Summary
With antibiotic resistance increasing at alarming rates, targets for new antimicrobial therapies must be identified. A particularly promising target is the bacterial two-component system. Two-component systems allow bacteria to detect, evaluate and protect themselves against changes in the environment, such as exposure to antibiotics, and also to trigger production of virulence factors. Drugs that target the response regulator portion of two-component systems represent a potent new approach so far unexploited. Here we focus efforts on the highly virulent bacterium Francisella tularensis tularensis. Francisella contains only three response regulators, making it an ideal system to study. In this study, we initially present the structure of the N-terminal domain of QseB, the response regulator responsible for biofilm formation. Subsequently, using binding assays, computational docking and cellular studies, we show that QseB interacts with 2-aminoimidazole based compounds that impede its function. This information will assist in tailoring compounds to act as adjuvants that will enhance the effect of antibiotics.
Abbreviated Summary
Targets for new antimicrobial therapies must be identified to combat rising antibiotic resistance worldwide. The 2-aminoimidazole-based adjuvant small molecules resensitize bacteria to conventional antibiotics. To elucidate one potential mechanism of action, we provide evidence that these compounds interact with a key transcriptional regulator from Francisella sp. The knowledge gained form these studies will inform future compound development.
Introduction
Tularemia is a devastating infectious disease, caused by the bacterium Francisella tularensis tularensis, which the Centers for Disease Control and Prevention list as a Category A select agent (Sjöstedt, 2007). Unnervingly, F. tularensis tularensis is considered to be an extremely viable biological warfare agent thanks to its ease of aerosolization, high infectivity, and quick incapacitation of those infected. Astonishingly, as few as ten cells can lead to an infection, making it one of the most pathogenic bacterial species in existence (Sjöstedt, 2007).
The ability of F. tularensis tularensis to form protective biofilms significantly contributes to its environmental persistence, pathogenicity, and resistance to antimicrobials (Durham-Colleran et al., 2010; Sutera et al., 2014). Biofilms are communities of bacteria that adhere to surfaces and one another, and bacteria are believed to spend an estimated 80% of their time in this state. Within the biofilm, they are enclosed in an extremely protective extracellular matrix (Donlan, 2002), exhibit differential gene expression, and can be up to 1,000-fold more resistant to antibiotics and the host immune response than while in a free-swimming, planktonic state (Donlan and Costerton, 2002; Rasmussen and Givskov, 2006; Percival et al., 2011; Foreman et al., 2011). To address this serious infection, we have been working towards better understanding of the formation of Francisella biofilms and subsequently eradicating them via small molecule intervention.
Biofilms in Francisella are regulated by only a small number of proteins (Durham-Colleran et al., 2010). The regulatory transcription factor, QseB (formerly referred to as PmrA), is a DNA-binding protein that is critical for Francisella biofilm formation (Durham-Colleran et al., 2010; van Hoek, 2013). Two additional transcription factors, MglA and SspA, may also play a role in biofilm regulation by forming a ternary complex with QseB (Bell et al., 2010). In addition, QseB has also been shown to play a critical role in controlling virulence and resistance in Francisella sp. (Mohapatra et al., 2007; Durham-Colleran et al., 2010; van Hoek, 2013).
QseB is a response regulator protein that works in combination with a sensor kinase to form the well-known and ubiquitous communication module referred to as a two-component system (Stock et al., 2000). In a prototypical two-component system, the sensor kinase is a transmembrane histidine kinase that detects an external signal, and responds by autophosphorylating. The phosphate group is then transferred to the response regulator, changing it from an “inactive” to an “active” state. The activated response regulator then propagates the signal through transcriptional regulation. Structurally, response regulators are composed of an N-terminal receiver domain (which contains the phosphorylation site) and a C-terminal DNA-binding domain. A highly flexible linker region of varying length connects the two domains. Commonly, upon activation, the receiver domain forms a head-to-head dimer, bringing two DNA-binding domains more proximal in order to better bind the two half sites of the cognate promoter (Gao and Stock, 2009). To date, no high-resolution structural information for any QseB domain has been available. Such information, of course, can greatly assist in informing the design of new therapeutics.
Only three response regulators have been identified in Francisella species: QseB, KdpE, and FTN1452 (in F. tularensis novicida; FTT1543 in F. tularensis tularensis, Larsson et al., 2005). KdpE and FTN1452 have not yet been well characterized. QseB has been more extensively studied and is known to be involved in biofilm formation, virulence, and intracellular growth. Sixty-five genes are under the control of QseB in F. tularensis novicida (a common surrogate for F. tularensis tularensis). Nearly a fifth of these genes are located on the Francisella pathogenic island, which is essential for the disease state (Mohapatra et al., 2007; Hare and Hueffer, 2014). Deletion of QseB from the genome results in no loss of cellular growth, but two main phenotypes are associated with QseB, biofilm formation and intramacrophage replication (Mohapatra et al., 2007; Durham-Colleran et al., 2010). Studies have confirmed that QseB is a DNA binding protein, able to bind its own promoter and the promoter of a protein from the Francisella pathogenic island (Bell et al., 2010). It has also been established that QseB can be phosphorylated by sensor kinases, and that removal of the phosphorylation site impacts gene regulation (Bell et al., 2010). These findings are consistent with the functionality generally observed for a response regulator.
A cell-permeable family of 2-aminoimidazole (2-AI) derivatives are known to interact with response regulators (Thompson et al., 2012). 2-AIs have been widely shown to inhibit and disperse biofilms and also to work synergistically with traditional antibiotics to re-sensitize multidrug-resistant bacteria (Richards, Ballard, et al., 2008; Richards, Huigens III, et al., 2008; Rogers and Melander, 2008; Ballard et al., 2008; Brackett et al., 2014). The specific mechanism by which 2-AIs interact with response regulators remains mostly unknown. It has been suggested that 2-AIs bind at the interface between the N-terminal receiver and C-terminal DNA-binding domains of response regulators, as a 2-AI compound has been shown to interact with each domain separately (along with the full-length protein) (Thompson et al., 2012).
Here, we report the high-resolution structure elucidation of the N-terminal receiver domain of QseB via X-ray crystallography, and provide evidence that QseB binds 2-AI compounds. We have also optimized a thermal shift assay that quickly identifies 2-AIs from a library that bind QseB with a high affinity. Molecular modeling further suggests a QseB/2-AI interaction, specifically in the domain interface as proposed for other response regulators. Finally, we provide confirmation that specific 2-AIs are able to inhibit biofilm formation, reduce intracellular macrophage survival, and lower F. tularensis novicida minimal inhibitor concentrations (MICs) in adjuvant studies with broad spectrum antibiotics. The combination of structural information, the high-throughput in vitro assay, and in silico docking should greatly help assist in the design of new and more potent 2-AI compounds.
