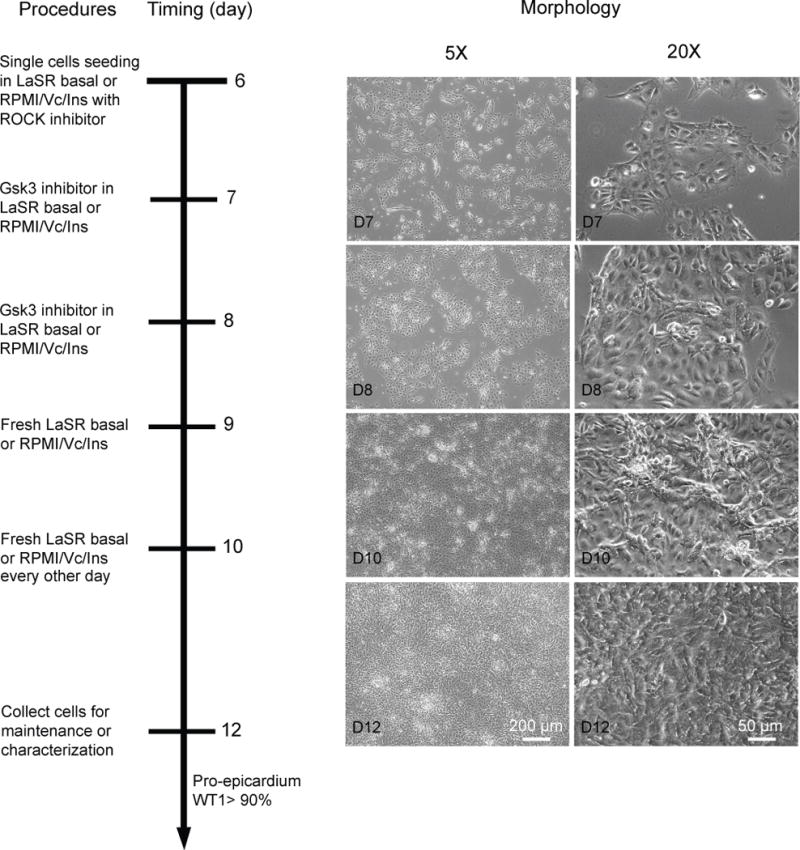
Schematic of protocol to differentiate epicardial cells by treatment of hPSC-derived cardiac progenitor cells with the Gsk3 inhibitor CHIR99021 (Steps 9–14). Bright field images of the typical morphology on day 7, day 8, day 10, and day 12 cells differentiated from H13 hESCs are shown. Scale bars, 50 or 200 μm, as indicated.
Long-term maintenance of hPSC-derived epicardial cells (Steps 15 A-C)
A summary of the maintenance protocol is shown in Fig. 3. To expand these hPSC-derived epicardial cells, confluent cells on day 12–16 following initiation of differentiation or day 4 after passage are split 1:3 to 1:9 using either Versene or Accutase (Fig. 3). Although Versene and Accutase work similarly at early passages, we found that Versene passaging leads to more efficient expansion of the epicardial cells after 8 passages. When cultured in LaSR basal medium without any treatment, these cells will spontaneously undergo EMT to form smooth muscle-like cells after 2 or 3 passages. Therefore, to promote cell self-renewal, a TGFβ inhibitor (e.g. A83-01 or SB431542) is added into the culture medium, resulting in long-term self-renewing epicardial cells. Even after 2 months and 25 doublings in LaSR basal medium, these cells still retain molecular markers and morphological and functional characteristics of primary epicardial cells (Figs. 4 and 5). hPSC-derived epicardial cells can also be maintained for about 24 days in albumin-free RPMI/Vc/Ins medium.
Figure 3.
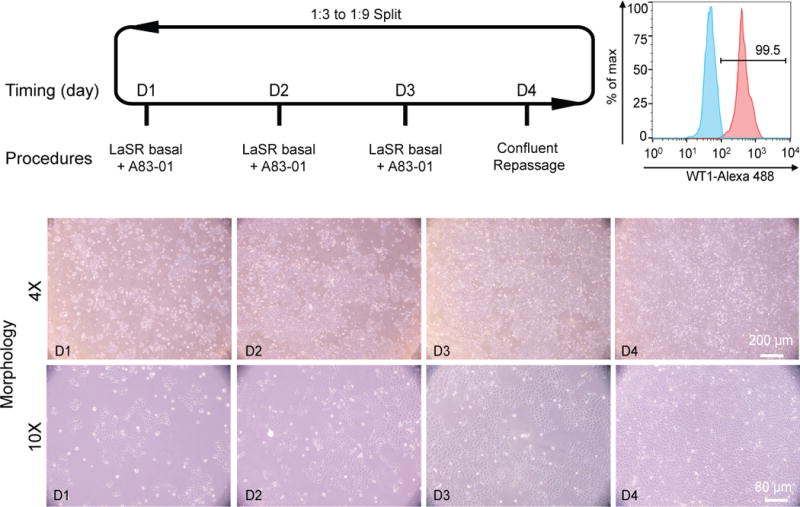
Schematic of protocol to maintain self-renewing hPSC-derived epicardial cells (Step 15A). Bright field images of the typical morphology of day 1, day 2, day 3, and day 4 epicardial cells differentiated from H13 hESCs are shown. Scale bars, 80 or 200 μm, as indicated. 10–50k cells were analyzed from each flow cytometry sample with a WT1+ percentage of 97.1± 3.3. Data are presented as mean ± S.D. from three independent experiments: WT1 (98.5%, 99.5%, 93.3%).
Figure 4.
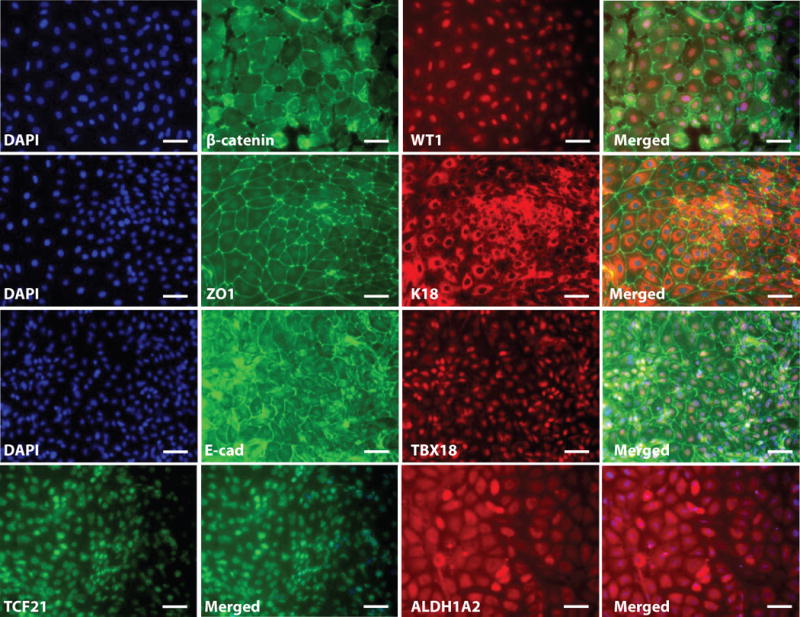
Immunostaining analysis of epicardial cells differentiated from hPSCs via small molecule modulation of Wnt signaling (Step 15B). Epicardial cells were generated from H13 hESCs as described in Fig. 2 and maintained as described in Fig. 3. At day 48, cells were individualized and replated on 0.1% gelatin-coated coverslips. Immunostaining for β-catenin, WT1, ZO1, K18, E-cadherin (E-cad), TBX18, TCF21 and ALDH1A2 are shown. The antibodies used are given in Table 1. Scale bars, 50μm.
