Abstract
Aging-related brain diseases consist of a number of important neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Huntington’s diseases, all of which have become more prevalent as the life expectancy of humans is prolonged. Age-dependent brain disorders are associated with both environmental insults and genetic mutations. For those brain disorders that are inherited, gene editing is an important tool for establishing animal models to investigate the pathogenesis of disease and identify effective treatments. Here we focus on the tools for gene editing, especially CRISPR/Cas9, and discuss their application for generating animal models that can recapitulate the brain pathology seen in human diseases. We also highlight the advantages and disadvantages of establishing genetically modified animal models. Finally, we discuss recent findings to resolve technical issues related to the use of CRISPR/Cas9 for generating animal models of brain diseases.
Keywords: CRISPR/Cas9, gene editing, neurodegeneration, animal models
Introduction
A number of important brain disorders are associated with aging, and their prevalence continues to climb as the life expectancy of humans is extended. These brain disorders include Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), Huntington’s disease (HD), and frontotemporal dementia (FTD). The common features of these neurological diseases are late-onset neurological symptoms and degeneration that preferentially affects neuronal cells in the brain. Although most cases of neurodegenerative diseases are sporadic, a number of genetic mutations are known to cause age-dependent neurodegeneration. For example, In HD, an expanded CAG repeat (>36 repeats) in the huntingtin gene causes neurodegeneration in an autosomal dominant manner (Gusella et al., 1993). In AD, mutations in the genes for presenilin, amyloid precursor protein (APP), or tau contribute critically to selective neurodegeneration (Hutton et al., 1997; Kang et al. 1987; Alzheimer et al. 1995). In PD, mutations in alpha-synuclein or Parkin and PINK1 can cause autosomal dominant or recessive neurodegeneration (Polymeropoulos et al., 1997; Dodson et al., 2007). Familial ALS can be caused by mutations in one of at least 32 known genetic loci, including superoxide dismutase 1 (SOD1), TAR DNA-binding protein 43 (TDP-43), fused in sarcoma (FUS), and C9ORF72 (Joyce et al., 2011; Neumann M, et al., 2006; DeJesus-Hernandez, et al., 2011). Identification of the genetic mutations in these diseases has enabled the generation of a variety of animal models via genome modifications.
Although genetically modified animal models have been widely used to investigate the mechanisms of different brain disorders and to develop therapeutic strategies, the ways to produce animal models determine the nature of the disease models and their utility for different purposes. In general, genetically modified animals models can be classified as transgenic or gene-targeted animal models. In transgenic models, the transgene is randomly inserted into the genome as an exogenous gene and is expressed under the exogenous promoter. In gene-targeted animal models, the specific locus or region of an endogenous gene is altered to mimic a specific genetic mutation in humans. Obviously, gene-targeted (knock-in or knockout) animal models are intended to recapitulate the same genetic defects seen in human beings and are expected to more faithfully replicate patient pathology. In this review, we focus on genomic editing tools that enable the generation of animal models of brain diseases by modifying endogenous genomes.
1. Genomic editing tools
The classic method for manipulating the endogenous genome involves use of a gene targeting vector to replace a specific region of endogenous genome via homologous recombination. This method relies on embryonic stem (ES) cells that allow for gene targeting in vitro, and then transplantation in vivo for differentiation into different types of cell. Because mouse ES cells are available, this approach has been widely used to generate germline knockout or knock-in mouse models. However, gene targeting via ES cells is both time consuming and costly. Moreover, lack of appropriate ES cells from other species prevents the use of knock-in and knockout approaches to generate disease models of different species, especially large animals including non-human primates. Thus, although precise manipulation of the genome is very important for generating animal models that can faithfully recapitulate genetic mutations and phenotypes of human diseases, it has been very difficult to generate large animal models that carry the same genetic mutations in the endogenous genes to mimic genetic defects in humans.
