Abstract
The future of precision medicine is heavily reliant on the use of human tissues to identify the key determinants that account for differences between individuals with the same disorder. This need is exemplified by the neurofibromatosis type 1 (NF1) neurogenetic condition. As such, individuals with NF1 are born with a germline mutation in the NF1 gene, but may develop numerous distinct neurological problems, ranging from autism and attention deficit to brain and peripheral nerve sheath tumors. Coupled with accurate preclinical mouse models, the availability of NF1 patient-derived induced pluripotent stem cells (iPSCs) provides new opportunities to define the critical factors that underlie NF1-associated nervous system disease pathogenesis and progression. In this review, we discuss the generation and potential applications of iPSC technology to the study of NF1.
Keywords: NF1, neurodevelopment, neurofibroma, iPSC, stem cell, optic pathway glioma, retinal ganglion cell, microglia, tumor
1. Introduction
Neurofibromatosis type 1 (NF1) is a complex multisystem cancer predisposition syndrome, with a birth incidence between 1 in 2,500 and 1 in 3,000 individuals worldwide [1, 2]. The condition is caused by autosomal dominantly-inherited or de novo loss-of-function mutations in the NF1 gene located on chromosome 17q11.2 [3]. Affected individuals present with a wide range of clinical manifestations, including pigmentary abnormalities (café-au-lait macules, skinfold freckling, Lisch nodules), peripheral (neurofibromas, malignant peripheral nerve sheath tumors) and central (optic pathway and brainstem gliomas) nervous tumors, bone abnormalities, vasculopathy and other cancers [4]. In addition to these medical problems, over 80% of children with NF1 have learning disabilities, social perception deficits (autism spectrum disorder), and attention deficits [5].
While there has been enormous progress over the past 25 years since the identification of the NF1 gene in 1990, there are still a limited number of molecular targets for therapeutic drug design, and few of these molecularly-targeted therapies have been effective when evaluated in human clinical trials. In this regard, oral imatinib mesylate successfully reduced plexiform neurofibroma size and metabolic activity in a preclinical Nf1 mouse model [6], but resulted in variable reductions in tumor volume in a phase 2 study [7]. Lovastatin was also successful in a preclinical Nf1 mouse model by normalizing long-term potentiation (LTP) deficits and reversing spatial learning and attention impairments [8]. However, several randomized placebo-controlled lovastatin and simvastatin clinical trials produced no detectable improvements in measures of attention [9–13].
One potential reason for this apparent lack of preclinical translation is the inherent differences between rodents and humans. Although they share substantial genomic homology, there are significant dissimilarities to consider when using animal models to inform about human disorders. Anatomically, rodent brains are unlike human brains in that they are lissencephalic, meaning that their cerebral cortices do not undergo gyrification during development like their human counterparts [14]. In addition, cerebral progenitor zone complexity and organization differs between rodents and humans [15]. Furthermore, specific cell types, like microglia, exhibit striking interspecies differences in proliferation in vitro, immune system receptor expression and response to immune stimuli [16].
For these reasons, it would be desirable to complement Nf1 mouse models with preclinical experiments using actual human biospecimens. One such approach entails the use of patient-derived xenografts (PDX), in which patient tumor tissues are transferred into immunodeficient mice, allowing for preservation of tumor histology, genetic composition, and drug sensitivity. This platform has been highly successful for high-grade brain tumors, such as glioblastoma [17], but has been problematic for low-grade gliomas and neurofibromas due to premature senescence and low clonogenic frequencies. Another approach employs pathologic specimens, which maintain intact tissue architecture and gene expression patterns. However, the dynamic changes inherent in these tissues are reduced to a static image, and much of the information in these biospecimens regarding cell-cell interactions, stromal contributions, or the impact of germline genetics on disease development and progression is lost.
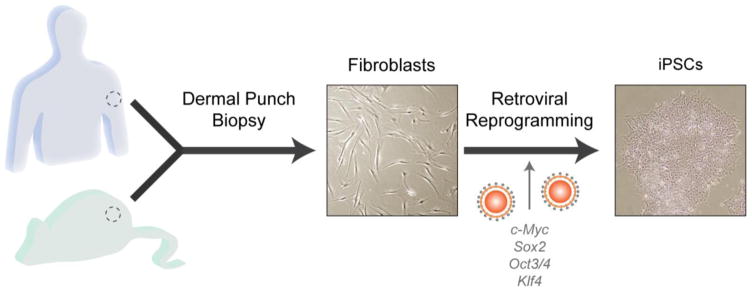
These limitations support the pressing need for an in vitro human system amenable to genetic engineering, as well as dynamic molecular and functional analyses. The discovery of somatic cell reprogramming to a pluripotent state by Shinya Takahashi and colleagues in 2006 [18] ushered in an era of in vitro human disease modeling. The work that Dr. Yamanaka received the Nobel Prize for in 2012 involved retroviral delivery of transcription factors Oct3/4, Sox2, c-Myc and Klf4 into mouse embryonic fibroblasts, generating induced pluripotent stem cells (iPSCs) with the capacity to differentiate into any cell type in the body (Fig. 1) [18]. Within the last ten years, refinements in reprogramming and differentiation techniques have resulted in the generation and application of human-derived iPSCs [19] to model complex genetic disorders, such as Rett syndrome [20], Fragile X syndrome [21], schizophrenia [22], and bipolar disorder [23]. In this review, we discuss the current capabilities of somatic cell reprogramming, iPSC differentiation and the potential of iPSC technology to provide multidimensional models of neurodevelopment and tumorigenesis in NF1. In addition, we will highlight potential applications of iPSC technology to therapeutic delivery and screening, as well as discuss the inherent limitations of this approach.
Fig. 1.
iPSCs can be generated from somatic cells by transcription factor-mediated reprogramming. Mouse fibroblasts and human dermal fibroblasts were originally reprogrammed by Shinya Takahashi and colleagues via retrovirus-mediated transfection of transcription factors Oct3/4, Sox2, c-Myc, and Klf4.
2. iPSC sources and reprogramming
Induced pluripotent stem cells, like most stem cells, are capable of generating more iPSCs (self-renewal), but also can give rise to cell types from any of the three germinal layers formed during embryogenesis (ectoderm, mesoderm, and endoderm). In this regard, they are similar to human embryonic stem cells (hESCs), but do not carry the ethical concerns associated with the use of embryos for hESC isolation. Importantly, iPSCs do not derive from embryonic tissues, and are instead generated by genetic reprogramming of non-germ cells (somatic cells).
