Abstract
The first two differentiation events in the embryo result in three cell types – epiblast, trophectoderm (TE), and hypoblast. The purpose here was to identify molecular markers for each cell type in the bovine and evaluate differences in gene expression among individual cells of each lineage. The cDNA from 67 individual cells of dissociated blastocysts was used to determine transcript abundance for 93 genes implicated as cell lineage markers in other species or potentially involved in developmental processes. Clustering analysis indicated that the cells belonged to two major populations (clades A and B) with two subpopulations of clade A and four of clade B. Use of lineage-specific markers from other species indicated that the two subpopulations of clade A represented epiblast and hypoblast, respectively while the four subpopulations of clade B were TE. Among the genes upregulated in epiblast were AJAP1, DNMT3A, FGF4, H2AFZ, KDM2B, NANOG, POU5F1, SAV1 and SLIT2. Genes overexpressed in hypoblast included ALPL, FGFR2, FN1, GATA6, GJA1, HDAC1, MBNL3, PDGFRA and SOX17 while genes overexpressed in all four TE populations were ACTA2, CDX2, CYP11A1, GATA2, GATA3, IFNT, KRT8, RAC1 and SFN. The subpopulations of TE varied amongst each other for multiple genes including the prototypical TE marker IFNT. New markers for each cell type in the bovine blastocyst were identified. Results also indicate heterogeneity in gene expression among TE cells. Further studies are needed to confirm whether subpopulations of TE cells represent different stages in development of a committed TE phenotype.
INTRODUCTION
Preimplantation embryonic development involves a series of cleavage divisions and cell differentiation processes that lead to the formation of the three primary cell types in the embryo: epiblast (precursor of fetus), hypoblast (precursor of extraembryonic endoderm including yolk sac) and trophectoderm (precursor of placenta) (Arnold & Robertson 2009). Upon compaction of the morula, the outer cells become polarized and mechanisms for tight junction formation are triggered (Levy et al. 1986; Nikas et al. 1996; Van Soom et al. 1997). Consequently, the outer cells begin to differentiate into trophectoderm (TE) cells while the cells of the inner cell mass (ICM) maintain pluripotency and cell-to-cell communication through gap junctions (Ducibella & Anderson 1975; Lo & Gilula 1979; Togashi et al. 2015). In the mouse, this first lineage differentiation event is regulated in part by the Hippo signaling pathway which maintains pluripotency of the cells when turned on (i.e., in the ICM) or favors differentiation of the cells when turned off (i.e., in the TE cells) (Paramasivam et al. 2011; Hirate et al. 2013). The presence of gap junctions in the ICM allows activation of a phosphorylation cascade that activates Nf2, Mst1 and Lats1/2. The last molecule in this cascade phosphorylates and inactivates Yap1to favor transcription of pluripotency factors such as Pou5f1 (Lian et al. 2010) and Sox2 (Lorthongpanich et al. 2012). In the absence of gap junctions in the outer cells of the embryo, the phosphorylation cascade is inactive, Yap1 remains dephosphorylated and the protein undergoes nuclear translocation and interaction with Tead4 to promote transcription of differentiation promoting factors such as Cdx2 and Gata3 (Nishioka et al. 2008; Ralston et al. 2010). An additional player of the Hippo pathway is Amot, which inactivates Yap1 by interacting with Yap1 and membrane bound Cdh1 to prevent nuclear translocation of Yap1. In the outer cells of the embryo, Amot is unable to inactive Yap1 because it is restricted to the apical domain of cells by binding to F-actin (Hirate et al. 2013; Leung & Zernicka-Goetz 2013). Transcription factors involved in later TE differentiation include Elf5 (Ng et al. 2008; Pearton et al. 2011) and Eomes (Russ et al. 2000).
As the blastocyst continues to develop, the ICM differentiates into the epiblast and hypoblast. This second differentiation event in the mouse is accomplished in part through activation of the Fgf4-Fgfr2 pathway which regulates transcription of Gata6 and Nanog (Kang et al. 2013). The first group of cells to enter the ICM express Fgf4 but lack Fgfr2 so that expression of Nanog is maintained (Morris et al. 2010, 2013). The cells from the second round of cell division in the ICM begin to express Fgfr2 while Fgf4 from neighboring cells activates Fgf4-Fgfr2 signaling to induce expression of Gata6 and repression of Nanog. Thus, the first group of cells become precursors of the epiblast and the second group are precursors of the hypoblast (Morris et al. 2010, 2013).
The processes for formation and early differentiation of the blastocyst have been been well described in species other than the mouse. Indeed, mechanisms for development during the preimplantation period are not completely conserved between mammalian species. There are large-scale species differences in orthologous gene expression patterns during the preimplantation period caused in large part to mutations in transcription factor binding sites and insertion of transposons containing cis-regulatory elements (Xie et al. 2010). Among the resultant differences in regulatory processes between species are the use of the transcription factor CDX2 to downregulate the pluripotency factor Oct4 (i.e., Pou5f1) in ICM (Berg et al. 2011) and the role of CCL24 in movement of hypoblast cells (Negrón-Pérez et al. 2017).
Understanding the mechanisms for differentiation and function of cell lineages in the bovine embryo has been hampered by the limited repertoire of markers for each cell type. The only definitive marker identified for epiblast is NANOG (Kuijk et al. 2012; Denicol et al. 2014). One marker for ICM in mouse, Pou5f1, is expressed by both ICM and TE in bovine (Berg et al. 2011; Ozawa et al. 2012). GATA6 can serve as a marker for hypoblast in bovine (Kuijk et al. 2012; Denicol et al. 2014) but interpretation of differences in gene expression for this transcription factor is complicated by the fact that expression can also occur for TE (Kuijk et al. 2012; Denicol et al. 2014). The most commonly used marker for TE in bovine is CDX2 (Dobbs et al. 2013; Schiffmacher & Keefer 2013; Denicol et al. 2014) but the observation that outer cells of the blastocyst only gradually become committed to the TE lineage (Berg et al. 2011) is suggestive that there is heterogeneity within this cell population that could be elucidated if additional markers were available.
Here we took advantage of recently-developed microfluidics-based technology for isolation of RNA from individual cells to identify markers of epiblast, hypoblast and TE in the bovine blastocyst. The approach was to assess expression of 93 genes including those that are used as markers for epiblast, hypoblast and TE in other species (Chazaud et al. 2006; Guo et al. 2010; Yan et al. 2013; Hermitte & Chazaud 2014; Blakeley et al. 2015; Boroviak et al. 2015), genes differentially expressed between ICM and TE in the bovine (Ozawa et al. 2012; Nagatomo et al. 2013; Brinkhof et al. 2015; Hosseini et al. 2015; Zhao et al. 2016), and genes involved in important processes for the developing embryo including the Hippo signaling pathway, epigenetic regulation, tight junction formation, cell polarity and chemokine signaling. The result was identification of markers for each cell lineage of interest, evidence that the TE is a heterogeneous tissue in the developing blastocyst, and identification of markers that distinguish between subtypes of TE.