Recently, new approaches have emerged that allow investigators to manipulate virtually any gene in any cell type or organism. The crucial technology, referred to as “genome editing,” is based on the use of engineered nucleases that can first target DNA, and then cut the targeted DNA. These nucleases, which include zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR), have the same capacity to induce double-strand breaks (DSBs) on the targeted DNA; these DSBs then stimulate the cellular DNA repair mechanisms, including error-prone non-homologous end joining (NHEJ) and homology-directed repair (HDR). The versatility of ZFNs, TALENs, and CRISPR/Cas9 arises from their ability to bind DNA in a sequence-dependent manner. These nucleases, especially CRISPR/Cas9, have greatly accelerated the creation of many new disease models, from small animals to non-human primates (Tu et al., 2015; Yang et al., 2016). These animal models have proved that CRISPR/Cas9 is an efficient tool to directly modify or repair genomic DNA in somatic and germline cells, without the need to establish embryonic stem cells for genomic manipulation. Gao et al found that Natronobacterium gregoryi Argonaute (NgAgo), can modify genome by DNA-guided in mammalian cells (Gao et al., 2016), however, the efficiency and application still need to be verification.
2. Development of CRISPR/Cas9 technology
The CRISPR/Cas9 system is derived from a prokaryotic RNA-guided defense system. CRISPR repeats were first discovered as an unusual repeat site located downstream of the iap gene in Escherichia coli (Ishino et al., 1987). There are also other repeat elements in the genomes of different bacterial and archaeal strains. By 2002, these repeat elements were named CRISPR, and CRISPR-associated (Cas) genes were found to be well conserved and adjacent to the repeat elements (Jansen et al., 2002). CRISPR and Cas proteins are able to use small crRNA molecules to target and destroy the DNAs or RNAs of invading viruses and plasmids (Barrangou et al., 2007; Brouns et al., 2008; Jansen et al., 2002; Karginov and Hannon, 2010). The type II prokaryotic CRISPR/Cas9 from Streptococcus pyogenes (SpCas9) is one of three distinct types of CRISPR/Cas immune systems. There are at least three components (Cas9, the CRISPR RNA, and trans-activating crRNA) that are essential for reconstituting the type II CRISPR nuclease system. The CRISPR/Cas9 system is equipped with two cleavage nuclease domains: the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like nuclease domain cleaves the non-complementary strand. Thus, the nuclease Cas9 can cut both strands of DNA precisely, and the damaged DNA is repaired via NHEJ or HDR, thereby resulting in gene disruptions and inactivation of the targeted gene. Since these initial studies, Cas9 has been used widely for genome editing applications in a variety of experimental model systems (Jinek et al., 2012; Mali, Esvelt, & Church, 2013).
Of many variants of the CRISPR/Cas9 system, the most commonly used one consists of a small Cas9 (1,368-residues) protein from S. pyogenes (SpCas9) (Haft et al., 2005). A smaller form from Staphylococcus aureus (Sa) Cas9 analog (SaCas9) was identified later, which requires a more-complex protospacer adjacent motif (PAM) of NNGRRT for gene targeting (Ran et al., 2015; Friedland et al., 2015). More recently, an additional Cas9 nuclease (Cpf1, CRISPR from Prevotella and Francisella 1) was uncovered and found to be a single-RNA-guided nuclease that can be turned into nCas9 to generate single-strand breaks (SSBs) (Zetsche et al., 2015). Compared with other genome editing tools, such as ZFN and TALEN, the Cas9 system works based on DNA-RNA, but not DNA-protein, recognition and is easier and simpler to design and generate.