Results and Discussion
Structure of the QseB N-terminal receiver domain
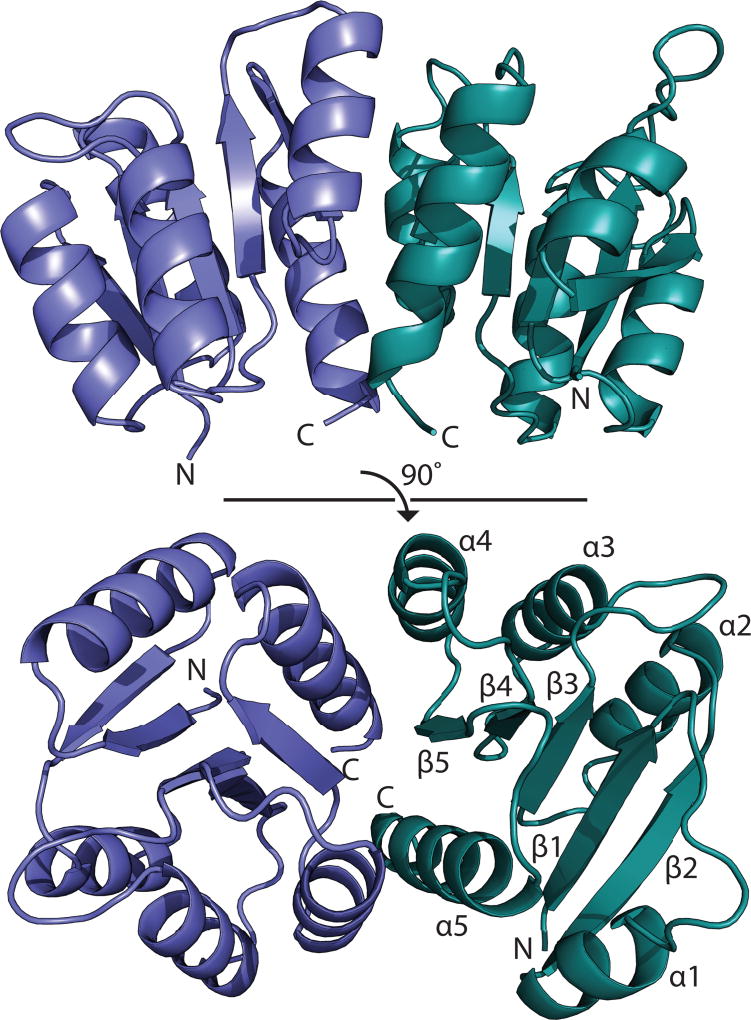
Response regulators are highly flexible proteins, and their structures are frequently characterized by separately probing the individual N- and C-terminal domains. In the first part of these studies, the structure of the N-terminal receiver domain of QseB (QseBN) was determined to a resolution of 2.5 Å using X-ray crystallography (Table S1). This domain becomes activated when phosphorylated, causing a change in the DNA-binding propensity of the C-terminal domain. The asymmetric unit contains two copies of QseBN in the form of a biologically relevant dimer (Fig. 1). The QseBN construct is composed of residues Met1-Ser126 with fifteen additional N-terminal residues remaining after cleavage of the affinity tag. Only residues Asn0 to Ile119 of chain A and Asn0 to Asp120 of chain B could be traced in the electron density.
Fig. 1.
Structure of Francisella response regulator, QseBN. The N-terminal domain of QseB is composed of two monomers, blue and green. Secondary structure elements are labeled.
Structurally, QseBN adopts an αβ doubly-wound, or (βα)5, fold that is characteristic of the OmpR/PhoB-type response regulators (Bourret, 2010). Five β-strands (β1-β5) are arranged in parallel across the central core. These β-strands are flanked by two α-helices (α1 and α5) on one side and three helices (α2, α3, α4) on the other to form the a β1-α1-β2-α2-β3-α3-β4-α4-β5-α5 topology (Fig. 1) observed for other response regulators (Buckler et al., 2002; Robinson et al., 2003). The two monomers of QseBN are very similar with an RMSD of 0.49 Å over 85 Cα atoms. The largest discrepancies between the two monomers occur in the helix α1 and the loop connecting sheet β4 and helix α4. Movement of the β4-α4 loop is known to be involved in a phosphorylation-mediated conformational change (Bourret, 2010; Bobay et al., 2012). It is suggested that QseB is sampling a range of conformations prior to activation, and upon activation, the β4-α4 loop is stabilized (Bobay et al., 2010; Bobay et al., 2012). In chain B, part of helix α1 is uncoiled into a disordered loop. This suggests high flexibility in regions of QseBN that are involved in regulation of binding activity or protein-protein interactions (Tzeng and Hoch, 1997; Tzeng et al., 1998; Jiang et al., 1999).
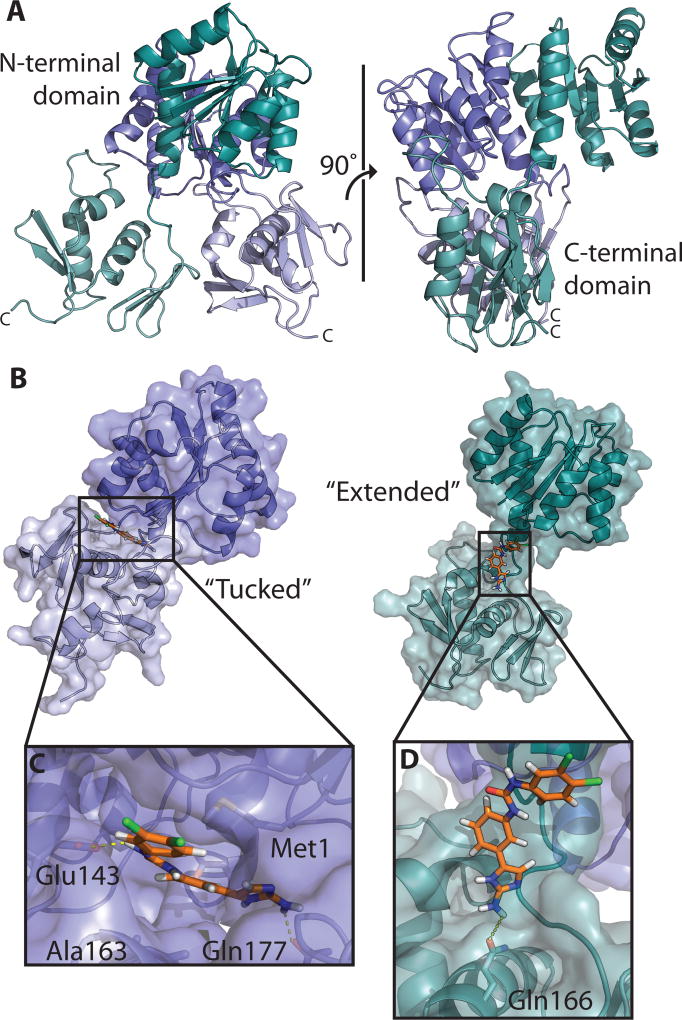
OmpR/PhoB-type response regulators also contain a C-terminal winged helix-turn-helix DNA-binding domain, connected to the receiver domain through a variable length flexible linker. This linker allows for a wide range of mobility between the two domains. It is broadly accepted that response regulators can occupy two extreme states, likely based on the linker length. As an example, the recent structure of Klebsiella pneumoniae PmrA bound to DNA (PDB ID 4S04 and 4S05 (Lou et al., 2015)) is composed of an asymmetrical dimer containing both “tucked” and “extended” conformations (Lou et al., 2015). The “tucked” state represents the N- and C-terminal domains being in closer proximity than in the “extended” state. Similar conformations are observed in the structure of Escherichia coli KdpE bound to DNA (PDB ID 4KFC and 4KNY (Narayanan et al., 2014)). The “tucked” and “extended” states likely represent the extremes of movement that can occur between the receiver and DNA-binding domains. Since the putative binding site for 2-AIs lies at the interface between the N- and C-termini, understanding the distribution and characteristics of these conformations must be considered during the development of possible therapeutics.