MATERIALS AND METHODS
In vitro production of embryos
Production of embryos involved use of culture media described elsewhere (Ortega et al. 2017). Oocytes and sperm were from cattle representing B. taurus and admixtures of B. taurus and B. indicus. Oocytes were obtained from ovaries collected at an abattoir by bisecting follicles 3–8 mm in diameter using a scalpel and, after follicles on an ovary were bisected, vigorously washing the ovary in oocyte collection medium (BoviPRO ™, MOFA Global, Verona, WI, USA) to dislodge cumulus-oocyte complexes (COC). Fluid containing the COC was then filtered and COC rinsed with fresh collection medium. The COC were then retrieved using a wiretrol pipette (Drummond Scientific Company, Broomall, PA, USA) while visualizing under a dissecting microscope. The COCs were washed another three times in fresh medium. An average of 12–15 COCs (having at least one layer of cumulus cells and containing homogeneous cytoplasm) were collected from each ovary. The COCs were pooled in groups of 10 and matured for 22–24 h in oocyte maturation medium (Tissue Culture Medium-199 with Earle’s salts supplemented with 2% (v/v) bovine steer serum, 100 U/ml penicillin-G, 0.1 mg/ml streptomycin, and 1 mM glutamine) in 50 μl microdrops covered with mineral oil (Sigma-Aldrich, St. Louis, MO, USA) and in an atmosphere of 5% (v/v) CO2 in humidified air at 38.5°C.
Matured COC were pooled, washed three times in HEPES-buffered synthetic oviduct fluid (HEPES-SOF) and placed in 1.7 ml synthetic oviduct fluid for fertilization (SOF-FERT) in groups of up to 300 COCs. For fertilization, frozen-thawed semen from three bulls was pooled, purified by centrifugation, at 1000 × g for 10 min, over a bilayer of Isolate® [Irvine Scientific, Santa Ana, CA, USA; 50% (vol/vol) and 90% (vol/vol) Isolate], washed in HEPES-SOF and diluted with SOF-FERT. Then, 120 μl of semen plus 80 μL of 0.5 mM penicillamine, 0.25 mM hypotaurine, and 25 μM epinephrine solution were added to the COC for a final sperm concentration of 1×106 sperm/ml. Fertilization dishes were incubated in an atmosphere of 5% (v/v) CO2 in humidified air at 38.5°C for 16–18 h.
Presumptive zygotes were collected and exposed to hyaluronidase (1000 U/ml in approximately 0.5 ml HEPES-SOF) to remove cumulus cells and washed three times in HEPES-SOF prior to culture. Embryos were pooled in groups of 25–30 and cultured at 38.5°C in 50 μl microdrops of BBH7 (Cooley Biotech, Gainesville, Florida, USA) covered with mineral oil in a humidified environment consisting of 5% (v/v) O2, 5% (v/v) CO2 and the balance nitrogen.
The proportion of embryos that cleaved was assessed at day 3 after fertilization and the proportion that became blastocysts assessed at day 7. At day 8.75 (207–209 hours post-insemination), all blastocysts (including non-expanded, expanded, hatching and hatched blastocysts) were collected and subjected to blastomere dissociation. Embryos were collected in a total of three replicates. A replicate was defined as a single in vitro fertilization procedure consisting of insemination of 900–1,200 COC. A total of eight bulls were used in the three replicates. The cleavage rate for the three replicates averaged 80% and the percent of inseminated oocytes becoming blastocysts averaged 20% on day 7 and 29.8% on day 8.75.
Preparation of cDNA from single blastomeres
cDNA was prepared from individual blastomeres using the C1 Single-Cell Auto Prep IFC (integrated fluidic circuit) system from Fluidigm (South San Francisco, CA, USA) using manufacturer instructions.
For each replicate, single-cell suspensions were prepared from the blastocysts collected at 207–209 h post-insemination. The number of blastocysts processed for each replicate ranged from 227–336. Blastocysts were washed three times in Dulbecco’s phosphate-buffered saline (DPBS) containing 0.1% (w/w) polyvinylpyrrolidone (DPBS-PVP; Kodak, Rochester, NY, USA), incubated in 0.1% (w/v) protease from Streptococcus griseus (Sigma-Aldrich, St. Louis, MO, USA) in DPBS until the zonae dissolved, and then washed another three times in fresh DPBS-PVP. Embryos were then incubated in 50 μl drop of TrypLE Select Enzyme 10× (ThermoFisher Scientific, Waltham, MA, USA) for 15 min at 38.5°C to disaggregate cells. Finally, blastomeres were transferred to a 1.7 ml tube, vortexed for 2 min, resuspended in 500 μl DPBS-PVP and centrifuged for 5 min at 6000 × g. The supernatant was removed and the cells were resuspended in 10 μl DPBS-PVP.
Following blastocyst disaggregation, cell viability and concentration were measured using the Countess Automated Cell Counter (Life Technologies, Carlsbad, CA, USA) and the concentration was adjusted to 300–400 cells/μl. Viability in each replicate ranged from 33%–82%. A subset of the cells was subjected to a buoyancy test as part of the quality control process for the C1 system to optimize probability of cell capture probability. The cells were mixed with C1 Cell Suspension Reagent at a ratio of 7:1, 7:2 and 7:3 (v:v) and placed in a well of the C1 IFC plate. The buoyancy check was performed under 10× magnification immediately and after 10 min waiting time. Buoyancy was determined by looking at three planes of the well (top, middle and bottom) under the microscope and noting where the majority of the cells were at time 0. This was repeated after the 10 min incubation period. The goal was to have the majority of cells suspended in the middle plane for better capture. The best ratio of suspension reagent to cell suspension was found to be 7:2; at this ratio 78% of cells were in the middle plane.
The C1 Single-Cell Auto Prep IFC for Preamp (10–17 μm) was primed following the manufacturer’s instructions. Because the cells were 13 μm in diameter on average, the plate and protocol used for priming was C1 DNA Seq IFC: Prime (1773×). The cell suspension was loaded into the C1 plate for individual cell capture, lysis for RNA extraction and cDNA pre-amplification. A viability cell staining was performed for one of the replicates using the LIVE/DEAD Viability/Cytotoxicity kit from Fluidigm to assess the proportion of live and dead cells that had been captured. In this replicate, 54 cells were captured including 11 dead cells and 43 live cells. Only the cDNA from the 43 viable cells was used for further analysis. Overall, the success of the system for capturing individual cells was low. The number of cells collected for each replicate were 6, 19 and 42 for a total of 67 cells analyzed. The C1 plates containing single-cell cDNA were stored at −20°C until gene expression analysis.
Gene expression analysis
The Fluidigm® qPCR microfluidic device Biomark™ HD system was used for gene expression assays. Primers were designed by Fluidigm® Delta Gene™ assays (Fluidigm Co., San Francisco, CA, USA) and validated by Miami Center for AIDS Research (CFAR) at the University of Miami Miller School of Medicine. A description of the genes analyzed, the primers used, and the rationale for including in the analysis are in Supplemental File S1. Primers were designed for 2 housekeeping genes (ACTB and GAPDH), 9 epiblast markers from one or more of 3 species; 9 hypoblast markers; 11 trophectoderm markers, 16 genes involved in chemokine signaling, 9 genes involved in Hippo signaling, 13 genes involved in epigenetic modification, 11 genes involved in tight junction formation and cell polarity, and, another 16 genes of interest. Initially, three genes, SDHA, H2AFZ and HPRT1, were also selected as housekeeping genes but they were not used as such because of non-uniform expression.