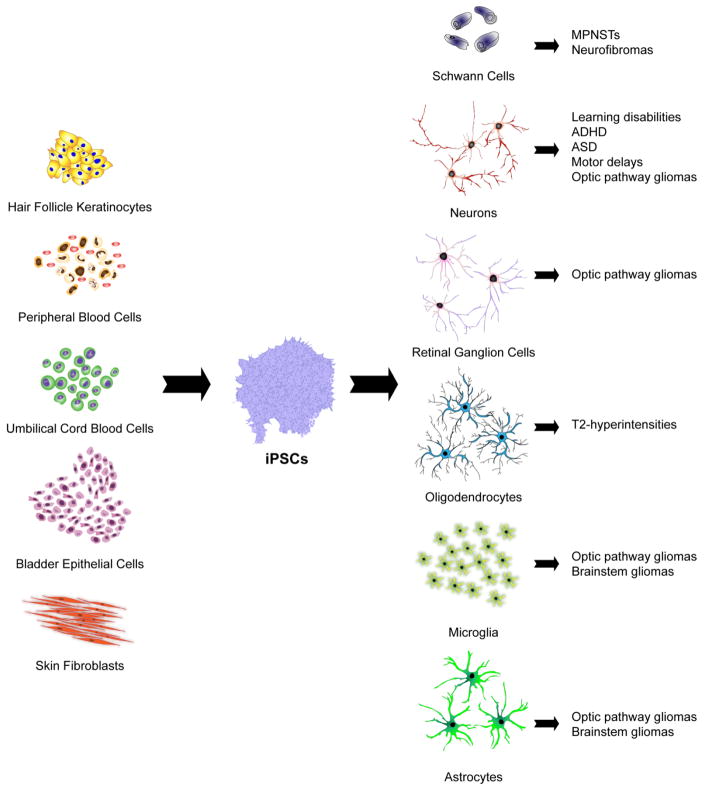
There are multiple somatic cell types that can be reprogrammed to generate iPSCs (Fig. 2), each with unique advantages and disadvantages. Dermal fibroblasts from skin punch biopsies were the first source of human-derived iPSCs [19, 24], and are the most frequently used cell type for reprogramming. iPSC sources have since been expanded to include stem cells from adult peripheral blood and umbilical cord blood collected after birth [25]. Exfoliated renal tubular epithelial cells isolated from urine [26] and keratinocytes from hair [27] are also viable reprogramming sources for the generation of iPSCs. There is currently no consensus regarding the ideal tissue from which to harvest cells for reprogramming, but the cell type of origin has been shown to affect programming efficiency [28, 29].
Fig. 2.
Patient-derived iPSCs can be generated by direct or indirect reprogramming of hair follicle keratinocytes, skin fibroblasts, urine-derived bladder epithelial cells, umbilical cord blood cells and peripheral blood cells. iPSCs can then be efficiently differentiated into the many distinct cell types that contribute to NF1 pathology using appropriate growth factors, cytokines, and extracellular matrix molecules. In this manner, iPSC-derived Schwann cells can be used to model malignant peripheral nerve sheath tumors (MPNSTs) and neurofibromas, while reprogrammed neurons can be employed to investigate the molecular etiologies of learning disabilities, autism spectrum disorder (ASD), attention-deficit-hyperactivity disorder (ADHD) and motor delays. Neurons, microglia and astrocytes comprise tumor microenvironments, and can enable modeling of optic pathway and brainstem gliomas, whereas reprogrammed retinal ganglion cells (RGCs) may be useful to evaluate the pathogenesis of optic glioma-associated RGC loss. Lastly, iPSC-derived oligodendrocytes can be employed to investigate the pathogenesis of the T2-weighted hyperintensities commonly found on magnetic resonance imaging (MRI) scans in young people with NF1.
For example, reprogramming of primary human keratinocytes using conventional retroviral transduction with OCT4, SOX2, KLF4 and MYC achieves 100-fold more efficient and 2-fold faster reprogramming compared to human fibroblasts [27]. Differences in efficiency and timing of reprogramming are likely attributable to more robust endogenous expression of stem cell-related genes in keratinocytes; however, the mechanisms underlying these differences have not been fully elucidated. Donor cells may also retain DNA methylation signatures of the original tissue after reprogramming, which predisposes to lineage-specific differentiation, suggesting that reprogramming of cells from the same germ layer as the desired differentiated cell population may be advantageous. As proof of concept, human beta cell-derived iPSCs have been shown to maintain an open chromatin structure at key beta cell genes, as well as an increased ability to differentiate into insulin-producing cells relative to ESCs and isogenic non-beta cell-derived iPSCs [30]. This retained epigenetic memory varies with the specific reprogramming method, and can be reset with chromatin modifying compounds in situations where donor cell epigenetic memory is disadvantageous for future differentiation [31]. Ultimately, the reprogramming method and efficiency, ease of obtaining cells, and differentiation capacity must all be considered when selecting a somatic cell source for reprogramming. For this reason, the incorporation of control, non-disease-bearing, normal cell lines matched to the somatic cell of origin is important for providing relevant comparators.
In addition, genome editing tools, like CRISPR/Cas9, allow for the correction or generation of specific genetic mutations on the same genomic background. Using this approach, germline NF1 gene mutations can introduced into the same iPSC line, allowing the original non-engineered line to serve as a control. Similarly, patient-derived iPSC lines can have the causative NF1 gene mutation corrected to a wild-type functional allele. In both cases, this method eliminates the genetic/genomic variation observed between different patients, and thus facilitates more accurate comparative analyses in situations where genomic differences may influence disease susceptibility or severity [32].
2.1 Indirect reprogramming methods
The first vectors used to generate iPSCs were gamma retroviruses, specifically murine leukemia retroviruses, chosen for their favorable long-term transgene expression (Fig. 3A) [18, 19]. Retroviral vectors consist of an RNA genome which is converted to DNA by virally-encoded reverse transcriptase once inside the target cell. The viral DNA is then integrated into the recipient genome during cell division, providing a persistent template for viral replication and gene expression. Replication-defective retroviruses engineered with insertion of transgenes in the place of viral protein-coding genes allow for delivery and integration of genes encoding reprogramming transcription factors. However, retroviruses are limited by their inability to transduce non-dividing and slow-growing cells, as well as the potential increased cancer risk associated with the integration of viral vectors near proto-oncogenes [33].
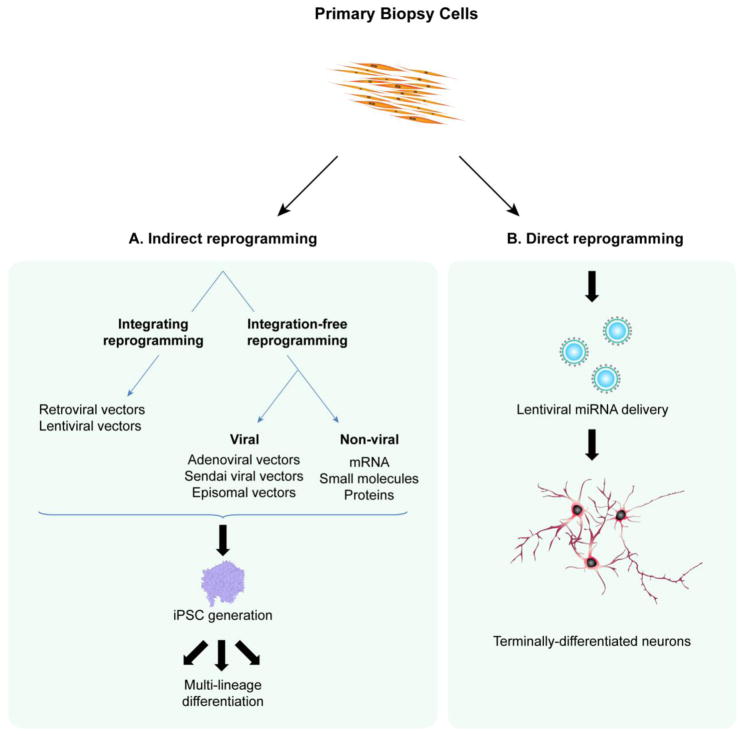
Fig. 3.
