Abstract
Mutations in MECP2 cause Rett syndrome (RTT), an X-linked neurological disorder characterized by regressive loss of neurodevelopmental milestones and acquired psychomotor deficits. However, the cellular heterogeneity of the brain impedes an understanding of how MECP2 mutations contribute to RTT. Here we developed a Cre-inducible method for cell type-specific biotin tagging of MeCP2 in mice. Combining this approach with an allelic series of knockin mice carrying frequent RTT mutations (T158M and R106W) enabled the selective profiling of RTT-associated nuclear transcriptomes in excitatory and inhibitory cortical neurons. We found that most gene expression changes are largely specific to each RTT mutation and cell type. Lowly expressed cell type-enriched genes are preferentially disrupted by MeCP2 mutations, with upregulated and downregulated genes reflecting distinct functional categories. Subcellular RNA analysis in MeCP2 mutant neurons further reveals reductions in the nascent transcription of long genes and uncovers widespread post-transcriptional compensation at the cellular level. Finally, we overcame X-linked cellular mosaicism in female RTT models and identified distinct gene expression changes between neighboring wild-type and mutant neurons, altogether providing contextual insights into RTT etiology that support personalized therapeutic interventions.
Introduction
RTT is a progressive X-linked neurological disorder that represents one of the most common causes of intellectual disability among young girls. Patients experience a characteristic loss of acquired social and psychomotor skills and develop stereotyped hand movements, breathing irregularities, and seizures after 6–18 months of normal development1. Approximately 95% of RTT cases are mapped to the X-linked gene encoding methyl-CpG binding protein 2 (MeCP2), a ubiquitously expressed protein that is highly enriched in postmitotic neurons2,3. The majority of RTT-associated mutations cluster within two functionally distinct domains of MeCP2. The Methyl-CpG Binding Domain (MBD) allows MeCP2 to bind to methylated cytosines4. The Transcriptional Repression Domain (TRD) mediates protein-protein interactions with histone deacetylase-containing co-repressors, such as the NCoR-SMRT and mSin3A complexes5–7. These domains support MeCP2 as a chromatin factor that mediates transcriptional repression7,8, although transcriptional activation by MeCP2 is also reported9–11.
Different mutations in MECP2, together with random X-chromosome inactivation (XCI), underlie a spectrum of clinical severity in RTT patients12. Among the most frequent RTT mutations, three are missense mutations in the MeCP2 MBD, including R106W (2.76% of RTT patients), R133C (4.24%), and T158M (8.79%)13. Typical RTT patients bearing the R133C mutation display milder clinical symptoms, whereas patients carrying the T158M or R106W mutation exhibit moderate or severe symptoms, respectively12. Although the clinical severity of these mutations scales with their effects on MeCP2 binding affinity to methylated DNA14–17, this relationship is not fully understood on a molecular level. Mouse models carrying RTT mutations can recapitulate this phenotypic variability, but most studies are limited to hemizygous male mice17–21. Despite that RTT predominantly affects heterozygous females, an experimental strategy to selectively identify gene expression changes from Mecp2 mutant-expressing cells in a mosaic female brain has not yet been developed.
Given that MeCP2 is a chromatin-bound nuclear protein, the identification of MeCP2 transcriptional targets in the brain remains key towards illuminating RTT etiology22. However, target identification is confounded by the cellular heterogeneity of the brain, which contains multiple intermixed cell types that differ in morphology, function, electrophysiological properties, and transcriptional programs23–25. Analyses using heterogeneous brain tissues obscure cell type-specific gene expression changes, impeding the assessment of MeCP2 function at the transcriptional level26. The identification of transcriptional targets is further complicated by the widespread binding patterns of MeCP2 to methylated cytosines (mCpG and mCpA)8,27,28 or unmethylated GC-rich regions29 throughout the genome.
In this study, we addressed the confounding effects of cellular heterogeneity by engineering genetically modified mice whereby MeCP2 is labeled with biotin using Cre-Lox recombination. To understand the molecular impact of RTT mutations on cell type-specific gene expression in vivo, we also developed an allelic series of knockin mice bearing one of two frequent RTT missense mutations, T158M and R106W. When combined with Fluorescence-Activated Cell Sorting (FACS), this strategy allows for the isolation of neuronal nuclei from targeted cell types, effectively circumventing cellular heterogeneity in the mouse brain and X-linked mosaicism in female mice. Our findings support a contextualized model by which cell autonomous and non-autonomous transcriptional changes in different cell types contribute to the molecular severity of neuronal deficits in RTT, providing new directions for therapeutic development.
Results
Engineering a System to Genetically Biotinylate MeCP2 In Vivo
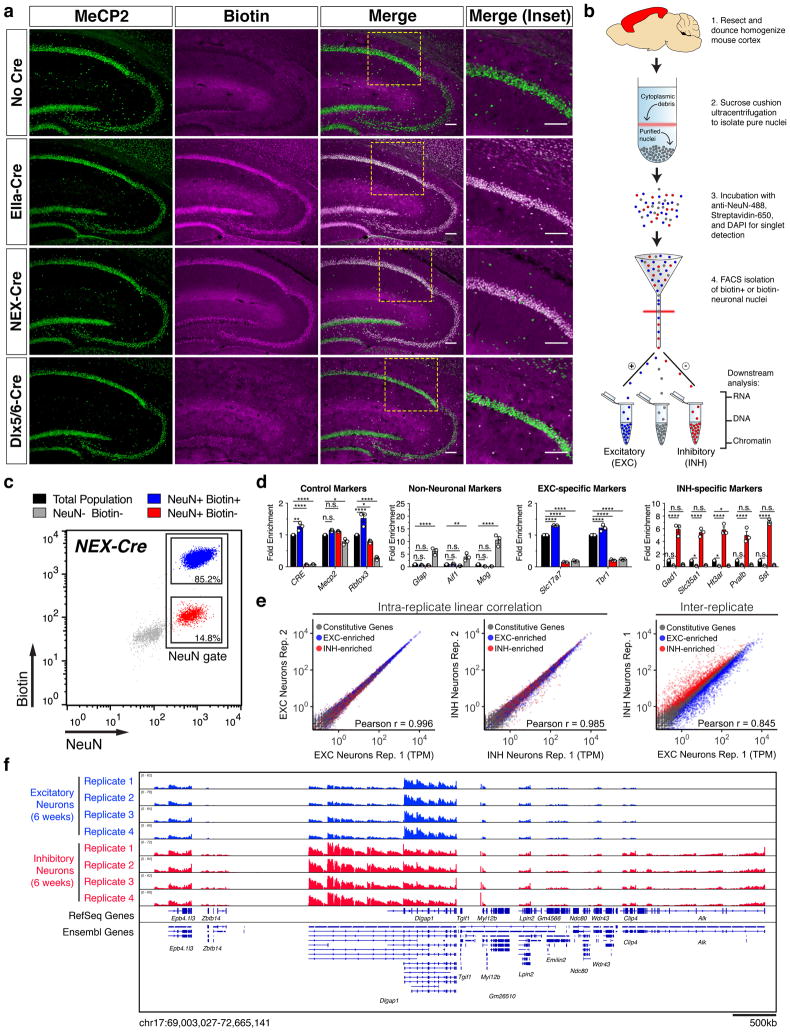
Biotin-mediated affinity tagging has been widely utilized in cell and animal models for multiple experimental approaches because of the strong (Kd = 4×10−14M) and specific interaction between biotin and avidin protein30. We exploited this approach to investigate MeCP2 function by using homologous recombination to insert a short 23-amino acid affinity tag immediately upstream of the Mecp2 stop codon (Fig. 1a and Supplementary Fig. 1a). This tag comprises a TEV protease cleavage site and a 15-amino acid biotinylation consensus motif (termed Tavi, TEV and Avidin-binding) that can be post-translationally labeled with biotin by the E. coli biotin ligase, BirA. To biotinylate the tag in cell types of interest, we also generated Cre-dependent BirA transgenic mice (herein R26cBirA; Supplementary Fig. 1b). Therefore, upon crossing these mice to a cell type-specific Cre line, BirA is expressed and subsequently biotinylates MeCP2-Tavi (Fig. 1b). We used EIIa-Cre31 to ubiquitously express BirA (R26BirA) and confirmed that MeCP2 is specifically biotinylated in vivo only when BirA is expressed and the Tavi tag is present (Fig. 1c and Supplementary Fig. 1c).
Figure 1.
Utilization and characterization of Mecp2Tavi mice and associated RTT variants. (a) Diagram of wild-type and tagged MeCP2 showing R106W or T158M missense mutations. MBD, Methyl-CpG Binding Domain; TRD, Transcriptional Repression Domain. (b) Breeding strategy to biotinylate the Tavi tag in a Cre-dependent manner. (c) Representative western blot showing the conditions in which the Tavi tag is biotinylated using whole brain nuclear extracts. Blot is probed with streptavidin for biotin detection and antibodies against MeCP2 N-terminus, Tavi tag, and NeuN. (d) Representative images showing immunofluorescent detection of biotinylated MeCP2 and mutant variants in hippocampal sections of untagged (WT) and tagged (TAVI, T158M, R106W) male mice at 6 weeks of age. Tissue is probed with streptavidin for biotin detection and antibody against the MeCP2 C-terminus. Scale bars represent 10 μm. (e) Quantification and representative western blot comparing MeCP2 protein expression levels between TAVI and mutant (T158M, R106W) male mice at 6 weeks of age. Blot is probed with antibodies against the MeCP2 C-terminus and TBP (nreplicates = 3, One-way ANOVA). (f) Quantification of salt-extracted MeCP2 from chromatin using 200mM (left) and 400mM (right) NaCl, normalized to extracts using 500mM NaCl (see Supplemental Fig. 2e; nreplicates = 4–5, One-way ANOVA). (g) Box-and-whisker plot of brain weights from untagged (WT, KO (Mecp2-null)) and tagged (TAVI, T158M, R106W) male mice at 6 weeks of age (nWT = 20, nTAVI = 11, nKO = 6, nT158M = 6, nR106W = 12; One-way ANOVA). Box limits denote 25th and 75th percentiles, center line denotes median, ‘+’ denotes mean, and whiskers denote data max and min. Each genotype is indicated with a different color. (h) Body weight over postnatal age in untagged (WT, KO) and tagged (TAVI, T158M, R106W) male mice. Data points consist of at least 6 observations each. Total number of mice assessed: nWT = 31, nTAVI = 23, nKO = 15, nT158M = 14, nR106W = 28. (i) RTT-like phenotypic score across postnatal development in untagged (WT, KO) and tagged (TAVI, T158M, R106W) male mice. Data points over time consist of at least 6 observations each. Total number of mice assessed: nWT = 31, nTAVI = 23, nKO = 15, nT158M = 14, nR106W = 28. (j) Kaplan-Meier survival curve for untagged (WT, KO) and tagged (TAVI, T158M, R106W) male mice (nWT = 31, nTAVI = 23, nKO = 17, nT158M = 39, nR106W = 26). *P < 0.5, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant; all pooled data depicts mean ± SEM unless otherwise stated. See also Supplementary Figs. 1 and 2.
To examine the possibility that tagging MeCP2 adversely affects its function, we assessed MeCP2 expression levels, DNA binding, and protein-protein interactions in 20-week old Mecp2Tavi/y (herein TAVI) and littermate Mecp2+/y (WT) mice. We found that total MeCP2 protein, but not RNA, is significantly reduced by ~40% in TAVI mice, and a similar trend towards ~40% reduction is also observed among soluble and chromatin-bound protein fractions (Supplementary Fig. 1d–f). However, Tavi-tagged and untagged MeCP2 both exhibit comparable levels of chromatin binding at high and low affinity genomic sites, including highly methylated major satellite repeats and IAP elements, and MeCP2-Tavi remains associated with the NCoR-SMRT co-repressor (Supplementary Fig. 1g–h). Although a 50% reduction in MeCP2 expression is sufficient to cause hypoactivity and subtle behavioral phenotypes in mice32, TAVI mice appear similar to WT mice and do not display overt RTT-like features using phenotypic scoring33 over an observational period of 20 weeks (Fig. 1g–j, Supplementary Fig. 1i, and data not shown/B.S.J).
MeCP2 Missense Mutations Recapitulate RTT-like Phenotypes in Mice
To examine the molecular relationship between MeCP2 affinity for methylated DNA and phenotypic severity, we generated independent Mecp2T158M-Tavi (herein T158M) and Mecp2R106W-Tavi (R106W) knock-in mice in parallel with TAVI mice (Fig. 1a). Relative to TAVI controls, we found that T158M and R106W mice both display a ~70–80% reduction in MeCP2 protein expression despite equivalent levels of mRNA at 6 weeks of age (Fig. 1e, Supplementary Fig. 1j), similar to other mouse models bearing MeCP2 MBD mutations17,18,21. However, there is a trend towards relatively higher MeCP2 protein levels in T158M than R106W mice across development (Supplementary Fig. 2a). Immunofluorescent (IF) staining of hippocampal sections from T158M and R106W mice revealed diffusely distributed MeCP2 throughout the nucleus that accompanied a loss of localization to heterochromatic foci, supporting the impaired binding of mutant MeCP2 to mCpGs in vivo (Fig. 1d). Streptavidin IF, which is noticeably lower in Mecp2 mutant mice, also confirmed a loss of mutant MeCP2 localization to heterochromatic foci and further illustrated a redistribution of mutant MeCP2 to the nucleolus (Fig. 1d and Supplementary Fig. 2b–c), a property reminiscent of GFP-tagged MeCP2 lacking its MBD34.