The procedure for quantitative PCR (qPCR) using the Biomark™ system (Dominguez et al. 2013) was as follows. Primer-probe sets and samples were loaded on an IFC plate and placed into a controller that prepares the nanoliter reactions. The plate was then transferred into the Biomark™ machine which includes a thermocycler for real-time qPCR. A total of 40 PCR cycles were performed using the 96.96 dynamic array IFC developed by the manufacturer. Cycle threshold (Ct) values were calculated by the Fluidigm Real-Time PCR analysis software. The cutoff for detectable genes was those with Ct >27. The geometric mean of the two housekeeping genes was calculated and used to obtain the delta Ct (dCt) values of the other 93 genes of interest. Fold changes were calculated as 2−dCt relative to housekeeping genes.
The parameters for the validation of primers for qPCR using the Biomark™ HD system were as follows. Validation was achieved using two pools of 40 embryos each (day 5 morulae and day 7 and day 8 blastocysts), endometrium, fetal liver and leukocytes. RNA was extracted from all samples using the RNeasy micro kit (Qiagen, Valencia, CA, USA); a DNase treatment was included as part of the protocol. Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) following manufacturer’s instructions. The cDNA was pre-amplified following the guidelines for the Ambion® Single Cell-to-CT™ kit (ThermoFisher Scientific) and diluted-serially in 2 fold dilutions to a single cell equivalent. Primer-probe sets and samples were loaded on an IFC plate and placed into a controller that prepares the nanoliter reactions. The plate was then transferred into the Biomark™ machine for real-time qPCR. Standard curves and cycle threshold (Ct) values were calculated by the Fluidigm Real-Time PCR analysis software. The quality control criteria for passing a primer were an R2≥0.97, efficiency of 0.8–1.3 and a slope of −3.92 to −2.76. All 96 primer pairs met these criteria except for the primer set for KLF2 (epiblast marker). Thus, results for this gene were not used for further analysis.
Clustering and Statistical Analysis
Cells were classified based on patterns of gene expression using unsupervised cluster analysis with Gene Cluster 3.0 clustering software (de Hoon et al., 2004). Clustering analyses were performed using six separate conditions of distance and linkage: Spearman correlation and centroid linkage, Spearman correlation and complete linkage, correlation center and centroid linkage, correlation center and complete linkage, Euclidian distance and centroid linkage, and Euclidian distance & complete linkage. Java Treeview 3.0 (https://doi.org/10.5281/zenodo.160573) was used to visualize and group the clustered data. Two or more populations of cells were identified for each analysis. For each clustering analysis, differences among populations of cells clusters in expression of each of the 93 genes of interest was determined by least-squares analysis of variance using the generalized linear models procedure (PROC GLM) of SAS v. 9.4 (SAS Institute Inc., Cary, NC, USA). Data analyzed were values for fold change relative to housekeeping genes. When the main effect of treatment was significant, differences between individual populations were determined using the pdiff statement of PROC GLM.
The effectiveness of each clustering analysis to identify epiblast, hypoblast and TE was evaluated by evaluating differences among clusters in expression of selected gene markers considered characteristic of epiblast (FGF4, NANOG and POU5F1), hypoblast (FGFR2, GATA6 and PDGFRA) and TE (CDX2, IFNT and KRT8). Using these criteria, the most effective clustering analysis was that using Euclidian distance and complete linkage. Subsequent analyses of differences in gene expression were based on populations of cells identified with this analysis. Discrimination of cells in the 6 populations identified by this cluster analysis was further illustrated by principal component analysis using the PROC PRINCOMP procedure of SAS v 9.4.
RESULTS
Identification of cell populations using cluster analysis
Unsupervised clustering of gene expression resulted in identification of 6 cell populations in two large clades (A and B) that were subdivided into smaller subclades (Figure 1). Clade A contained two subclades termed A1 (n=7 cells) and A2 (4 cells). Clade B was subdivided into two subclades [B1 (13 cells) and B2 (43 cells), with clade B2 consisting of three smaller subpopulations termed B2.1 (8 cells), B2.2 (19 cells) and B2.3 (16 cells). Note that cells in clade B were in the all three replicates of the cell separation procedure whereas cells in clade A were present in the third replicate only.
FIGURE 1.
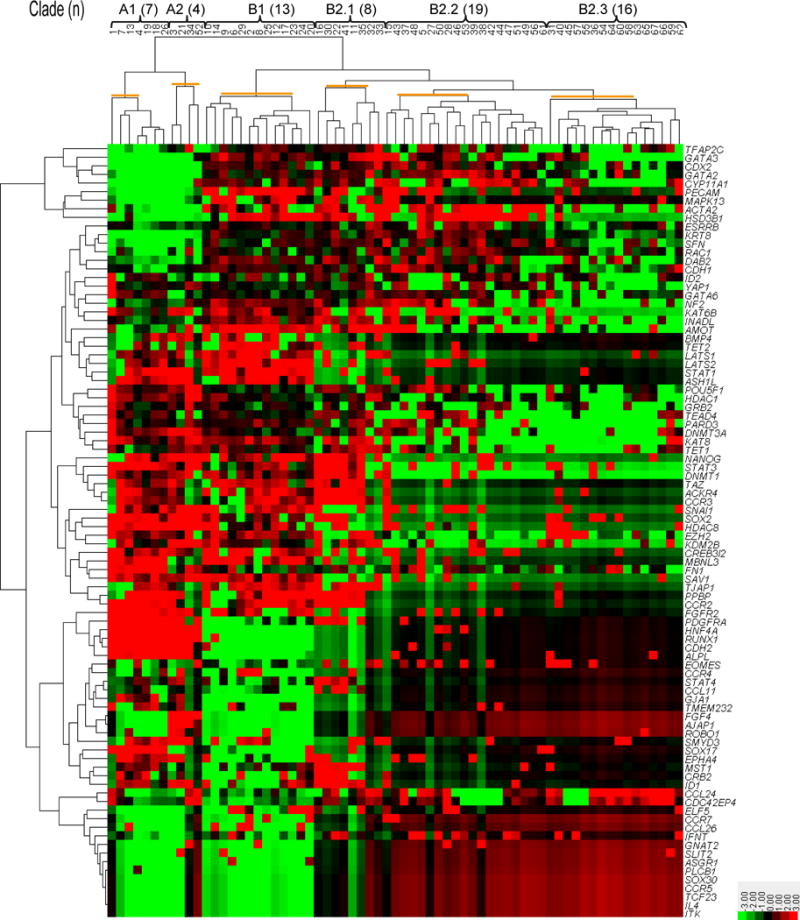
Identification of embryonic cell populations by unsupervised clustering analysis of gene expression. Heatmaps were generated for expression levels of 90 genes from 67 individual blastomeres after normalizing to housekeeping genes, ACTB and GAPDH. The output yielded two major clades (A and B) that were subdivided into six smaller clusters (highlighted with orange lines): A1, A2, B1, B2.1, B2.2 and B2.3. The numbers in parentheses are the number of cells in each group. The color key is at the bottom right (−3=bright green, 0=black, 3=bright red).