(A) Indirect reprogramming of human biopsy samples requires the generation of an intermediate pluripotent population and can leverage both transgenic and transgene-free methods. Retroviral and lentiviral vectors can be used for the delivery and integration of reprogramming factors, whereas adenoviral vectors, Sendai vectors, episomal vectors, mRNA, protein and small molecules utilize transgene-free cellular reprogramming approaches. (B) Direct reprogramming bypasses a pluripotent intermediate stage, and can produce post-mitotic neurons from terminally-differentiated somatic cells using lentiviral microRNA transduction.
Lentiviral vectors, a retroviral subclass, can transduce both non-dividing and rapidly-dividing cells with efficiencies comparable to other retroviruses, thus expanding potentially reprogrammable biomaterials to include cells with low division rates, like neurons and macrophages [34]. iPSC conversion with virally-delivered transgenes is a highly efficient reprogramming method [35], but transgene systems have inherent drawbacks: Insertional mutagenesis and transgene reactivation in iPSC-derived differentiated cells confers the capacity for malignant transformation, making iPSCs generated with these reprogramming technologies potentially unsafe for transplant therapies. Transplanted pluripotent stem cell-derived neural precursors can induce tumor formation in animal models, despite the removal of pluripotent cells prior to transplantation [36, 37]. To circumvent these risks, excisable lentiviral vectors have been developed using Cre/loxP technology [38], while transposon-derived vectors have emerged as another approach [39]. However, even excisable transgene technologies still pose a risk for non-specific recombination events and genomic instability during gene excision.
The clinical application of iPSCs to cell transplantation therapies necessitates the reduction of oncogenic potential and genetic instability associated with genome-integrating reprogramming methods. Adenoviral vectors are considered a non-integrative option for reprogramming factor delivery, with the caveat that they require extremely high viral titers for genomic integration [40]. Despite this limitation, adenoviral vectors have been used to successfully reprogram human fibroblasts [41]. Sendai viral vectors are another non-integrative reprogramming alternative [42–44]. Since these are RNA viruses with exclusively cytoplasmic replication, they do not integrate into the target cell genome. The derivation of iPSCs free of transgene sequences has also been achieved using episomal vectors to deliver reprogramming factors. In this regard, human fibroblasts have been successfully reprogrammed after a single transfection with oriP/EBNA1 (Epstein-Barr nuclear antigen-1)-based episomal vectors expressing OCT4, SOX2, MYC, KLF4, NANOG, LIN28 and Simian Virus 40 Large T (SV40LT) [45]. Alternative approaches have been reported, involving the repeated application of cell-penetrating peptide-anchored reprogramming proteins [46] and small molecule cocktails to replace Yamanaka reprogramming factors [47, 48]. These transgene-free methods are promising, but remain challenging due to low reprogramming efficiencies. Repeated administration of modified synthetic messenger RNA cocktails encoding Yamanaka factors plus LIN28 has also been employed to reprogram differentiated human cells to pluripotency without genomic integration. Conversion efficiencies were substantially superior using modified RNAs as compared to reprogramming with established retroviral protocols [49]. Future research would benefit from the optimization and standardization of reprogramming technologies capable of reproducibly and efficiently preserving iPSC genomic integrity.
2.2 Direct reprogramming methods
The discovery of iPSCs confirmed that somatic cells are not restricted to a particular differentiated state, thus inspiring the use of transdifferentiation methods as a complementary approach to the aforementioned dedifferentiation technique. The first direct lineage conversion of post-mitotic neurons from mouse embryonic fibroblasts (MEFs) and tail-tip fibroblasts (TTFs) was performed by Thomas Vierbuchen and colleagues [50]. Functional excitatory neurons were efficiently induced from MEFs and TTFs via lentiviral transduction of three transcription factors Achaete-scute family bHLH transcription factor 1 (Ascl1), POU class 3 homeobox 2 (Brn2), and myelin transcription factor 1 (Myt1l) identified from a pool of candidate genes with documented roles in neural development. The resulting induced neuronal cells expressed neuron-specific proteins and formed functional synapses, thus demonstrating that somatic lineage conversion can bypass a pluripotent intermediate and occur between lineages from different germ layers [50]. To confirm that the induced neuronal population was not derived from rare neural crest cell derivatives that potentially contaminate fibroblast cultures, the conversion of differentiated mouse hepatocytes into functional neurons was achieved using the same three factors [51]. Using lentiviral transduction of these same factors plus NEUROD1, functional neurons have also been induced from human fibroblasts [52].
Small noncoding RNA molecules, called microRNAs, have similarly been utilized to promote transdifferentiation of functional excitatory neurons from terminally-differentiated somatic cells (Fig. 3B). Since induction of the neural-progenitor-specific BAF (nBAF) chromatin-remodeling complex represents an essential switch in neuronal differentiation and function, microRNAs miR-9* and miR-124 have been used to repress BAF function and facilitate neuronal induction [53]. As such, human fibroblasts have been successfully converted to neurons by miR-9* and miR-124 lentiviral transduction, which is accelerated by the addition of the NEUROD2, ASCL1 and MYT1L transcription factors [54]. Human fibroblasts have also been reprogrammed into functional neurons using the combination of miR-124 and the MYT1L and BRN2 transcription factors [55]. In addition, small molecule strategies with no risk of transgene integration have also been used to directly reprogram human fibroblasts into neurons [56–58].
To date, various populations of neurons, including inhibitory, spinal motor, dopaminergic, and excitatory neurons, have been generated by direct conversion [59]. The direct reprogramming method is a promising alternative to methods involving an iPSC intermediate, and researchers have many advantages and limitations of each to consider before choosing an approach. In bypassing a pluripotent intermediate, the direct reprogramming method has the capacity to generate cell populations with a safer profile for transplantation therapies. However, without a pluripotent intermediate, the direct platform is limited in expandability, as direct conversion produces post-mitotic cells which cannot be expanded in culture like iPSCs. Direct reprogramming also preserves putative cellular aging markers, which could be advantageous in the study of late-onset diseases where age is a significant risk factor [60]. In this regard, neurons generated by direct conversion of fibroblasts from older adult mice demonstrate comparable levels of oxidative stress and DNA damage as the starting fibroblasts, and exhibit higher expression of age-related genes [61].
3. iPSCs for neurodevelopmental disorders in NF1
Cognitive impairments in individuals with NF1 are characterized by extensive clinical variability and can involve executive function, attention, memory, visuospatial and visuo-perceptual abilities, as well as emotionality, behavioral and social competence [4, 62]. Comorbid neurodevelopmental disorders, such as autism spectrum disorder (ASD) and attention-deficit-hyperactivity disorder (ADHD), have been reported with increased prevalence in the NF1 population [63], and NF1 has been clinically associated with an ASD phenotype of elevated symptom burden [64].