QseB binds 2-AI compounds
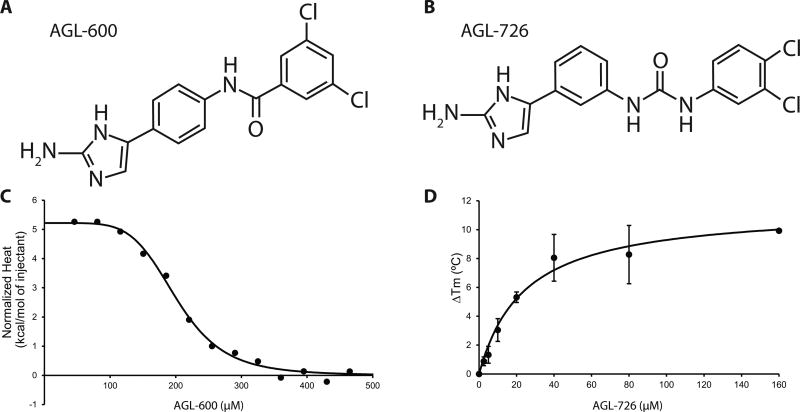
From a library of 2-AIs, the compound AGL-600 (Fig. 2A) was initially selected to study, as it demonstrates significant activity in cellular studies (Brackett et al., 2014). To characterize potential interactions between 2-AIs and QseB, it was first necessary to establish that AGL-600 does indeed bind to QseB. Isothermal titration calorimetry (ITC) was used to measure the energetics of AGL-600 binding to both QseB and “activated” beryllium fluorinated QseB (QseB-BeF3−). Beryllium fluoride is a widely accepted mimic for this process as it stabilizes the active state in the absence of a phosphorylation event (Wemmer and Kern, 2005). Activation of OmpR/PhoB response regulators is designated by the phosphorylation of a conserved aspartate located in the receiver domain, which influences downstream events such as DNA binding (Gao and Stock, 2009). The ITC experiments revealed that AGL-600 binds QseB with a Kd of 200.6 ± 9.8 µM, and QseB-BeF3− with a Kd of 228.6 ± 1.8 µM. Affinities were determined using an averaged dataset from two replicates and fitting to a single-site binding model (Fig. 2C). The affinities and stoichiometry of AGL-600 for QseB and QseB-BeF3− are similar, resulting in a 1:1 molar ratio of the protein-ligand complex. These data indicate that QseB binds AGL-600 with a high micromolar affinity, and that the binding affinity is not significantly impacted by activation of the protein.
Fig. 2.
QseB binds 2-AI compounds. A) Chemical structure of AGL-600. B) Chemical structure of AGL-726. C) Simulated ITC data showing AGL-600 binds QseB with a Kd of 200.6 ± 9.8 µM. D) Thermal shift data showing dose-dependent change in Tm of QseB with addition of AGL-726.
To determine binding potential of a library of 2-AI compounds, we developed a higher throughput assay that could more quickly screen for compounds that interact with QseB. A fluorescence-based thermal shift assay was employed to evaluate binding based on a shift in melting temperature (Tm) of QseB (Niesen et al., 2007). The increase in the Tm correlates to an increase in protein stability, and suggests the formation of a protein-ligand complex. The change in QseB melting temperature was evaluated in the presence of twenty-nine different 2-AI compounds (Fig S1), twelve of which resulted in an increase in Tm above background (Fig S2A). The compound AGL-726 (Fig. 2B) showed the largest shift in temperature from apo-QseB with a ΔTm of 6.7 ± 1.2°C at 25 µM compound. Dose-dependent analysis showed an increase in Tm with an increase in compound concentration. From this, a Kd of 24.0 ± 7.4 µM was determined (Fig. 2D). AGL-600, which was confirmed through ITC to interact with low affinity with QseB, did not have a significant impact on melting temperature. This suggests that the thermal shift assay under these conditions is only able to detect compounds with moderate to high binding affinities in a high throughput manner.
The potential binding interactions between QseB’s receiver domain, QseBN, and AGL-726 and AGL-600 were also investigated using the thermal shift assay. QseBN requires a significantly higher concentration of protein to obtain a reliable signal for analysis, 100 µM compared to the 5 µM needed for full length QseB. QseBN has a Tm of 82.1 ± 0.6°C, indicating that it is significantly more stable than full length QseB (Tm of 65.4 ±0.2°C). To verify that the drastic differences in Tms are due to the inherent properties of QseB constructs, and not because of the increase in concentration, full length QseB was evaluated at 100 µM. A 20-fold increase in QseB concentration yielded only a ~3°C raise in Tm. Since response regulators are highly flexible proteins, the intact C-terminal domain is likely adding significant movement to the full-length protein. This flexible nature is likely responsible for reduction in the overall melting temperature compared to the more rigid N-terminal domain alone.
Focusing on AGL-726 and AGL-600, the compounds were added to QseBN in increasing amounts, and the change in Tm measured (Fig S2B). AGL-726 raises the Tm of QseBN slightly at a protein:compound ratio of 1:1, resulting in a 1.6°C increase. Under the same conditions, an increase of 1.3°C is observed for AGL-600, suggesting that there is some interaction between the 2-AIs and the N-terminal domain. These results support previous pulldown assays that showed that the N-terminal domain of response regulators can bind 2-AIs (Thompson et al., 2012). As more 2-AI is added, the change in melting temperature decreases. Beyond a 1:1 and 1:1.5 protein:ligand ratio, neither compound significantly interacts with QseBN. Binding of AGL-726 increases the Tm of full length QseB by more than 6°C. If AGL-726 exclusively interacts with the N-terminal domain, then we would expect to see a similar raise in Tm with the N-terminal alone. The Tm should also increase in a concentration dependent manner as observed with full length QseB (Fig 2D). Since neither of these trends were observed, the results suggest that 2-AIs do not interact exclusively with the N-terminal domain. These data give further support to our model that the 2-AIs bind to the interface between the N- and C-terminal domains.
Molecular docking of 2-AIs to QseB
In the absence of a full length structure of QseB, computational modeling was employed to provide information about potential protein-ligand binding interactions. A model of full length QseB (Fig. 3A) was built from the structure of PmrA (PDB ID 4S05 (Lou et al., 2015)) using MODELLER (Eswar et al., 2007). PmrA shares 61% sequence homology with QseB, and structural alignment of the N-terminal domain Cα backbone of PmrA and our solved QseBN structure has a Cα RMSD of 1.805 Å, validating this PmrA model. An ensemble of full-length QseB models generated from GROMACS (Abraham et al., 2015) were input into HADDOCK (van Zundert et al., 2016) for docking with compounds.
Fig. 3.
Molecular docking of AGL-726 to a model of QseB. A) Full-length model of QseB based off of homologous crystal structures. One monomer (blue) takes on the “tucked” conformation with N-terminal domain in dark blue and C-terminal domain in light blue. The second monomer (green) is in the “extended” conformation with N-terminal domain in dark green and C-terminal domain in light green. B) Surface representation of AGL-726 (orange) docked to the “tucked” (blue) and “extended” (green) conformations. C) Close-up view of binding pocket of the “tucked” conformation. Yellow dashes representing H-bonding distances between AGL-726 and side chains. D) Close-up view of “extended” conformation binding site with yellow dashes representing potential H-bonding.
Not surprisingly, during preparation for docking, it became apparent that the putative 2-AI binding sites between QseB’s “tucked” and “extended” states showed differences, and compound binding could be greatly impacted. Due to this, both the “extended” and “tucked” conformations (Fig. S3) were used in the docking protocol to identify any preferences a 2-AI may have for one extreme conformation over the other. As we have seen with other response regulator/2-AI combinations, the top docking results show the 2-AI clustering in the interface between the N- and C-terminal domains (Fig 3B), (Thompson et al., 2012).
Comparison of the docking results for the “extended” and “tucked” conformations of the full-length two-domain protein suggests that AGL-726 preferentially binds (lower energy) the “tucked” conformation. The mean FireDock scores for the top-ranking clusters are −24.97 kJ/mol and −18.26 kJ/mol for “tucked” and “extended”, respectively (Fig. S4). Inspection of the AGL-726 docking site shows potential interactions between the compound and the N- and C-terminal domain residues. In the “tucked” conformation, the 2-AI ring and a nitrogen of the meta-urea are in position for forming H-bonds with Glu143 and Gln178 (Fig. 3C). The remainder of the binding pocket is predominately lined with hydrophobic residues, allowing the 2-AI to fit snugly between the two domains. The “extended” conformation exhibited fewer potential binding interactions and a less defined binding pocket. Here, AGL-726 is positioned along the linker region, spanning the space between the two domains (Fig. 3D). The 2-AI ring is in position to form a hydrogen bond with Gln166. All other observed interactions are van der Waals interactions. Docking was repeated for AGL-600 and resulted in similar binding conformations (Fig. S5) and a mean scoring of −28.24 kJ/mol and −17.69 kJ/mol for “tucked” and “extended”, respectively (Fig. S4). The formation of a clear binding pocket in the “tucked” conformation parallels the binding energies calculated by FireDock, and suggests that the “tucked” state provides tighter binding potential for 2-AIs and is preferred.