Principal component analysis also resulted in clustering of cells into two major populations, with one population corresponding to clade A of the unsupervised clustering analysis and the other population corresponding to clade B (Figure 2). Principal component analysis also indicated discrimination between subclades identified by the clustering analysis.
FIGURE 2.
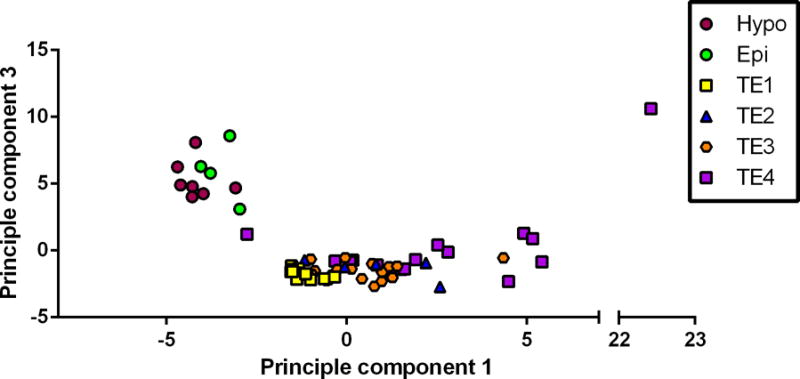
Principal component analysis of 67 cells subjected to gene expression analysis. Note that cells labeled as Hypo and Epi correspond to cells in Clade 1 from the unsupervised clustering analysis while cells labeled as TE1, TE2, TE3 and TE4 correspond to Clade 2.
Identification of cell subpopulations as epiblast, hypoblast, and TE
Expression patterns of genes known to be markers for specific cell lineages in the bovine or other species were used to identify each subclade of cells. Expression patterns of nine genes considered as markers of epiblast in bovine (Kuijk et al. 2012; Denicol et al. 2014)], mouse (Guo et al. 2010; Boroviak et al. 2015), or human (Yan et al. 2013; Blakeley et al. 2015) are shown in Figure 3. Three of these genes, FGF4, NANOG, and POUF51, were more highly expressed in subclade A2 than other subclades. In addition, another marker, HNF4A, was more highly expressed in subclades A1 and A2 than for cells in clade B; FN1 was upregulated in clade A1 as compared to the other five subclades. Based on these patterns of gene expression, subclade A2 was considered to represent epiblast. Note that several markers of epiblast in the mouse (Guo et al. 2010; Boroviak et al. 2015) were not preferentially expressed in subclade A2 including BMP4, ESRRB, and SOX2.
FIGURE 3.
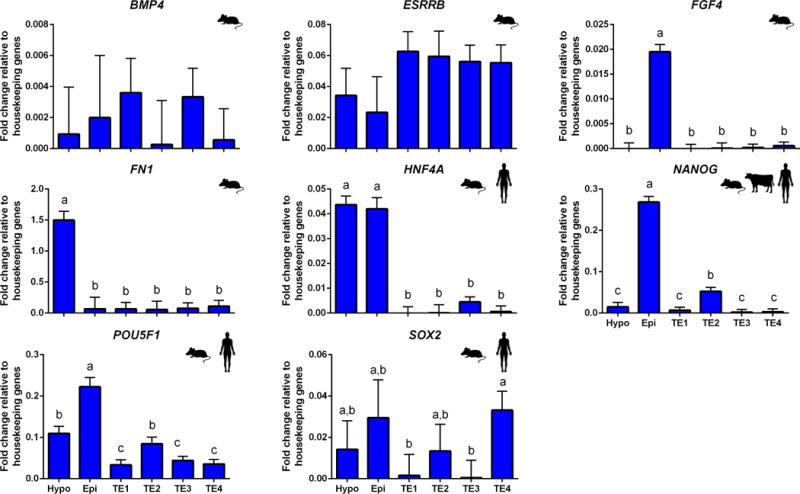
Expression of genes considered as prototypical epiblast markers. Data were normalized to the geometric mean of ACTB and GAPDH and are presented as least-squares means ± SEM. The number of cells within each subgroup were as follows: epiblast (subclade A2) n=4, hypoblast (subclade A1) n=7, TE1 (subclade B1) n=13, TE2 (subclade B2.1) n=8, TE3 (subclade B2.2) n=19 and TE4 (Subclade B2.3) n=16. Bars labeled with different letters are different from each other (P<0.05). Species symbols are used to denominate upregulation of the gene in epiblast of mouse (Guo et al. 2010; Boroviak et al. 2015), human (Yan et al. 2013; Blakeley et al. 2015) or bovine (Denicol et al. 2014).
Expression of nine genes that are characteristic of hypoblast in either bovine (Kuijk et al. 2012; Denicol et al. 2014), mouse (Chazaud et al. 2006; Guo et al. 2010; Boroviak et al. 2015) or human (Yan et al. 2013; Blakeley et al. 2015) are shown in Figure 4. Four of these genes (FGFR2, GATA6, PDGFRA and SOX17) were more expressed in subclade A1 than other populations. Accordingly subclade A1 was labeled as hypoblast. Note that this subclade, like the epiblast subclade, also expresses high amounts of HNF4A (Figure 3) and RUNX1, which is a hypoblast marker in mouse (Guo et al. 2010). Other markers of hypoblast in mouse, including CREB3L2, GRB2, SNAI1, and TCF23 (Chazaud et al. 2006; Guo et al. 2010) were not overexpressed in the hypoblast clade.
FIGURE 4.
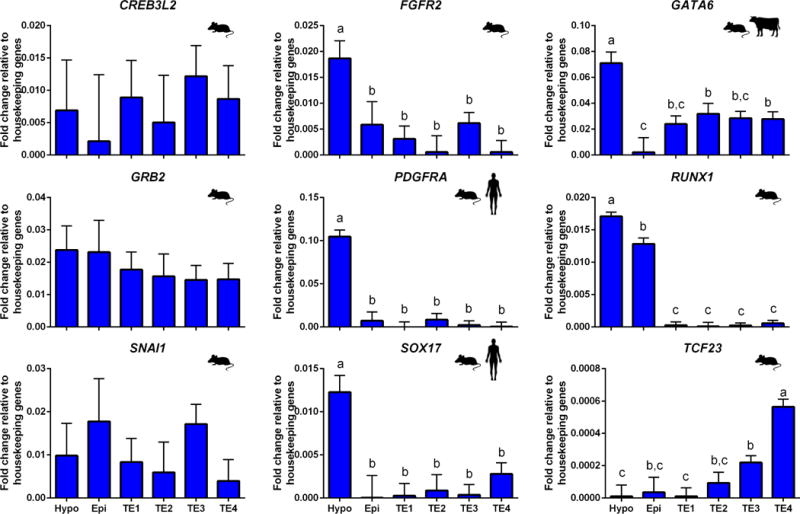
Expression of genes considered as prototypical hypoblast markers. Species symbols are used to denominate upregulation of the gene in hypoblast of mouse (Chazaud et al. 2006; Guo et al. 2010; Boroviak et al. 2015), human (Yan et al. 2013; Blakeley et al. 2015) or bovine (Kuijk et al. 2008; Denicol et al. 2014). Refer to Figure 3 for additional information.