Figure 5.
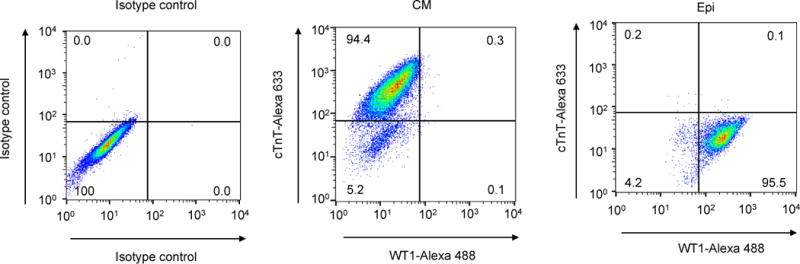
Quantitative analysis of marker expression in epicardial cells differentiated from hPSCs via small molecule modulation of Wnt signaling (Step 15C). Epicardial cells (Epi) were generated from H13 hESCs as described in Fig. 2. At day 48, 10–50 k cells were analyzed for cTnT/WT1 expression by flow cytometry with percentages of cTnT: 0.3±0.1 and WT1: 96.3±0.8, compared to cTnT: 94.6 ± 1.5 and WT1: 0.2 ± 0.1 of cardiomyocytes (CMs). Data are presented as mean ± S.D. from three independent experiments: epicardial cell WT1 (95.6%, 96.1%, 97.2%) and cTnT (0.3%, 0.2%, 0.4%) and cardiomyocyte WT1 (0.4%, 0.1%, 0.2%) and cTnT (94.7%, 96%, 93%).
Differentiation of hPSC-derived epicardial cells to fibroblasts and SMCs (Steps 15 D)
A summary of this differentiation protocol is shown in Fig. 6A. To further confirm the functional identity of these hPSC-derived epicardial cells, singularized WT1+ cells are seeded at a density of 0.01 to 0.06 million cells per cm2 in LaSR basal medium supplemented with 5 μM Y-27632 (Fig. 3). After 24 hr, the medium is changed to LaSR basal medium and treated with bFGF or/and TGFβ1 for 6 days. bFGF-treated cells adopt a fibroid spindle-like morphology typical of cultured fibroblasts, while TGFβ1− or TGFβ1+bFGF-treated cells display a fusiform-shaped appearance typical of smooth muscle cells (Fig. 6B). The expression of calponin and smooth muscle myosin heavy chain (SMMHC) in TGFβ1-induced cultures further support their smooth muscle cell identity, and vimentin (VIM) and CD90 expression support their fibroblast identity.
Figure 6.
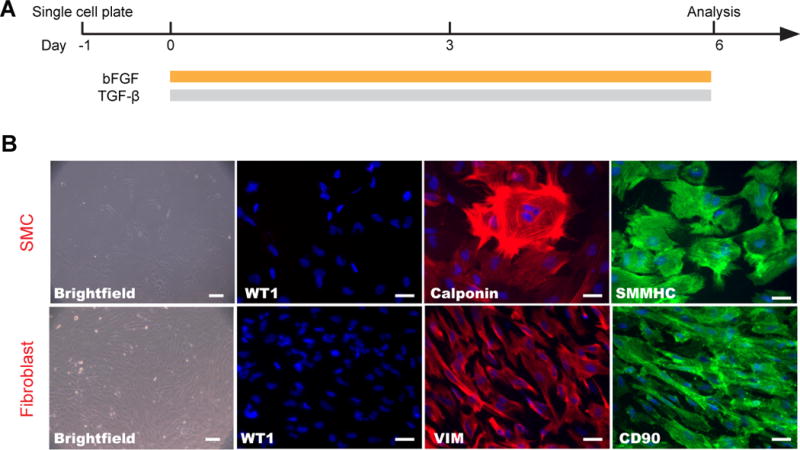
Differentiation of hPSC-derived epicardial cells to fibroblasts and smooth muscle cells (Step 15D). (A) Schematic of protocol for the differentiation of hPSC-derived epicardial cells. (B) Bright field images of the typical morphology of fibroblasts and smooth muscle cells and immunostaining images with correspondent markers are shown. The antibodies used are listed in Table 1. Scale bars, 50 μm.
MATERIALS
REAGENTS
Antibodies for characterization (Table 1)
2% gelatin solution (Sigma, cat. no. G1393)
16% formaldehyde (Polysciences, cat. no. 18814)
A83-01 (R&D systems, cat. no. 2939)
Accutase (Innovative Cell Technology, cat. no. AT104)
Advanced DMEM/F12 (Life Technologies, cat. no. 12634-028)
Albumin, human (Sigma, cat. no. A9713-5G)
bFGF (R&D systems, cat. no. 233-FB)
B27 supplement (Life Technologies, cat. no. 17504-044)
B27 supplement Minus Insulin (Life Technologies, cat. no. A1895601)
Bovine Serum Albumin (Sigma, cat. no. A9418)
CHIR99021 (Selleckchem, cat. no. S2924-25mg)
DMEM (Life Technologies, cat. no. 11965-092)
DMEM/F12 (Life Technologies, cat. no. 11330-057)
DMSO (Sigma, cat. no. D8418)
Essential 8™ Medium (Life Technologies, cat. no. A1517001)
FBS (Life Technologies, cat. no. 16000-044)
GlutaMAX (Life Technologies, cat. no. 35050-061)
Gold Anti-fade reagent with DAPI (Life Technologies, cat. no. P-36931)
HCl (Sigma, cat. no. 320331) !CAUTION HCl is corrosive and toxic in case of inhalation, ingestion and skin absorption. Wear proper personal protective equipment and handle carefully to avoid direct contact with skin or eye.
Hoechst 33342 (Life Technologies, cat. no. H3570) !CAUTION Hoechst 33342 is toxic in case of inhalation, ingestion and skin absorption. Wear proper personal protective equipment and handle carefully to avoid direct contact with skin or eye.
Insulin solution human (Sigma, cat. no. I9278)
IWP2 (Tocris, cat. no. 3533-10 mg)
IWP4 (Stemgent, cat. no. 04-0036)
L-ascorbic acid (Sigma, cat. no. A8960)
RPMI (Life Technologies, cat. no. 11875-119)
mTeSR1 (STEMCELL Technologies, cat. no. 05857)
Matrigel (BD Biosciences, cat. no. 354277)
Non Fat Dry Milk (Bio-rad, cat. no. 170-6404XTU)
Phosphate Buffered Saline (Sigma, cat. no. D8537)
SB431542 (R&D systems, cat. no. 1614)
Synthemax II-SC Substrate (Corning, cat. no. 3535)
TGFβ1 (R&D systems, cat. no. 240-B)
Triton X-100 (Sigma, cat. no. T8532) !CAUTION Triton X-100 is corrosive, toxic and hazardous to the aquatic environment. Wear proper personal protective equipment and handle carefully. After use, please dispose appropriately to avoid water contamination.