Given the absence of human experimental platforms for modeling the neurocognitive deficits in NF1, Nf1 mouse models have been employed to elucidate the potential structural and functional causes of these cognitive abnormalities. Mice with neuron-specific Nf1 loss have abnormal development of the cerebral cortex, with reduced cortical thickness and astrogliosis [65], while Cre-mediated Nf1 excision in neuroglial progenitors results in extensive astrogliosis and increased neuroglial progenitor proliferation [66, 67]. In addition, mice heterozygous for a mutation in the Nf1 gene (Nf1+/− mice) display increased GABA-mediated inhibition and deficits in LTP [68, 69], as well as reduced hippocampal dopamine levels, which each contribute to the observed behavioral abnormalities [70].
Despite the wealth of information imparted by Nf1 genetically-engineered mouse (GEM) models, it has been difficult to reconcile NF1 patient clinical findings with the pathophysiology exhibited by these models. For example, the assessments used to evaluate cognitive and behavioral phenotypes in rodents, such as the Morris Water Maze for spatial learning and memory, do not fully recapitulate the complexity of higher-order cognitive processes in humans. Consequently, normalization of cognitive dysfunction with drugs in these preclinical models has not served as reliable predictors of therapeutic efficacy in human clinical trials. In this respect, lovastatin, an HMG-CoA reductase inhibitor, reversed LTP deficits and impaired spatial learning in Nf1+/− mice, as evaluated by performance in the Morris water maze [8]. However, several randomized placebo-controlled lovastatin and simvastatin trials revealed no detectable improvement in measures of attention in children with NF1 [9–13]. Similarly, treatment of Nf1 mutant mice with methylphenidate (Ritalin) normalized striatal dopamine levels and improved the performance of mice on tests of attention-related behaviors and spatial learning [70, 71]. However, when examined in pediatric clinical trials, methylphenidate only showed short-term improvement in attention using a simplified Connors’ Parent Rating Scale, with no effect on learning and memory [72].
In addition, the specific patient germline NF1 gene mutation may also contribute to the phenotypic diversity observed in people with NF1 (see below), where it might be one factor underlying differences in autistic trait severity [64]. The number of GEM strains harboring patient-specific germline NF1 gene mutations is limited, and this underrepresentation of mutation heterogeneity may be another barrier to preclinical translation. The ability to engineer iPSCs representative of the spectrum of patient-specific mutations and differentiate them into virtually any cell type renders the iPSC platform a powerful technology to examine neurodevelopmental abnormalities in NF1 (Fig. 2).
3.1 iPSCs for modeling brain development
Cultures of homogenous human-derived cell populations are extremely useful for evaluating cell-specific molecular signaling pathways in disease, but these two-dimensional (2D) cultures do not permit a detailed examination of heterogeneous cell-cell interactions, patterning, and circuit abnormalities seen in the brains of patients with neurodevelopmental disorders. Recently, three-dimensional (3D) culture systems have been developed to support the evolution of complex, self-organizing cerebral tissues. These 3D structures, termed brain organoids, can be established from patient-derived iPSCs (Fig. 4), and replicate many aspects of cellular diversity, connectivity, brain and regional identity found in the developing human brain [73–75]. The first cerebral organoids were generated to model microcephaly [76], where skin fibroblasts from a patient with severe microcephaly and truncating CDK5RAP2 mutations were reprogrammed using lentiviral OCT4, SOX2, MYC and KLF4 delivery. Following aggregation into embryoid bodies and embedding in Matrigel, these cells were maintained under conditions permissive to neuroectodermal differentiation. Reverse transcriptase PCR (RT-PCR) and immunohistochemistry confirmed the presence of discrete brain regions, including midbrain, hindbrain, forebrain and hippocampal structures within the tissues, as well as the sophisticated organization of dorsal cortical regions. Moreover, the organoids derived from these patient iPSCs had premature neural differentiation and failure of radial glial stem cell expansion, demonstrating their ability to serve as an in vitro model of a human neurodevelopmental disorder which had been difficult to recapitulate in mice [76].
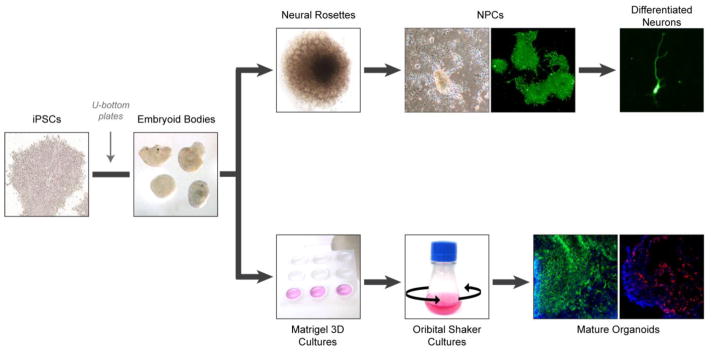
Fig. 4.
Schematic of systems for culturing patient-derived cerebral tissues. iPSCs are aggregated into embryoid bodies to allow for deposition of matrix and the formation of dense tissues. The generation of differentiated neurons involves the transfer of embryoid bodies to adhesive plates to allow for neural rosette formation. Neural rosettes are dissociated and differentiated into neural progenitor cells (NPCs), which can be further differentiated into various types of post-mitotic neurons. Representative cultured NPCs and post-mitotic neuron express TUJ1 (green). The generation of cerebral organoids involves the transfer of embryoid bodies to Matrigel, and then neural induction in stationary culture. After sufficient expansion of the resulting neuroepithelium, samples are transferred to a rotating suspension culture to promote nutrient diffusion and further organoid maturation. Mature organoids are subsequently collected for molecular and cellular analyses. Representative organoids express VIMENTIN (green) and FOXG1 (red). Nuclei are counterstained with DAPI (blue).
Cerebral organoids have since been used to recapitulate some of the intricacies of human progenitor zone organization, where they develop well-defined outer subventricular zone (oSVZ)-like layers containing specialized outer radial glia cell (oRGC)-like regions [77]. This progenitor zone expansion is significantly reduced in rodents, and is believed to be important for the increased size and complexity of the human cortex [78, 79]. Reproducibility and homogeneity of cerebral organoid generation has been optimized by the development of miniaturized spinning bioreactors and the pre-patterning of embryoid bodies to specific brain regions [77]. In this respect, telencephalic organoids have been developed from patient-derived iPSCs to model ASD. At the RNA level, organoids best reflect the expression patterns seen in the dorsal telencephalon during the first trimester of human fetal development, as well as altered GABA/glutamate levels, which was attributable to dysregulated FOXG1 expression in iPSC-derived organoids from ASD patients with macrocephaly [80]. Using this versatile technology, new insights into the pathogenic mechanisms underlying Zika virus exposure [77] and Alzheimer’s disease have emerged [81]. Further refinements in organoid generation may permit modeling of long-range functional connectivity, and ultimately identify potential convergence patterns of neuronal circuit dysfunction in complex disorders like NF1.