Results for expression of 11 genes considered as TE markers in bovine (Ozawa et al. 2012; Nagatomo et al. 2013; Brinkhof et al. 2015; Hosseini et al. 2015; Zhao et al. 2016), mouse (Guo et al. 2010; Boroviak et al. 2015), or human (Yan et al. 2013; Blakeley et al. 2015) are shown in Figure 5. The genes CDX2, GATA3, IFNT and KRT8 exhibited, or tended to exhibit, higher expression in the subpopulations of subclade B than for cells of subclade A. Accordingly, clade B was considered to represent TE and subclades were renamed as follows: B1=TE1, B2.1=TE2; B2.2=T3 and B2.3=T4. Expression of GATA3 tended to be higher for TE1 and TE2 than TE3 and TE4, IFNT was highest in TE3 and lowest in TE1, and KRT8 was lowest for TE4. Also, the mouse TE marker, EOMES, was more highly expressed in TE4 than in TE1 and tended to be more expressed than in other TE subpopulations or in epiblast or hypoblast. There was no difference in gene expression between subpopulations for DAB2, MBNL3, PECAM1, TEAD4 or TFAP2C. Also, the mouse TE marker, MBNL3, was most highly expressed in hypoblast (significantly different from epiblast and TE2 and TE4).
FIGURE 5.
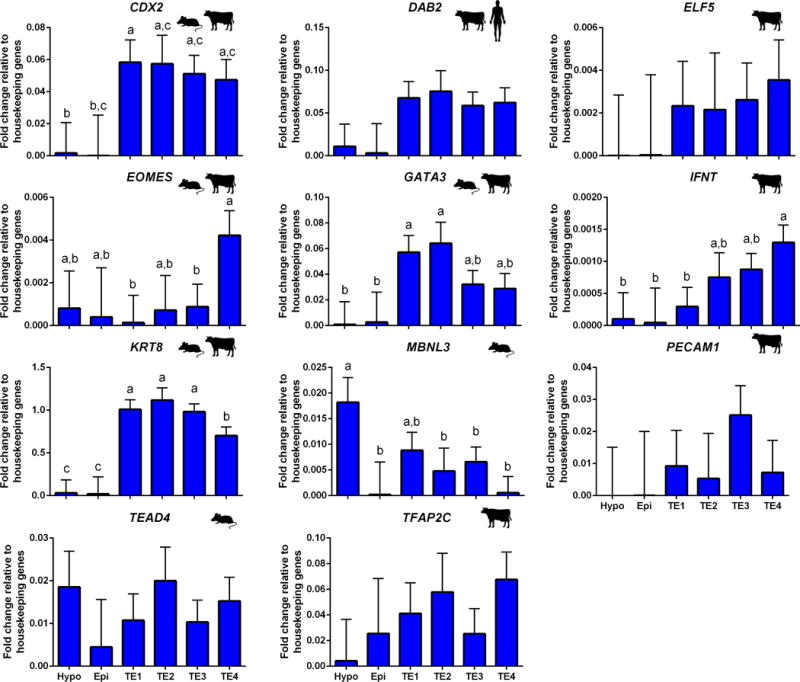
Expression of genes considered as prototypical trophectoderm markers Species symbols are used to denominate upregulation of the gene in TE of mouse (Guo et al. 2010; Boroviak et al. 2015), human (Yan et al. 2013; Blakeley et al. 2015) or bovine (Ozawa et al. 2012; Nagatomo et al. 2013; Brinkhof et al. 2015; Hosseini et al. 2015; Zhao et al. 2016). Refer to Figure 3 for additional information.
Note that expression of GATA6, which is a marker of hypoblast in bovine, but which also shows moderate immunolabeling in TE and low immunolabeling in epiblast (Kuijk et al. 2012; Denicol et al. 2014), exhibited a similar pattern of transcript abundance – high in hypoblast, low in epiblast and moderate in TE populations (Figure 4).
Other genes overexpressed in epiblast and hypoblast
In addition to genes used to define epiblast (Figure 3) and hypoblast (Figure 4), there were six additional genes that were most highly expressed in epiblast (AJAP1, DNMT3A, H2AFZ, KDM2B, SAV1, and SLIT2), two genes most highly expressed in hypoblast (ALPL and GJA1) and three genes (CDH2, HDAC1, and HDAC8) that were more expressed in both epiblast and hypoblast than the TE populations (Figure 6). Of these latter three genes, CDH2 was expressed equally in epiblast and hypoblast, HDAC1 was more expressed in hypoblast than epiblast, and HDAC8 was more expressed in epiblast than hypoblast (Figure 6). One gene, ID2, was expressed more in hypoblast than epiblast, with amounts in TE being intermediate (Figure 6).
FIGURE 6.
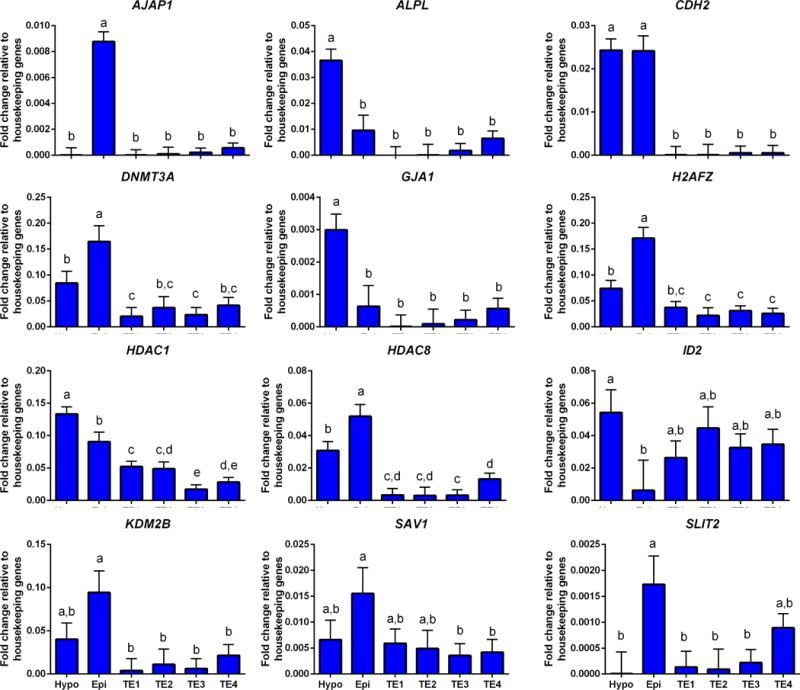
Additional genes that were differentially expressed in epiblast and hypoblast. Refer to Figure 3 for additional information.
Other genes overexpressed in all subpopulations of TE
Besides the genes used to define TE populations, there were an additional five genes that were expressed in all four TE subpopulations (Figure 7). Of these, expression of GATA2, RAC1 and SFN was relatively uniform between TE subpopulations, AMOT was higher for TE1 than TE4 (with amounts for TE2 and TE3 being intermediate), and expression of CYP11A1 tended to be higher for TE3 than other TE populations.
FIGURE 7.