Versene (Life Technologies, cat. no. 15040-066)
Y27632 (Tocris, cat. no. 1254)
Table 1.
Antibodies used for characterization of hPSC-derived epicardial cells.
| Antibody | Source/Isotype/clone/cat. no. | Concentration |
|---|---|---|
| Cardiac troponin T | Lab Vision/Mouse IgG1/13-11/ms-295-p1 | 1:200 (FC) |
| ISL1 | DSHB/Mouse IgG2b/39.4D5-s | 1:20 (FC) |
| NKX2-5 | Santa Cruz/Rabbit IgG/sc-14033/H-114 | 1:100 (FC) |
| WT1 | Abcam/Rabbit IgG/ab89901 | 1:250 (IF/FC) |
| TCF21 | Sigma-Aldrich/Rabbit IgG/HPA013189 | 1:200 (IF) |
| TBX18 | Sigma-Aldrich/Rabbit IgG/HPA029014 | 1:200 (IF) |
| ALDH1A2 | Sigma-Aldrich/Rabbit IgG/HPA010022 | 1:50 (IF) |
| Brachyury | R&D Systems/Goat IgG/AF2085 | 1:200 (FC) |
| ZO1 | Invitrogen/Rabbit IgG/402200 | 1:200 (IF) |
| β-catenin | Cell Signaling/Mouse IgG1/2698/L87A12 | 1:200 (IF) |
| E-cadherin | BD Biosciences/Mouse IgG2a/560061 | 1:200 (IF) |
| Cytokeratin 18 (K18) | Thermo Scientific/Mouse IgG1/DC10/ms-142 | 1:200 (IF) |
| Oct-3/4 | Santa Cruz/Rabbit IgG/H-134/sc-9081 | 1:40 (FC) |
| Vimentin | Sigma-Aldrich/Mouse IgG1/V6630/V9 | 1:200 (IF) |
| SMMHC | Abcam/Rabbit IgG/ab82541 | 1:800 (IF) |
| Calponin | Abcam/Mouse IgG1/ab700/CALP | 1:200 (IF) |
| CD90 | BD Pharmingen/Mouse IgG1/559869 | 1:200 (IF) |
| Rabbit IgG Control | Abcam/Rabbit IgG/ab172730 | 1:200 (IF/FC) |
| Secondary Antibody | Alexa 488 Chicken anti-Gt IgG/A-21467 | 1:1,000 (IF/FC) |
| Secondary Antibody | Alexa 488 Chicken anti-Rb IgG/A-21441 | 1:1,000 (IF/FC) |
| Secondary Antibody | Alexa 488 Goat anti-Ms IgG1/A-21121 | 1:1,000 (IF/FC) |
| Secondary Antibody | Alexa 488 Goat anti-Ms IgG/A-11001 | 1:1,000 (IF/FC) |
| Secondary Antibody | Alexa 488 Goat anti-Rb IgG/A-11008 | 1:1,000 (IF/FC) |
| Secondary Antibody | Alexa 594 Goat anti-Ms IgG2b/A-21145 | 1:1,000 (FC) |
| Secondary Antibody | Alexa 594 Goat anti-Rb IgG/A-11012 | 1:1,000 (FC) |
| Secondary Antibody | Alexa 633 Goat anti-Ms IgG1/A-21126 | 1:1,000 (IF/FC) |
| Secondary Antibody | Alexa 647 Goat anti-Ms IgG2b/A-21242 | 1:1,000 (FC) |
| Secondary Antibody | Alexa 647 Goat anti-Rb IgG/A-21244 | 1:1,000 (FC) |
IF: Immunofluorescence; FC: Flow cytometry.
Feeder-free hPSC lines
We’ve used hESC lines H9, H13, ES03, and iPSC lines 19-9-7, 19-9-11 (WiCell, Madison, WI, USA). ES03 were engineered with CRISPR/Cas9 to make ES03 WT1-eGFP. !CAUTION The cell lines used should be regularly checked to ensure they are authentic, have not acquired an abnormal karyotype, and are not infected with mycoplasma.
EQUIPMENT
15- and 50-ml conical tubes (BD Biosciences, cat. nos. 352095 and 352073)
Synthemax plates (Corning, cat. no. 3979)
6-, 12-, 24- and 96-well plates (Nunc, cat. nos. 140675, 150628, 142475 and 165306)
CL2 Centrifuge (Thermo Scientific, part no. 004260F)
Mr. Frosty™ freezing container (Thermo Scientific, cat. no. 5100-001)
Polypropylene conical tubes (15 ml; BD Biosciences, cat. no. 352097)
Polypropylene conical tubes (50 ml; BD Biosciences, cat. no. 352098)
Sterile biosafety cabinets
Liquid waste disposal system
Flow cytometry FACSCalibur (Becton Dickinson)
FlowJo v10 software (FlowJo LLC)
Sterilized pasteur pipettes (Fisher, 13-678-20D)
Humidified tissue culture incubator (37 °C, 5% CO2)
Hemocytometer (Hausser Scientific, cat. no. 02-671-52)
Inverted phase contrast microscope (Nikon, ECLIPSE TS100)
Microcentrifuge tube (1.5 ml, Fisher Scientific, cat. no. 05-408-129)
VWR Scientific 1205 Dual Heated Water Bath Incubator (VWR, cat. no. 14405)
Serological pipettes 5-, 10- and 25-ml (Fisher Scientific, cat. nos. 13-678-11D, 13-678-11E and 13-678-11)
Stericup filtration system (Millipore, cat. no. SCGPU05RE, 500 ml)
Stericup filtration system (Millipore, cat. no. SCGPU02RE, 250 ml)
Steriflip filtration system (Millipore, cat. no. SCGP00525, 50 ml)
0.45 μm Steriflip-HV Filter Unit (Millipore, cat. no. SE1M003M00, 50 ml)
Coverslips (18mm) (Fisher Scientific, 12-545-100 and 12-545-101)
Flow round-bottom tube (5 ml) (BD Biosciences, cat. no. 352052)
Flow round-bottom tube caps (BD Biosciences, cat. no. 352032)
REAGENT SETUP
10% FBS DMEM medium
In a sterile environment, mix 50 ml FBS and 450 ml DMEM. Filter the medium with a 500 ml Stericup filtration system. The medium can be stored at 4 °C for up to 2 months.
1% formaldehyde
Add 62.5 μl 16% formaldehyde into 1 ml PBS. We do not recommend storing this solution.
4% formaldehyde
Add 1 ml 16% formaldehyde into 3 ml PBS. We do not recommend storing this solution.
90% methanol
Add 5 ml MilliQ water to 45 ml pure methanol and store at −20 °C for up to 6 months.
0.1% BSA
Add 1 g BSA into 1000 ml PBS and filter it using Stericup filtration systems. Store at 4 °C for up to 6 months.
A83-01 (10mM)
Add 2.37 ml DMSO to 10 mg A83-01. Aliquot and store at −20 °C for up to 1 year.
bFGF (25μg/ml)
Add 1 ml sterile PBS to 25 μg bFGF. Aliquot and store at −20 °C for up to 3 months.
CHIR99021 (36 mM)
Add 1.49 ml DMSO to 25 mg CHIR99021. Aliquot and store at −20 °C for up to 1 year.
CPC Freezing medium (50 ml)
In a sterile environment, mix 30 ml DMEM, 15 ml FBS, 5 ml DMSO and 50 μl Y27632. The medium can be stored at 4 °C for up to 2 months.