3.2 iPSCs for modeling patient-specific mutations
Learning and behavioral deficits vary greatly in phenotype and severity among individuals with NF1, and this clinical variability is thought to reflect, in some part, the specific patient germline NF1 gene mutation. In this regard, children with large NF1 locus microdeletions exhibit more severe clinical manifestations, such as dysmorphic facial features, developmental delay, and an elevated risk of malignant peripheral nerve sheath tumors (MPNSTs) relative to the general NF1 population [82]. We have recently generated the first collection of NF1 patient-derived iPSCs to examine the impact of the germline NF1 gene mutation on cognitive disability [83].
NF1 patient iPSCs were generated by Sendai virus reprogramming of primary fibroblasts and subsequently differentiated into neural progenitor cells (NPCs) (Fig. 4). All of the differentiated NPCs exhibited increased RAS activity and reduced cAMP levels relative to age and sex-matched controls. Interestingly, these changes in intracellular signaling pathway activity were independent of the levels of NF1 protein (neurofibromin) expression. In contrast, there was a striking positive correlation between neurofibromin expression and the levels of dopamine. This neurofibromin dose-dependent effect offers a potential explanation for the variability in cognitive disabilities detected among Nf1 genetically-engineered mice and the limited clinical success of therapeutics targeting dopamine homeostasis. Future research exploring genotype-phenotype correlations in NF1 would benefit greatly from the use of patient-derived iPSCs to further dissect the molecular repercussions of unique germline mutations on deregulated signaling pathways in specific human cell populations.
4. iPSCs for tumor modeling and drug screening
4.1 Brain tumor modeling
Optic pathway gliomas (OPGs) are seen in 15–20% of children with NF1, which are almost never biopsied as part of routine medical care [4]. NF1-OPG formation requires biallelic NF1 gene inactivation, in which the germline NF1 gene mutation is coupled with somatic loss of the one remaining functional NF1 allele in the presumed cell of origin for these tumors [84, 85].
Since these tumors are rarely biopsied, the majority of our understanding of NF1-OPG pathogenesis derives from the use of Nf1 GEM models. Mice heterozygous for a germline Nf1 gene mutation [86], in which somatic Nf1 loss occurs in neuroglial progenitors, develop optic gliomas [87]. Importantly, Nf1 loss in neuroglial progenitor cells alone does not result in gliomagenesis: Tumor formation requires the presence of cells heterozygous for an inactivating Nf1 gene mutation [66]. This finding suggests that Nf1+/− stromal cells are important participants in tumor formation and maintenance [88]. One of these non-neoplastic cell types in these tumors are microglia, immune system-like cells that mature within the developing brain [89]. Formal proof for the critical role for microglia in murine optic glioma formation and maintenance derives from studies in which pharmacologic or genetic inhibition of microglial function is sufficient to delay tumorigenesis and reduce tumor proliferation, respectively [90–92].
While these mouse tumors share many of the histologic and biologic features of their human counterparts, there are important differences. One difference relates to the level of microglial enrichment in gliomas, which is much smaller in mice (~10–15%) relative to human NF1-low grade gliomas (35–50%) [93]. Recent advances in iPSC reprogramming enable the generation of human microglia-like cells, with expression profiles similar to both primary fetal and adult microglia [94]. Future studies using in vitro co-cultures containing NF1-deficient human neuroglial progenitor cells in combination with NF1 patient-derived iPSC-microglia may reveal new targets for future stroma-directed low-grade glioma treatments.
4.2 Neurofibroma modeling
Neurofibromas are benign peripheral nerve sheath tumors (PNSTs) that are thought to arise from Schwann cell progenitors. NF1-deficient Schwann cells isolated and cultured from patient surgical specimens [95, 96] have been grown as explants in immunocompromised mice to model tumorigenesis [2, 32, 97, 98]. In addition, Nf1 GEM models of cutaneous and plexiform neurofibromas have been developed [99–101]. However, these first-generation models do not fully recapitulate many of the features of the intact human neurofibromas.
To develop potential future human neurofibroma models, an in vitro 3D skin raft culture system was recently established in mice, allowing for more detailed examination of neoplastic and non-neoplastic cell interactions [102]. In this approach, 3D skin rafts were constructed from collagen type I and dermal fibroblasts supplemented with Nf1+/− nerve tissue or non-nerve tissue controls. Nf1-deficient skin-derived precursors (SKPs) gave rise to tumors with many of the classic features of human plexiform neurofibromas [102].
In vitro tissue modeling is a scalable and easily manipulated technology. Given the limited proliferative capacity of primary neurofibroma tumor cells in vitro, it would be ideal to incorporate the disease relevant cell types derived from expandable patient iPSCs into in vitro tumor models. Schwann cells have been successfully generated from human-derived induced pluripotent stem cells [98, 103], and 3D human iPSC systems for neurofibroma modeling are currently under development by Dr. Lu Le (UT-Southwestern, personal communication). These systems will potentially allow for more detailed characterization of host-specific tumor pathophysiology and could serve as a tractable preclinical platform for drug development and screening.
5. iPSCs for neurorestorative therapy
Nearly half of children with NF1-OPG develop decreased visual acuity [104], which has been effectively modeled in Nf1 optic gliomas models [105–107]. However, mice rely more on smell than vision to navigate their environment [108], and the human retina and visual system is more complex than it is in mice [109, 110]. For this reason, future efforts devoted to therapies that aim to restore vision or prevent further declines in visual acuity might consider the use of reprogrammed pluripotent stem cells.
These reprogrammed cells could be used in one of two major ways. First, NF1 patient iPSC-derived retinal ganglion cells (RGCs) could serve as drug screening platforms. As proof of concept, RGCs derived from a patient with genetically-inherited glaucoma exhibited a significant reduction in caspase-3 activation, an apoptotic indicator, upon treatment with neuroprotective factors BDNF or PEDF [111]. High-throughput screening assays have also been developed for iPSC-derived retinal pigment epithelium (RPE) cultured in 96- and 384-well microtiter plates. These screening assays facilitate the monitoring of developmental, functional and disease-relevant gene expression in response to small molecule application, which could aid in the identification of therapeutic drugs for RPE-associated degenerative diseases in the future [112]. Second, pluripotent stem cell-derived RGCs could be used to replace degenerated retinal cells through autologous cell therapy. In proof-of-concept studies, human stem cell-derived RGCs were differentiated and transplanted into the retina by direct vitreous injection following mouse optic nerve injury. These transplanted RGCs were able to undergo terminal maturation, integrate into the retina, and lead to improvements in visual function [113, 114].
The use of autologously-transplanted retinal cells would require correction of the germline NF1 gene mutation prior to transplantation. Efficient site-specific gene editing technologies, such as TALENs [115], CRISPR/Cas9 [116], and zinc-finger nucleases (ZFNs) [117], have emerged as efficient methods to correct genetic defects. The correction of mutations in human iPSCs using gene editing technologies has already been reported for several diseases, including β-thalassemia [118], Duchenne muscular dystrophy [119], and Niemann-Pick Type C [120] and many more [121]. Moreover, these “repaired” iPSC-RGCs should not elicit an immune response when reintroduced, resulting in more successful engraftment.