3. Delivering CRISPR/Cas9 into the brain
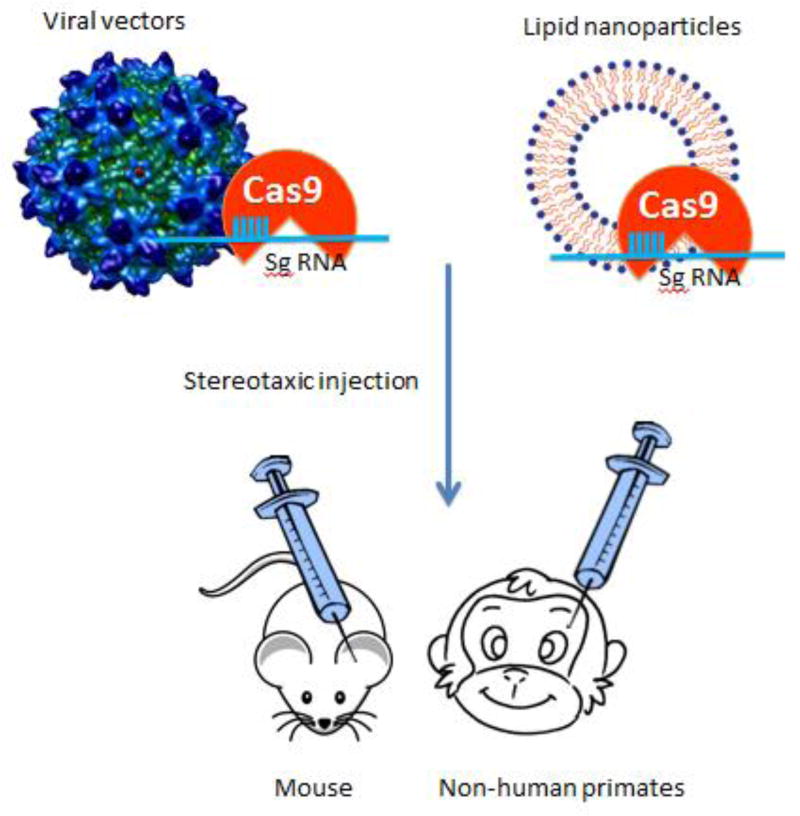
Because of its ability to modify genomes in any type of cell, CRISPR/Cas9 has been used to modify genes in neuronal cells in the brain. Delivering genetic modifiers into the brain is difficult, however. Viral vectors are one suitable choice for delivering a variety of transgenes, including CRISPR/Cas9 (Figure 1). Lentiviruses can infect non-dividing cells, and their packaging limit is 8.5 kb, which is sufficient to package most Cas9 genes, guide RNA, and specific promoters (Kumar et al., 2001). Lentiviral Cas9 has been delivered successfully into mice to control the progression of lung cancer (Sánchez-Rivera et al., 2014). Adenoviruses can infect both replicating and non-replicating cells but also elicit a strong immune response (Wang et al., 2004). Adeno-associated viruses (AAVs) are the most attractive gene delivery vectors as they do not lead to genomic integration and also show a safe non-immune response. AAV has been approved by the Food and Drug Administration (FDA) for use in human clinical trials. Smaller Cas9 orthologs, such as spCas9 (4.2kb) or saCas9 (3.2 kb), are more attractive because they can be packaged in a single AAV vector for in vivo gene editing in the brain (Ran et al., 2015). By injecting viral vectors expressing CRISPR/Cas9 in the mouse brain, several groups have successfully modified specific genes in neurons (Incontro et al., 2014; Platt et al., 2014; Straub et al., 2014; Heidenreich et al., 2016; Walters et al., 2016).
Figure 1. Direct expression of CRISPR/Cas9 in the brain.
Viral vectors (lentiviral, adenoviral, and AAV) or lipid-nanoparticles can be used to deliver CRISPR/Cas9 into a specific brain region in animals (e.g., mouse, non-human primates, and other species) via stereotaxic injection, resulting in gene editing in specific brain regions.
Compared with viral vectors, non-viral vectors possess low immunogenicity and do not lead to genomic DNA integration. The non-viral delivery methods use polyethylenimine (PEI), bioreducible lipid, liposome-mediated transfection, and in utero electroporation. Wang and colleagues used bioreducible lipid-packaged Cas9 protein and directly delivered it into the mouse brain for treatment of neurological disorders (Figure 2). They also directly injected the bioreducible lipid-packaged (−27) GFP-Cre protein into the mouse brain to successfully achieve Cre-mediated gene recombination (Wang et al., 2016). Hydrodynamic injection (HDI) of plasmid DNA is convenient, and investigators have directly injected DNA plasmids encoding Cas9 and sgRNA into the livers of mice to inhibit the hepatitis B virus (HBV) (Ramanan et al., 2015). Liposome Cas9/gRNAs complexes can also be delivered into the genome in mouse inner ear hair cells and neurons (Zuris et al., 2015; Wang et al., 2016). Whether these non-viral systems can be used to directly deliver CRISPR/Cas9 into the brain, however, remains to be investigated. Also, these non-viral delivery systems may not be able to produce stable expression, and the safety of injecting them into the brain remains unclear.