Pro-epicardial Freezing medium (50 ml)
In a sterile environment, mix 30 ml LaSR basal medium,15 ml FBS, 5 ml DMSO, 2.5 μl A83-01 and 50 μl Y27632. The medium can be stored at 4 °C for up to 2 months.
L-ascorbic acid (100 mg/ml)
In a sterile environment, add 50 ml sterile MilliQ water to 5 g L-ascorbic acid. Aliquot into 1 ml samples and store at −20°C for up to 1 year.
LaSR basal medium (500 ml)
In a sterile environment, mix 500 ml advanced DMEM/F12 medium, 6.5 mL GlutaMAX and 500 μl 100 mg/ml ascorbic acid solution. The medium can be stored at 4 °C for up to 2 months.
LaSR basal medium + 3 μM CHIR99021 (24 ml)
Add 2 μl of 36 mM CHIR99021 into 24 ml LaSR basal medium. We do not recommend storing this medium.
RPMI/B27-insulin (510 ml)
In a sterile environment, mix 500 ml of RPMI and 10 ml B27 supplement Minus Insulin. The medium can be stored at 4 °C for up to 1 month.
RPMI/B27 (510 ml)
In a sterile environment, mix 500 ml of RPMI and 10 ml of B27 supplement. The medium can be stored at 4 °C for up to 1 month.
RPMI20 (250 ml)
In a sterile environment, mix 200 ml of RPMI and 50 ml FBS, then filter through a 250 ml Stericup filtration system. The medium can be stored at 4 °C for up to 1 month.
RPMI/Vc/Ins (500 ml)
In a sterile environment, mix 500 ml of RPMI, 1 ml of 100 mg/ml L-ascorbic acid and 500 μl of insulin solution. The medium can be stored at 4 °C for up to 2 months.
RPMI/Vc/Ins + 3 μM CHIR99021 (24 ml)
Add 2 μl of 36 mM CHIR99021 into 24 ml RPMI/Vc/Ins medium. We do not recommend to storing this medium.
IWP2 (5 mM)
Add 4.28 ml DMSO to 10 mg IWP2. Incubate the mixture at 37 °C for 10 min to dissolve the IWP2. Aliquot 100 μl samples into 1.5 ml tubes and store at−20 °C for up to 1 year.
IWP4 (5 mM)
Add 0.805 ml DMSO to 2 mg IWP4. Incubate the mixture at 37 °C for 10 min to dissolve the IWP4. Aliquot 100 μl samples into 1.5 ml tubes and store at−20 °C for up to 1 year.
5 mM Y27632
In a sterile environment, add 6.24 ml PBS to 10 mg Y27632. Aliquot 100 μl samples into 1.5 ml tubes and store at−20 °C for up to 1 year.
mTeSR1 + 5 μM Y27632
Add 50 μl of 5 mM Y27632 to 50 ml mTeSR1 (final concentration of Y27632 is 5 μM). Store at 4 °C for up to 2 weeks.
0.1% Gelatin (30 ml)
Add 1.5 ml 2% gelatin solution into 30 ml sterile MilliQ water. We do not recommend storing this solution.
FlowBuffer-1 (500 ml)
Add 2.5 g BSA into 500 ml PBS and filter using a 500 ml Stericup filtration system. The medium can be stored at 4 °C for up to 6 months.
1% Triton X-100 solution (500 ml)
Add 5 ml Triton X-100 into 495 ml PBS and shake the bottle to dissolve the Triton. The medium can be stored at 4 °C for up to 6 months.
FlowBuffer-2 (550 ml)
Add 2.5 g BSA and 50 ml 1% Triton X-100 solution into 500 ml PBS and filter using a 500 ml Stericup filtration system. The medium can be stored at 4 °C for up to 6 months.
10% Triton X-100 solution (50 ml)
Add 5 ml Triton X-100 into 45 ml PBS and shake the tube to dissolve the Triton. The medium can be stored at 4 °C for up to 6 months.
5% non fat dry milk, 0.4 % Triton X-100
Add 0.5 g non fat dry milk and 4 ml 1% Triton X-100 solution into 6 ml PBS. We do not recommend storing this solution.
Hoechst staining solution (5 μg/ml)
Add 2.5 μl Hoechst 33342 stock solution (10 mg/ml) into 5 ml PBS. We do not recommend storing this solution.
SB431542 (10 mM)
Add 2.60 ml DMSO to 10 mg SB431542. Aliquot and store at −20 °C for up to 1 year.
Synthemax II-SC stock solution (1 mg/ml)
Add 10 ml of sterile water to the vial of 10 mg Corning Synthemax II-SC substrate, and mix well to make a homogenous 1 mg/ml stock solution. The solution can be stored at 4°C for up to 6 months.
TGFβ1 (20 μg/ml)
Add 100 μl sterile 4 mM HCl containing 0.1% bovine serum albumin to 2 μg TGFβ1. Aliquot and store at −20 °C for up to 3 months.
EQUIPMENT SETUP
Matrigel-coated plates
Matrigel-coated plates can be made according to our previous protocol6. Briefly, in a sterile hood, add 30 ml of cold (4 °C) DMEM/F12 to a 50 ml conical tube and keep it cold by placing it on ice. Remove one Matrigel aliquot (2.5 mg) from the freezer, and add 1 ml of cold DMEM/F12 to it. Gently pipette the Matrigel solution with a P1000 tip to thaw and dissolve the Matrigel. Immediately transfer the Matrigel solution to the 50 ml conical tube that contains 30 ml cold DMEM/F12. Immediately add 1 ml/well Matrigel in DMEM/F12 for 6-well plates, 0.5 ml/well for 12-well plates, 250 μl/well for 24-well plates, or 100 μl/well for 96-well plates. Allow the Matrigel to set for at least 30 minutes at room temperature before use. The Matrigel-coated plates can be stored at 4 °C for up to 3 weeks. CRITICAL: We recommend dissolving 0.5 mg Matrigel into 6 ml cold DMEM/F12. Use Matrigel lots qualified by BD Biosciences for hESC/iPSC culture. Some Matrigel lots do not support hPSC self-renewal.
Synthemax II-SC coated plates
In a sterile hood, add 6 ml of room temperature sterile water to a 12 ml conical tube and add 150 μl of 1 mg/ml Synthemax II-SC stock solution into it. Gently pipette the Synthemax solution and mix well. Immediately add 1 ml/well Synthemax in water for 6-well plates, 0.5 ml/well for 12-well plates, 250 μl/well for 24-well plates, or 100 μl/well for 96-well plates. Allow the Synthemax-coated plates to sit at room temperature for at least 2 hours. Aspirate all remaining solution and the Synthemax-coated plates are ready to use, or can be stored at 4°C for up to 3 months.
0.1% gelatin coated plates
In a sterile hood, add 30 ml of sterile water and 1.5 ml of 2% gelatin solution to a 50 ml conical tube. Gently pipette the gelatin solution and mix well. Immediately add 1 ml/well gelatin solution for 6-well plates, 0.5 ml/well for 12-well plates, 250 μl/well for 24-well plates, or 100 μl/well for 96-well plates. Allow the gelatin to sit for at least 30 min at 37°C before use. The gelatin-coated plates can be stored at 4 °C for up to 3 weeks.