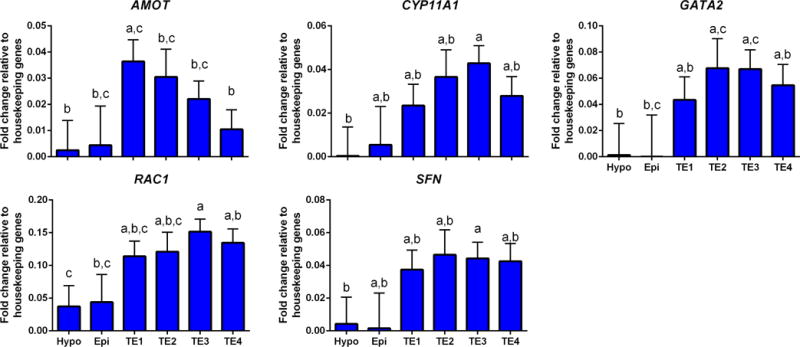
Additional genes upregulated in trophectoderm that were expressed in all four TE subpopulations. Refer to Figure 3 for additional information.
Other genes overexpressed in some TE subpopulations
There were several genes that were differentially expressed in one or more TE subpopulations as compared to other cell subpopulations. Cells in TE1 expressed higher amounts of transcripts for CCL26, LATS1 and LATS2 than for all or some subpopulations of cells (Figure 8). Expression of LATS1 was lowest for TE3 and TE4. Expression of TET2 was also higher than for other subpopulations except TE2 (Figure 8). Cells in TE2 expressed higher amounts of transcript for ACKR4, CCL11, CCR2, CCR3, CRB2, DNMT1, EPHA4, HPRT1, ID1, MST1, PPBP, TAZ, and TJAP1 than other TE subpopulations and, except for HPRT1 (high expression also in hypoblast), higher than epiblast or hypoblast (Figure 8). Expression of INADL was higher for TE2 than all cell subpopulations than TE1, KAT6B was higher for TE2 than TE4, STAT3 was higher for TE2 than all populations except hypoblast, and STAT4 (Figure 8) was higher for TE2 than all populations except epiblast (Figure 8). SOX2 was highly expressed in epiblast, hypoblast, TE2 and TE4 but lowly expressed in TE1 and TE3 (Figure 3).
FIGURE 8.
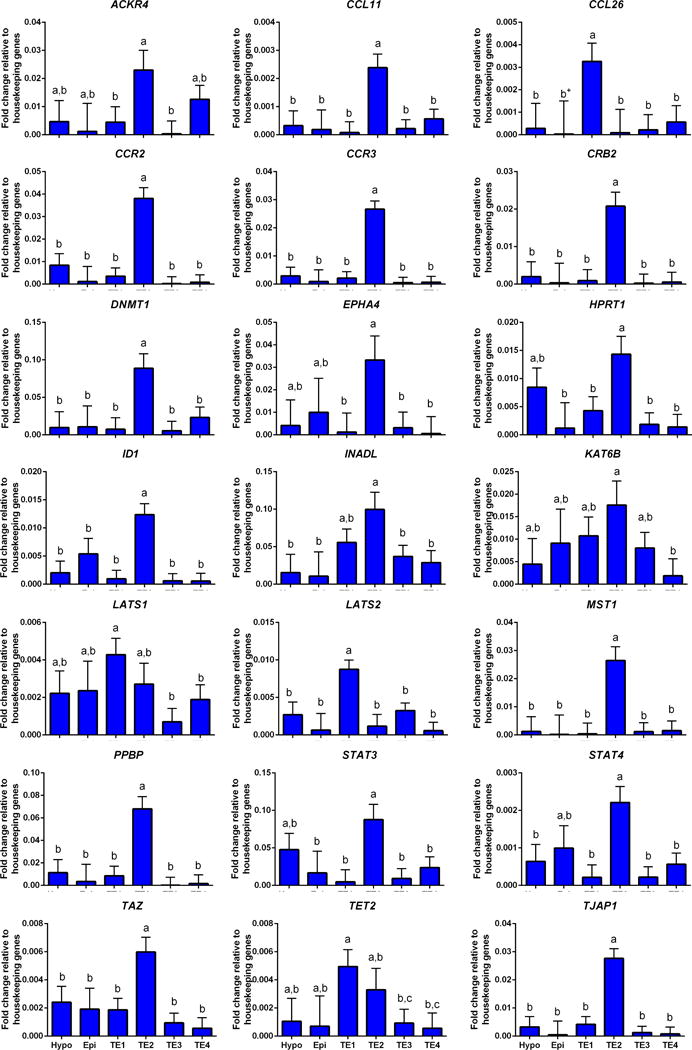
Genes that were differentially expressed in TE1 and TE2 cell populations. Refer to Figure 3 for additional information.
As compared to other cell populations, expression of ACTA2 was higher for TE3 than other TE populations and cells in TE4 expressed higher amounts of ASGR1, CCL24, CCR5, CDC42EP4 (difference with epiblast not significant), IL4, ITK, PLCB1 and SOX30 than other cells; cells in TE3 expressed lower amounts of transcripts for EZH2, cells in TE3 and TE4 expressed lower amounts of LATS1 than TE1, cells in TE3 and TE4 expressed lower amounts of SDHA than for other populations, and cells in TE4 expressed the lowest amount of HSD3B1 (Figure 9). Moreover, expression of TET1 was highest in TE2 and lowest in TE3 (Figure 9).
FIGURE 9.
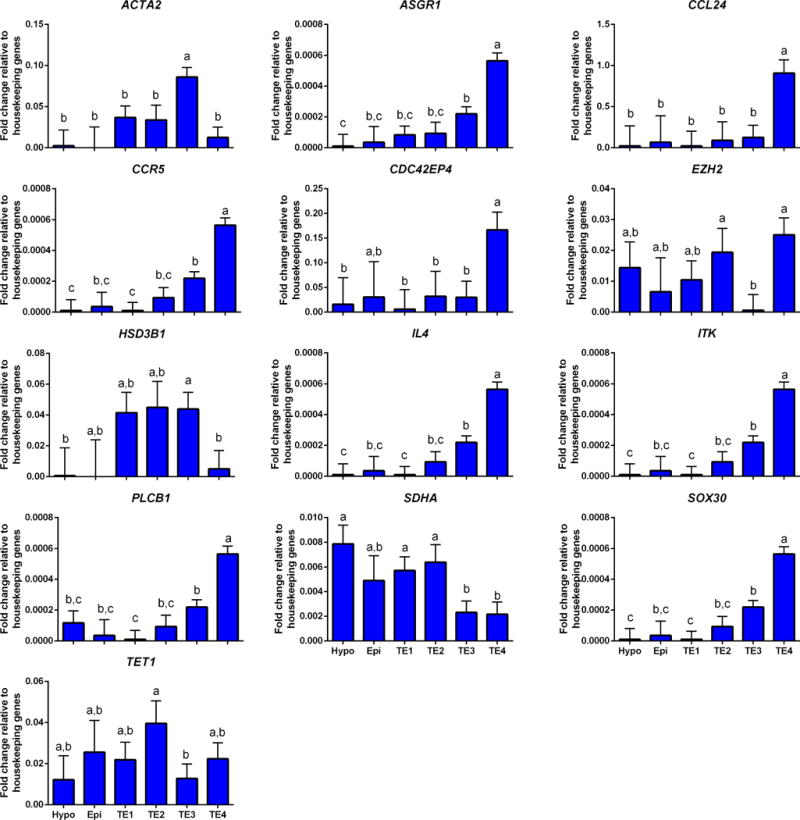
Genes that were differentially expressed in TE3 and TE4 cell populations. Refer to Figure 3 for additional information.