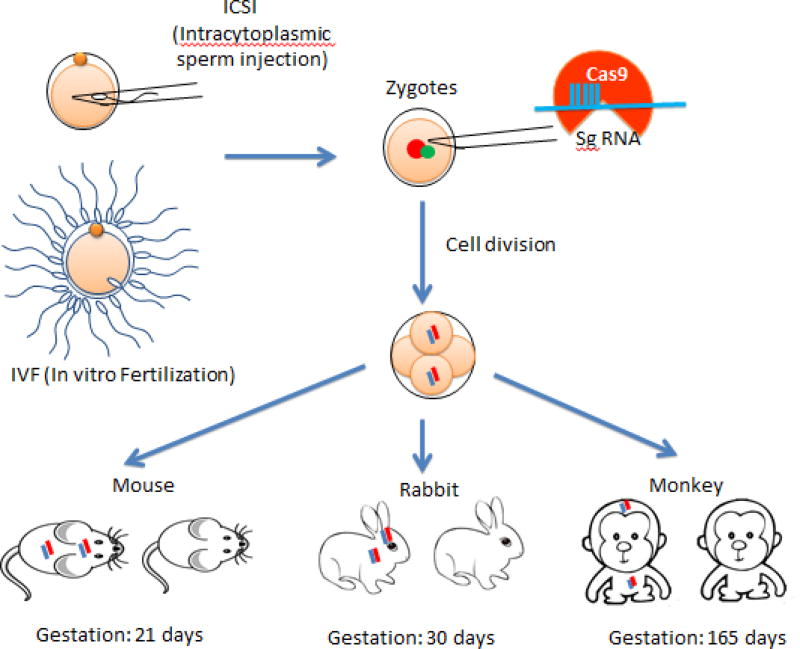
Figure 2. Expression of CRISPR/Cas9 in zygotes.
CRISPR/Cas9 can be injected directly into fertilized eggs to target specific genes, resulting in genetically modified animals, including mice, rabbits, and non-human primates, which have modified genomes throughout the whole body, including the brain. Color lines represent modified genes.
Another approach is expressing CRISPR/Cas9 in zygotes to generate live animals that will express CRISPR/Cas9 in their brain tissues. For this approach, Cas9 mRNA and sgRNA are injected into zygotes to modify genomes in early embryos. The resulting animals carry the genetically modified genes in different types of cells, including neuronal cells in the brain, derived from the early embryos. This approach has been used successfully to generate small animal models, including zebrafish (Hruscha et al., 2013; Hwang et al., 2013a; Hsu et al., 2013), flies (Bassett et al., 2013), frogs (Nakayama et al., 2013), mice (Wang et al., 2013; Li et al., 2013a), rats (Li et al., 2013b), and rabbits (Lv et al., 2016; Song et al., 2016). Since the genetic mutations created occur at the one-cell stage or early embryos, this approach is believed to generate genetic mutations similar to those genetic defects in humans.
The power of CRISPR/Cas9 is its ability to target any gene in any type of cell, including early embryos. This allows the use of CRISPR/Cas9 to directly modify genomes in embryos of large animals whose germline transmission of modified genes requires considerably prolonged breeding times. Although small animal models provide valuable tools for investigating neurological disorders, their brain anatomy, circuitry, aging processes, and behaviors are quite different from those in humans. Thus, the use of large animal models to better understand brain disorders is imperative. Via injection of CRISPR/Cas9 into animal zygotes, genetically modified larger animals, including dogs (Zou et al., 2015), pigs (Tang et al., 2014), and non-human primates (Niu et al., 2014; Chen et al., 2015), have been recently established. The genetically modified non-human primates are especially valuable to help us understand the higher cognitive functions and provide important animal models for investigating brain diseases with psychiatric symptoms.