Using the Gibbs free energy equations, FireDock scores for AGL-726 and AGL-600 were used to estimate corresponding Kd values. Docking predicts that AGL-726 binds with a 32.7 µM affinity for the “tucked” state and 633.7 µM affinity for the “extended” state. The binding energy from the molecular modeling to the “tucked” conformation is similar to the measured binding affinity of AGL-726 determined through the thermal shift assay (Fig. 2D). This suggests that AGL-726 either binds to QseB when QseB is sampling the “tucked” conformation, or when AGL-726 induces a conformational change and traps QseB in the “tucked” state. On the other hand, AGL-600 docking scores suggest binding affinities of 11.3 µM for “tucked” and 795.9 µM for “extended”. Since the affinity measured by ITC lies between these two models, it is likely that AGL-600 binds QseB in a manner that still allows for some movement in the DNA binding domain, with the “extended” conformation being sampled more frequently. Comparable trends were seen for AGL-778 and AGL-774, which had similar thermal shift results to AGL-726 and AGL-600, respectively (Fig. S2). Most importantly, in cases where a significant change in Tm was seen upon QseB/2-AI binding, the affinities determined by the thermal shift assay agree well with predicted energies for the 2-AI preferentially binding the “tucked” conformation. In contrast, compounds that do not change the Tm notably have experimental binding affinities more consistent with predicted energies for binding a confirmation that is between “tucked” and “extended” (Fig 2C and data not shown).
By combining experimentally-measured binding affinity data with molecular modeling information, the following features about QseB:2-AI complexes can be suggested. The change in melting temperature of QseB through the addition of 2-AIs may correlate to the trapping of QseB in a particular conformational state. An increase in melting temperature may signify that the compound preferentially targets the “tucked” conformation of QseB. By doing this, the population of the “extended” conformation is reduced, and the relative motion of the DNA binding domain with respect to the N-terminal domain is mitigated to some degree. Compounds that do not alter the Tm of QseB could be preferentially binding a population of more “extended” conformations and do not affect relative motions as much, since there would be fewer stabilizing interactions between the domains. It is likely that affecting the mobility of the DNA-binding domain has significant consequences to QseB’s ability to identify and bind its target DNA, or interact with other protein partners, such as MglA and SspA.
2-AIs inhibit biofilm formation
QseB plays a significant role in biofilm formation. Deletion of QseB results in a 60% reduction in biofilm growth compared to wild type F. tularensis novicida (Durham-Colleran et al., 2010). AGL-726 and AGL-600 both appear to inhibit biofilm formation in a dose-dependent manner. AGL-600 has an IC50 of 57.64 ± 15.12 µM for biofilm inhibition. AGL-726 is a more potent inhibitor with an IC50 of 15.03 ± 1.99 µM (Table 1). The reduction in biofilms suggests that QseB is a target of the compounds.
Table 1.
Growth and biofilm inhibition of F. tularensis novicida by AGL-600 and AGL-726
| Compound | IC50 Cellular Growth | IC50 Biofilm Inhibition |
|---|---|---|
| AGL-600 | > 125 µM | 57.64 ± 15.12 µM |
|
|
||
| AGL-726 | 15.58 ± 1.83 µM | 15.03 ± 1.99 µM |
|
|
||
QseB is not needed for cellular survival. Deletion of QseB results in a slight defect in growth (~86%) compared to the wild-type rate (Durham-Colleran et al., 2010). At high concentrations, 2-AIs do appear to have an impact of cellular growth. This concentration is dependent on the 2-AI. F. tularensis novicida cells that are treated with 25 µM of AGL-726 do not appear to grow. When the cells are then transferred to fresh media with no 2-AI, the cells grow normally (data not shown). This suggests that AGL-726 is causing the cells to go into a persistent state and is not toxic. Similar results were observed for an early 2-AI derivative, RA-2AI-1 (Stowe et al., 2015). We hypothesize that the 2-AIs are binding response regulators and impacting bacterial response. In a dormant state, cells are waiting for environmental cues to begin growth again. If 2-AIs are blocking these signals through their influence on response regulators, the cells will remain in the persistent state.
The production of a biofilm is directly related to the overall level of cellular growth (Durham-Colleran et al., 2010). Reduction in growth, in our case by entering a persistent state, may mask an effect on biofilm formation. Unfortunately, this makes the accurate determination of the influence of 2-AIs on the production of biofilms difficult. To evaluate the effect of 2-AIs on cellular growth, the optical density of each biofilm plate was taken prior to staining with crystal violet. Comparison of the IC50 of cellular growth and the IC50 for biofilm formation allows a more precise determination of the impact of 2-AIs on biofilms. AGL-600 has significantly higher IC50 for growth versus biofilm formation, while AGL-726 has a nearly identical IC50 (Table 1). AGL-600 is clearly able to inhibit biofilm production.
Since AGL-726 had significantly reduced cellular growth, samples were removed from the wells and streaked out on agar plates. Growth levels of cells exposed to higher concentrations of 2-AIs were compared to untreated cells after 24 and 48 h. Cells exposed to 12.5 µM compound showed no noteworthy reduction in growth in the biofilm assay, 76.2 ± 9.4% growth compared to untreated. Increasing the concentration of AGL-726 to 25 µM and 50 µM significantly decreased the percentage of growth, ~10%, but cells grew at the same rate as wild type once the compound was removed. Exposure to 100 µM of AGL-726 resulted in a delay in cell recovery, but growth was clearly visible after 48 h. Thus, AGL-600 can reduce biofilm formation, which suggests that QseB is a cellular target. Since AGL-726 pushes cells into a persistent state, its impact exclusively on biofilm production could not be evaluated as accurately.
2-AIs impact intramacrophage survival
To further confirm is QseB is a target of 2-AIs, intramacrophage survival assays were performed. QseB has also been shown to be necessary for cellular replication. Deletion of QseB prevents F. tularensis novicida survival within macrophages (Mohapatra et al., 2007; Bell et al., 2010). Exposing F. tularensis novicida to AGL-600 and AGL-726 prior to exposure to J774A.1 macrophages results in a decrease in intramacrophage survival (Fig. 4). Since QseB is needed for survival, this suggests that QseB activity is impacted by the 2-AIs. These results are not as extreme as the 73-fold difference between wild type and a QseB deletion mutant observed by Mohapatra et al, but it is clear that the compounds are having an impact on intracellular survival. The discrepancy is likely due to the compounds not completely inhibiting QseB activity.
Fig. 4.
Intramacrophage survival assay. F. tularensis novicida that was exposed to AGL-600 and AGL-726 prior to infection show a decrease in survivability within J774A.1 macrophages after a 12 h incubation. Assays were conducted at a multiplicity of infection of about 50:1, bacteria to macrophages.
2-AIs induce antibiotic re-sensitization
To investigate the cellular impact of treatment with 2-AIs, the MIC lowering activity of the adjuvant compounds were tested in F. tularensis novicida. By comparing the MIC values of an antibiotic alone to the combination of 2-AI and the antibiotic, it is possible to determine the effect that the 2-AI has on re-sensitizing the bacteria to that antibiotic. AGL-600 is known to cause MIC lowering for both meropenem and imipenem in Acinetobacter baumannii ATCC-1605 and ceftazidime and meropenem in Pseudomonas aeruginosa PA53 (Brackett et al., 2014). The MIC lowering ability of AGL-600 was measured in F. tularensis novicida against seven antibiotics from different classes (Table 2). Meropenem, from the carbapenem class of antibiotics, was impacted the most, with AGL-600 significantly reducing the MIC in a dose dependent manner. With no 2-AI present, meropenem has an MIC value > 1024 µg/mL. The addition of 50, 75, and 100 µM AGL-600 reduced the MIC by 32, 64, and 128-fold, respectively. These results are particularly important as thirty-eight strains of F. tularensis tularensis isolated from both humans and animals in Finland were all resistant to meropenem (Ikäheimo et al., 2000). In addition, thirty-six isolates from Japan also showed notably reduced susceptibility to meropenem (Hotta et al., 2013). The ability of 2-AIs to re-sensitize Francisella to meropenem may once again allow for treatment with broad spectrum antibiotics that have long been ineffectual.