Genes whose expression did not differ between subpopulations
There were 14 genes in which there was no significant difference in expression between cell populations (Figure 10).
FIGURE 10.
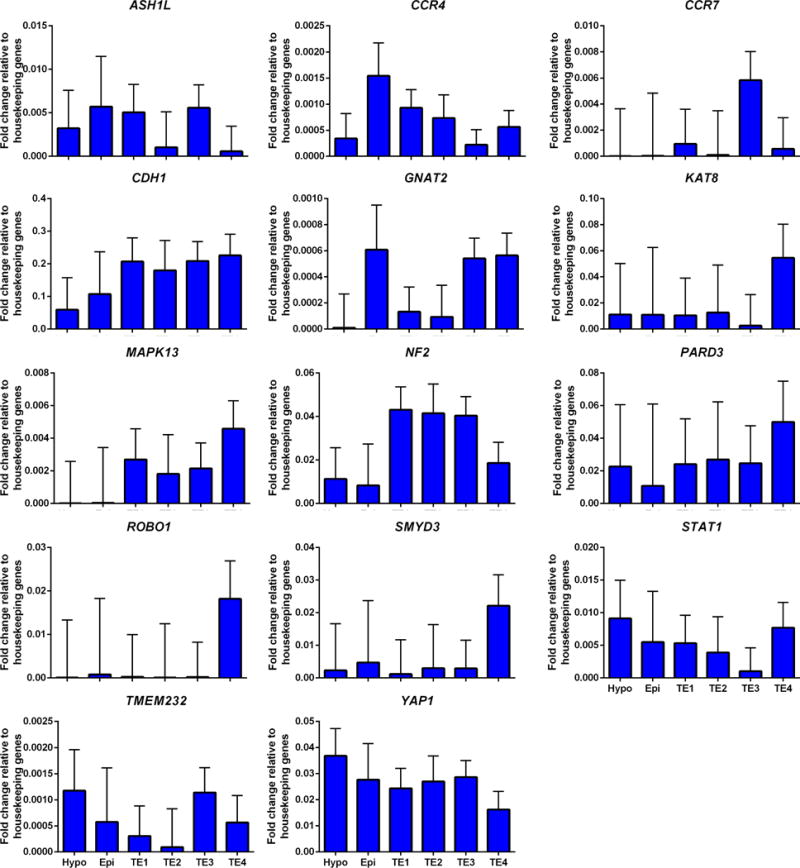
Transcripts whose expression did not differ significantly among cell populations. Refer to Figure 3 for additional information.
DISCUSSION
In the present experiment, analysis of gene expression at the single-cell level has revealed several heretofore unknown characteristics of the bovine blastocyst. This accomplishment was achieved despite the inefficiency of the procedures for isolation of individual cells using the C1 Single-Cell Auto Prep IFC. Only 67 usable cells were captured in three replicates although the theoretical number of cells that could be captured was 288. The reasons for the low capture rate are not known. Contributing factors could have included cell aggregation or suboptimal buoyancy conditions for cell capture. Clearly, more work is required to optimize the use of the C1 technology for isolation of individual cells from the bovine blastocyst. Alternative approaches for isolation of individual cells could include micromanipulation to pluck individual cells from disaggregated embryos, as has been performed for the mouse and human embryo (Guo et al. 2010; Yan et al., 2013; Blakely et al. 2015; Zhou et al., 2015).
One consequence of the low number of cell numbers analyzed is that there could be some differences in gene expression between populations that did not approach statistical significance because of the small degree of replication. It is also possible that the selection of cells was not random but that cells of certain characteristics were more likely to be captured than others. Nonetheless, the experiment was successful in the identification of molecular markers for epiblast, hypoblast and TE lineages in the bovine blastocyst, including some markers that allow discrimination of 4 subpopulations of TE. These are summarized in Figure 11. It is proposed that mRNA for these genes, or the protein products encoded by those mRNA, can be used to identify and study the function of specific lineages in the bovine blastocyst. Some of the molecular markers for epiblast, hypoblast and TE were expressed almost exclusively by one cell lineage. Examples include AJAP1, FGF4 and NANOG in epiblast, FGFR2, FN1, and SOX17 in hypoblast, and CDX2, GATA2, GATA3, IFNT, KRT8 and SFN in TE. Others, while more expressed in one cell subpopulation than others, were expressed to variable degrees by more than one cell subpopulation. Examples include HDAC1 and POU5F1, which were highest in epiblast but which were also expressed in hypoblast and TE cells, GATA6, in which transcript abundance was highest for hypoblast but which was also expressed in TE cell subpopulations, and SLIT2, most expressed in epiblast but also moderately expressed in the TE4 subpopulation. Gene markers identified in Figure 11 that allow discrimination between the four subpopulations of TE also varied in the degree to which expression was limited to one subpopulation. Thus, some genes, like ACKR4, CCL11, CCL26, CCR2, CCR3, CRB2, DNMT1, EOMES, EPHA4, ID1, MST1, PPBP, STAT3, STAT4, TAZ, and TJAP1, were much more expressed for one TE subpopulation than for other subpopulations whereas others, like AMOT, CYP11A1 LATS1, and TET2, while being more expressed in one or more cell subpopulations than others, were expressed more uniformly among the subpopulations.
FIGURE 11.
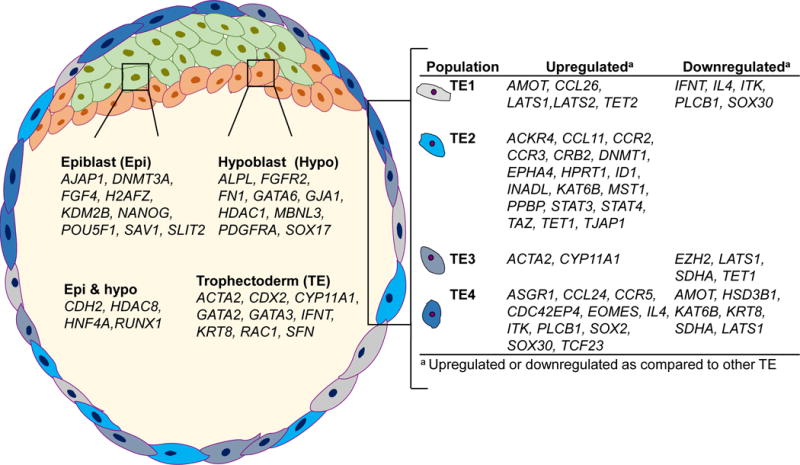
Model of genes whose expression differs between cell populations in the bovine blastocyst. Genes listed under Epiblast, Hypoblast, Epi & hypo and Trophectoderm were upregulated in that cell type as compared to other cell populations: For the four populations of trophectoderm cells, two types of genes are listed: those that were upregulated relative to one or more other populations of trophectoderm cells and those that were downregulated.