Using sub-nuclear fractionation, we confirmed that a greater proportion of MeCP2 T158M or R106W protein occupies the soluble fraction when compared to WT or TAVI protein (Supplementary Fig. 2d), consistent with the loss of chromatin binding in mutant mice (Fig. 1d). By further extracting chromatin-bound MeCP2 with different salt concentrations, we found that MeCP2 R106W is more readily released at lower salt concentrations (200mM NaCl) than MeCP2 WT, TAVI, or T158M protein, suggesting that MeCP2 R106W has the lowest binding affinity for chromatin (Fig. 1f and Supplementary Fig. 2e). Phenotypic comparisons revealed that T158M and R106W mice both exhibit RTT-like features similar to that of Mecp2-null mice, including decreased brain and body weight, and an age-dependent increase in phenotypic score (Fig. 1g–i). Although lifespan is significantly reduced in all three Mecp2 mutant mice, the median survival of R106W mice more closely resembles that of Mecp2-null than T158M mice (Fig. 1j). Both mutations demonstrated a significant difference in survival curves (T158M median survival = 14 weeks; R106W median survival = 10 weeks; Mantel Cox P = 0.012). Together, these data suggest that T158M and R106W mutations represent a partial and complete loss-of-function, respectively.
Genetic Biotinylation Permits Cell Type-specific Transcriptional Profiling
We next devised a biotinylation-based strategy for cell type-specific nuclei isolation and transcriptional profiling (Fig. 2a–b). We used the NeuroD6/NEX-Cre line35 to drive BirA expression and MeCP2-Tavi biotinylation in forebrain excitatory neurons (Fig. 2a and Supplementary Fig. 3a–g). Quantification of pan-neuronal (NeuN), pan-inhibitory (GAD67), and inhibitory-specific (parvalbumin, somatostatin and calretinin) neuronal markers in the somatosensory cortex of Mecp2Tavi/y;R26cBirA/+;NEXCre/+ (herein NEX-Cre) mice demonstrated that biotinylation occurs in ~80% of NeuN+ cortical neurons devoid of inhibitory markers (Supplementary Fig. 3h). FACS using stained cortical nuclei from NEX-Cre mice identified three distinct nuclear populations (Fig. 2c). RT-PCR for cell type-specific markers confirmed that NeuN+Biotin+ nuclei reflect excitatory neurons, whereas NeuN+Biotin− nuclei represent a mixture of inhibitory interneuron subtypes (Fig. 2c–d). Astrocytic, microglial and oligodendrocytic markers are restricted to the third, non-neuronal population of NeuN−Biotin− nuclei (Fig. 2c–d). We also used the Dlx5/6-Cre line36 to drive BirA expression in forebrain GABAergic neurons and obtained results inverse to that of NEX-Cre (Fig. 2a and Supplementary Fig. 3a–j), confirming that MeCP2-Tavi is reliably biotinylated in Cre-defined cell types.
Figure 2.
Cell type-specific transcriptional profiling of neuronal nuclei. (a) Representative images showing immunofluorescent detection of biotinylated MeCP2-Tavi protein in Cre-specified neuronal populations of the mouse hippocampus. Probed using streptavidin for biotin detection and antibody against the MeCP2 C-terminus. Scale bars represent 100μm. (b) Schematic of cortical nuclei preparation and FACS isolation. (c) FACS analysis of labeled cortical nuclei populations. Data shown is representative of nine independent experiments using NEX-Cre mice. Percentages indicate the mean distribution of neurons that are NeuN+Biotin+ (excitatory; 85.2% ± 0.35) or NeuN+Biotin− (inhibitory; 14.8% ± 0.35). (d) RT-PCR validation of FACS-isolated populations depicted in (c) (nreplicates = 3, Two-way ANOVA). (e) Pearson correlation of biological replicate nuclear RNA-seq libraries within (intra-replicate) and across (inter-replicate) FACS-isolated populations depicted in (c). Colors correspond to EXC-enriched (blue) and INH-enriched (red) genes identified through differential expression analysis of excitatory and inhibitory neurons. Note lower Pearson correlation and clear dispersal of cell type-enriched genes across FACS populations. (f) IGV browser snapshot of Dlgap1 genomic locus in excitatory and inhibitory neurons of TAVI male mice at 6 weeks of age. RefSeq and Ensembl gene annotations are both shown. *P < 0.5, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant; all pooled data depicts mean ± SEM. See also Supplementary Figs. 3 and 4.
Because MeCP2 is known to modulate transcription22, nuclear RNA-seq would afford an unique opportunity to study the primary effects of RTT mutations on gene expression. We thus performed transcriptional profiling in 6-week old male mice near the onset of RTT-like phenotypes. We employed the NEX-Cre driver and isolated cortical excitatory and inhibitory nuclei from T158M, R106W and TAVI mice via FACS, followed by total RNA-seq (Supplementary Table 1). Biological replicates were well-correlated (Fig. 2e), and ~74% of total reads mapped to introns, serving as a proxy for chromatin-associated transcriptional activity37,38. Unsupervised hierarchical clustering shows that replicate transcriptomes from cortical excitatory and inhibitory neurons in TAVI mice are highly correlated by cell type, and genic-mapped reads illustrate selectively expressed genes in each cell type (Fig. 2e–f and Supplementary Fig. 4a). We identified 9,379 differentially expressed genes (DEGs, FDR < 0.05) between excitatory and inhibitory neurons, the majority (86.9%) of which comprise protein-coding genes (Table 1 and Supplementary Fig. 4b). Among the protein-coding fraction of cell type-enriched DEGs, 3,958 genes (0.15 – 4.70 fold change) display Gene Ontology (GO) functions consistent with glutamatergic pyramidal cell types (EXC-enriched; Supplementary Fig. 4c), whereas the remaining 4,194 genes (0.17 – 7.77 fold change) exhibit GO functions consistent with metabolically active GABAergic interneurons (INH-enriched; Supplementary Fig. 4d).
Table 1.
Summary of Differentially Expressed Genes (DEGs) identified in the study
| Experiment | Cell Type | Genotype | Differentially Expressed Genes (DEGs) |
Upregulated/ Downregulated (Protein-coding) |
|Log2 Fold Change| (Protein-coding) |
Gene Length (kb) (Protein-coding) |
Proportion of Cell Type Enriched Genes (Protein-coding) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Coding | Non- coding |
Median [IQR] | Median [IQR] | Constitutive | Excitatory (EXC) |
Inhibitory (INH) |
|||||
| 6 WEEKMales | Actively Expressed Genes | Excitatory Neurons | TAVI (Control) | 13877 | 10926 | 2951 | - | - | 24.9 [9.9 – 61.3] | 46.5% | 28.5% | 24.9% |
| Inhibitory Neurons | TAVI (Control) | 10369 | 8319 | 2050 | - | - | 25.4 [10.3 – 61.5] | 46.8% | 24.3% | 28.8% | ||
| Cell Type-enriched Gene Expression | Excitatory Neurons | TAVI (Control) | 4593 | 3958 | 635 | - | 0.75 [0.41 – 1.46] | 50.6 [21.5 – 110.1] | - | 100% | - | |
| Inhibitory Neurons | TAVI (Control) | 4783 | 4194 | 589 | - | 0.73 [0.45 – 1.47] | 21.2 [8.8 – 53.0] | - | - | 100% | ||
| MeCP2-dependent Gene Expression | Excitatory Neurons | T158M | 197 | 177 | 20 | 63.8% DOWN | 0.45 [0.33 – 0.62] | 88.6 [33.5 – 173.1] | 16.4% | 49.2% | 34.5% | |
| R106W | 425 | 386 | 39 | 61.7% DOWN | 0.44 [0.34 – 0.61] | 82.5 [36.7 – 146.0] | 17.6% | 51.3% | 31.1% | |||
| Shared DEGs | 75 | 69 | 6 | 65.2% DOWN | see Figure 3D | 109.8 [49.9 – 184.1] | 14.5% | 52.2% | 33.3% | |||
| Inhibitory Neurons | T158M | 146 | 143 | 3 | 62.9% UP | 0.45 [0.29 – 0.55] | 107.8 [41.4 – 188.7] | 20.3% | 44.8% | 35.0% | ||
| R106W | 758 | 697 | 61 | 56.2% UP | 0.41 [0.33 – 0.56] | 40.5 [19.1 – 102.9] | 29.4% | 35.3% | 35.3% | |||
| Shared DEGs | 109 | 107 | 2 | 64.5% UP | see Figure 3D | 112.8 [42.9 – 202.8] | 18.7% | 46.7% | 34.6% | |||
| 18 WEEK Females | MeCP2-dependent Gene Expression | Excitatory Neurons | T158MWT | 42 | 28 | 14 | 64.3% DOWN | 0.52 [0.49 – 0.79] | 54.6 [27.2 – 112.6] | - | - | - |
| R106WWT | 346 | 294 | 52 | 62.2% UP | 0.64 [0.47 – 0.93] | 51.4 [21.0 – 140.4] | - | - | - | |||
| T158MMUT | 585 | 516 | 69 | 67.2% DOWN | 0.49 [0.40 – 0.73] | 51.2[20.4 – 113.2] | - | - | - | |||
| R106WMUT | 634 | 569 | 65 | 52.2% UP | 0.54 [0.43 – 0.83] | 65 [31.7 – 142.1] | - | - | - | |||
| SharedMUT | 207 | 185 | 22 | 65.9% DOWN | see Figure 5I | 77.5 [36.2 – 143.6] | - | - | - | |||
Protein-Coding Genes are More Severely Affected in R106W Mice
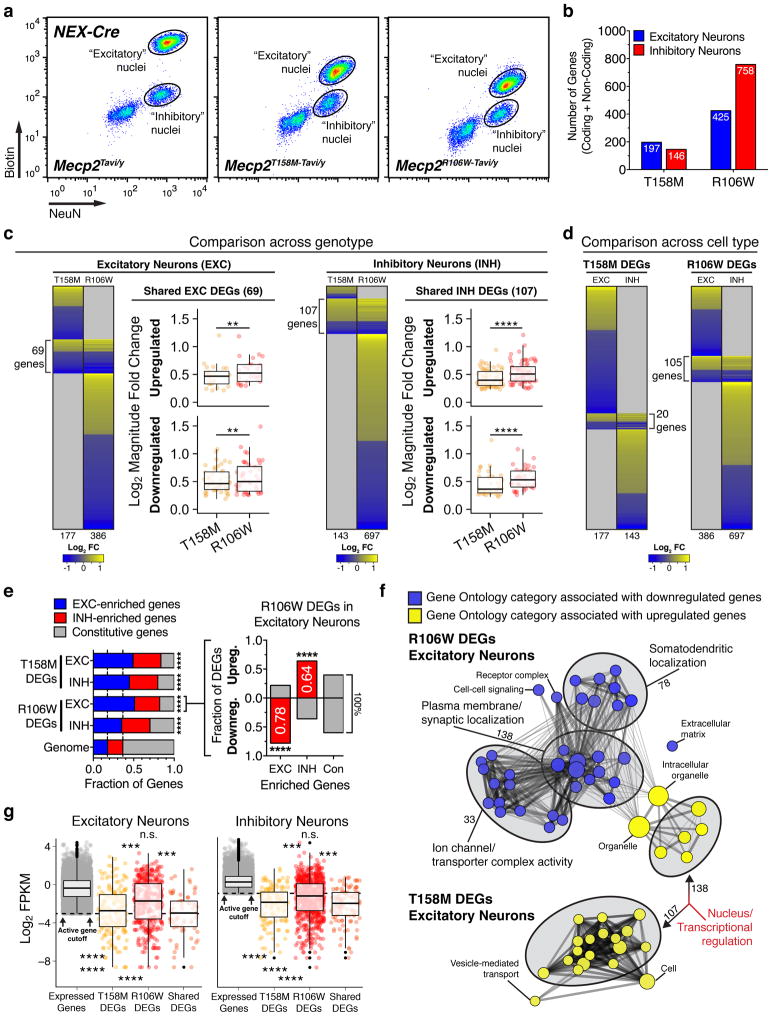
We next compared nuclear gene expression profiles in excitatory and inhibitory neurons between mutant (T158M, R106W) and control (TAVI) mice to identify and characterize DEGs associated with the appearance of RTT-like phenotypes (Fig. 3a). We identified more DEGs in excitatory and inhibitory neurons of R106W than T158M mice, indicating that the number of misregulated genes positively scales with the severity of the Mecp2 mutation (Fig. 3b and Table 1). More than 90% of MeCP2 DEGs are protein-coding genes (Supplementary Fig. 5a), significantly higher than the percentage of protein-coding genes in the genome (60.4%), or among actively expressed (77.7–78.3%) and cell type-enriched (86.2–87.7%) genes (Supplementary Fig. 4b). We therefore excluded non-coding genes from further analyses. We note that the number and percentage of protein-coding DEGs overlapping between T158M and R106W genotypes is greater in inhibitory (107 genes) than excitatory neurons (69 genes). Moreover, overlapping DEGs tend to be misregulated in the same direction (Fig. 3c).