0.1% gelatin coated coverslips
Gelatin-coated coverslips can be made according to our previous protocol6. Briefly, autoclave the coverslips at 121°C, 15 psi for 30 minutes and place one sterile coverslip in each well of a 12-well plate. Add 1 ml of 0.1% gelatin per well and incubate at 37 °C overnight. Store the gelatin coated coverslips at 4 °C for up to 2 months.
Flow cytometry data analysis
Data are collected on a FACSCaliber flow cytometer and analyzed using FlowJo v10 as illustrated in Fig. S1. Briefly, the intact cell population, without cell debris, is gated from forward and side scatter for further analysis (Fig. S1A). Change the X and Y-parameters (in this case, Histogram and Alexa-488) on the Graph Window to gate the cell population of interest in both isotype and primary antibody stained samples with a ranged tool (Fig. S1B). Use the Layout Editor to create an overlay histogram report (Fig. S1C).
PROCEDURE
Cardiac progenitor differentiation with Gsk3 inhibitor and Wnt inhibitor (GiWi2 protocol)
1| Culture the hPSCs on Matrigel-coated or Synthemax-coated 6-well plates in mTeSR1or E8 medium to 80–90% confluence using instructions provided in our previous protocol to generate cardiomyocytes6. Aspirate the medium and add 1ml of room temperature Accutase to each well. Put the plate in a 37°C, 5% CO2 incubator and wait for 5 min.
2| Add 0.5 ml of mTeSR1 or E8 into each well of the 6-well plate and pool all of the cells into a 15 ml conical. Mix well and count the total cell number with a hemocytometer. Centrifuge the cells at 200 × g for 5 min.
3| Aspirate the supernatant, resuspend the cells in mTeSR1 or E8 + 5 μM Y27632 at a cell density of 2 million cells/ml, and plate 0.5 – 2.0 million cells/well in each well of a 12-well Matrigel- or Synthemax-coated plate. Add mTeSR1 or E8 + 5 μM Y27632 medium to each well to make a final volume of 1 ml in each well of the 12-well plate. This time point corresponds to day -3.
CRITICAL STEP: The starting seeding cell density is very critical for efficient cardiac differentiation. The initial plating density and/or the time of expansion prior to initiation of differentiation may require optimization for different cell lines or expansion conditions. We recommend plating 0.5 – 2.0 million cells per well of 12-well plate and expanding the cells for 3 days prior to initiation of differentiation.
4| Day -2 and day -1, aspirate the medium and replace with 2 ml room temperature mTeSR1 or E8 per well of the 12-well plate. For cells cultured in E8, pre-condition with 0.1–0.6 μM CHIR99021 on day -1 for cardiac differentiation.
5| Day 0, prepare RPMI medium containing 6 μM CHIR99021. You will need 2 ml RPMI for each well of the 12 well plate, so 24 ml to differentiate cells in all 12 wells. Add 4 μl of 36 mM CHIR99021 into 24 ml RPMI basal medium to make RPMI medium containing 6 μM CHIR99021. Aspirate the old medium and then add 2 ml RPMI medium containing 6 μM CHIR99021 to each well of the 12-well plate and record the time.
CRITICAL STEP: Recording the time when you finish adding RPMI medium containing CHIR99021 is important since exactly 24 hr later you need to change the medium. Although we identified 6 μM CHIR99021 as the optimal concentration for the hPSC lines that we tested, other lines may respond to CHIR99021 treatment differently. We have also found significant lot-to-lot variability in CHIR99021 activity. Thus, optimization of CHIR99021 concentration may be required, especially when changing lots or suppliers. We recommend testing 3–12 μM CHIR99021.
6| Day 1, aspirate the medium from each well of the 12-well plate and replace with 2 ml room temperature RPMI basal medium. Put the plate back into the 37°C, 5% CO2 incubator.
7| Day 3 (72 hr post addition of CHIR99021), prepare combined medium: use a 5 ml pipette to collect 1ml medium from the 12-well plate and mix it with 1 ml of fresh RPMI basal medium in a 15 ml conical tube. This 2 ml medium is called combined medium. Add 1 μl of 5 mM IWP2 (final concentration is 2.5 μM) into the 2 ml combined medium. Prior to aspirating, gently rock the plate back and forth to suspend any cell debris, ensuring that the cell debris will be removed after aspiration. Aspirate the remaining 1ml medium from each well of the 12-well plate, then add 2 ml/well of the combined medium containing IWP2 to each well.
CRITICAL STEP: Though we identified 2.5 μM IWP2 as the optimal concentration for the hPSC lines that we tested, other lines may respond to IWP2 treatment differently. Thus, optimization of the IWP2 concentration may be required. We recommend testing 2–8 μM IWP2.
8| Day 5, aspirate the medium from each well of the 12-well plate and add 2 ml/well room temperature RPMI basal medium. Return the plate to the 37°C, 5% CO2 incubator.
CRITICAL STEP: Though we identified day 6 cells as the optimal starting cell population for epicardial cell generation, day 5 or 7 cells also efficiently generated WT1+ epicardial cells after 2 days of CHIR99021 treatment (day 6 to day 8, or day 8 to day 10). If day 5 cells are used, it’s not necessary to change the medium on day 5. Instead, singularize the cells and seed according to steps 9–12.
Directed differentiation of cardiac progenitors into pro-epicardial cells
9| Day 6 of GiWi25 or GiWi6 protocol, aspirate the medium, add 1 ml Accutase per well in a 12-well plate, and incubate in a 37°C, 5% CO2 incubator for 5 min. For a quick passage without centrifugation (skipping Steps 10–12), aspirate Accutase, gently scrape cells with a glass pipette into corresponding plating medium, and seed cells at a split ratio of 1:3 to 1:12. If you have more time however proceed instead to step 10.
CRITICAL STEP: Although we found centrifugation is not necessary for cell passaging, 5 min treatment of Accutase might dissociate the cells from the plate surface, resulting in cell loss. In that case, decrease the Accutase incubation time or use a more gentle dissociating buffer (e.g. Versene).
10| Pipette 5–10 times with a P1000 tip to singularize the cells and then transfer the 1 ml cell mixture into a 15 ml conical containing 2 ml RPMI20 medium.
11| Count the cells with a hemocytometer, centrifuge the cells at 200 × g for 5 min, and aspirate the supernatant.
12| Resuspend the cell pellet in albumin-containing LaSR basal or albumin-free RPMI/Vc/Ins medium + 5 μM Y27632 at a concentration of 100,000 cells/ml, and then seed onto a gelatin- or Synthemax-coated cell culture dish at a density of 20,000 – 40,000 cells/cm2 (or a 1:6 to 1:12 split) in LaSR basal medium or 40,000 – 80,000 cells/cm2 (or a 1:3–1:6 split) in RPMI/Vc/Ins medium. Incubate the plate at 37°C, 5% CO2 overnight without medium change to allow cell attachment.
CRITICAL STEP: Although albumin (bovine serum albumin or human recombinant albumin) and FBS are not required here, the addition of 1% albumin or FBS may improve cell attachment and viability. Day 6 cell pellets can also be resuspended in cardiac progenitor cell (CPC) freezing medium at a density of 1 million cells/ml and frozen for long-term storage following instructions in Box 1.
BOX 1 |Cryostorage and thawing of cardiac progenitors and pro-epicardial cells.