Table 2.
MIC activity of F. tularensis novicida by AGL-600
| AGL-600 | |||||
|---|---|---|---|---|---|
|
| |||||
| Antibody | 0 µM | 50 µM | 75 µM | 100 µM | |
| Streptomycin | MIC (µg/mL) | 4 | 4 | 2 | 2 |
|
|
|||||
| Fold reduction | 0 | 2 | 2 | ||
|
|
|||||
| Colistin | MIC (µg/mL) | 1024 | 512 | 512 | 512 |
|
|
|||||
| Fold reduction | 2 | 2 | 2 | ||
|
|
|||||
| Gentamicin | MIC (µg/mL) | 1 | 2 | 1 | 1 |
|
|
|||||
| Fold reduction | 0.5 | 0 | 0 | ||
|
|
|||||
| Azithromycin | MIC (µg/mL) | 2 | 1 | 1 | 0.5 |
|
|
|||||
| Fold reduction | 2 | 2 | 4 | ||
|
|
|||||
| FR 900098 | MIC (µg/mL) | 16 | 16 | 16 | 16 |
|
|
|||||
| Fold reduction | 0 | 0 | 0 | ||
|
|
|||||
| Meropenem | MIC (µg/mL) | >1024 | 32 | 16 | 8 |
|
|
|||||
| Fold reduction | 32 | 64 | 128 | ||
|
|
|||||
| Polymyxin B | MIC (µg/mL) | 1024 | 256 | 128 | 128 |
|
|
|||||
| Fold reduction | 4 | 8 | 8 | ||
It is important to note that it is not currently known which of the three response regulators (QseB, KdpE, and FTN1452) is responsible for meropenem resistance in Francisella. Nevertheless, we have shown that 2-AIs are able to interact with a variety of response regulators (Thompson et al., 2012). Consequently, the following studies on QseB are likely applicable to KdpE and FTN1452 also.
PmrA from Salmonella typhimurium is known to be responsible for polymyxin resistance (Gunn et al., 1998; Tamayo et al., 2002). Since there is high sequence homology between S. typhimurium PmrA and QseB, we propose that QseB is likewise responsible for polymyxin B resistance in Francisella. As an excellent benchmark for our studies, Gunn and coworkers have shown that deletion of QseB causes F. tularensis novicida to be 32-fold more susceptible to polymyxin B than wild type (Mohapatra et al., 2007). In the absence of 2-AIs, polymyxin B has an MIC of 1024 µg/ml. The addition of AGL-600 reduced the MIC in a dose-dependent manner, with 100 µM of compound lowering the MIC by 8-fold (Table 2). While the presence of AGL-600 has an impact on the MIC of polymyxin B, the fold reduction is not as potent as the deletion of QseB. This suggests that AGL-600 lowers QseB activity but does not completely inhibit it. AGL-726 also has a slight dose-dependent MIC lowering activity. The addition of 1, 2, and 3 µM AGL-726 reduced the MIC by 2, 2, and 4-fold, respectively. Considerably lower doses of compound had to be used due to AGL-726 having a significantly lower MIC value than AGL-600, 12.5 µM and 200 µM, respectively. Since AGL-726 causes cells to enter a persistent state, higher concentrations could not be tested. Regardless, if we are correct in our hypothesis that 2-AIs are causing the bacteria to go into a dormant state, then 2-AIs are ideal for adjuvant treatment with current wide spectrum antibiotics.
Conclusions
The X-ray structure of QseBN confirms that it adopts a fold similar to other response regulator receiver domains. This is the first structural information for QseB. Cellular studies in the presence of AGL-600 confirm that 2-AIs can resensitize F. tularensis spp. to broad spectrum antibiotics. Using the thermal shift assay, 2-AI compounds that bind with high affinity to QseB can be identified quickly. Molecular modeling allows for visualization of the potential interaction, and is guiding future drug design in the absence of complete structural information and the lack of a compound-bound structure. Computational docking of QseB provides strong support for the suggestion that QseB samples two confirmations - “tucked” and “extended”, with the N- and C-terminal domains being respectively proximal and more removed. Using this knowledge, along with experimental data, we predict that AGL-726 preferentially targets and traps QseB in the “tucked” conformation. Since 2-AIs are known to target other response regulators with variable affinities, it is likely that the compounds may also affect KdpE and FTN1452 somewhat.
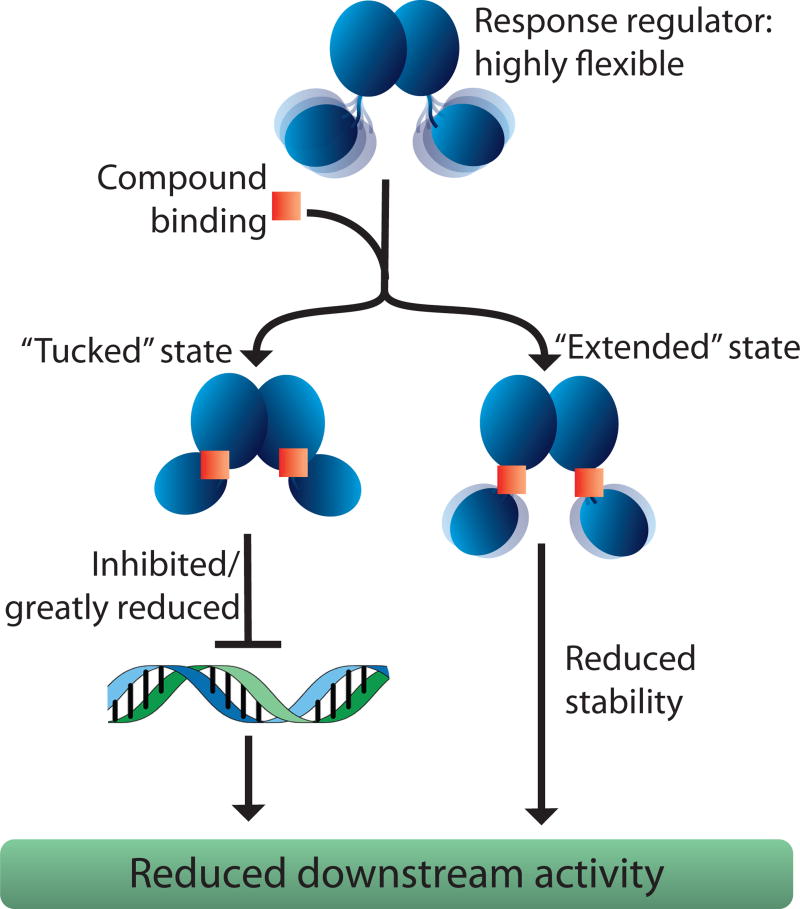
Preferentially targeting response regulators in the “tucked” state, and/or pushing them towards adopting this conformation, can impede their ability to interact with target DNA. Such a functional down-regulation would impede the initiation of downstream events such as the formation of biofilms, protective responses, and virulence factors. A better understanding of the binding interactions between QseB and 2-AI compounds will provide necessary information for further developing potent drugs that target response regulators. Based on our data, we propose two possible mechanisms by which 2-AIs interact with response regulators: (i) impeding DNA binding in the “tucked” conformation, and (ii) reducing movement of the response regulator, causing it to be held open in a more “extended” conformation (Fig 5). These mechanisms strongly support the “tucked”/”extended” model proposed by others (Narayanan et al., 2014; Lou et al., 2015). Knowing how tightly a 2-AI binds, and which conformational state it preferentially binds to, impacts drug design. A better understanding of how response regulators interact with 2-AIs and how those interactions manifest within virulent bacteria will inform development of future compounds.