Figure 3.
Analysis of T158M and R106W differentially expressed genes. (a) FACS isolation of cortical excitatory and inhibitory neuronal nuclei from TAVI, T158M, or R106W male mice at 6 weeks of age. (b) Total number of protein coding and non-coding differentially expressed genes (DEGs) identified in excitatory or inhibitory neurons of Mecp2 mutant mice. (c) Heatmaps display log2 fold changes among protein-coding DEGs in excitatory and inhibitory neurons of Mecp2 mutant mice, compared across genotypes. Excitatory DEGs nshared = 69 genes, Hypergeometric P = 3.15e−77. Inhibitory DEGs nshared = 107 genes, Hypergeometric P = 5.33e−134. Boxplots compare log2 median fold changes among overlapping DEGs between T158M and R106W neurons (One-tailed Wilcoxon Signed Rank). (d) Heatmap displaying log2 fold changes among protein-coding DEGs in excitatory and inhibitory neurons of Mecp2 mutant mice, compared across cell types. (e) Left graph, Distribution of constitutive, EXC- or INH-enriched genes among T158M and R106W protein-coding DEGs, compared against genomic distribution (Chi-square Goodness-of-Fit). Right graph, Bar plot summarizing R106W DEGs in excitatory neurons, partitioned by cell type-enriched or constitutive genes, and which are preferentially upregulated or downregulated. Red indicates statistical significance (One-tailed Fisher’s Exact Test). (f) Enrichment map of pre-ranked Gene Set Enrichment Analysis (GSEA) functional network associations. Data represents DEGs from R106W (top) and T158M (bottom) excitatory neurons (P-value < 0.01, Q-value < 0.1). Nodes denote functional categories, colored by Normalized Enrichment Score (NES). Line weight denotes extent of gene overlap between connected nodes. Red text highlights the similarity in functional annotations between both genotypes. (g) Boxplots comparing the log2 FPKM distribution of actively expressed genes against T158M, R106W, and shared DEGs for each cell type (Pairwise Wilcoxon Rank Sum P displayed). *P < 0.5, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant. See also Supplementary Figs. 5 and 6.
The median fold change of T158M and R106W DEGs is small in mutant neurons compared to overall differences in gene expression between excitatory and inhibitory neurons (Supplementary Fig. 5b and Table 1). We further compared fold changes between T158M and R106W DEGs, limiting our analysis to protein-coding genes that overlap between genotypes to account for disproportionate numbers of DEGs. Within this subset, we found that the median fold change among upregulated and downregulated DEGs is consistently higher in both cell types of R106W mice than those of T158M mice (Fig. 3c), consistent with a more severe phenotype in R106W mice.
Transcriptional Features of T158M and R106W DEGs
We next compared MeCP2 DEGs across excitatory and inhibitory neurons and found limited overlap between the two cell types (6.2% of T158M DEGs, 10.7% of R106W DEGs; Fig. 3d), indicating that most DEGs reflect cell type-specific transcriptional changes. Indeed, EXC/INH-enriched genes are significantly overrepresented among MeCP2 DEGs in each cell type, comprising ~70–80% of genes (Fig. 3e and Table 1). Moreover, EXC- and INH-enriched genes are preferentially downregulated and upregulated, respectively, in each cell type (Fig. 3e and Supplementary Fig. 5c).
We next performed a pre-ranked Gene Set Enrichment Analysis (GSEA, FDR < 0.1) to determine whether upregulated and downregulated DEGs represent functionally distinct categories. Upregulated DEGs in T158M and R106W mice are both primarily associated with transcriptional regulation. These include DNA-binding transcriptional activators, repressors, and chromatin remodelers, most of which tend to be INH-enriched genes (Fig. 3f and Supplementary Fig. 5d). Significant functional categories associated with downregulated DEGs, however, are specifically detected in R106W excitatory neurons and enriched for post-synaptic membrane proteins, including various ion channels, synaptic scaffolding proteins, and ionotropic glutamate receptors (Fig. 3f). Although significant gene functions were not identified among downregulated DEGs in inhibitory neurons using our GSEA FDR cutoff, gene functions associated with upregulated DEGs in R106W inhibitory neurons are related to cellular metabolism and signal transducer activity (Supplementary Fig. 5e).
Upon examining the relative expression levels of MeCP2 DEGs using Fragments Per Kilobase of transcript per Million mapped reads (FPKM), we noticed that T158M, R106W, and overlapping DEGs display significantly lower FPKM values relative to total expressed genes in each cell type (Fig. 3g). To exclude the possibility of gene filtering biases associated with RNA-seq, we randomly selected 12 low-expressing DEGs that overlap between both mutations and independently measured their primary and mature RNA transcripts in excitatory neuronal nuclei using RT-PCR (Supplementary Table 2). We found that 10 out of 12 genes show significant gene expression changes that resemble those using RNA-seq (83.3% positive validation rate; Supplementary Fig. 6a–b), confirming that genes with low transcriptional activity are indeed affected by MeCP2 mutations. To examine whether lowly expressed genes are selectively enriched for MeCP2 DEGs, we binned actively expressed genes from each cell type into four percentiles (Q1–Q4) according to expression level. EXC- and INH-enriched genes served as reference distributions across percentiles for each cell type (Supplementary Fig. 6c). In comparison, T158M and R106W DEGs are preferentially enriched in Q1, the bottom 25th percentile of actively expressed genes, in both excitatory and inhibitory neurons (Fisher Exact one-tailed P, T158M: EXC = 1.11e-07, INH = 2.03e-04; R106W: EXC = 4.04e-08, INH = 1.50e-02; Supplementary Fig. 6c). Between the two mutations, T158M DEGs are more likely to be lowly expressed than R106W DEGs (Fisher Exact Odds Ratio (OR) for Q1, T158M: EXC = 3.1, INH = 3.2; R106W: EXC = 2.0, INH = 1.3; Supplementary Fig. 6c). Accordingly, R106W-specific DEGs have significantly higher FPKM values than T158M DEGs in both cell types and are predominantly downregulated in R106W neurons (Fig. 3g and Supplementary Fig. 6d). This preferential downregulation of high-expressing genes appears consistent with the specific loss of synaptic gene functions in R106W excitatory neurons (Fig. 3f–g and Supplementary Fig. 6d).
Subcellular RNA Pools Reveal Global Transcriptional and Post-transcriptional Changes
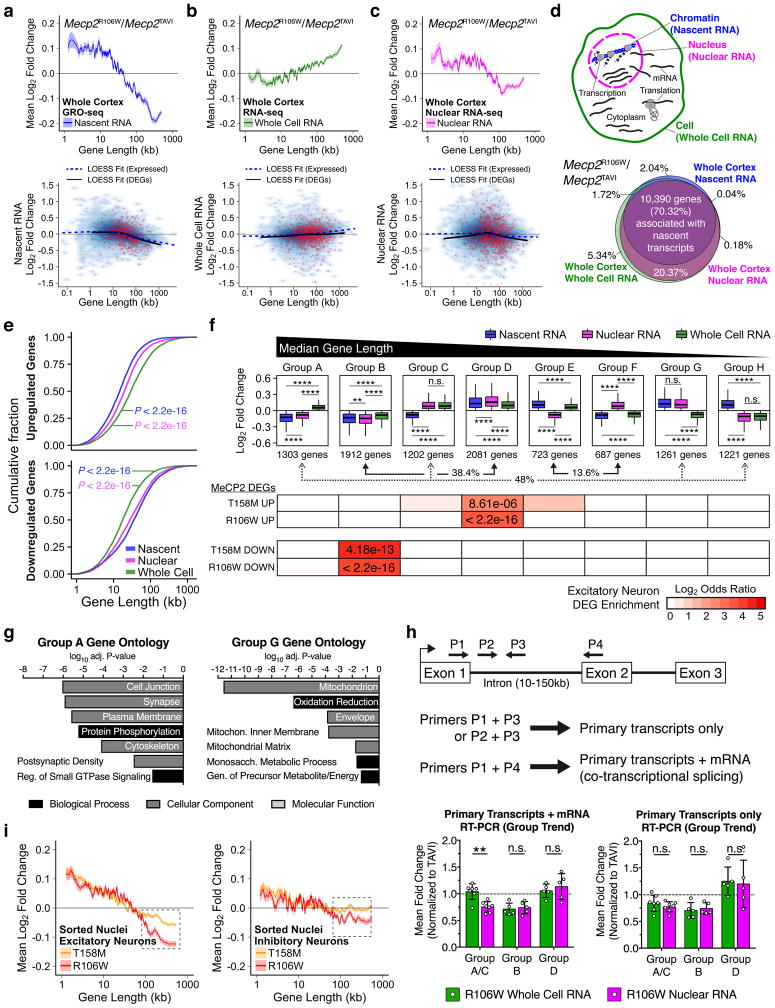
Two recent reports implicate MeCP2 in the transcriptional repression of long genes, which are preferentially upregulated in the neurons of multiple RTT models27,39. We therefore examined the possibility that genome-wide transcriptional changes may correlate with T158M and R106W phenotypic and molecular severity. Similar to those studies, we sorted and binned expressed protein-coding genes according to gene length and measured the mean fold change in Mecp2 mutant neurons. Notably, nuclear transcriptomes revealed a striking inversion of previously reported gene expression changes whereby short (≤ 100kb in gene length) and long (> 100kb in gene length) genes trend towards upregulation and downregulation, respectively, in a length-dependent manner (Supplementary Fig. 7a).
Although most nuclear RNAs comprise intron-containing pre-mRNA transcripts on chromatin, the presence of processed mRNA transcripts awaiting nuclear export may potentially confound the assessment of transcriptional events40. We therefore performed global nuclear run-on with high-throughput sequencing (GRO-seq41) to directly assess de novo transcriptional activity by RNA polymerase in cortical nuclei of TAVI and R106W mice. Similar to sorted nuclear RNA, the nascent transcription of short and long genes in R106W neurons is predominantly increased and decreased, respectively (Fig. 4a). LOESS local regression of DEGs that were identified in R106W excitatory and inhibitory neurons also revealed a similar overall trend towards the preferential downregulation of long genes (Fig. 4a). The genome-wide trend we observe in sorted nuclear RNA thus represents a primary effect at the transcriptional level, prompting us to further investigate if the length-dependent upregulation of long genes that was previously reported may represent an indirect effect of MeCP2-dependent transcriptional deregulation. To test this, we resected cortical tissue from TAVI and R106W mice, and subjected each cortical half to whole cell or nuclear RNA isolation in parallel, followed by sequencing. Cortical whole cell RNA from mutant mice displayed a length-dependent increase in the mean expression of long genes (Fig. 4b), similar to what was previously described27,39. In contrast, cortical nuclear RNA isolated from the same TAVI and R106W mice exhibited a length-dependent upregulation of short genes and downregulation of long genes (Fig. 4c), corroborating the transcriptional changes we observed from nascent RNA (Fig. 4a) and nuclear RNA from sorted nuclei (Supplementary Fig. 7a). Using the 10,390 expressed genes associated with de novo transcription by GRO-seq (Fig. 4d), we observed that genes upregulated in nascent and nuclear RNA pools are cumulatively shorter in length relative to those upregulated in whole cell RNA, and the inverse was observed among downregulated genes (Fig. 4e). Thus, gene expression changes in Mecp2 mutant neurons appear to be substantially different between subcellular compartments.
Figure 4.