1. Following dissociation of cardiac progenitor cells or pro-epicardial cells as described in the main PROCEDURE, resuspend cardiac progenitors (from Step 11) or pro-epicardial cells (from Step 15 A (iii)) at a density of 2×106 cells per ml in CPC or pro-epicardial cell freezing medium.
2. Aliquot 1 ml of the cell suspension into each cryovial, and freeze in a Mr. Frosty™ freezing container at −80°C overnight.
3. The next day, transfer the cryovials to liquid nitrogen for long-term storage.
PAUSE POINT Cells can be stored in liquid nitrogen for at least 1 year.
Thawing cells
4. Incubate the vial in a 37°C water bath until almost all of the ice crystals thaw.
5. Gently transfer the thawed cardiac progenitor cells or pro-epicardial cells to a 15-ml conical tube containing 5 ml of DMEM with 10% FBS.
6. Centrifuge the cells at 200 × g for 5 min then aspirate the supernatant.
7. Gently resuspend the cells in 1 ml of LaSR basal medium + 20% FBS with 5 μM Y27632 and transfer into a gelatin-or Synthemax-coated 12-well plate at a density of 0.1–0.5 million cells per cm2. For pro-epicardial cell thawing, addition of 0.5 μM A83-01 or 2 μM SB431542 to the medium will significantly increase cell attachment and viability.
8. The next day, aspirate the medium in each well and replace with 2 ml fresh room temperature LaSR basal medium. Depending on the step at which you froze the cells either follow Steps 13 of the main PROCEDURE to make epicardial cells or step 15 A (v) of the main PROCEDURE for long-term maintenance.
13| Day 7 and day 8, aspirate the medium and replace with 1 ml room temperature LaSR basal or RPMI/Vc/Ins medium + 3 μM CHIR99021 per well of the 12-well plate.
CRITICAL STEP: Although we identified 3 μM CHIR99021 as the optimal concentration for the hPSC lines that we tested, other lines may respond to CHIR99021 treatment differently. Thus, optimization of CHIR99021 concentration may be required. We recommend testing 1–6 μM CHIR99021.
14| Day 9, day 10, and day 11, aspirate the medium and replace with 1 ml room temperature LaSR basal or RPMI/Vc/Ins medium per well of 12-well plate. On day 12, more than 90% WT1+ cells can be collected for long-term maintenance (see step 15 A), characterization (see steps 15 B and C), (or further differentiation into fibroblasts and SMCs (see step 15 D). Day 12 pro-epicardial cells can also be frozen for long-term storage following instructions in Box 1.
Downstream assays
15| Cells from Steps 15 onward should express hallmark markers of epicardial cells. If you wish to passage hPSC-derived epicardial cells long term, follow Option A. To analyze these WT1+ cells, perform immunostaining (Option B) and flow cytometry analysis (Option C). We recommend flow cytometry for quantitative analysis of the purity of hPSC-derived epicardial cells. Antibody combinations of cTnT/WT1, β-catenin/WT1 and E-cadherin/TBX18 are recommended for double staining. To differentiate cells to fibroblasts and SMCs follow Option D.
-
(A) Long-term maintenance of hPSC-derived epicardial cells
-
(i) On day 12, aspirate the medium, add 1 ml Accutase per well in a 12-well plate, and incubate in a 37°C, 5% CO2 incubator for 5 min. For a quick passage without centrifuge (skipping Steps 15 A (ii)-(iv)), aspirate Accutase, gently scrape cells with a glass pipette into corresponding plating medium, and seed cells at a split ratio of 1:3 to 1:9. Alternatively proceed to next step.
CRITICAL STEP: Although we found centrifugation is not necessary for cell passaging, 5 min treatment of Accutase might dissociate the cells from the plate surface, resulting in cell loss. In that case, decrease the Accutase incubation time or use a more gentle dissociating buffer (e.g. Versene).
(ii) Pipette 5–10 times with a P1000 tip to singularize the cells and then transfer the 1 ml cell mixture into a 15 ml conical containing 2 ml RPMI20 medium.
(iii) Count the cells with a hemocytometer, centrifuge the cells at 200 × g for 5 min, and aspirate the supernatant.
(iv) Resuspend the cell pellet in LaSR basal medium or RPMI/Vc/Ins medium + 0.5 μM A83-01 or 2 μM SB431542 supplemented with 5 μM Y27632 and 1% human recombinant albumin or FBS at a concentration of 100,000 cells/ml, and then seed onto a gelatin- or Synthemax-coated cell culture dish at a density of 40,000–80,000 cells/cm2. Incubate the plate at 37°C, 5% CO2 overnight without medium change to allow cell attachment.
(v) The next day and every day following, aspirate the medium from each well of the 12-well plate and add 2 ml/well room temperature LaSR basal or RPMI/Vc/Ins medium with 0.5 μM A83-01 or 2 μM SB431542. Return the plate to the 37°C, 5% CO2 incubator.
-
(vi) Once the cells are confluent, dissociate the cells with Accutase or Versene, and then repeat steps ii–v until analysis (Options B and C). This passage has been successfully repeated 15 times.…Although both Accutase and Versene work similarly at early passages, we found that Versene passaging resulted in greater cell attachment after 8 passages. To passage with Versene, treat cells with Versene for 3 to 4 mins, aspirate the Versene, gently scrape cells with a glass pipette into plating medium, and seed cells at a split ratio of 1:3 to 1:9.
CRITICAL STEP During the first round of maintenance the cells transition from ALDH1A2- cells to ALDH1A2+ cells. It’s highly recommended to use Accutase to dissociate day 12 cells for passage since Versene does not adequately separate the cells at this stage.
-
-
(B) Immunostaining analysis
(i) Wash the differentiated cells from Step 14 or 15 A (vi) with 1 ml PBS per well in a 12-well plate. For immunostaining analysis without coverslips, directly go to Step 15 (B) (vii).
(ii) Aspirate the PBS, add 1 ml Accutase or Versene per well, and incubate in a 37°C, 5% CO2 incubator for 5 min.
(iii) Pipette 5–10 times with a P1000 tip to singularize the cells and then transfer the 1 ml cell mixture into a 15 ml conical tube containing 2 ml of 10% FBS DMEM medium.
(iv) Count the cells with a hemocytometer, centrifuge the cells at 200 × g for 5 min, and aspirate the supernatant.
(v) Resuspend the cell pellet in RPMI20 + 5 μM Y27632 at a concentration of 100,000 cells/ml. Plate 1 ml of the resuspended cell solution in each well of a 12-well plate containing a gelatin-coated coverslip. Incubate the plate at 37°C, 5% CO2 overnight without medium change to allow cell attachment.
(vi) The next day, aspirate the medium and add 1 ml of PBS per well to wash the cells.
(vii) Aspirate the PBS, add 1 ml of 4% formaldehyde per well, and incubate for 15 min at room temperature to fix the cells. Aspirate the formaldehyde solution and then add 1 ml of PBS per well and aspirate to rinse the cells. Repeat the PBS rinse step twice.
PAUSE POINT Cells in PBS after rinsing twice can be stored at 4°C for up to 3 months.
(viii) Add 300 μl 5% non-fat dry milk, 0.4% Triton X-100 in PBS per well and then add primary antibodies into individual wells according to Table 1. Incubate at room temperature for 1 hr or at 4°C overnight on a shaker. Antibodies include but are not restricted to WT1, Ki67, E-cadherin, TBX18 and ALDH1A2.