6. Future directions
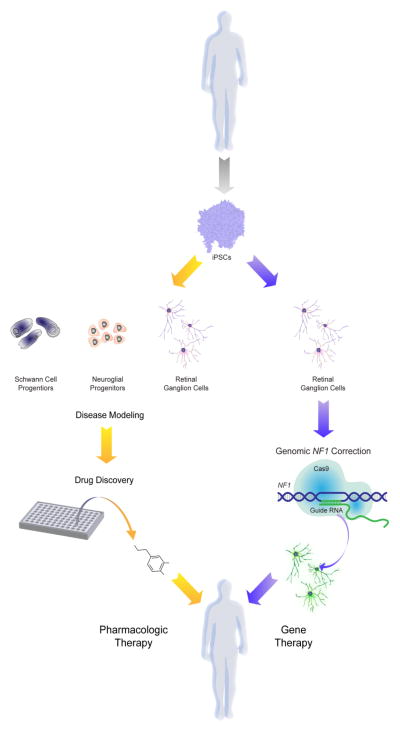
The availability of renewable sources of human cells has the potential to transform both research investigation and clinical practice for individuals with genetic disorders (Fig. 5). This is particularly relevant for disorders in which disease phenotypes involve organs and tissues that are not typically biopsied (e.g., brain) or where cells that underlie the particular medical problem cannot be easily propagated in culture. As such, patient-derived iPSCs are excellent complementary platforms, in conjunction with credentialed small-animal disease models, for studies aimed at disease modeling, drug discovery and cellular transplantation.
Fig. 5.
iPSCs can be differentiated into NF1-relevant cell types (neuroglial progenitors, Schwann cell progenitors and retinal ganglion cells) for disease modeling and drug discovery. In addition, CRISPR/Cas9 gene editing technology can be utilized to correct germline mutations in retinal ganglion cells for gene therapy in patients with visual impairment.
In addition, NF1 patient-derived iPSCs offer tools for precision medicine, in which investigations focused on specific germline NF1 gene mutations in the context of human genomic variation can be performed. These types of studies will provide important insights into the impact of specific germline patient mutations on NF1 disease pathogenesis [81], as well as the effect of genomic differences in people that potentially contribute to clinical variability. For example, only when using isogenic stem cells engineered to harbor a mutant LRRK2 allele did researchers find that reduced alpha-synuclein (αSYN) levels were observed in derivative neurons. In striking contrast, no difference in αSYN levels was detected between LRRK2-mutant Parkinson’s disease patient-derived lines and non-isogenic, healthy controls, which reflected the high variability of αSYN expression in healthy neurons [122]. The use of site-specific nucleases to correct mutations in NF1 patient-derived iPSCs or introduce known patient NF1 gene mutations into control (isogenic) lines represent important approaches to understanding the contributions of germline genetics and background genomics to disease pathogenesis and response to therapy.
Another potential future application of iPSC technology is in the area of risk assessment. As the number of NF1 patient-derived iPSCs grows, one could envision the creation of an international repository for high-throughput cellular and molecular phenotyping. The use of RNA-sequencing, immunophenotyping with cell type-specific antibodies, and functional characterization with electrophysiology offer unprecedented opportunities to discover subgroups of patients most likely to exhibit specific NF1 clinical features or respond to specific therapies. This “fingerprinting” approach might facilitate the identification of individuals at greatest risk for particular problems in NF1 (biomarkers), as well as the development of personalized drug development strategies.
Similarly, the refinement of 3D organoid cultures for specific NF1 clinical features will be valuable for dissecting the complex interplay between different cell types relevant to disease pathogenesis and progression. The clear co-dependency between neoplastic and non-neoplastic cells in the tumor microenvironment can be efficiently examined in human organoid cultures, and therapies that target the tumor ecosystem identified. Similarly, in the brain, where numerous neuronal populations contribute to learning and behavioral phenotypes in NF1, the use of “brain in a dish” approaches may reveal new targets for therapeutic drug design.
While the promise of iPSC technology shines bright, there remain unresolved issues. One significant challenge to the widespread application of iPSC technology involves the time investment and cost of establishing and differentiating patient-derived iPSC lines. Differentiation protocols for deriving various cell populations are numerous and often involve culturing in media supplemented with numerous, expensive growth factors, cytokines, and extracellular matrix molecules. Production of disease-relevant cell types in large quantities for drug screening and cell transplantation therapies necessitates standardization of practical protocols. In addition, genome instability of iPSCs during the reprogramming process [123], prolonged cell culture [124], or gene correction is another important concern, especially in populations destined for cell transplantation where genetic variations could lead to malignancy. Nonetheless, continual improvements in genome editing specificity [125], quality control and efficient reprogramming methodologies will undoubtedly provide new insights into the molecular and cellular etiologies of NF1, and accelerate the development and translation of personalized and targeted therapeutics for patients with this condition.
Highlight statements.
iPSCs provide a unique experimental platform for investigating the cellular and molecular etiologies of human neurodevelopmental abnormalities in NF1.
iPSCs enable mechanistic studies aimed at understanding the effects of patient-derived NF1 gene mutations and genomic variations on human nervous system cell dysfunction.
3-dimensional organoid systems incorporating patient iPSCs may serve as tractable human models of NF1-associated brain and nerve pathology.
iPSCs have the potential to enable improved preclinical drug identification and evaluation relevant to specific patients (personalized medicine), as well as serving as platforms for genome-edited cell transplantation therapy.