Genome-wide length-dependent transcriptional changes in RTT mutant mice. (a) Genome-wide log2 fold changes in R106W mice (n = 2) compared to TAVI mice (n = 2) at 6 weeks of age using GRO-seq. Top, Lines represent mean fold change in expression for genes binned according to gene length (200 gene bins, 40 gene step) as described in27. Ribbon represents SEM of genes in each bin. Bottom, Smoothed scatterplot depicting LOESS correlation between gene length and log2 fold change for all individual protein-coding genes detected in GROseq. Genes in red highlight R106W DEGs identified from sorted excitatory and inhibitory neuronal nuclei. (b,c) Same as in (a), but using total RNA-seq analysis of whole cell (b) or nuclear (c) RNA isolated from left or right cortex of the same mice at 6 weeks of age (n=2). (d) Top, Diagram of RNA distribution across subcellular compartments. Bottom, Area proportional Venn diagram comparing overlap in gene expression changes between nuclear RNA, whole cell RNA, and nascent RNA. (e) Cumulative distribution function of gene lengths for all upregulated and downregulated protein-coding genes among nascent, nuclear, and whole cell RNA pools (n = 10,390 genes, Kolmogorov-Smirnov). (f) Top, Boxplots depicting median log2 fold changes in R106W mice between nascent, nuclear, and whole cell RNA pools, classified by the direction of gene misregulation (n = 10, 390 genes, Pairwise Wilcoxon Rank Sum P displayed). Gene groups are arranged by median gene length (black bar on top). Arrows highlight the percentage of 10,390 genes that display similar (38.4% of expressed genes), opposite (48%), or dynamic changes (13.6%) across subcellular RNA pools. Bottom, Heatmap displaying statistical enrichment of T158M and R106W DEGs in excitatory neurons among gene groups (One-tailed Fisher’s Exact Test). (g) DAVID Gene ontology terms (Benjamini P < 0.01, FDR < 0.05) for Group A and Group G sets of genes defined in (f). (h) Top, Diagram of RT-PCR primer design to measure mature and primary RNA transcripts. Bottom, Data shows overall trend in gene expression mean fold changes using primers against primary and mature RNA transcripts (left) or primary transcripts only (right)_across individual genes from Group A/C (n = 7 genes), Group B (n = 5 genes), and Group D (n = 5 genes) in R106W compared to TAVI mice (Two-way ANOVA). Data depicts mean ± S.D. (i) Mean log2 fold change in 6-week R106W (red; n = 4) and T158M (orange, n = 4) sorted excitatory (left) and inhibitory neurons (right) using genes that are also detected in GRO-seq. *P < 0.5, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant. See also Supplementary Figs. 7–9.
To directly compare individual genes across subcellular compartments, we next classified all 10,390 expressed genes into eight subgroups that reflect the total number of arrangements by which a gene can be misregulated across three given RNA pools (23 = 8). Groups B and D comprise 38.4% of expressed genes which are involved in neuronal projection and cellular stress, respectively, and represent expression changes that are misregulated in the same direction across nascent, nuclear, and whole cell RNA pools (Fig. 4f and Supplementary Fig. 7b). Among these groups of genes, log2 fold changes measured from the whole cell are significantly smaller than that in the nuclear compartment, suggesting that gene expression changes in the nucleus are post-transcriptionally minimized in the cell (Fig. 4f and Supplementary Fig. 7c). The majority of genes (48%), however, exhibit expression changes in nascent RNAs that are reversed in the whole cell compartment (Groups A,C,G,H; Fig. 4f). Groups A and C consist of relatively long, EXC-enriched genes that are transcriptionally downregulated in nascent RNA but post-transcriptionally upregulated in whole cell RNA (Fig. 4f and Supplementary Fig. 7d–e). DAVID gene ontology revealed that Group A genes are associated with synaptic functions and intracellular signaling (Fig. 4g). Groups G and H consist of considerably shorter, INH-enriched genes that are transcriptionally upregulated in nascent RNA but post-transcriptionally downregulated in whole cell RNA (Fig. 4f and Supplementary Fig. 7d–e), and Group G genes are functionally associated with cellular energy and metabolism in mitochondria (Fig. 4g). RT-PCR validation of primary and mature RNA transcripts for several genes from Groups A/C, B, and D recapitulated these apparent expression differences between subcellular compartments, particularly when genes were analyzed as a collective in their respective Group (Fig. 4h, Supplementary Fig. 8 and Supplementary Table 2). Notably, upon analyzing de novo transcriptional activity derived from GRO-seq for genes expressed in sorted excitatory or inhibitory neurons (Supplementary Fig. 7f), we found a trend towards long genes being more severely downregulated in both cell types bearing the R106W mutation than the T158M mutation (Fig. 4i and Supplementary Fig. 7g), consistent with a more severe phenotype in R106W mice.
To gain insight into the apparent switch in gene misregulation between subcellular compartments, we next used publically available HITS-CLIP datasets from the mouse brain to examine genes whose transcripts are typically bound and regulated by RNA-binding proteins (RBPs), and tested for associations with distinct subcellular gene expression changes in Mecp2 mutant neurons. K-means clustering of 10,390 cortically expressed genes across HITS-CLIP data for 12 RBPs (MBNL1-2, TDP43, FUS, TAF15, FMR1, HuR, APC, RBFOX1-3, and AGO2) identified 5 major gene clusters (Supplementary Fig. 9a). We found one subset of genes whose transcripts display significantly higher levels of HuR binding, but lower levels of AGO2 binding (RBP Clusters 1 and 4), and another subset showing significantly higher levels of AGO2 binding but lower levels of HuR binding (RBP Clusters 2, 3 and 5; Supplementary Fig. 9b–c). HuR binds to the 3′UTR of mRNA transcripts (Supplementary Fig. 9d) and is known to increase mRNA stability42. Conversely, AGO2 functions to promote mRNA degradation through AGO2-bound miRNAs43. Both HuR and AGO2 genes are also actively expressed in neurons at 6-weeks of age (Supplementary Fig. 9e). By comparing gene Groups A-H, which summarize subcellular gene expression changes in Mecp2 mutant neurons (Fig. 4f), to functionally-distinct RBP clusters (Supplementary Fig. 9a–c), we found that many downregulated nascent RNA transcripts from Groups A, B, and C are significantly associated with RBP Clusters 1 and 4 and are post-transcriptional targets of HuR (Supplementary Fig. 9f). By contrast, upregulated nascent RNA transcripts in Mecp2 mutant neurons, particularly from Groups G and H, show associations with RBP Clusters 2, 3, and 5, and are targets of AGO2-bound miRNAs (Supplementary Fig. 9f). The opposite functions of HuR and AGO2 in the post-transcriptional regulation of mRNA stability likely alter the abundance of cellular RNAs in a group- or cluster-specific manner. Therefore, gene expression differences between subcellular compartments in Mecp2 mutant mice could be post-transcriptionally mediated in part by RBPs (Supplementary Fig. 9g).
Female RTT Mouse Models Reveal Cell and Non-Cell Autonomous DEGs
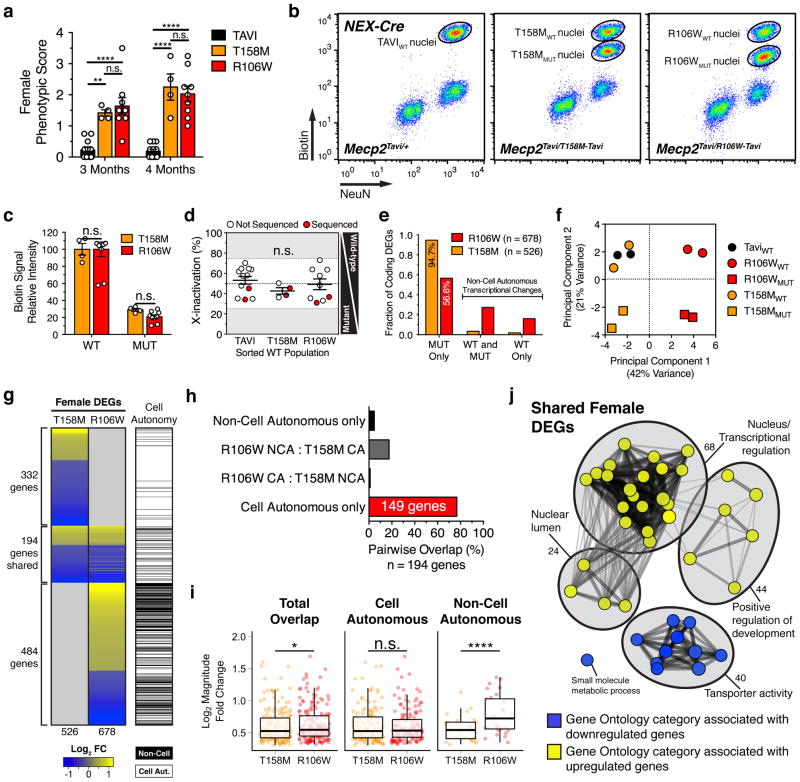
RTT is an X-linked disorder that primarily affects heterozygous females. However, the extent to which intermixed Mecp2 WT and mutant (MUT) neurons in cellular mosaic RTT females affect each other at the level of gene expression remains unknown. The reduced expression level of T158M and R106W mutant protein allowed us to use the same tagging and sorting strategy in male mice to isolate and profile WT (denoted by subscript: T158MWT, R106WWT) and MUT (T158MMUT, R106WMUT) cells from mosaic female mice. These include TAVI (Mecp2Tavi/+;R26cBirA/+;NEXCre/+), T158M (Mecp2Tavi/T158M-Tavi;R26cBirA/+;NEXCre/+), and R106W (Mecp2Tavi/R106W-Tavi;R26cBirA/+;NEXCre/+) heterozygous females that each carry a Tavi-tagged WT allele and a Tavi-tagged T158M, R106W, or untagged WT allele. Upon aging these mice to ~18 weeks, when T158M and R106W females both exhibit RTT-like phenotypes (Fig. 5a), cortical excitatory nuclei were isolated for FACS (Fig. 5b–c and Supplementary Fig. 10a). From the number of females sampled, we did not detect skewed XCI (> 75%) among excitatory neurons in TAVI, T158M, or R106W mice (Fig. 5d).
Figure 5.
T158M and R106W differentially expressed genes in mosaic female mice. (a) RTT-like phenotypic score in TAVI (n = 12), T158M (n = 4), and R106W (n = 9) heterozygous female mice (Two-way ANOVA). Data depicts mean ± SEM. (b) FACS isolation of excitatory neuronal nuclei from the cortex of heterozygous TAVI, T158M, or R106W female mice. (c) Biotin signal intensity from FACS-isolated populations depicted in (b) (nT158M = 4, nR106W = 9, Two-way ANOVA). Data depicts mean ± SEM. (d) X-inactivation ratios among cortical excitatory neurons in all sorted female mice, displayed as a percentage of the FACS-sorted WT population (nTAVI = 12, nT158M = 4, nR106W = 9, One-way ANOVA). Data points in red indicate samples used for RNA-seq. Data depicts mean ± SEM. (e) Bar graph showing the cell and non-cell autonomous distribution of total protein-coding DEGs identified from T158M and R106W female mice. (f) Principal component analysis of WT and MUT cell populations isolated from TAVI, T158M, and R106W female mice. (g) Heatmap displaying log2 fold changes among the total number of protein-coding DEGs detected in both WT and MUT populations from T158M or R106W female mice. Note genes that overlap across genotype (n = 194). (h) Proportion of cell autonomous (CA) and non-cell autonomous (NCA) genes that overlap between T158M and R106W female excitatory neurons (One-tailed Fisher’s Exact Test). (i) Boxplots comparing absolute log2 fold change between cell autonomous and non-cell autonomous shared DEGs (n = 185) between T158M and R106W female mice (One-tailed Wilcoxon Signed Rank). (j) Enrichment map of pre-ranked GSEA functional network associations (P-value < 0.01, Q-value < 0.1). Data represents DEGs that overlap between T158M and R106W mice (n = 185). Nodes denote functional categories, colored by Normalized Enrichment Score (NES). Line weight denotes extent of gene overlap between connected nodes. *P < 0.5, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = not significant. See also Supplementary Fig. 10.
By comparing the gene expression profiles of WT or MUT neurons from heterozygous mutant mice to those from control mice (TAVIWT), a total of 526 and 678 unique protein-coding DEGs in T158MWT and MUT and R106WWT and MUT neurons were identified, respectively (Fig. 5e, Supplementary Fig. 10b and Table 1). Most DEGs represent cell autonomous gene expression changes that occur in mutant neurons alone (Fig. 5e). However, a larger proportion of R106W DEGs are also found in R106WWT neurons (43.4%; Fig. 5e), revealing a mutation-specific susceptibility of WT neurons to non-cell autonomous gene expression changes in heterozygous females. Using principal component analysis (PCA) to plot the first two major axes of transcriptome variation, we found that PC2 separates neuronal populations by Mecp2 allele status (WT vs. MUT neurons) irrespective of the T158M or R106W mutation (Fig. 5f), indicating that Mecp2 mutations induce cell autonomous changes that are transcriptionally distinct from neighboring wild-type neurons. However, PC1 accounts for nearly twice the variation as PC2 and clusters R106WWT and MUT populations away from other genotypes, likely due to the extensive number of indirect DEGs associated with this mutation. In contrast, against PC1 and PC2, T158MWT neurons closely resemble TAVIWT (Fig. 5f), indicating that the non-cell autonomous DEGs observed in R106WWT neurons specifically arise due to the increased severity of R106W mutation in R106WMUT neurons.