(ix) Aspirate the antibody solution. Add 1 ml of PBS to each well and then aspirate the PBS. Repeat this wash twice.
(x) Dilute the secondary antibodies specific to the primary IgG subtype at 1:1000 in 5% milk, 0.4% Triton X-100. Add 300 μl of secondary antibody solution to each well and then incubate at room temperate for 30 min or at 4°C overnight with gentle shaking in the dark.
(xi) Aspirate the secondary antibody solution in the dark, add 1 ml of PBS to each well and then aspirate the PBS. Repeat this wash twice.
(xii) Seal the coverslips to glass slides with Gold Anti-fade reagent with DAPI. Examine the slides with an epifluorescence microscope. For immunostaining analysis without coverslips, incubate the cells with Hoechst staining solution (5 μg/ml) for 5 min in dark and then wash the cells with PBS twice before imaging. Typical β-catenin, ZO1, K18, E-cadherin (E-cad), TCF21, ALDH1A2, WT1 and TBX18 expression patterns are shown in Fig. 4.
-
(C) Flow cytometry analysis
(i) Follow Steps 15 B (i)-(iv) using Accutase, then add 1 ml of 1% formaldehyde to resuspend the cell pellet and then incubate at room temperature for 20 min.
(ii) Centrifuge the cells at 200 × g for 5 min, aspirate the supernatant and then resuspend the fixed cells in 1 ml of 90% (vol/vol) cold methanol per tube. Incubate the mixture at −20°C for one hr or overnight and calculate the cell density based on the cell count obtained in step 15 B (iv).
PAUSE POINT Cells in methanol can be stored at −20°C for up to 3 months.
(iii) Add 1 million cells into a 15-ml tube containing 2 ml of FlowBuffer-2, centrifuge the cells at 200 × g for 5 minutes at room temperature, and aspirate the supernatant. Repeat this wash twice to completely remove the methanol.
(iv) Resuspend the cell pellet in 100 μl of FlowBuffer-2 with the appropriate dilution of primary antibody according to Table 1. Antibody combinations of cTnT/WT1 and Ki67/WT1 are recommended for double staining. Incubate the mixture for 1hr at room temperature or overnight at 4°C.
(v) Wash the cells with 2 ml of FlowBuffer-2 twice and resuspend the cell pellet in 100 μl of FlowBuffer-2 containing 1:1000 dilution of secondary antibody. Incubate the mixture for 30 min at room temperature or overnight at 4°C in dark.
(vi) Wash the cells with 2 ml of FlowBuffer-2 twice, resuspend the cell pellet in 300 μl of FlowBuffer-1, and transfer the cell suspension into flow round-bottom tubes. Place the flow tubes on ice and perform flow cytometric analysis with a BD FACSCalibur™ or similar flow cytometer. Fig. 5 provides representative results of cTnT/WT1 double staining of day 48 H13 hESC-derived epicardial cells differentiated via the GiWiGi protocol.
-
(D) Differentiation of hPSC-derived epicardial cells to fibroblasts and smooth muscle cells
-
i. Follow steps i to iii of Option A. Then resuspend the epicardial cells and seed onto a gelatin-coated cell culture dish at a density of 10,000–60,000 cells per cm2 in LaSR basal medium supplemented with 5 μM Y-27632. Incubate the plate at 37°C, 5% CO2 overnight without medium change to allow cell attachment. This time point corresponds to day -1.
CRITICAL STEP: Although FBS or TGFβ inhibitors are not required here, the addition of 1% FBS and 0.5 μM A83-01 may improve cell attachment and viability.
ii. The next day and every day following, aspirate the medium from each well of the 48-well plate and add 400 μl/well room temperature LaSR basal medium with 10 ng/ml bFGF or 5 ng/ml TGFβ1 for fibroblast and SMC differentiation (Fig. 6A), respectively. For SMC differentiation, 10 ng/ml bFGF can be used instead of 5 ng/ml TGFβ1 from day 3 to 6. Return the plate to the 37°C, 5% CO2 incubator.
iii. On day 6, follow Options B and C for immunostaining and flow cytometry analysis for CD90, vimentin (VIM), SMMHC, and calponin. Typical morphology of fibroblasts and SMCs and correspondent immunostaining images are shown in Fig. 6B.
-
TIMING
Steps 1–3, passaging hPSCs: 10 min
Step 4–8, cardiac progenitor differentiation with Gsk3 inhibitor and Wnt signaling inhibitor: 6 days
Steps 9–14, pro-epicardial differentiation with Gsk3 inhibitor: 6 days
Step 15 A, long-term maintenance of hPSC-derived epicardial cells: 4 days
Steps 15 B, immunostaining analysis of hPSC derived epicardial cells: 2 days
Steps 15 C, flow cytometric analysis of hPSC derived epicardial cells: 2 days
Steps 15 D, differentiation of hPSC-derived epicardial cells to fibroblasts and SMCs: 7 days
Box 1, Cryostorage and thawing of cardiac progenitors and pro-epicardial cells: 2 days.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting
| Step | Problem | Possible reasons | Solution |
|---|---|---|---|
| 4 | Poor attachment of hPSCs on Matrigel- or Synthemax-coated plates | No ROCK inhibitor included in step 3 or coating substrate of low quality used | Include a ROCK inhibitor, (e.g. Y27632) in thawing medium; use qualified substrate |
| 6 | Cell death or detachment of hPSCs after exposure to CHIR99021 for 24 hr | Initial cell seeding density is not optimal or too high of a CHIR99021 concentration is used | Optimize initial cell seeding density in Step 3 or pretreat cells with low dose CHIR99021 (e.g. 1 μM) on day -1 in Step 4; optimize CHIR99021 (3–12 μM) for your specific hPSC lines or CHIR99021 lot. |
| 13, 15 A (v), 15 D (ii), Box 1 | Detachment of cardiac progenitors or epicardial cells | Cell seeding density is too low or cells need more albumin or FBS | Increase cell seeding density in Steps 12, 15 A (iv), and Box 1; Increase concentration of albumin or FBS (up to 20%) |
| 14 | Low percentage of WT1+ cells at day 12 | Initial cardiac progenitor purity is too low or cell seeding density is not optimal, or CHIR99021 addition dose and/or time is not optimal | Make sure Isl1+Nkx2.5+ cells are over 50% on day 6 or optimize initial cardiac progenitor cell seeding density in Step 9; Optimize CHIR99021 concentration on day 7–9 in Step 13 |
| 15 A (vi) | Slow proliferation or differentiation of WT1+ cells after passage | The concentration of TGFβ inhibitor is not optimal; the initial cell seeding density is too low | Optimize TGFβ inhibitor concentration; Increase the initial cell seeding density. |
ANTICIPATED RESULTS
This protocol presents a rapid and efficient (>90% WT1+ cells after two weeks) method for the generation of self-renewing epicardial cells from multiple hPSC lines. Before starting the differentiation protocol, well-maintained hPSCs should have high levels (more than 95%) of pluripotency markers including Oct4 (Fig. 1). 24 hours post-addition of CHIR99021, more than 90% of the differentiated culture should express brachyury, a mesendoderm marker (Fig. 1). A differentiation producing less than 90% brachyury-positive cells might be due to the poor quality of the initial hPSCs. After 5–6 days of differentiation, more than 50% of the cells should express cardiac progenitor specific markers including Isl1 and Nkx2.5 (Fig. 1). Cardiac progenitor differentiation below 20% will result in low yield of WT1+ cells.