Acknowledgments
Financial support: D.H.G. is supported by a R35 grant from the National Institute of Neurological Disorders and Stroke. M.L.W. is supported by the MSTP Training Grant (5T32 GM007200). We also acknowledge and appreciate generous private support from the Walk Family, whose early investment in the Washington University NF Center iPSC initiative was critical.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lammert M, et al. Prevalence of neurofibromatosis 1 in German children at elementary school enrollment. Arch Dermatol. 2005;141(1):71–4. doi: 10.1001/archderm.141.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Evans DG, et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A. 2010;152A(2):327–32. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 3.Uusitalo E, et al. Neurofibromatosis type 1 gene mutation analysis using sequence capture and high-throughput sequencing. Acta Derm Venereol. 2014;94(6):663–6. doi: 10.2340/00015555-1843. [DOI] [PubMed] [Google Scholar]
- 4.Jett K, Friedman JM. Clinical and genetic aspects of neurofibromatosis 1. Genet Med. 2010;12(1):1–11. doi: 10.1097/GIM.0b013e3181bf15e3. [DOI] [PubMed] [Google Scholar]
- 5.Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65(7):1037–44. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- 6.Yang FC, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/−- and c-kit-dependent bone marrow. Cell. 2008;135(3):437–48. doi: 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson KA, et al. Imatinib mesylate for plexiform neurofibromas in patients with neurofibromatosis type 1: a phase 2 trial. Lancet Oncol. 2012;13(12):1218–24. doi: 10.1016/S1470-2045(12)70414-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15(21):1961–7. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 9.Bearden CE, et al. A randomized placebo-controlled lovastatin trial for neurobehavioral function in neurofibromatosis I. Ann Clin Transl Neurol. 2016;3(4):266–79. doi: 10.1002/acn3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krab LC, et al. Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: a randomized controlled trial. JAMA. 2008;300(3):287–94. doi: 10.1001/jama.300.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne JM, et al. Randomized placebo-controlled study of lovastatin in children with neurofibromatosis type 1. Neurology. 2016;87(24):2575–2584. doi: 10.1212/WNL.0000000000003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Vaart T, et al. Simvastatin for cognitive deficits and behavioural problems in patients with neurofibromatosis type 1 (NF1-SIMCODA): a randomised, placebo-controlled trial. Lancet Neurol. 2013;12(11):1076–83. doi: 10.1016/S1474-4422(13)70227-8. [DOI] [PubMed] [Google Scholar]
- 13.van der Vaart T, et al. Behavioral and cognitive outcomes for clinical trials in children with neurofibromatosis type 1. Neurology. 2016;86(2):154–60. doi: 10.1212/WNL.0000000000002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semple BD, et al. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;106–107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molnar Z, Vasistha NA, Garcia-Moreno F. Hanging by the tail: progenitor populations proliferate. Nat Neurosci. 2011;14(5):538–40. doi: 10.1038/nn.2817. [DOI] [PubMed] [Google Scholar]
- 16.Smith AM, Dragunow M. The human side of microglia. Trends Neurosci. 2014;37(3):125–35. doi: 10.1016/j.tins.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Joo KM, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3(1):260–73. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mor-Shaked H, Eiges R. Modeling Fragile X Syndrome Using Human Pluripotent Stem Cells. Genes (Basel) 2016;7(10) doi: 10.3390/genes7100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–5. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HM, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry. 2014;4:e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 25.Loh YH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113(22):5476–9. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou T, et al. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol. 2011;22(7):1221–8. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26(11):1276–84. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 28.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(12):1117–9. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3(3):340–5. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar-Nur O, et al. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9(1):17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HS, et al. Genomic editing tools to model human diseases with isogenic pluripotent stem cells. Stem Cells Dev. 2014;23(22):2673–86. doi: 10.1089/scd.2014.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kvaratskhelia M, et al. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014;42(16):10209–25. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu K. All roads lead to induced pluripotent stem cells: the technologies of iPSC generation. Stem Cells Dev. 2014;23(12):1285–300. doi: 10.1089/scd.2013.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayart E, Cohen-Haguenauer O. Technological overview of iPS induction from human adult somatic cells. Curr Gene Ther. 2013;13(2):73–92. doi: 10.2174/1566523211313020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nori S, et al. Long-term safety issues of iPSC-based cell therapy in a spinal cord injury model: oncogenic transformation with epithelial-mesenchymal transition. Stem Cell Reports. 2015;4(3):360–73. doi: 10.1016/j.stemcr.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DR, et al. PSA-NCAM-negative neural crest cells emerging during neural induction of pluripotent stem cells cause mesodermal tumors and unwanted grafts. Stem Cell Reports. 2015;4(5):821–34. doi: 10.1016/j.stemcr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soldner F, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136(5):964–77. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–70. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harui A, et al. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73(7):6141–6. doi: 10.1128/jvi.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27(11):2667–74. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- 42.Ban H, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108(34):14234–9. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fusaki N, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki T, Yuasa S, Fukuda K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc. 2012;7(4):718–28. doi: 10.1038/nprot.2012.015. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–6. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin T, Wu S. Reprogramming with Small Molecules instead of Exogenous Transcription Factors. Stem Cells Int. 2015;2015:794632. doi: 10.1155/2015/794632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7(6):651–5. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marro S, et al. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9(4):374–82. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476(7359):220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoo AS, et al. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–6. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ambasudhan R, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9(2):113–8. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu W, et al. Direct Conversion of Normal and Alzheimer’s Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell. 2015;17(2):204–12. doi: 10.1016/j.stem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 57.Dai P, Harada Y, Takamatsu T. Highly efficient direct conversion of human fibroblasts to neuronal cells by chemical compounds. J Clin Biochem Nutr. 2015;56(3):166–70. doi: 10.3164/jcbn.15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu ML, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanabe K, Haag D, Wernig M. Direct somatic lineage conversion. Philos Trans R Soc Lond B Biol Sci. 2015;370(1680):20140368. doi: 10.1098/rstb.2014.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mertens J, et al. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat Rev Neurosci. 2016;17(7):424–37. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y, et al. Enhanced Rejuvenation in Induced Pluripotent Stem Cell-Derived Neurons Compared with Directly Converted Neurons from an Aged Mouse. Stem Cells Dev. 2015;24(23):2767–77. doi: 10.1089/scd.2015.0137. [DOI] [PubMed] [Google Scholar]
- 62.Payne JM, et al. Theory of mind in children with Neurofibromatosis Type 1. Neuropsychology. 2016;30(4):439–48. doi: 10.1037/neu0000262. [DOI] [PubMed] [Google Scholar]
- 63.Garg S, et al. Autism and other psychiatric comorbidity in neurofibromatosis type 1: evidence from a population-based study. Dev Med Child Neurol. 2013;55(2):139–45. doi: 10.1111/dmcn.12043. [DOI] [PubMed] [Google Scholar]