We further found 194 DEGs that overlap between T158M and R106W female mice, most of which are misregulated in the same direction (Fig. 5g). Among these genes, cell autonomous transcriptional changes (149 genes, 76.8%) are more likely to be shared across independent Mecp2 mutations than non-cell autonomous changes (9 genes, 4.6%; Fig. 5h). These overlapping DEGs also show higher fold changes in R106W than T158M female mice, but this difference is mainly driven by indirect DEGs in R106W neurons (Fig. 5i).
In R106W female mice, we found that non-cell autonomous DEGs are predominantly upregulated (~60%) in contrast to cell autonomous DEGs (~48%; Supplementary Fig. 10b), and display significantly higher fold changes than cell autonomous DEGs (Supplementary Fig. 10c). Furthermore, cell autonomous DEGs are considerably longer in gene length, specifically among upregulated genes (Supplementary Fig. 10d). To determine if cell and non-cell autonomous DEGs represent distinct biological processes, we also performed pre-ranked GSEA (FDR < 0.1) and found that non-cell autonomous gene expression changes primarily affect cell-to-cell signaling and negative regulation of protein phosphorylation (Supplementary Fig. 10e). These DEGs include several immediate early and late response genes that are induced by neuronal activity and modulate signaling pathways associated with synaptic plasticity44. In contrast, cell autonomous DEGs are significantly associated with transcriptional regulation (Supplementary Fig. 10f). These functional categories demonstrate a marked resemblance to those observed in excitatory neurons of male T158M and R106W mice (Fig. 5j). The striking consistency with which these functional annotations characterize Mecp2 mutant neurons, despite apparent differences in age and sex, supports the cell autonomous disruption of these functions as a key, contributing factor to RTT pathogenesis.
Discussion
The complexity of MeCP2 molecular function, coupled with the cellular heterogeneity of the brain, confounds the study of transcriptional changes in RTT. We thus combined in vivo biotinylation with Cre-Lox technology to label both wild-type and mutant MeCP2 from different neuronal populations and examined RTT-associated transcriptomes in mice. Notably, the 23AA Tavi tag can be readily used to target any given protein using CRISPR-Cas9 technology45 for cell type-specific biochemical purifications, molecular profiling, and imaging applications.
By using an allelic series of RTT mutations to perform a transcriptome analysis of cortical neurons, we identified similarities and differences in gene expression features that couple impairments in MeCP2’s ability to bind DNA to RTT phenotypic severity. We found that lowly-expressed, cell type-enriched genes are sensitive to the effects of both T158M and R106W mutations, which likely contributes to the specificity of MeCP2-mediated gene expression changes among different neuronal cell types. Both mutations also display conserved transcriptional features among upregulated DEGs in male and female neurons, which include genes encoding INH-enriched transcription factors and chromatin remodelers. The upregulation of transcriptional regulators could contribute to the shared RTT etiology between T158M and R106W mice, as well as the genome-wide trend towards increased transcription of shorter, INH-enriched genes associated with cellular respiration and energy metabolism. This provides transcriptional insight into clinical features among both mildly and severely affected RTT patients that resemble mitochondrial and metabolic disorders46.
However, the greater impairment in MeCP2 R106W binding to neuronal chromatin associates with increased RTT phenotypic severity, and notably correlates with the larger number and degree of misregulated genes that are more highly expressed and predominately downregulated relative to the T158M mutation. These transcriptional differences extend to most long genes throughout the genome, which are highly expressed in neurons47. Our datasets are in partial agreement with global reductions in Ser5-phosphorylated RNA polymerase in Mecp2-null neuronal nuclei48, supporting MeCP2 as a global modulator of gene transcription. Loss of MeCP2 occupancy may either alter local chromatin organization, which could decrease the efficiency of transcriptional elongation49 and lead to the downregulation of long genes, or may abrogate HDAC3-mediated recruitment of transcription factors required for long gene transcriptional activation50. Because downregulated genes associate with synaptic morphology and function, and R106W mice have reduced lifespans compared to T158M mice, reductions in long gene transcription may act as modifiers to worsen RTT-like phenotypes. RTT patients with mutations that preserve MeCP2 binding do exhibit milder features than patients for whom binding is disrupted12. Transcriptional assessments with mutations preserving MeCP2 binding are thus necessary to further refine these genotype-phenotype correlations.
Because MeCP2 is a DNA-binding nuclear protein22, nuclear and nascent RNA pools provide additional insights into the primary effects of RTT mutations on transcriptional activity37,38,51 that complement the reported whole cell upregulation of long genes in RTT27,39. Whole cell RNA is subject to post-transcriptional regulation40,52,53, being notably enriched for cytoplasmic mRNAs that are bound by various RBPs to modulate their steady-state abundance and turnover. We found that many downregulated nascent RNA transcripts are targets of HuR, which increases mRNA stability in the brain42 and may post-transcriptionally upregulate these transcripts in whole cell RNA. Upregulated nascent RNA transcripts tend to associate with miRNA-bound AGO2, which may post-transcriptionally mitigate their upregulation by increasing rates of mRNA decay43. These post-transcriptional mechanisms may abate cellular consequences arising from global alterations in synaptic, mitochondrial, and metabolic gene transcription. Whole cell gene expression changes in RTT may thus be compensatory and not entirely representative of transcriptional activity, questioning the therapeutic benefit of decreasing long gene transcription for treating RTT. Identifying RBPs that contribute to cellular compensation may yield a novel class of interventional therapies administered prior to or during the initial phases of RTT, minimizing its pathological impact.
Finally, our approach allows for the molecular profiling of mosaic neurons from female mice that represent accurate preclinical RTT models, revealing non-cell autonomous changes in WT neurons that depend on mutation severity. However, to better elucidate direct and indirect contributions to RTT, further investigation requires examination of females with a wide range of XCI ratios across multiple ages, cell types, and Mecp2 mutations. Non-cell autonomous DEGs include genes induced by neuronal activity to reduce synaptic responsiveness to excessive neuronal stimuli44. Nuclear RNA transcripts of two late-response genes in particular, Bdnf and Igf1, were found to be transcriptionally upregulated in WT and MUT neuronal nuclei of 18-week old R106W female mice. As Bdnf and Igf1 encode neuroprotective peptides that ameliorate RTT symptoms20, the selective upregulation of non-cell autonomous DEGs in R106W mice may be a protective response to increased neuronal activity or stress among severely affected mosaic neurons. Currently, BDNF and IGF-1 peptides are being tested in clinical trials54,55. Further study of molecular pathways associated with non-cell autonomous DEGs may thus reveal additional RTT therapeutic targets and avenues.
Data availability
All sequencing data reported in this study has been deposited in the NCBI Gene Expression Omnibus (GSE83474). Mouse lines generated from this study have been deposited at The Jackson Laboratory (Bar Harbor, ME) under the following stock numbers: R26cBirA (Stock #030420), Mecp2Tavi (Stock #030422), Mecp2T158M-Tavi (Stock #029642), and Mecp2R106W-Tavi (Stock #029643).
Online Methods
Generation of Mouse Lines
The targeting construct used for homologous recombination at the Mecp2 locus in murine ES cells was cloned in two arms by PCR amplification of sv129 genomic DNA. The 5′ arm was PCR amplified with 5′-AGGAGGTAGGTGGCATCCTT-3′ and 5′-CGTTTGATCACCATGACCTG-3′ primers, whereas the 3′ arm was PCR amplified with 5′-GAAATGGCTTCCCAAAAAGG-3′ and 5′-AAAACGGCACCCAAAGTG-3′ primers. Restriction sites at the ends of each arm were created using nested primers for cloning into a vector containing a loxP-flanked neomycin cassette (Neo) and a diphtheria toxin A negative-selection cassette. QuikChange (Stratagene) insertional mutagenesis was used to generate the Mecp2Tavi targeting construct by inserting the Tavi tag immediately upstream of the Mecp2 stop codon within the 5′ arm. The portion of the Tavi tag containing the biotinylation consensus sequenced flanked by 5′ NaeI and 3′ BspHI restriction sites was inserted through two rounds of mutagenesis:
Round 1 Forward: 5′-GACCGAGAGAGTTAGCGCCGGCCTGAACGACATCTTCGAGTCATGACTTTACATAGAGCG-3′
Round 1 Reverse: 5′-CGCTCTATGTAAAGTCATGACTCGAAGATGTCGTTCAGGCCGGCGCTAACTCTCTCGGTC-3′
Round 2 Forward: 5′-CTGAACGACATCTTCGAGGCTCAGAAAATCGAATGGCACGAATCATGACTTTACATAGAG-3′
Round 2 Reverse: 5′-CTCTATGTAAAGTCATGATTCGTGCCATTCGATTTTCTGAGCCTCGAAGATGTCGTTCAG-3′
The portion of the tag containing the TEV protease cleavage site was inserted upstream of the NaeI restriction site with a third round of mutagenesis:
Round 3 Forward: 5′-GACCGAGAGAGTTAGCGAAAACCTGTATTTTCAGGGCGCCGGCCTGAACGACATC-3′
Round 3 Reverse: 5′-GATGTCGTTCAGGCCGGCGCCCTGAAAATACAGGTTTTCGCTAACTCTCTCGGTC-3′
To generate Mecp2Tavi targeting constructs bearing independent RTT-associated point mutations, QuikChange site-directed mutagenesis was used to mutate MeCP2 arginine 106 to tryptophan and MeCP2 threonine 158 to methionine within the 3′arm and 5′arm, respectively. A single nucleotide at codon T160 also underwent site-directed mutagenesis for a silent mutation to introduce a BstEII restriction site to correctly identify targeted ES cells.
To generate conditional R26cBirA transgenic mice, PCR primers containing AscI restriction sites and a Kozak consensus sequence were used to subclone the BirA coding sequence and insert it downstream of both a CAG promoter and a floxed transcriptional attenuator, Neo-STOP, within pROSA26-1, a transgenic targeting vector that has previously been characterized56.
After confirmation by Sanger sequencing and linearization with NotI (Mecp2Tavi targeting construct and its mutant variants) or SgfI (cBirA targeting construct), the constructs were electroporated into sv129-derived murine ES cells. Correctly targeted ES cells were independently injected into C57BL/6 blastocysts and subsequently implanted into pseudopregnant females. Agouti offspring were screened by southern blot and PCR genotyping to confirm germline transmission of the Mecp2Tavi, Mecp2T158M-Tavi, Mecp2R106W-Tavi, and R26cBirA alleles. In the case of the Mecp2Tavi allele and its mutant variants, the resulting offspring were mated with C57BL/6 EIIa-cre mice to ensure germline deletion of the floxed Neo cassette between Mecp2 exons 3 and 4.
Additional Mouse lines
Dlx5/6-Cre (Stock #008199) and EIIa-Cre (Stock #003724) mice were obtained from The Jackson Laboratory (Bar Harbor, ME)31,36. NeuroD6/NEX-Cre mice were obtained with permission from the Nave Laboratory35.
Animal Husbandry
Experiments were conducted in accordance with the ethical guidelines of the US National Institutes of Health and with the approval of the Institutional Animal Care and Use Committee of the University of Pennsylvania. All of the experiments described were performed using mice on a congenic sv129:C57BL/6J background with the knock-in/transgenic alleles backcrossed to C57BL/6J mice (The Jackson Laboratory) for at least five generations, unless otherwise stated. Mice were housed in a standard 12h light/12h dark cycle with access to ample amounts of food and water. Mice bearing the Tavi tag were genotyped using a bipartite primer PCR-based strategy to detect the Tavi tag at the 3′-end of the endogenous Mecp2 gene (Forward: 5′-CACCCCGAAGCCACGAAACTC-3′, Reverse: 5′-TAAGACTCAGCCTATGGTCGCC-3′) and give rise to a 318-bp product from the wild-type allele and a 388-bp product from the tagged allele. Mice bearing the BirA transgene were genotyped using a tripartite primer PCR-based strategy to detect the presence or absence of the CAG promoter at the Rosa26 locus (Forward:5′-TGCTGCCTCCTGGCTTCTGAG-3′, Reverse #1: 5′-GGCGTACTTGGCATATGATACAC-3′, Reverse #2: 5′-CACCTGTTCAATTCCCCTGCAG-3′) and give rise to a 173-bp product from the wild-type allele and a 477-bp product from the transgene-bearing allele. Mice bearing Cre-recombinase (either NeuroD6/NEX-Cre or Dlx5/6-Cre) were genotyped using PCR-based strategies as previously described35,36.
Phenotypic Assessment
For tagged Mecp2 knock-in mice, phenotypic scoring was performed on a weekly basis for the presence or absence of overt RTT-like symptoms as previously described33. Investigator was blinded to genotypes during phenotypic assessment of mice. For BirA transgenic mice, no formal scoring was performed. However, R26BirA heterozygous and homozygous mice are viable, fertile, and devoid of any gross abnormalities, consistent with previously engineered transgenic mice that express BirA either ubiquitously or within restricted tissues using cell type-specific promoters57,58.