Fig. 5.
Proposed mechanisms for 2-AI binding. Response regulator shown as a blue cartoon and 2-AI as an orange square. 2-AIs either coerce the response regulator to favor a “tucked” conformation (left-hand path), hindering DNA-binding activity, or they bind in a manner that pulls the domains apart into the “extended” conformation (right-hand path), reducing protein stability.
Experimental Procedures
Cloning, expression, and purification
The coding region of qseb (FTN1495) was cloned from F. tularensis novicida genomic DNA (100% homology to FTT1557 in F. tularensis tularensis) provided by the van Hoek lab (George Mason University). QseB and the QseB N-terminal domain construct (QseBN, residues 1–126) were cloned into pET28a (Novagen) using EcoRI and XhoI restriction sites to generate a protein construct containing a cleavable N-terminal His6 affinity tag. Proteins were over-expressed in BL21(DE3)pLysS cells at 37°C in LB. At an OD600 of 0.6–0.8, cells were induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 25°C, overnight. Harvested cell pellets were stored at −80°C for later use. Pellets were resuspended in lysis buffer (20 mM sodium phosphate pH 7.3, 400 mM NaCl, 10% v/v glycerol, 0.02% w/v sodium azide, and 0.1 mM of AEBSF) at 10 mL g−1 of pellet. Cells were sonicated at 40% amp for 20 min and the resulting lysate clarified at 20,400 × g for 15 min. The clarified lysate was loaded onto 10 mL of Ni-NTA resin (QIAGEN) pre-equilibrated in lysis buffer. Ni-NTA-bound protein was subsequently washed with 40 column volumes of lysis buffer. The protein was eluted with a linear gradient from lysis buffer to elution buffer (20 mM sodium phosphate pH 7.3, 400 mM NaCl, 10% v/v glycerol, and 300 mM imidazole). Fractions containing protein were pooled and dialyzed into 20 mM sodium phosphate pH 7.3, 400 mM NaCl, 10% v/v glycerol, 0.5 mM EDTA, and 0.02% w/v sodium azide. The sample was then dialyzed into 20 mM HEPES pH 6.5, 300 mM NaCl, 0.5 mM EDTA, and 0.02% w/v sodium azide for 4 h. The affinity tag was then cleaved by 100 units of thrombin and allowed to dialyze for 2 h at room temperature. Cleavage was quenched with 0.1 mM AEBSF and the sample was dialyzed into the final buffer of 20 mM HEPES pH 6.5, 100 mM NaCl, 5 mM MgCl2, and 0.02% w/v sodium azide.
Crystallization and structure determination
QseBN crystals were grown through hanging-drop vapor diffusion against 1 M potassium citrate, 0.1 M glycine, and 0.05 M bis-tris propane (BTP) pH 7.0 at room temperature. Crystals grew at 9 mg mL−1 with a protein to cocktail ratio of 1:1. Crystals were cryoprotected using perfluoropolyether and data was collected at a wavelength of 1.000 Å at APS beamline 22-ID (SER-CAT) on a 300 mm MAR Mosaic CCD detector. Diffraction was indexed, merged, and scaled using HKL-2000 (Otwinowski and Minor, 1997). The MRage (Krissinel and Henrick, 2004) molecular replacement component of PHENIX (Adams et al., 2010) was used to solve the structure of QseBN using OmpR/PhoB (PDB ID: 1KGS, (Buckler et al., 2002)) as a starting model. The resulting solution was further refined using COOT (Emsley et al., 2010) and phenix.refine (Adams et al., 2010). Statistics for data collection and refinement are located in Table S1.
Activation by BeF3−
Sample was activated as previously described (Yan et al., 1999; Cho et al., 2001; Bachhawat et al., 2005). In short, BeCl2 was added to the sample at a final concentration of 5 mM. NaF was then added to a final concentration of 20 mM. The solution was mixed by gentle pipetting and incubated undisturbed at room temperature for at least 1 h.
Isothermal titration calorimetry (ITC)
ITC experiments were performed on a low volume NanoITC (TA Instruments) at the North Carolina State University Biological Core Facility. Stock samples of QseB were quantified by UV-Vis spectroscopy, then supplemented with 100% PEG 400 to a final concentration of 4% v/v, resulting in working QseB concentrations between 160–180 µM for experiments. Samples of AGL-600 were prepared from a 50 mM master stock dissolved in 100% PEG 400 by diluting in buffer to a working concentration of 2 mM compound in 4% v/v PEG 400. Titrations were performed in duplicate with 13 injections of 4 µL and a 0.5 µL initial injection separated by 300 sec spacing with continuous 250 rpm stirring at 25°C. Isotherms were processed with Nano Analyze (TA Instruments) and extracted to GraphPad Prism 6 for simulated curve fitting of a “one-site specific-binding” model to derive experimental binding constants and thermodynamic parameters. Errors are reported based on the goodness-of-fit (R2 = 0.9944) to an averaged binding isotherm generated from two separate representative titrations. Plots of the raw data and simulated isotherms were made in Kaleidagraph, version 4.52 (Synergy Software).
Thermal shift assay (TSA)
QseB and QseBN were dialyzed into reaction buffer containing 20 mM MES pH 7.0, 100 mM NaCl, and 10% v/v glycerol. AGL compounds were dissolved in 100% PEG 400 to a final concentration of 1 mM. For QseB, reactions were carried out using final concentrations of 5 µM QseB, 25 µM compound, 10% v/v PEG 400, and 10× SYPRO orange (ThermoFisher Scientific). Samples were prepared in three technical replicates on a CFX384 Touch Real-Time PCR Detection System (BioRad). Samples were heated from 25° to 95°C in 0.5°C increments, holding for 30 sec at each step. Fluorescence was detected using the default HEX wavelengths. Data was fit to a Boltzmann curve using SigmaPlot. For QseBN, reactions were carried out using final concentrations of 100 µM QseBN, 100 to 400 µM compound, 10% PEG 400, and 8× SYPRO orange (ThermoFisher Scientific). Samples were prepared in technical replicates on a QuantStudio 3 (Applied Biosystems). Samples were headed from 25° to 99° C in 0.05°C/s. Fluorescence was detected using ROX as the reporter. Data was analyzed using Protein Thermal Shift Software (Applied Biosystems) and fit to a Boltzmann curve. Assays were repeated in triplicate.
Homology model construction of QseB
MODELLER v10.1 was used to construct a model of QseB from F. tularensis using PmrA as the template (PDB entry: 4S05) (Eswar et al., 2007; Lou et al., 2015). An alignment between QseB and PmrA provided an E-value of 1.97115e−38 and 43% sequence identity, indicating good alignment and template choice. The 4S05 structure also afforded the opportunity to construct a dimeric response regulator protein PDB with one DNA binding C-terminal domain immediately adjacent to the N-terminal response domain, while the other monomer is composed of two domains in which the N-terminal and C-terminal domains are extended and not immediately adjacent. A total of 500 models were generated between an alignment of QseB and 4S05 and scored by the internal MODELLER scoring method Discrete Optimized Protein Energy (DOPE, (Shen and Sali, 2006)). DOPE is a statistical potential used to assess homology models in protein structure prediction. DOPE is based on an improved reference state that corresponds to non-interacting atoms in a homogeneous sphere with the radius dependent on a sample native structure; thus, it accounts for the finite and spherical shape of the native structures. The structure with the lowest DOPE score was subsequently run through PROCHECK (Laskowski et al., 1993) and WHATCHECK (Hooft et al., 1996) (checks the stereochemical quality of a protein structure) for structural quality.