Subsequently, the cardiac progenitor cells will be directed towards epicardial cells upon Gsk3 inhibitor treatment. The cobblestone-like morphology can be observed as early as day 8 (Fig. 2), which is different from spindle-shaped cardiomyocytes4. The cell number of epicardial cells is significantly higher than the cardiomyocytes with a same seeding density of cardiac progenitor cells on day 6. More than 90% of cells should express epicardial cell markers, including WT1 (Fig. 3). The resulting WT1+ putative epicardial cells are highly proliferative upon TGFβ inhibitor treatment, and can be passaged every 4 days, displaying epicardial characteristics for more than 2 months in LaSR basal medium. Epithelial cell markers, such as β-catenin, ZO1, K18 and E-cadherin, as well as epicardial cell markers, such as WT1, TBX18, TCF21 and ALDH1A2, should be always highly expressed during long-term maintenance (Fig. 4, 5).
The hPSC-derived epicardial cells can further differentiate into fibroblasts or SMCs upon bFGF or TGFβ1 treatment for 6 days. bFGF- and TGFβ1-treated cells display a fibroid spindle-like and a fusiform-shaped appearance typical of cultured fibroblasts and SMCs, respectively (Fig. 6). The expression of calponin and SMMHC in TGFβ1-induced cultures further support their smooth muscle cell identity, and VIM and CD90 expression in bFGF-treated cells support their fibroblast identity.
Supplementary Material
Figure S1. Flow cytometry data analysis of WT1+ cells. (A) The intact cell population is gated using forward (FWD) and side scatter to exclude cell debris. The numbers in the pink polygon show the percentage of total events that are intact cells in the pink gated region. (B) The WT1+ gated region is identified using both isotype control (blue) and WT1 antibody stained samples (red) with a ranged tool. The numbers above black line show the percentage of WT1+ cells in the gated population from (A). (C) Overlay histogram showing percentage of WT1+ cells in the gated population from (A).
Editorial summary.
This protocol differentiates human pluripotent stem cells to self-renewing epicardial cells by appropriate differentiation stage-specific application of Gsk3 inhibitor, Wnt inhibitor, then Gsk3 inhibitor in a completely defined, xeno-free system.
Acknowledgments
This study was supported by NIH grant EB007534 (S.P.P.), NSF grant 1547225 (S.P.P.), and a fellowship from the University of Wisconsin Stem Cell and Regenerative Medicine Center (X.B.).
Footnotes
AUTHOR CONTRIBUTIONS
X.B. and S.P.P. designed this study and prepared the manuscript. X.B. undertook experimentation and data analysis. T.Q., X.L., V.J.B., and T.H. contributed to the development of this protocol.
COMPETING FINANCIAL INTERESTS
All authors declare no competing financial interests.
References
- 1.Ashton RS, Keung AJ, Peltier J, Schaffer DV. Progress and prospects for stem cell engineering. Annu Rev Chem Biomol Eng. 2011;2:479–502. doi: 10.1146/annurev-chembioeng-061010-114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson JA. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science (80-) 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Brenner C, Franz W-M. Pluripotent-stem-cell-derived epicardial cells: a step toward artificial cardiac tissue. Cell Stem Cell. 2014;15(5):533–4. doi: 10.1016/j.stem.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Lian XJ, Hsiao C, Wilson G, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian X, Bao X, Zilberter M, et al. Chemically defined, albumin-free human cardiomyocyte generation. Nat Methods. 2015;12(7):595–596. doi: 10.1038/nmeth.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian X, Zhang J, Azarin SM, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–75. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kattman SJ, Witty AD, Gagliardi M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Minami I, Yamada K, Otsuji TG, et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012;2(5):1448–60. doi: 10.1016/j.celrep.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Bao X, Lian X, Dunn KK, et al. Chemically-defined albumin-free differentiation of human pluripotent stem cells to endothelial progenitor cells. Stem Cell Res. 2015;15(1):122–129. doi: 10.1016/j.scr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao X, Lian X, Palecek SP. Directed Endothelial Progenitor Differentiation from Human Pluripotent Stem Cells Via Wnt Activation Under Defined Conditions. Methods Mol Biol. 2016;1481:183–96. doi: 10.1007/978-1-4939-6393-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahara M, Hansson EM, Wernet O, Lui KO, Später D, Chien KR. Manipulation of a VEGF-Notch signaling circuit drives formation of functional vascular endothelial progenitors from human pluripotent stem cells. Cell Res. 2014;24(7):820–41. doi: 10.1038/cr.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuel R, Daheron L, Liao S, et al. Generation of functionally competent and durable engineered blood vessels from human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110(31):12774–9. doi: 10.1073/pnas.1310675110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lian X, Bao X, Al-Ahmad A, et al. Efficient Differentiation of Human Pluripotent Stem Cells to Endothelial Progenitors via Small-Molecule Activation of WNT Signaling. Stem Cell Reports. 2014;3(5):804–16. doi: 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung C, Bernardo AS, Trotter MWB, Pedersen RA, Sinha S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin–dependent disease susceptibility. Nat Biotechnol. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang A, Tang Z, Li X, Jiang Y, Tsou DA, Li S. Derivation of Smooth Muscle Cells with Neural Crest Origin from Human Induced Pluripotent Stem Cells. Cells Tissues Organs. 2012;195:5–14. doi: 10.1159/000331412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Hu J, Jiao J, et al. Engineering vascular tissue with functional smooth muscle cells derived from human iPS cells and nanofibrous scaffolds. Biomaterials. 2014;35:8960–8969. doi: 10.1016/j.biomaterials.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brade T, Pane LS, Moretti A, Chien KR, Laugwitz K-L. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med. 2013;3(10):a013847. doi: 10.1101/cshperspect.a013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Männer J, Ruiz-Lozano P. Development and Function of the Epicardium. Adv Dev Biol. 2007;18:333–357. [Google Scholar]
- 19.Witty AD, Mihic A, Tam RY, et al. Generation of the epicardial lineage from human pluripotent stem cells. Nat Biotechnol. 2014;32(10):1026–1035. doi: 10.1038/nbt.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer D, Gambardella L, Bernard WG, et al. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development. 2015;142(8):1528–41. doi: 10.1242/dev.119271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Tuyn J, Atsma DE, Winter EM, et al. Epicardial cells of human adults can undergo an epithelial-to-mesenchymal transition and obtain characteristics of smooth muscle cells in vitro. Stem Cells. 2007;25:271–278. doi: 10.1634/stemcells.2006-0366. [DOI] [PubMed] [Google Scholar]
- 22.Bao X, Lian X, Hacker TA, et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat Biomed Eng. 2016;1:3. doi: 10.1038/s41551-016-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braitsch CM, Combs MD, Quaggin SE, Yutzey KE. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol. 2012;368(2):345–57. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flow cytometry data analysis of WT1+ cells. (A) The intact cell population is gated using forward (FWD) and side scatter to exclude cell debris. The numbers in the pink polygon show the percentage of total events that are intact cells in the pink gated region. (B) The WT1+ gated region is identified using both isotype control (blue) and WT1 antibody stained samples (red) with a ranged tool. The numbers above black line show the percentage of WT1+ cells in the gated population from (A). (C) Overlay histogram showing percentage of WT1+ cells in the gated population from (A).