Immunofluorescence and Microscopy
Mice were anesthetized with 1.25% Avertin (wt/vol), transcardially perfused with 4% paraformaldehyde (wt/vol) in 0.1M sodium-potassium phosphate buffered saline and postfixed overnight at 4°C. Brains were coronally or sagittally sectioned at 20μm using a Leica CM3050 S cryostat. Immunofluorescence on free-floating sections was performed as previously described18, except sections were permeabilized with 0.5% Triton without methanol for 20 minutes, and sections were blocked overnight with 10% Normal Goat Serum and 1:100 unconjugated goat anti-mouse IgG (Sigma M5899). The following primary antibodies were incubated at 4°C overnight: rabbit anti-MeCP2 C-terminus (1:1000, in house), rabbit anti-nucleolin (1:1000, Abcam ab22758), mouse anti-parvalbumin (1:500, Millipore MAB1572), rabbit anti-calretinin (1:1000, Swant 7699/3H), mouse anti-GAD67 (1:500, Millipore MAB5406), mouse anti-NeuN (1:500, Millipore MAB377). For rat anti-somatostatin (1:250, Millipore MAB354MI), primary incubation was performed for 48 hours at 4°C. Fluorescence detection of primary antibodies was performed using Alexa 488-conjugated goat anti-rabbit (1:1000, Invitrogen A11008), Alexa 488-conjugated goat anti-mouse (1:1000, Invitrogen A11029), or Alexa 488 goat anti-rat (1:1000, Invitrogen A11006). Fluorescence detection of biotin was performed simultaneously with Streptavidin Dylight 650 (1:1000, Fisher 84547) for fluorescence microscopy and Streptavidin Dylight 550 (1:1000, Fisher 84542) for confocal microscopy. Sections were counterstained with DAPI (1:1000, Affymetrix 14564) to visualize DNA before mounting with Fluoromount G (SouthernBiotech). Images were acquired using a Leica DM5500B fluorescent microscope with a Leica DFC360 FX digital camera (region-specific biotinylation, quantification of neuronal cell type-specific markers) or a Leica TCS SP8 Multiphoton confocal microscope (representative images of neuronal cell type specific markers, subcellular localization of MeCP2). Images were acquired using identical settings for laser power, detector gain amplifier offset and pinhole diameter in each channel. Image processing was performed using ImageJ and Adobe Photoshop, including identical adjustments of brightness, contrast, and levels in individual color channels and merged images across genotypes.
Quantitative Western analysis
Quantitative Western blot was performed using Odyssey Infrared Imaging System (Licor). Primary antibodies used in this study include rabbit anti-MeCP2 C-terminus (1:4000, in house), mouse anti-MeCP2 N-terminus (1:4000, Sigma M7433), mouse anti-NeuN (1:500, Millipore MAB377), rabbit anti-Avi tag (1:5000, Abcam ab106159, listed as anti-Tavi in main text, detects the minimal peptide substrate of biotin ligase BirA regardless of biotinylation status), rabbit anti-HDAC3 (1:1000, Santa Cruz sc-11417), rabbit anti-TBLR1 (1:1000, Bethyl A300–408A), rabbit anti-Sin3A, (1:500, Thermo Scientific PA1-870), rabbit anti-Histone H3 (1:1000, Abcam ab1791), and rabbit anti-TBP (1:1000, Cell Signaling #8515). Secondary antibodies include anti-rabbit IRDye 680LT (1:10,000, Licor), anti-mouse IRDye 800CW (Licor), Streptavidin Dylight 650 (1:10,000, Fisher 84547) and Streptavidin Dylight 800 (1:10,000, Fisher 21851). Quantification of protein expression levels was carried out following Odyssey Infrared Imaging System protocols. Scans of full-length Western blot membranes are provided in Supplementary Figs. 11–13.
Co-immunoprecipitation using nuclear extracts
Tissues were mined on ice and homogenized in ice cold lysis buffer (10 mM HEPES pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.5% NP-40, 0.2mM EDTA, protease inhibitors). Nuclei were pelleted, washed and resuspended in nuclear extract (NE) buffer (20mM HEPES pH 7.9, 1.5mM MgCl2, 500mM KCl, 0.2mM EDTA, 10% glycerol, protease inhibitors). Nuclei were incubated in NE buffer at 4°C for two hours with rotation. Samples were cleared by ultracentrifugation with a TLA 100.3 rotor (Beckman Optima TL) at 4°C for 30 minutes and the supernatant taken for nuclear extract. Protein concentration was quantified using a modified Bradford assay (Bio-Rad). 1mg of nuclear extract was adjusted to 300μl total volume with NE buffer to perform IP in duplicate. Protein G Dynabeads or Streptavidin M-280 Dynabeads (Life Technologies) were washed three times in PBS with 0.1% Tween-20 and 0.1% BSA. Nuclear extracts were cleared for 30 minutes at 4°C with 25μl Protein G Dynabeads. For streptavidin pulldown, 50μl of Streptavidin M-280 Dynabeads were added to the nuclear extract and incubated at 4°C for two hours with rotation. To test if the Tavi tag was required for streptavidin pulldown, nuclear extracts were split and incubated with or without 200U TEV protease (Invitrogen) in the absence of a reducing agent and without agitation at 4°C for ≥ 4 hours prior to IP. For antibody immunoprecipitations, 5μg of antibody was added to the nuclear extract and incubated overnight at 4°C with rotation. Protein G beads were blocked in wash buffer overnight at 4°C with rotation. Blocked beads were then incubated with antibody-bound nuclear extract for two hours at 4°C with rotation. Beads were washed four times in PBS with 0.1% Tween-20 and split into two equal volumes. Each sample was resuspended in 25μl loading buffer with 50mM DTT and boiled for 10 minutes at 95°C prior to loading on a 4–12% Bis-Tris NuPage gel (Life Technologies).
Chromatin immunoprecipitation
Forebrain tissues from male mice at 20 weeks of age were homogenized in cross-linking buffer (1% formaldehyde (wt/vol), 10mM HEPES (pH 7.5), 100mM NaCl, 1mM EDTA, 1mM EGTA) and cross-linked for 5 minutes at RT. After quenching with 125mM glycine, cross-linked tissue was washed with ice-cold PBS and dounced with 16 strokes in lysis buffer (50mM HEPES (pH 7.5), 140mM NaCl, 1mM EDTA, 1mM EGTA, 10% glycerol (vol/vol), 0.5% NP-40 (vol/vol), and 0.25% Triton X-100 (vol/vol) with protease inhibitors). Nuclei were pelleted, washed and resuspended in chromatin buffer (10mM Tris-HCl (pH 8.0), 1mM EDTA, and 0.5mM EGTA with protease inhibitors). Chromatin was sonicated using a Diagenode Bioruptor, and salt and detergent were added to adjust the chromatin buffer to 0.5% Triton X-100, 150mM NaCl, 10mM EDTA, and 0.1% sodium deoxycholate (DOC, vol/vol), and precleared at 4°C with Protein A Dynabeads (Invitrogen). For immunoprecipitation, 3μg of purified rabbit anti-MeCP2 IgG (in house) or non-specific rabbit IgG control (Millipore NI01) was incubated with 45μg of chromatin for 4 hours, followed by an overnight incubation with pre-blocked Protein A Dynabeads, at 4°C with rotation. Bead-bound chromatin was washed with low salt buffer (50mM HEPES pH 7.5, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% DOC), high salt buffer (50mM HEPES pH 7.5, 500mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% DOC), LiCl buffer (50mM Tris-HCl pH 8.0, 150mM NaCl, 1mM EDTA, 0.5% NP-40, 0.5% DOC) and TE buffer (10mM Tris-HCl pH 8.0, 1mM EDTA). Chromatin was eluted with elution buffer (50mM Tris-HCl pH 8.0, 10mM EDTA, and 1% SDS (wt/vol)), digested with proteinase K (0.5mg ml−1), and reversed crosslinked at 65°C overnight. After RNase A treatment, DNA fragments were extracted with phenol/chloroform and ethanol-precipitated.
Quantitative real-time PCR (qPCR) analysis was carried out using SYBR green detection (Life Technologies) on an ABI Prism 7900HT Real-Time PCR System (Applied Biosystems). The percent input for each amplicon was determined by comparing the average threshold cycle of the immunoprecipitated DNA to a standard curve generated using serial dilutions of the input DNA and interpolating the “fraction of input” value for this sample.
Sub-nuclear Fractionation
To prepare nucleoplasm-enriched proteins, cortices were dounce homogenized in 5 ml NE10 buffer (20 mM HEPES, pH 7.5, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, and 15 mM β-mercaptoethanol) 30 times using a loose pestle. The resulting nuclei were washed with NE10 buffer and rotated in NE300 buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, and 15 mM β-mercaptoethanol) for 1 hour at 4°C. Samples were centrifuged at 500 g for 5 minutes, and the supernatant, which represents the nucleosolic fraction, was collected and saved. The insoluble pellet, consisting of the chromatin-bound fraction, was washed in NE150 buffer and incubated with 500 units of benzonase (Sigma-Aldrich) for 5 minutes at room temperature. The pellet was then resuspended in 50 μl NE150 buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, and 15 mM β-mercaptoethanol) and rotated for 1 hour at 4°C. Samples were centrifuged at 16,000 g, and the supernatant was collected as the chromatin-bound fraction.
FACS Isolation of Neuronal Nuclei for RT-PCR and RNA-seq
Nuclei were isolated from fresh cortical tissue for FACS under ice-cold and nuclease-free conditions. Mouse cortices were rapidly resected on ice and subjected to dounce homogenization in homogenization buffer (0.32M sucrose, 5mM CaCl2, 3mM MgAc2, 10mM Tris-HCl pH 8.0, 0.1% Triton, 0.1mM EDTA, Roche Complete Protease Inhibitor without EDTA). Homogenates were layered onto a sucrose cushion (1.8M sucrose, 10mM Tris-HCl pH 8.0, 3mM MgAc2 Roche Complete Protease Inhibitor without EDTA) and centrifuged in a Beckman Coulter L7 Ultracentrifuge at 25,000 rpm at 4°C for 2.5 hours using a Beckman Coulter SW28 swinging bucket rotor. Nuclei were resuspended & washed once in blocking buffer (1× PBS, 0.5% BSA (Sigma A4503), RNasin Plus RNase Inhibitor (Promega)) and pelleted using a tabletop centrifuge at 5000 RCF at 4°C for 10 minutes. Nuclei were resuspended in blocking buffer to a concentration of ~6×106 nuclei/ml, blocked for 20 minutes at 4°C with rotation, then incubated with Streptavidin Dylight 650 (1:1000, Fisher 84547) and Alexa 488-conjugated anti-NeuN antibody (1:1000, Millipore MAB377X) for 30 minutes at 4°C with rotation. After a 5-minute incubation with 1:1000 DAPI to enable singlet detection during FACS, labeled nuclei were washed for an additional 30 minutes at 4°C with blocking buffer, pelleted and resuspended in blocking buffer with 1% BSA. A BD Biosciences Influx cell sorter at the University of Pennsylvania Flow Cytometry and Cell Sorting Facility was used to identify cell type-specific populations of nuclei, and 1.2 – 2.5 ×105 singlet nuclei from specified populations were directly sorted into Qiagen Buffer RLT Plus for immediate lysis and stabilization of RNA transcripts. Total nuclear RNA was processed using the Qiagen AllPrep DNA/RNA mini kit according to manufacturer instructions, with exception to the on-column DNaseI treatment. RNA was eluted from RNeasy mini spin columns and treated with DNaseI (Qiagen 79254) for 25 minutes at room temperature, then precipitated with glycogen/NaOAc and stored in ethanol at −80°C. Ethanol precipitation of nuclear RNA was carried out to completion prior to initiating RT-PCR or RNA-seq library construction.
For RNA-seq, total RNA was prepared from FACS-isolated cortical nuclei of male mice at 6 weeks (TAVI, T158M, R106W, 2–3 mice pooled per biological replicate, 4 independently-sorted biological replicates total) and female mice at 18 weeks (TAVI, T158M, R106W, 1 single mouse per biological replicate, 2 independently-sorted biological replicates total). No method of randomization was used to determine how animals were allocated to experimental groups, which was determined by genotype with matching age and sex. The numbers of biological replicates used for differential gene expression analysis are in compliance with ENCODE consortium long RNA-seq recommendations (≥2 replicates). Furthermore, the total amount of RNA isolated from 120,000–250,000 sorted nuclei was used as input for library construction; hence differential gene expression comparisons between FACS-isolated Mecp2 control and mutant neurons are performed using RNA from equivalent numbers of neuronal nuclei. Total RNA was depleted of ribosomal RNAs, subjected to 5 minutes of heat fragmentation, and converted to strand-specific cDNA libraries using the TruSeq Total RNA library prep kit with RiboZero depletion (Illumina). Multiplexed libraries were submitted for 100 paired-end sequencing on the Illumina HiSeq 2000/2500 platform at the University of Pennsylvania Next Generation Sequencing Core facility, yielding approximately 30–40M total reads per library. 90–95% of total reads were uniquely mapped to the mouse Ensembl GRCm38/mm10 mouse genomic assembly.