- 64.Morris SM, et al. Disease Burden and Symptom Structure of Autism in Neurofibromatosis Type 1: A Study of the International NF1-ASD Consortium Team (INFACT) JAMA Psychiatry. 2016 doi: 10.1001/jamapsychiatry.2016.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15(7):859–76. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Y, et al. Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005;132(24):5577–88. doi: 10.1242/dev.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hegedus B, et al. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1(4):443–57. doi: 10.1016/j.stem.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 68.Costa RM, et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415(6871):526–30. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 69.Cui Y, et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135(3):549–60. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diggs-Andrews KA, et al. Dopamine deficiency underlies learning deficits in neurofibromatosis-1 mice. Ann Neurol. 2013;73(2):309–15. doi: 10.1002/ana.23793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown JA, et al. Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis-1 mutant mice. Hum Mol Genet. 2010;19(22):4515–28. doi: 10.1093/hmg/ddq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lion-Francois L, et al. The effect of methylphenidate on neurofibromatosis type 1: a randomised, double-blind, placebo-controlled, crossover trial. Orphanet J Rare Dis. 2014;9:142. doi: 10.1186/s13023-014-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huch M, Koo BK. Modeling mouse and human development using organoid cultures. Development. 2015;142(18):3113–25. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 74.Quadrato G, Brown J, Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med. 2016;22(11):1220–1228. doi: 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- 75.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–40. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qian X, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165(5):1238–54. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31(10):3683–95. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mariani J, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162(2):375–90. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raja WK, et al. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS One. 2016;11(9):e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mautner VF, et al. Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J Med Genet. 2010;47(9):623–30. doi: 10.1136/jmg.2009.075937. [DOI] [PubMed] [Google Scholar]
- 83.Anastasaki C, et al. Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum Mol Genet. 2015;24(12):3518–28. doi: 10.1093/hmg/ddv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gutmann DH, et al. Molecular analysis of astrocytomas presenting after age 10 in individuals with NF1. Neurology. 2003;61(10):1397–400. doi: 10.1212/wnl.61.10.1397. [DOI] [PubMed] [Google Scholar]
- 85.Gutmann DH, et al. Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res. 2013;23(3):431–9. doi: 10.1101/gr.142604.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gutmann DH, et al. Haploinsufficiency for the neurofibromatosis 1 (NF1) tumor suppressor results in increased astrocyte proliferation. Oncogene. 1999;18(31):4450–9. doi: 10.1038/sj.onc.1202829. [DOI] [PubMed] [Google Scholar]
- 87.Bajenaru ML, et al. Astrocyte-specific inactivation of the neurofibromatosis 1 gene (NF1) is insufficient for astrocytoma formation. Mol Cell Biol. 2002;22(14):5100–13. doi: 10.1128/MCB.22.14.5100-5113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bajenaru ML, et al. Natural history of neurofibromatosis 1-associated optic nerve glioma in mice. Ann Neurol. 2005;57(1):119–27. doi: 10.1002/ana.20337. [DOI] [PubMed] [Google Scholar]
- 89.Ginhoux F, et al. Origin and differentiation of microglia. Front Cell Neurosci. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daginakatte GC, Gutmann DH. Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth. Hum Mol Genet. 2007;16(9):1098–112. doi: 10.1093/hmg/ddm059. [DOI] [PubMed] [Google Scholar]
- 91.Pong WW, et al. Reduced microglial CX3CR1 expression delays neurofibromatosis-1 glioma formation. Ann Neurol. 2013;73(2):303–8. doi: 10.1002/ana.23813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Solga AC, et al. RNA Sequencing of Tumor-Associated Microglia Reveals Ccl5 as a Stromal Chemokine Critical for Neurofibromatosis-1 Glioma Growth. Neoplasia. 2015;17(10):776–88. doi: 10.1016/j.neo.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simmons GW, et al. Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth. J Neuropathol Exp Neurol. 2011;70(1):51–62. doi: 10.1097/NEN.0b013e3182032d37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22(11):1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wallace MR, et al. Culture of cytogenetically abnormal schwann cells from benign and malignant NF1 tumors. Genes Chromosomes Cancer. 2000;27(2):117–23. [PubMed] [Google Scholar]
- 96.Li H, et al. Immortalization of human normal and NF1 neurofibroma Schwann cells. Lab Invest. 2016;96(10):1105–15. doi: 10.1038/labinvest.2016.88. [DOI] [PubMed] [Google Scholar]
- 97.Li H, et al. Analysis of steroid hormone effects on xenografted human NF1 tumor schwann cells. Cancer Biol Ther. 2010;10(8):758–64. doi: 10.4161/cbt.10.8.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang A, et al. Induced pluripotent stem cells for neural tissue engineering. Biomaterials. 2011;32(22):5023–32. doi: 10.1016/j.biomaterials.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu Y, et al. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296(5569):920–2. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Z, et al. Cells of origin in the embryonic nerve roots for NF1-associated plexiform neurofibroma. Cancer Cell. 2014;26(5):695–706. doi: 10.1016/j.ccell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Le LQ, et al. Susceptible stages in Schwann cells for NF1-associated plexiform neurofibroma development. Cancer Res. 2011;71(13):4686–95. doi: 10.1158/0008-5472.CAN-10-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liao CP, et al. The role of nerve microenvironment for neurofibroma development. Oncotarget. 2016 doi: 10.18632/oncotarget.11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kreitzer FR, et al. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am J Stem Cells. 2013;2(2):119–31. [PMC free article] [PubMed] [Google Scholar]
- 104.Listernick R, et al. Optic pathway gliomas in children with neurofibromatosis 1: consensus statement from the NF1 Optic Pathway Glioma Task Force. Ann Neurol. 1997;41(2):143–9. doi: 10.1002/ana.410410204. [DOI] [PubMed] [Google Scholar]
- 105.Hegedus B, et al. Optic nerve dysfunction in a mouse model of neurofibromatosis-1 optic glioma. J Neuropathol Exp Neurol. 2009;68(5):542–51. doi: 10.1097/NEN.0b013e3181a3240b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim KY, et al. Ultrastructural characterization of the optic pathway in a mouse model of neurofibromatosis-1 optic glioma. Neuroscience. 2010;170(1):178–88. doi: 10.1016/j.neuroscience.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brown JA, Gianino SM, Gutmann DH. Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J Neurosci. 2010;30(16):5579–89. doi: 10.1523/JNEUROSCI.3994-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Latham N, Mason G. From house mouse to mouse house: the behavioural biology of free-living Mus musculus and its implications in the laboratory. Applied Animal Behavior Science. 2004;86(3–4):261–289. [Google Scholar]
- 109.Bennis A, et al. Comparison of Mouse and Human Retinal Pigment Epithelium Gene Expression Profiles: Potential Implications for Age-Related Macular Degeneration. PLoS One. 2015;10(10):e0141597. doi: 10.1371/journal.pone.0141597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Volland S, et al. A comparison of some organizational characteristics of the mouse central retina and the human macula. PLoS One. 2015;10(4):e0125631. doi: 10.1371/journal.pone.0125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohlemacher SK, et al. Stepwise Differentiation of Retinal Ganglion Cells from Human Pluripotent Stem Cells Enables Analysis of Glaucomatous Neurodegeneration. Stem Cells. 2016;34(6):1553–62. doi: 10.1002/stem.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ferrer M, et al. A multiplex high-throughput gene expression assay to simultaneously detect disease and functional markers in induced pluripotent stem cell-derived retinal pigment epithelium. Stem Cells Transl Med. 2014;3(8):911–22. doi: 10.5966/sctm.2013-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Satarian L, et al. Engrafted human induced pluripotent stem cell-derived anterior specified neural progenitors protect the rat crushed optic nerve. PLoS One. 2013;8(8):e71855. doi: 10.1371/journal.pone.0071855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hambright D, et al. Long-term survival and differentiation of retinal neurons derived from human embryonic stem cell lines in un-immunosuppressed mouse retina. Mol Vis. 2012;18:920–36. [PMC free article] [PubMed] [Google Scholar]
- 115.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 118.Xie F, et al. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24(9):1526–33. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li HL, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4(1):143–54. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maetzel D, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports. 2014;2(6):866–80. doi: 10.1016/j.stemcr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Merkert S, Martin U. Site-Specific Genome Engineering in Human Pluripotent Stem Cells. Int J Mol Sci. 2016;17(7) doi: 10.3390/ijms17071000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reinhardt P, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12(3):354–67. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 123.Sugiura M, et al. Induced pluripotent stem cell generation-associated point mutations arise during the initial stages of the conversion of these cells. Stem Cell Reports. 2014;2(1):52–63. doi: 10.1016/j.stemcr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471(7336):63–7. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]