PDB and parameter construction of 2-AI ligand
PDB and parameter file construction of each 2-AI ligand was achieved using a combination of PRODRG (Schüttelkopf and van Aalten, 2004) and CHARMM General Force Field (CGenFF) interface (https://cgenff.paramchem.org). PRODRG was supplied a text or .mol file description of each 2-AI ligand. Standard PRODRG parameters were used for PDB and parameter construction with full charges and chirality. CHARMM was then provided the .mol2 file from PRODRG output to determine partial charges for each atom. After visual inspection of the PRODRG DRGFIN.PDB file, partial charges from the CGenFF output were inserted into the DRGCNS.TOP parameter file for subsequent molecular docking. Also in this topology file, hydrogen bond donor (DONO) and acceptor (ACCE) flags were manually added for the appropriate atoms (H, O, N, and F).
Molecular dynamics (MD) simulations
MD simulations were performed on the QseB PDB derived model from MODELLER with the GROMACS (Pronk et al., 2013) 4.4.5 software package, using the OPLS/AA force field and the flexible SPC water model. This was performed to provide a more realistic solution PDB model of QseB. In addition, this provided the high ambiguity driven docking (HADDOCK (van Zundert et al., 2016)) protocol with an ensemble of structures of multiple conformations as to not bias the subsequent docking. The initial structures were immersed in a periodic water box of cube shape (1 nm thickness) and neutralized with counter ions. Electrostatic energy was calculated using the particle mesh Ewald method. Cutoff distances for the calculation of the Coulomb and van der Waals interactions were 1.0 nm. After energy minimization using a steepest decent method, the system was subject to equilibration at 300 K and normal pressure for 100 ps under the conditions of position restraints for heavy atoms and LINCS constraints for all bonds. The system was coupled to the external bath by Parrinello–Rahman pressure and temperature coupling. The final MD calculations were performed under the same conditions, except the position restraints were removed and the simulation was run for 100 ns. The last 40 ns of the simulation were taken as representative protein conformers and uploaded into HADDOCK. Cα root mean square deviation (r.m.s.d.) levels off to ~0.75 nm, indicating that the structure is very stable, while the radius of gyration remains reasonably invariant, indicating a compact (folded) stable conformation over the course the last 40 ns.
HADDOCK molecular docking
Default HADDOCK parameters were used throughout all docking procedures except as noted below. Residues with solvent accessibility >50% calculated by NACCESS (http://www.bioinf.manchester.ac.uk/naccess/) within the QseB dimer PDB model were denoted as active residues. Passive residues were automatically defined by HADDOCK. The entire 2-AI small molecule was defined as active. Partial charges for the 2-AI were determined as stated above. Non-crystallographic and C2 symmetry restraints were used throughout the docking protocol to ensure symmetry in the N-terminal dimerization domain. The C-terminal domain of each monomer was allowed to move independently through the assumption that these domains do not contribute to dimerization (i.e., no C2 symmetry restraints; however, non-crystallographic restraints were employed to ensure C-terminal structural identity). Hydrogen bond restraints were incorporated for defined secondary structure elements as well. Finally, intermolecular Cα distance restraints between the two N-terminal domains were incorporated to ensure dimerization retention throughout the docking process. One thousand structures were generated for the first rigid docking iteration, the top 20% (200) models were then further refined through a semi-flexible vacuum protocol, and the 200 lowest energy structures were subsequently water refined. Each docking attempt was performed 10 times, and the solution with the lowest HADDOCK score was retained. The carbon atom r.m.s.d. values of the complexes were calculated using the McLachlan algorithm as implemented in ProFit (McLachlan, 1982). A cluster analysis was performed on the final docking solutions using a minimum cluster size of four.
Scoring
All clusters for each compound were visually inspected for intuitive results, then analyzed according both XSCORE (Wang et al., 2002) (HPScore, HMScore, HSScore, -log(Kd), and AiG), and FireDock (Mashiach et al., 2008) (global, attractive VdW, repulsive VdW, ACE, and hydrogen bond). RStudio and SigmaPlot subsequently provided statistical analysis of the docking scores, resulting in a box plot ranked by median scoring value (excluding statistical outliers), where lower scores indicate higher affinity.
Bacterial strains, media, and antibiotics
F. tularensis novicida Utah-112 was obtained from BEI Resources as BEI # (U112). Cells were grown in tryptic soy broth supplemented with 0.1% cysteine (TSB-C) at 37°C. Antibiotics were purchased from Sigma-Aldrich.
Biofilm inhibition assay
Assays were performed as previously described (Durham-Colleran et al., 2010). Cells were grown in an overnight culture of TSB-C. Prior to the start of the assay, cells were diluted with fresh media to an optical density of 0.1. 2-AI compounds were diluted in 100% DMSO such that the addition of 1 µL of compound per 100 µL of cells gave the desired final compound concentration. 100 µL of sample containing diluted cells and 2-AI was added to 96-well flat bottom polystyrene microtiter plate. A no cell, no compound, and 1% DMSO only control was included on each plate and technical triplicates were performed for each compound concentration. Plates were covered with plastic wrap and incubated for at least 24 h at 37°C under stationary conditions in a humid incubator. Media was gently aspirated, wells were gently washed with dH2O, and allowed to air dry. 200 µL of 99% methanol was added to each well for 15 min to fix the biofilm to the plate. Methanol was then removed and the plate was allowed to dry. Wells were then stained with 120 µL of 1% crystal violet for 5 min. Wells were washed three times with dH2O and dried. The stain was then dissolved in 160 µL of 33% acetic acid for 15 min. Plates were read for absorbance at 600 nm and 540 nm. Growth was normalized to the 1% DMSO control. Assay was performed in triplicate.
Intramacrophage survival assay
Assays were performed as previously described (Mohapatra et al., 2007; Bell et al., 2010). J774A.1 murine macrophages (ATCC) were infected with F. tularensis novicida Utah-112.Wells were seeded with ~4 × 104 macrophages in Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum (FBS) and ~2×106 bacteria per well, a ~50:1 multiplicity of infection. After incubation at 37°C and 5% CO2 for 2 hours, cells were washed to remove extracellular bacteria. Macrophages were lysed with 0.1% sodium dodecyl sulfate (SDS) at various time points post infection. Lysate was diluted in phosphate buffer saline (PBS) and then plated on TBS-C agar plates. Cell viability counts were then determined.
Minimum inhibitory concentration (MIC) assay
Overnight bacterial cultures (~16 h) were subcultured to 5 × 105 CFU mL−1 in TSB-C. AGL-600 from a 100 mM stock solution in DMSO was added to cells to give the desired concentration to be tested (25% of the compound MIC value). Antibiotics were added to small culture tubes to give the desired starting concentration (2–2048 µg mL−1). Rows 2–12 of a 96-well microtiter plate were filled with 100 µL per well of the bacterial subculture. The wells of the first row were filled with 200 µL each of the compound-containing sample. Row 1 wells were mixed a minimum of five times, followed by a 100 µL aliquot to row 2. The process was repeated until the final row, in which the last 100 µL aliquot was discarded. The 96-well plate was then sealed with a plastic lid and incubated under stationary conditions at 37°C for 48 h. The compound MIC was recorded as the lowest concentration at which no bacterial growth was observed.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Monique van Hoek for supplying the F. tularensis novicida genomic DNA, Dr. Ninell Mortensen for assistance with macrophage culturing, and the North Carolina State University Structural Biology Resource. Crystallographic data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201500010C, grant R01-GM055769, and by the V Foundation for Cancer Research.
Footnotes
Accession Codes
Coordinates and structure factors have been deposited with the Protein Data Bank under accession code 5UIC.
Author Contributions
MEM, CLA, EAF, and BGB designed research; EAF and CLA expressed and purified proteins; CLA collected and processed diffraction data and solved the structure; MEM refined structures; EAF performed ITC experiments; MEM performed thermal shift experiments; BGB and KET performed molecular dynamics simulations; MEM performed biofilm inhibition assays; MEM and RJT performed intramacrophage survival studies; MDS, RJM, and MEM performed antibiotic MIC screen; DLJ designed and synthesized AGL-726; DZ provided all synthesized compounds; MEM analyzed data; RJT, CM, and JC supervised research; MEM and CLA wrote the paper.