Real-Time PCR (RT-PCR)
For RT-PCR of FACS-isolated cortical nuclei, total RNA was prepared (as described in preceding section) from 120,000 sorted nuclei of TAVI or R106W male mice at 6 weeks of age (2–3 mice pooled per biological replicate, 3 independently-sorted biological replicates total). For remaining RT-PCR assays, total RNA was isolated from whole tissue or unsorted cortical nuclei of WT, TAVI, T158M, or R106W mice as specified in figure legends (1 mouse per biological replicate, 3 biological replicates total). Total RNA was converted to cDNA with random hexamers using the SuperScript III First-Strand Synthesis System (Invitrogen). RT-PCR was performed on a ABI Prism 7900HT Real-Time PCR System (Applied Biosystems). To validate cell type-specific cortical nuclei populations (Fig. 2d and Supplementary Fig. 3i–j), exon-spanning Taqman gene expression assays to detect mRNA transcripts for the following genes: CRE (Mr00635245_cn), Mecp2 (Mm01193537_g1), Rbfox3 (Mm01248771_m1), Gfap (Mm01253033_m1), Aif1 (Mm00479862_g1), Mog (Mm00447824_m1), Slc17a7 (Mm00812886_m1), Tbr1 (Mm00493433_m1), Gad1 (Mm04207432_g1), Slc35a1 (Mm00494138_m1), Ht3ar (Mm00442874_m1), Pvalb (Mm00443100_m1), Sst (Mm00436671_m1), Pgk1 (Mm00435617_m1), Actb (Mm00607939_s1), β2m (Mm00437762_m1). A geometric mean was calculated to normalize mRNA expression levels to multiple housekeeping genes (Actb, β2m, and Pgk1), and cell type-enrichment for each sorted population was determined relative to the total mixed population of DAPI+ nuclei. For RT-PCR validation of low expressing genes and subcellular gene expression changes (Fig. 4h and Supplementary Figure 8), primers against primary transcripts and mRNAs were used (listed in Supplementary Table 2), and geometric means were calculated to normalize mRNA expression levels to multiple housekeeping genes (Actb, β2m, and Pgk1).
GRO-seq
Nuclei were isolated from fresh cortical tissue of TAVI or R106W male mice at 6 weeks of age (2 mice pooled per biological replicate, 2 biological replicates total) under ice-cold and nuclease-free conditions as described in the preceding section. After ultracentrifugation, nuclei were resuspended & washed once in PBS (1× PBS, RNasin Plus RNase Inhibitor (Promega)) and pelleted using a tabletop centrifuge at 5000 RCF at 4°C for 10 minutes. Nuclei were resuspended in PBS, pipetted through a 0.22μm filter and counted using a hemocytometer. Nuclei were then pelleted, resuspended to a concentration of 5×106 – 10×106 nuclei/100μl in glycerol storage buffer (50mM Tris pH 8.3, 40% glycerol, 5mM MgCl2, 0.1 mM), and flash frozen in liquid N2 for storage until needed.
For each nuclear run-on (NRO), 100μl of nuclei was mixed with 46.5μl NRO Reaction Buffer (10mM Tris pH 8.0, 5mM MgCl2, 1 mM DTT, 300mM KCl), 3.5μl Nucleoside Mix (50μM ATP, 50μM GTP, 2μM CTP, 50μM Br-UTP, 0.4U/μl RNasin), and 50μl 2% Sarkosyl Nuclear Run On Stop Solution (20mM Tris pH 7.4, 10mM EDTA, 2% SDS). The NRO reaction was performed at 30°C for 5 minutes, then terminated by a 20 minute incubation with DNAse I at 37°C, followed by a hour-long incubation with 225μl NRO Stop Buffer (20mM Tris, pH 7.4, 10mM EDTA, 2% SDS) and Proteinase K at 55°C. Phenol-extracted RNA was fragmented with 0.2N NaOH, and BrdU-RNA was isolated three consecutive times with BrdU-antibody beads, treated with enzymatic tobacco acid pyrophosphatase (TAP) and T4 polynucleotide kinase (PNK) to remove the cap and 3’-phosphate and to add a 5’-phosphate, as well as Illumina TruSeq small RNA sample prep kit adapter ligations between BrU-RNA isolation steps as described41,59.
RNA-seq Mapping, Read Counting, and Differential Expression Analysis
The mouse mm10 genomic sequence (Mus_musculus.GRCm38.75.dna.primary_assembly.fa.gz) and gene information (Mus_musculus.GRCm38.75.gtf.gz) were downloaded from Ensembl release 75. The genome files used for mapping were built by STAR (version 2.3.0)60 using the parameters ‘STAR --runMode genomeGenerate --runThreadN 12 --genomeDir ./ --genomeFastaFiles Mus_musculus.GRCm38.75.dna.primary_assembly.fa.gz --sjdbGTFfile Mus_musculus.GRCm38.75.gtf --sjdbOverhang 100′. The FASTQ files were mapped to the mouse Ensembl GRCm38/mm10 genome assembly by STAR (version 2.3.0) using the parameters ‘--genomeDir ENSEMBL_75_mm10 --runThreadN 10 --outFilterMultimapNmax 1 --outFilterMismatchNmax 3′. Perl scripts generated in-house were used to count the number of read pairs that mapped to genic regions (exon + intron) for each gene. If one end of a read pair overlapped with the annotated genomic region of a given gene and the other did not, the read pair was included in the final count for that gene. The total number of read pairs that overlapped within a given gene represented the final read count for that gene. All intron and exon-mapped reads were used for differentially expressed gene comparisons, which were performed by using the R packages “edgeR” (v3.10.0)61 and “DEseq2” (v1.8.0)62. Genes exhibiting low expression due to a substantially low number of mapped reads and whose edgeR CPM values satisfied the condition ‘rowSums(cpm(data_y)) < 2′ were filtered out from differential gene expression analyses. Conversely, genes with ‘rowSums(cpm(data_y)) ≥ 2′ were retained for differential gene expression analyses. A false discovery rate < 0.05 was set to identify differentially expressed genes, and no fold change cutoff was applied. For each comparison, the results of both edgeR and DESeq2 analyses were merged into a final non-redundant and FDR-controlled list of genes to avoid method-specific biases. The mean fold change and the mean FDR generated from both methods were used for generating plots and heatmaps.
RNA Binding Protein (RBP) Data Pre-processing and Analysis
To determine the enrichment of neuronally expressed RNA-binding proteins on gene transcripts, raw HITS-CLIP reads derived from the mouse brain were obtained from publically available datasets in the GEO repository (listed below). The quality of raw reads were assessed with FastQC63 and contaminants were removed using Trimgalore64 with parameters ‘-q 15 --length 20 --stringency 5′. Remaining reads were aligned to a mouse reference genome derived from the Ensembl v75 archived assembly using STAR (version 2.5)60 with parameters ‘--outFilterMultimapNmax 1 --outFilterScoreMinOverLread 0 --outFilterMatchNminOverLread 0.5 --alignEndsType Local’. Replicates were merged and then subsampled to match the sample with the lowest library size. Genome annotation from Ensembl v75 and in-house developed scripts facilitated the calculation of RNA binding protein coverage in gene models. Genes not included in Groups A-H (Figure 4f) were filtered out. For k-means clustering of RBPs, raw read counts were first normalized using the variance stabilizing transformation function in “DESeq2”62, and the R packages “factoextra”, “cluster”, and “NbClust” were used to perform a Silhouette coefficient analysis and cluster genes based on the optimal k number of clusters. The Kruskal-Wallis test, followed by a post-hoc Wilcoxon Rank Sum Test with Holm’s correction for multiple comparisons, was used to identify statistically significant differences in RNA-binding protein enrichment within each gene cluster. RBP gene clusters were then used to perform One-tailed Fisher’s Exact Test with genes from Groups A-H to identify RBP enrichments that significantly associate with genes displaying subcellular differences in gene expression changes in R106W mutant mice.
| HITS-CLIP datasets used for RBP Analysis | ||
|---|---|---|
| AGO2 (Rep 1–6/9–12) | GSE73058 | PMID: 26602609 |
| APC (Rep 1–4) | SRP042131 | PMID: 25036633 |
| MBNL1 (Rep 1–2) | GSE39911 | PMID: 22901804 |
| MBNL2 (Rep 1–3) | GSE38497 | PMID: 22884328 |
| ELAVL1 (Rep 1–2) | GSE45148 | PMID: 21784246 |
| FMR1 (Rep 1–2) | GSE45148 | PMID: 21784246 |
| FUS (Rep 1–3) | GSE40651 | PMID: 23023293 |
| TAF15 (Rep 1–2) | GSE43294 | PMID: 23416048 |
| TDP43 | GSE40651 | PMID: 23023293 |
| RBFOX1 | SRP030031 | PMID: 24213538 |
| RBFOX2 | SRP030031 | PMID: 24213538 |
| RBFOX3 (Rep 1–5) | SRP039559 | - |
Functional Enrichment of Differentially Expressed Genes
For DAVID gene ontology, a list of differentially expressed protein-coding genes was compared to a background list of actively expressed protein-coding genes from their respective cell type. Statistically significant terms (Benjamini P < 0.01, FDR < 0.05) were plotted for Figures S3C-D. For Gene Set Enrichment Analysis (GSEA), we performed a seeded, pre-ranked GSEA from lists of differentially expressed protein-coding genes (ranked by fold change) using the September 2015 Mouse GO Gene Set Release (http://download.baderlab.org/EM_Genesets/September_24_2015/Mouse/). GSEA network associations (P-value < 0.1, Q-value < 0.1) were visualized using the Enrichment Map application (v2.0.1) in Cytoscape (v3.2.1)65,66, and clustered using gene set overlap coefficients.
Principal Components Analysis (PCA)
PCA analyses were performed with the top 500 genes exhibiting the highest row variance using the “plotPCA” function in the R package “DESeq2”. Principal components were plotted using Graphpad Prism version 6.0 for Mac (GraphPad Software, La Jolla California USA, www.graphpad.com).
Determination of Actively Expressed Genes
Actively expressed genes for excitatory and inhibitory neurons were determined by calculating the normalized FPKM (zFPKM) and using ZFPKM ≥ 3 for the active gene cutoff as previously described67.
Statistical Analyses
Statistical analyses were performed using Graphpad Prism version 6.0 for Mac (GraphPad Software, La Jolla California USA, www.graphpad.com) and R68. No statistical method was used to estimate sample size, as pre-specified effect sizes were not assumed. No animals or samples were excluded from analyses. Individual statistical tests are fully stated in the main text or figure legends. Comparisons of normally distributed data consisting of two groups with equal variances (F-test equality of variance P > 0.05) were analyzed using Student’s T-test, and unequal variances (F-test equality of variance P < 0.05) using Students T-test with Welch’s correction for unequal variance. Comparisons of normally distributed data consisting of three or more groups were analyzed using One-way ANOVA with the appropriate post-hoc test. Comparison of two or more factors across multiple groups was analyzed using a Two-way ANOVA with Sidak’s correction for multiple comparisons. Comparisons of non-normally distributed data were analyzed using the Mann-Whitney/Wilcoxon test (two groups) or the Kruskal-Wallis test (three or more groups) with the appropriate post-hoc test. For multiple comparisons, all p-values are adjusted using the Holm-Bonferroni correction unless otherwise indicated. Experimental design and analytical details are also listed in the Life Sciences Reporting Summary.
Main Figure Statistical Analyses
Figure 1 Utilization and characterization of Mecp2Tavi mice and associated RTT variants
(e) nreplicates = 3, One-way ANOVA [F = 25.55, P = 0.0012]; Tukey’s multiple comparisons correction applied. (f) Left, nWT = 4, nTAVI = 5, nT158M = 4, nR106W = 4; One-way ANOVA [F = 12.4, P = 0.0004]; Sidak’s multiple comparisons correction applied. Right, nWT = 4, nTAVI = 5, nT158M = 4, nR106W = 4;One-way ANOVA [F = 0.2977, P = 0.8264]; Sidak’s multiple comparisons correction applied. (g) nWT = 20, nTAVI = 11, nKO = 6, nT158M = 6, nR106W = 12; One-way ANOVA [F = 20.05, P < 0.0001]; Tukey’s multiple comparison correction applied. (j) nWT = 31, nTAVI = 23, nKO = 17, nT158M = 39, nR106W = 26, Mantel-Cox [χ2 = 109.3, df = 4, P < 0.0001].