4. Challenges in using CRISPR/Cas9
CRISPR/Cas9 can target to virtually any gene in a sequence-dependent manner, and its targeting efficiency is higher than other gene targeting approaches. However, because the genome editing by CRISPR/Cas9 relies on approximately 23 base pair matches (Hsu et al., 2014), off-target effects are considered an important issue. Many reports have focused on reducing these effects when using CRISPR/Cas9. Research has shown that Cas9 can tolerate mismatches, depending on their distribution and number (Hsu et al., 2013; Mali et al., 2013; Fu et al., 2014). Reducing Cas9 concentration and using specific gRNAs can also minimize off-target effects and improve gene targeting specificity (Hsu et al., 2013).
The second issue with CRISPR/Cas9 is mosaic mutations, which are different types of mutations in different types of cells in the same animals. For small animals with short breeding times, mosaic mutations can be diluted over generations, and specific mutations in DNA can be obtained by crossing animals for 4–5 generations. In contrast, for large animals like non-human primates, whose sexual maturation requires 4–5 years, mosaic mutations pose an obstacle to obtaining genetically modified animals that can faithfully mimic genetic mutations in human patients. More importantly, mosaicism can significantly affect the precision of gene therapy when using CRISPR/Cas9 technology. The mechanisms behind mosaic mutations remain unclear, but CRISPR/Cas9-mediated mosaic mutations likely result from random DNA breaks and repairs. Thus, reducing this random DNA cutting and repair is key to preventing the mosaicism in gene targeting by CRISPR/Cas9.
Another challenge facing CRISPR/Cas9 is the low rate of homologous recombination, which is important for gene replacement or knock-in with specific sequences. Gene replacement is the basis for gene therapy to correct genetic defects in human diseases. The nuclease Cas9 cuts both strands of DNA, and the damaged DNA is repaired via NHEJ or HDR. Generally, HDR takes place in the synthesis (S) and the premitotic (G2) phases (Heyer et al., 2010), whereas NHEJ occurs in the growth 1 (G1) and the mitotic (M) phases (Daley and Sung, 2014). It turns out that CRISPR/Cas9-mediated indel mutations via NHEJ have high efficiency, and the HDR rate is relatively low. Suppression of NHEJ key molecules is found to increase the HDR rate by CRISPR/Cas9 (Chu et al., 2015; Maruyama et al., 2015). Thus, increasing the HDR rate is expected to improve the efficiency of CRISPR/Cas9-mediated gene replacement.
5. Strategies to improve CRISPR/Cas9 targeting
As described, genetically modifying brain genomes in animals to model brain diseases can be achieved with CRISPR/Cas9. Direct injection of viral vectors expressing CRISPR/Cas9 into selective brain regions can rapidly generate animal models to mimic specific neuropathology in these brain areas. Expression of CRISPR/Cas9 in zygotes to target specific genes can generate animals that mimic genetic defects in the human brain and other organs. All these strategies, however, are compromised by the potential off-target effects and mosaic mutations created by CRISPR/Cas9. In terms of the off-target effects, many reports have shown that these can be minimized by using specific gRNAs and modified Cas9. For example, using a shorter sgRNA (17 or 18 nt) can greatly improve off-target specificity (Fu et al., 2014). Variants of monomeric SpCas9, such as SpCas9-HF1 (high-fidelity variant 1), are found to show improved specificity in gene targeting (Kleinstiver et al., 2016). Cpf1, another form of Cas9, is also reported to show no off-target effects (Kim et al., 2016). Despite these improvements, mosaic mutations remain a challenge to be resolved.
Mosaic mutations affect the efficiency and precision of CRISPR/Cas9’s ability to edit the genome, which can be an obstacle to obtaining animal models that faithfully mimic genetic mutations in human patients. More importantly, mosaicism can significantly affect the precision of gene therapy. Because expression of CRISPR/Cas9 in one-cell stage embryos (zygotes) can lead to mosaic mutations, it is possible that Cas9 mRNA may segregate into late embryo stages, such as the 4-cell to morula stages, to act on targeted DNAs, thus creating mosaicism in targeting. If so, reducing the prolonged expression of Cas9 in embryos could potentially reduce the mosaic mutation rate. To test this hypothesis, our group modified Cas9 activity by tagging the ubiquitin-proteasome degradation signal to the N-terminus of Cas9 to shorten its half-life. We injected this modified Cas9 into non-human primate embryos and found that the modified Cas9 with a shorter half-life apparently decreased mosaicism in DNA mutations in monkey embryos (Tu et al., 2017). Since modified Cas9 with reduced activity, such as nickase Cas9, is able to induce more specific cleavage sites (Hsu et al., 2013; Zetsche et al., 2015), modifying Cas9 activity or identifying new Cas9 proteins may help resolve the mosaic mutation issue.