References
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat P, Swapna GVT, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Struct Lond Engl 1993. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard TE, Richards JJ, Wolfe AL, Melander C. Synthesis and Antibiofilm Activity of a Second-Generation Reverse-Amide Oroidin Library: A Structure-Activity Relationship Study. Chem - Eur J. 2008;14:10745–10761. doi: 10.1002/chem.200801419. [DOI] [PubMed] [Google Scholar]
- Bell BL, Mohapatra NP, Gunn JS. Regulation of Virulence Gene Transcripts by the Francisella novicida Orphan Response Regulator PmrA: Role of Phosphorylation and Evidence of MglA/SspA Interaction. Infect Immun. 2010;78:2189–2198. doi: 10.1128/IAI.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay BG, Hoch JA, Cavanagh J. Dynamics and activation in response regulators: the β4-α4 loop. Biomol Concepts. 2012;3:175–182. doi: 10.1515/bmc-2011-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay BG, Thompson RJ, Hoch JA, Cavanagh J. Long range dynamic effects of point-mutations trap a response regulator in an active conformation. FEBS Lett. 2010;584:4203–4207. doi: 10.1016/j.febslet.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackett CM, Melander RJ, An IH, Krishnamurthy A, Thompson RJ, Cavanagh J, Melander C. Small-Molecule Suppression of β-Lactam Resistance in Multidrug-Resistant Gram-Negative Pathogens. J Med Chem. 2014;57:7450–7458. doi: 10.1021/jm501050e. [DOI] [PubMed] [Google Scholar]
- Buckler DR, Zhou Y, Stock AM. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Struct Lond Engl 1993. 2002;10:153–164. doi: 10.1016/s0969-2126(01)00706-7. [DOI] [PubMed] [Google Scholar]
- Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, et al. BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase. Proc Natl Acad Sci U S A. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM. Biofilms: Microbial Life on Surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham-Colleran MW, Verhoeven AB, Hoek ML van. Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb Ecol. 2010;59:457–465. doi: 10.1007/s00248-009-9586-9. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007 doi: 10.1002/0471140864.ps0209s50. Chapter 2: Unit 2.9. [DOI] [PubMed] [Google Scholar]
- Foreman A, Holtappels G, Psaltis AJ, Jervis-Bardy J, Field J, Wormald P-J, Bachert C. Adaptive immune responses in Staphylococcus aureus biofilm-associated chronic rhinosinusitis. Allergy. 2011;66:1449–1456. doi: 10.1111/j.1398-9995.2011.02678.x. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- Hare RF, Hueffer K. Francisella novicida Pathogenicity Island Encoded Proteins Were Secreted during Infection of Macrophage-Like Cells. PLOS ONE. 2014;9:e105773. doi: 10.1371/journal.pone.0105773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek ML van. Biofilms: an advancement in our understanding of Francisella species. Virulence. 2013;4:833–846. doi: 10.4161/viru.27023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RWW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272–272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Hotta A, Fujita O, Uda A, Sharma N, Tanabayashi K, Yamamoto Y, et al. In vitro antibiotic susceptibility of Francisella tularensis isolates from Japan. Jpn J Infect Dis. 2013;66:534–536. doi: 10.7883/yoken.66.534. [DOI] [PubMed] [Google Scholar]
- Ikäheimo I, Syrjälä H, Karhukorpi J, Schildt R, Koskela M. In vitro antibiotic susceptibility of Francisella tularensis isolated from humans and animals. J Antimicrob Chemother. 2000;46:287–290. doi: 10.1093/jac/46.2.287. [DOI] [PubMed] [Google Scholar]
- Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;33:389–395. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Larsson P, Oyston PCF, Chain P, Chu MC, Duffield M, Fuxelius H-H, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Lou Y-C, Weng T-H, Li Y-C, Kao Y-F, Lin W-F, Peng H-L, et al. Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA. Nat Commun. 2015;6:8838. doi: 10.1038/ncomms9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AD. Rapid comparison of protein structures. Acta Crystallogr A. 1982;38:871–873. [Google Scholar]
- Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, et al. Identification of an Orphan Response Regulator Required for the Virulence of Francisella spp. and Transcription of Pathogenicity Island Genes. Infect Immun. 2007;75:3305–3314. doi: 10.1128/IAI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Kumar S, Evrard AN, Paul LN, Yernool DA. An asymmetric heterodomain interface stabilizes a response regulator-DNA complex. Nat Commun. 2014;5:3282. doi: 10.1038/ncomms4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Percival SL, Hill KE, Malic S, Thomas DW, Williams DW. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regen Off Publ Wound Heal Soc Eur Tissue Repair Soc. 2011;19:1–9. doi: 10.1111/j.1524-475X.2010.00651.x. [DOI] [PubMed] [Google Scholar]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinforma Oxf Engl. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol IJMM. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Richards JJ, Ballard TE, Melander C. Inhibition and dispersion of Pseudomonas aeruginosa biofilms with reverse amide 2-aminoimidazole oroidin analogues. Org Biomol Chem. 2008;6:1356. doi: 10.1039/b719082d. [DOI] [PubMed] [Google Scholar]
- Richards JJ, Huigens RW, III, Ballard TE, Basso A, Cavanagh J, Melander C. Inhibition and dispersion of proteobacterial biofilms. Chem Commun. 2008:1698. doi: 10.1039/b719802g. [DOI] [PubMed] [Google Scholar]
- Robinson VL, Wu T, Stock AM. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J Bacteriol. 2003;185:4186–4194. doi: 10.1128/JB.185.14.4186-4194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SA, Melander C. Construction and screening of a 2-aminoimidazole library identifies a small molecule capable of inhibiting and dispersing bacterial biofilms across order, class, and phylum. Angew Chem Int Ed Engl. 2008;47:5229–5231. doi: 10.1002/anie.200800862. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, Aalten DMF van. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Shen M-Y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci Publ Protein Soc. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stowe SD, Thompson RJ, Peng L, Su Z, Blackledge M, Draughn GL, et al. Membrane-Permeabilizing Activity of Reverse-Amide 2-Aminoimidazole Antibiofilm Agents Against Acinetobacter baumannii. Curr Drug Deliv. 2015;12:223–230. doi: 10.2174/1567201811666140924125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutera V, Levert M, Burmeister WP, Schneider D, Maurin M. Evolution toward high-level fluoroquinolone resistance in Francisella species. J Antimicrob Chemother. 2014;69:101–110. doi: 10.1093/jac/dkt321. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Ryan SS, McCoy AJ, Gunn JS. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar typhimurium. Infect Immun. 2002;70:6770–6778. doi: 10.1128/IAI.70.12.6770-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RJ, Bobay BG, Stowe SD, Olson AL, Peng L, Su Z, et al. Identification of BfmR, a response regulator involved in biofilm development, as a target for a 2-Aminoimidazole-based antibiofilm agent. Biochemistry (Mosc) 2012;51:9776–9778. doi: 10.1021/bi3015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Feher VA, Cavanagh J, Perego M, Hoch JA. Characterization of interactions between a two-component response regulator, Spo0F, and its phosphatase, RapB. Biochemistry (Mosc) 1998;37:16538–16545. doi: 10.1021/bi981340o. [DOI] [PubMed] [Google Scholar]
- Tzeng YL, Hoch JA. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- Wemmer DE, Kern D. Beryllofluoride binding mimics phosphorylation of aspartate in response regulators. J Bacteriol. 2005;187:8229–8230. doi: 10.1128/JB.187.24.8229-8230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Cho HS, Hastings CA, Igo MM, Lee SY, Pelton JG, et al. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc Natl Acad Sci U S A. 1999;96:14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundert GCP van, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, et al. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.