Figure 2 Cell type-specific transcriptional profiling of neuronal nuclei
(d) nreplicates = 3, Two-way ANOVA, Control [Cell Type-Gene Interaction, F = 42.68, P < 0.0001; Cell Type, F = 222.0, P < 0.0001; Gene, F =80.03, P < 0.0001], Non-Neuronal [Cell Type-Gene Interaction, F = 12.47, P < 0.0001; Cell Type, F = 109.8, P < 0.0001; Gene, F = 7.655, P = 0.0027], EXC-specific [Cell Type-Gene Interaction, F = 4.376, P = 0.0198; Cell Type, F = 1227, P < 0.0001; Gene, F = 0.3267, P = 0.5756], INH-specific [Cell Type-Gene Interaction, F = 3.047, P = 0.0040; Cell Type, F = 646.5, P < 0.001; Gene, F = 2.916, P = 0.033]; Dunnett’s multiple comparisons correction applied.
Figure 3 T158M and R106W differentially expressed genes at 6 weeks of age
(c) One-tailed Wilcoxon Signed Rank, Excitatory PUpregulated = 4.357e-3, Excitatory PDownregulated = 7.345e-3, Inhibitory PUpregulated = 4.575e-09, Inhibitory PDownregulated = 1.684e-05. (e) Chi-square Goodness-of-Fit, Excitatory PT158M < 2.2e-16 [χ2 = 182.2, df = 2], Excitatory PR106W < 2.2e-16 [χ2 = 401.11, df = 2], Inhibitory PT158M < 2.2e-16 [χ2 = 119.94, df = 2], Inhibitory PR106W < 2.2e-16 [χ2 = 346.86, df = 2]. (f) Two-tailed Kruskal-Wallis Rank Sum, Excitatory P < 2.2e-16 [χ2 = 418.2, df = 3], Inhibitory P < 2.2e-16 [χ2 = 1026.9, df = 3]; Pairwise Wilcoxon Rank Sum P displayed.
Figure 4 Genome-wide length-dependent transcriptional changes in RTT mutant mice
(e) Top, n = 10,390 genes, Kolmogorov-Smirnov P < 2.2e-16 for each nascent or nuclear RNA versus whole cell RNA comparison, no correction for multiple comparisons. (f) n = 10, 390 genes, Kruskal-Wallis PGroup A < 2.2e-16 [χ2 = 2664.8, df = 2], PGroup B < 2.2e-16 [χ2 = 290.18, df = 2], PGroup C < 2.2e-16 [χ2 = 2403.3, df = 2], PGroup D < 2.2e-16 [χ2 = 319.36, df = 2], PGroup E < 2.2e-16 [χ2 = 1483.8, df = 2], PGroup F < 2.2e-16 [χ2 = 1385.8, df = 2], PGroup G < 2.2e-16 [χ2 = 2522.9, df = 2], PGroup H < 2.2e-16 [χ2 = 2442.7, df = 2]; Pairwise Wilcoxon Rank Sum P displayed. (h) Left, Primary Transcripts + mRNA RT-PCR (Group Trend), nGroup A/C = 7 genes, nGroup B = 5 genes, nGroup D = 5 genes, Two-way ANOVA, [Subcellular Compartment-Gene Group Interaction, F = 5.699, P = 0.0084; Subcellular Compartment, F = 1.419, P = 0.2436; Gene Group, F = 16.14, P < 0.0001]; Sidak’s multiple comparisons correction applied. Right, Primary Transcripts only RT-PCR (Group Trend), nGroup A/c = 7 genes, nGroup B = 5 genes, nGroup D = 5 genes, Two-way ANOVA, [Subcellular Compartment-Gene Group Interaction, F = 0.182, P = 0.8345; Subcellular Compartment, F = 0.1334, P = 0.7176; Gene Group, F = 15.12, P <0.0001]; Sidak’s multiple comparisons correction applied.
Figure 5 T158M and R106W differentially expressed genes in mosaic female mice
(a) Two-way ANOVA [Genotype-Time Interaction, F = 2.987, P = 0.0712; Genotype, F = 41.14, P < 0.0001; Time, F = 7.332, P = 0.0129; Subjects (matching), F = 1.873, P = 0.0744]. (b) FACS isolation of cortical mosaic excitatory neuronal nuclei from heterozygous TAVI, T158M, or R106W female mice. (c) nT158M = 4, nR106W = 9, Two-way ANOVA [Population-Genotype Interaction, F = 0.3320, P = 0.5703; Population, F = 111.1, P < 0.0001; Genotype, F = 0.332, P = 0.5703]. (d) nTAVI = 12, nT158M = 4, nR106W = 9, One-way ANOVA [F = 0.9376, P = 0.4067]. (h) One-tailed Fisher’s Exact Test [Odds Ratio = 19.3, P = 2.43e-05]. (i) One-tailed Wilcoxon Signed Rank, PTotal Overlap = 0.0331, PCell. Auto. = 0.5778, PNon-Cell Auto. = 8.825e-06.
Supplementary Material
Acknowledgments
We would like to thank the IDDRC Mouse Gene Manipulation Core at Children’s Hospital Boston (U54HD090255, M. Thompson), the Gene Targeting Core (P01DK049210, K. Kaestner) and the Transgenic and Chimeric Mouse Facility (J. Richa) at University of Pennsylvania for help in generating transgenic mice, the Flow Cytometry and Cell Sorting Resource Laboratory (H. Pletcher, W. DeMuth), and the Next Generation Sequencing Core (J. Schug) for technical assistance. B.S.J. is supported by a Cell and Molecular Biology Training Grant (TG32GM072290) and the UNCF/Merck Graduate Research Dissertation Fellowship. This work is supported by NIH grants K22AI112570 (G. V.), R21AI107067 and R01CA140485 (T.H.K.), R01MH091850 and R01NS081054 (Z.Z.), and a basic research grant from Rettsyndrome.org (Z.Z.). Z.Z is a Pew Scholar in the Biomedical Sciences.
Footnotes
Competing Financial Interest: The authors declare no competing financial interests.
Author Contributions: Conceptualization, B.S.J. and Z.Z.; Methodology, B.S.J., J.M.L., D.G., and Z.Z.; Investigation, B.S.J., Y.T.Z., M.F., J.M.L., K.H.W., Y.J.K., and D.B.; Formal Analyses, B.S.J., Y.T.Z., G.G., and T.H.K.; Validation, B.S.J., M.F., J.M.L., and G.V.; Resources, B.S.J., Y.T.Z. and Y.C.; Data Curation, Y.T.Z.; Writing – Original Draft, B.S.J.; Writing – Review & Editing, B.S.J., Y.T.Z., M.F. and Z.Z.; Visualization, B.S.J.; Project Administration and Funding Acquisition, Z.Z.
References
- 1.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–37. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 3.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 4.Lewis JD, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to Methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 5.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 6.Lyst MJ, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 8.Skene PJ, et al. Neuronal MeCP2 Is Expressed at Near Histone-Octamer Levels and Globally Alters the Chromatin State. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chahrour M, et al. MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Natl Acad Sci. 2015;112:5509–5514. doi: 10.1073/pnas.1505909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. Global Transcriptional and Translational Repression in Human-Embryonic-Stem-Cell-Derived Rett Syndrome Neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuddapah VA, et al. Methyl-CpG-binding protein 2 (MECP2) mutation type is associated with disease severity in Rett syndrome. J Med Genet. 2014;51:152–158. doi: 10.1136/jmedgenet-2013-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. [Accessed: 8th April 2016];RettBASE: Rett Syndrome Variation Database. Available at: http://mecp2.chw.edu.au/
- 14.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Gierasch LM, Woodcock CL. Rett syndrome-causing mutations in human MeCP2 result in diverse structural changes that impact folding and DNA interactions. J Biol Chem. 2008;283:20523–34. doi: 10.1074/jbc.M803021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho KL, et al. MeCP2 Binding to DNA Depends upon Hydration at Methyl-CpG. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 16.Ballestar E, Yusufzai TM, Wolffe AP. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry (Mosc ) 2000;39:7100–7106. doi: 10.1021/bi0001271. [DOI] [PubMed] [Google Scholar]
- 17.Brown K, et al. The molecular basis of variable phenotypic severity among common missense mutations causing Rett syndrome. Hum Mol Genet. 2015:ddv496. doi: 10.1093/hmg/ddv496. [DOI] [PMC free article] [PubMed]
- 18.Goffin D, et al. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat Neurosci. 2012;15:274–283. doi: 10.1038/nn.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker SA, et al. An AT-Hook Domain in MeCP2 Determines the Clinical Course of Rett Syndrome and Related Disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz DM, et al. Preclinical research in Rett syndrome: setting the foundation for translational success. Dis Model Mech. 2012;5:733–745. doi: 10.1242/dmm.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamonica JM, et al. Elevating expression of MeCP2 T158M rescues DNA binding and Rett syndrome–like phenotypes. J Clin Invest. 2017:127. doi: 10.1172/JCI90967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- 23.Fishell G, Heintz N. The Neuron Identity Problem: Form Meets Function. Neuron. 2013;80:602–612. doi: 10.1016/j.neuron.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Molyneaux BJ, et al. DeCoN: Genome-wide Analysis of In Vivo Transcriptional Dynamics during Pyramidal Neuron Fate Selection in Neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo A, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao YT, Goffin D, Johnson BS, Zhou Z. Loss of MeCP2 function is associated with distinct gene expression changes in the striatum. Neurobiol Dis. 2013;59:257–266. doi: 10.1016/j.nbd.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabel HW, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo JU, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rube HT, et al. Sequence features accurately predict genome-wide MeCP2 binding in vivo. Nat Commun. 2016;7:11025. doi: 10.1038/ncomms11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik HS, Henikoff S. A Simple Method for Gene Expression and Chromatin Profiling of Individual Cell Types within a Tissue. Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samaco RC, et al. A partial loss of function allele of Methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum Mol Genet. 2008;17:1718–1727. doi: 10.1093/hmg/ddn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–7. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, et al. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J Cell Sci. 2008;121:1128–1137. doi: 10.1242/jcs.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goebbels S, et al. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genes N Y N 2000. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- 36.Monory K, et al. The Endocannabinoid System Controls Key Epileptogenic Circuits in the Hippocampus. Neuron. 2006;51:455–466. doi: 10.1016/j.neuron.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatt DM, et al. Transcript Dynamics of Proinflammatory Genes Revealed by Sequence Analysis of Subcellular RNA Fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 39.Sugino K, et al. Cell-Type-Specific Repression by Methyl-CpG-Binding Protein 2 Is Biased toward Long Genes. J Neurosci. 2014;34:12877–12883. doi: 10.1523/JNEUROSCI.2674-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 41.Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci CMLS. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flavell SW, Greenberg ME. Signaling Mechanisms Linking Neuronal Activity to Gene Expression and Plasticity of the Nervous System. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, et al. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller M, Can K. Aberrant redox homoeostasis and mitochondrial dysfunction in Rett syndrome. Biochem Soc Trans. 2014;42:959–964. doi: 10.1042/BST20140071. [DOI] [PubMed] [Google Scholar]
- 47.Zylka MJ, Simon JM, Philpot BD. Gene Length Matters in Neurons. Neuron. 2015;86:353–355. doi: 10.1016/j.neuron.2015.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linhoff MW, Garg SK, Mandel G. A High-Resolution Imaging Approach to Investigate Chromatin Architecture in Complex Tissues. Cell. 2015;163:246–255. doi: 10.1016/j.cell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nott A, et al. Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat Neurosci. 2016 doi: 10.1038/nn.4347. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buxbaum AR, Yoon YJ, Singer RH, Park HY. Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol. 2015;25:468–475. doi: 10.1016/j.tcb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauger O, Lemoine F, Scheiffele P. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron. 2016;92:1266–1278. doi: 10.1016/j.neuron.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 54.Khwaja OS, et al. Safety, pharmacokinetics, and preliminary assessment of efficacy of mecasermin (recombinant human IGF-1) for the treatment of Rett syndrome. Proc Natl Acad Sci U S A. 2014;111:4596–4601. doi: 10.1073/pnas.1311141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lombardi LM, Baker SA, Zoghbi HY. MECP2 disorders: from the clinic to mice and back. J Clin Invest. 2015;125:2914–2923. doi: 10.1172/JCI78167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao C, et al. MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 57.de Boer E, et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 2003;100:7480–5. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Driegen S, et al. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14:477–82. doi: 10.1007/s11248-005-7220-2. [DOI] [PubMed] [Google Scholar]
- 59.Greer CB, et al. Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Rep. 2015;13:1444–1455. doi: 10.1016/j.celrep.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012:bts635. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrews S. [Accessed: 28th April 2017];Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 64.Krueger F. [Accessed: 28th April 2017];Babraham Bioinformatics - Trim Galore! Available at: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 65.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLOS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinforma Oxf Engl. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hart T, Komori HK, LaMere S, Podshivalova K, Salomon DR. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics. 2013;14:778. doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data reported in this study has been deposited in the NCBI Gene Expression Omnibus (GSE83474). Mouse lines generated from this study have been deposited at The Jackson Laboratory (Bar Harbor, ME) under the following stock numbers: R26cBirA (Stock #030420), Mecp2Tavi (Stock #030422), Mecp2T158M-Tavi (Stock #029642), and Mecp2R106W-Tavi (Stock #029643).