CRISPR/Cas9 causes double-strand breaks (DSBs) that can produce NHEJ and HDR. Some researchers found scr7 targeted the key enzyme in the NHEJ pathway to increase the efficiency of HDR (Maruyama et al., 2015). Post-translational Cas9-hGem (1/110) can lower its expression in G1, with increased expression in G2 and S, leading to a 1.87-fold increased rate of HDR compared to wild-type Cas9 (Gutschner et al., 2016). Komor et al. engineered fusions of CRISPR/Cas9 and a cytidine deaminase enzyme that retain the ability to be programmed with a guide RNA, do not induce dsDNA breaks, and mediate the direct conversion of cytidine to uridine (Komor et al., 2016). All these studies focus on the modification of Cas9 activity to improve its specificity for genome editing. Given that recent studies have identified many new species of Cas9 and their regulators (Pawluk et al., 2016; Burstein et al., 2016; Rauch et al., 2016), future studies to characterize these new Cas9 proteins hold out great promise of overcoming the mosaicism issue.
In summary, although the precision and knock-in efficiency of CRISPR/Cas9-mediated gene targeting need to be improved, this new genomic editing tool has been proved powerful to generate animal models for studying brain diseases. The ability of CRISPR/Cas9 to target genome in any type of cells allows for modeling brain diseases via delivering this system directly to the brain. Direct targeting the specific types of cells or regions in the brain is particularly useful for mimicking specific neuropathology in large animals whose prolonged breeding period does not allow for fast germline transmission of the targeted genes by CRISPR/Cas9.
Highlights.
Genomic editing has been used to generate animal models of human diseases to mimic neurological symptoms and to study disease pathogenesis and treatments. Of various genomic editing tools, CRISPR/Cas9 is powerful to modify genomes in all types of cells. We discuss the use of CRISPR/Cas9 to study brain diseases and the strategies to improve the specificity of its gene targeting.
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) grants (91332206, 91649115) and National Institutes of Health grants (NS041669). We thank Cheryl Strauss for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander H, Peter K, Alexandra R, Verena H, Jochen H, Christian H, Bettina S. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Alzheimer A, Stelzmann RA, Schnitzlein HN, Murtagh FR. An English translation of Alzheimer’s 1907 paper, “Uber eine eigenartige Erkankung der Hirnrinde”. Clin Anat. 1995;8:429–31. doi: 10.1002/ca.980080612. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ. Antiviral Defense in Prokaryotes. Genome Res. 2008;960:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. New CRISPR-Cas systems from uncultivated microbes. Nature. 2016 doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, Tu Z, Si C, Wang H, Xing R, Pu X, Yang SH, Li S, Ji W, Li XJ. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Daley JM, Sung P. 53BP1, BRCA1 and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 2014;34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NCA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GYR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr. Opin. Neurobiol. 2007 doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AV, Sousa A, Collins MA, Jayaram H, Cullen BR, Bumcrot D. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Shen XZ, Jiang F, Wu Y, Han C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol. 2016;34(7):768–73. doi: 10.1038/nbt.3547. Epub 2016 May 2. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME, Ambrose CM, Duyao MP. Molecular genetics of Huntington’s disease. Arch Neurol. 1993;50:1157–1163. doi: 10.1001/archneur.1993.00540110037003. [DOI] [PubMed] [Google Scholar]
- Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep. 2016;14:1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:0474–0483. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich M, Zhang F. Applications of CRISPR–Cas systems in neuroscience. Nat. Rev. Neurosci. 2015;17:36–44. doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–39. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Hardy J. The presenilins and Alzheimer’s disease. Hum. Mol. Genet. 1997;6:1639–1646. doi: 10.1093/hmg/6.10.1639. [DOI] [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JRJ. Heritable and Precise Zebrafish Genome Editing Using a CRISPR-Cas System. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, Complete Deletion of Synaptic Proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–33. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. 2839 [pii] [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. [Research Support, Non-U.S. Gov't] Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce PI, Fratta P, Fisher EMC, Acevedo-Arozena A. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: Recent advances in understanding disease toward the development of clinical treatments. Mamm. Genome. 2011;22:420–448. doi: 10.1007/s00335-011-9339-1. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire H-G, Unterbeck A, Salbaum JM, Masters CL, Grzeschik Karl-H, Multhaup G, Beyreuther K, Müller-Hill B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–6. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol. Cell. 2010 doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon S, Kim J-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863–8. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;61:5985–91. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12:1893–905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013a;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 2013b;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- Lv Q, Yuan L, Deng J, Chen M, Wang Y, Zeng J, Li Z, Lai L. Efficient Generation of Myostatin Gene Mutated Rabbit by CRISPR/Cas9. Sci. Rep. 2016;6:25029. doi: 10.1038/srep25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK1, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–42. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM-Y. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science (80-.) 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Pawluk A, Amrani N, Zhang Y, Sontheimer EJ, Maxwell KL, Davidson AR, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell. 2016;167:1–10. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL, Lazarrini AM, Golbe LI, Polymeropoulos MH, Uèda K, Chen X, Shitasaki Y, Iwai A, Weinreb PH, Jensen PH, Jensen PH, Maroteaux L, Scheller RH, Doty RL, Pearce RK, Hawkes CH, Daniel SE, George JM, Perez-Tur J, Sherrington R, Sorbi S, Wasco W, Levy-Lahad E, Rogaev EI, Hsiao K. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, Scot t Da, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott Da, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp Pa, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–190. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell. 2016;168(1–2):150–158. e10. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, Jacks T. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–31. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yuan L, Wang Y, Chen M, Deng J, Lv Q, Sui T, Li Z, Lai L. Efficient dual sgRNA-directed large gene deletion in rabbit with CRISPR/Cas9 system. Cell. Mol. Life Sci. 2016;73:2959–2968. doi: 10.1007/s00018-016-2143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Research. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Yang W, Yan S, Guo X, Li X-J. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol. Neurodegener. 2015;10:35. doi: 10.1186/s13024-015-0031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Yang W, Yan S, Yin An, Gao J, Liu X, Zheng Y, Zheng J, Li Z, Yang S, Li S, Guo X, Li X-J. Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci. Rep. 2017 doi: 10.1038/srep42081. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Azam AB, Gillon CJ, Josselyn SA, Zovkic IB. Advanced in vivo use of CRISPR/Cas9 and anti-sense DNA inhibition for gene manipulation in the brain. Front. Genet. 2016 doi: 10.3389/fgene.2015.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AY, Peng PD, Ehrhardt A, Storm Ta, Kay Ma. Comparison of adenoviral and adeno-associated viral vectors for pancreatic gene delivery in vivo. Hum. Gene Ther. 2004;15:405–413. doi: 10.1089/104303404322959551. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zuris Ja, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2016:10–15. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Wasiliew P, Kracht M. Interleukin-1 (IL-1) pathway. Sci. Signal. 3, cm1. 2010 doi: 10.1126/scisignal.3105cm1. [DOI] [PubMed] [Google Scholar]
- Yang W, Tu Z, Sun Q, Li X-J. CRISPR/Cas9: Implications for Modeling and Therapy of Neurodegenerative Diseases. Front. Mol. Neurosci. 2016;9:30. doi: 10.3389/fnmol.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Wang X, Liu Y, Ouyang Z, Long H, Wei S, Xin J, Zhao B, Lai S, Shen J, Ni Q, Yang H, Zhong H, Li L, Hu M, Zhang Q, Zhou Z, He J, Yan Q, Fan N, Zhao Y, Liu Z, Guo L, Huang J, Zhang G, Ying J, Lai L, Gao X. Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol. 2015;7(6):580–3. doi: 10.1093/jmcb/mjv061. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen Z-Y, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2014;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]