Many of the epiblast, hypoblast and TE markers identified in Figure 11 have been previously identified as cell lineage markers. Examples include Cdx2, Fgf4, Fgfr2, Gata3, Gata6, Hnf4a, Krt8, Nanog, Pdgfra, Pou5f1, Runx1, and Sox17 for mouse (Guo et al. 2010; Boroviak et al. 2015), HNF4A, NANOG, PDGFRA, POU5F1, and SOX17 for human (Yan et al. 2013; Blakeley et al. 2015) and CDX2, GATA3, GATA6, IFNT, KRT8, and NANOG for bovine (Kuijk et al. 2012; Ozawa et al. 2012; Nagatomo et al. 2013; Denicol et al. 2014; Brinkhof et al. 2015; Hosseini et al. 2015; Zhao et al. 2016). Others have not been previously described as cell lineage markers, for example, GATA2, MST1, SFN and TJAP1 for TE.
Several of the cell lineage markers in the mouse or human embryo displayed an expression pattern in the bovine embryo distinct from the expected pattern. Examples include BMP4, CREB3L2, ESRRB, FN1, GRB2,, MBNL3, SNAI1, TCF23, and TEAD4. Also, some TE markers in mice were only expressed in a subset of TE cells, most notably ELF5 and EOMES. Differences between the bovine and other species is not surprising because evolutionary analysis indicates that there is a large degree of heterogeneity in the control of gene expression in the early embryo among species (Xie et al. 2010; Berg et al. 2011; Niakan & Eggan 2013; Hosseini et al. 2015).
It remains to be determined whether differences in protein expression recapitulate differences in transcript abundance and whether markers that are useful for distinguishing between cell lineages at the mRNA level will also prove useful at the protein level. It is notable, however, that for the proteins previously examined, there is concordance with results on mRNA determined here. CDX2 is a widely-used protein marker for TE (Ross et al. 2009; Kuijk et al. 2012; Schiffmacher & Keefer 2013; Denicol et al. 2014) and was found to be highly expressed in all four TE populations in the present study. In addition, IFNT, whose transcript was more expressed in TE than epiblast and hypoblast, was immunolocalized to TE but not ICM (Johnson et al. 2006). Similarly, like for present findings regarding gene expression, GATA6 can be localized to both hypoblast and TE, with amounts of immunoreactive protein greater in hypoblast than TE (Kuijk et al. 2012; Denicol et al. 2014). The present finding that POU5F1 was most highly expressed in the epiblast but was also detected in populations of hypoblast and TE is consistent with earlier results that immunoreactive POU5F1 was present in ICM and TE, with more intense labeling in the former (Berg et al. 2011).
There was one case, however, where there is a lack of concordance between gene expression and protein accumulation. Immunoreactive YAP1 was greater in TE compared to ICM in day 7 bovine blastocysts (Tribulo et al. 2017) even though YAP1 mRNA was not different between cell lineages in the current study. Indeed, at least in the mouse, YAP1 is regulated post-translationally through control of phosphorylation and degradation (Basu et al. 2003; Verghese et al. 2012). It is also possible that differences in gene expression between cell lineages, seen here with blastocysts at day 8.75 of development, may be different at other stages in development. One example is CCL24, whose expression in the blastocyst peaks at about Day 7 of development (Negrón-Pérez et al. 2017). At Days 7–8, CCL24 is more expressed in ICM than TE (Ozawa et al. 2012; Brinkhof et al. 2015; Hosseini et al. 2015; Zhao et al. 2016). In the current experiment, however, CCL24 expression was largely limited to one TE subpopulation (TE4), suggesting a developmental switch in the site of expression of this chemokine gene.
Patterns of gene expression revealed in this study provides some insight into the processes leading to the formation and subsequent development of each cell lineage. As expected, the epiblast, which remains in a pluripotent state after formation of the TE and hypoblast (Nichols et al. 1998; Kirchhof et al. 2000; Avilion et al. 2003; Silva et al. 2009), exhibited upregulation of the pluripotency genes NANOG and POU5F1. The epiblast also overexpressed FGF4, which has been associated with high levels of SOX2 expression in the epiblast of mouse species (Yuan et al. 1995). Expression of two genes involved in epigenetic remodeling in the epiblast, DNMT3A and KDMB2, are also consistent with maintenance of epiblast cells. Studies using embryonic stem cells deficient for Dnmt1/Dnmt3a/Dnmt3b indicated that methyltransferases are important for allowing embryonic stem cells to contribute to embryonic lineages at day E10.5 but is not required for formation of extraembryonic tissues (Sakaue et al. 2010). KDM2B has also been implicated in maintenance of pluripotency by driving noncanonical Polycomb repressive complex 1 (ncPRC1) to CpG islands (Morey et al. 2015; Schoenfelder et al. 2015) and subsequent repression of differentiation promoting genes (Smith et al. 2016). Another gene upregulated in epiblast, H2AFZ, encodes for a variant of the histone octamer member H2A. The specific function of H2AFZ during early embryonic development is unknown but H2afz deficient mouse embryos die by E5.5 and have an abnormal ICM (Faast et al. 2001). A histone deacetylase, HDAC8, was upregulated in both epiblast and hypoblast, in agreement with a previous finding that the gene was upregulated in the ICM of the bovine embryo (Hosseini et al. 2015). The significance of this finding is unclear since HDAC8 is involved in silencing pluripotency genes (Saha et al. 2013). Another two genes upregulated in the epiblast may be involved in the Hippo signaling pathway that controls formation of the ICM. AJAP1 is an adherens junction associated protein. During formation of the ICM and TE, adherens junctions play a crucial role in promoting ICM formation by interacting with Cdh1 and Amot (Sasaki 2015). Another gene upregulated in epiblast, SAV1, is an upstream member of the phosphorylation cascade of the Hippo signaling pathway (Lee et al. 2008).
Differentiation of the hypoblast in mice involves actions of Ffg4 from neighboring cells on hypoblast precursors (Morris et al. 2010, 2013). The situation with respect to the bovine is unclear. Treatment of bovine embryos with FGF4 and heparin can lead to blastocysts where the ICM is composed entirely of hypoblast cells (Kuijk et al. 2012). In this same paper, inhibition of FGF4 signaling caused by addition of sodium chlorate caused no effect on number of hypoblast cells (Kuijk et al. 2012). Addition of FGF2 to bovine blastocysts increased the outgrowth of hypoblast cells expressing GATA4 and GATA6 (Yang et al. 2011). Present results evaluating gene expression are consistent with a role of FGF4 in the formation of the hypoblast. In particular, the epiblast subpopulation preferentially expressed FGF4 while cells of the hypoblast subpopulation preferentially expressed FGFR2. One downstream gene for FGF signaling in human pluripotent stem cells is HNF4A (Twaroski et al. 2015), which is also a hypoblast marker in the mouse (Hermitte & Chazaud 2014) and human (Yan et al. 2013). This gene was overexpressed in the epiblast and hypoblast subpopulations and mRNA was localized to ICM, particularly those lining the blastocoele, in the bovine blastocyst (Nagatomo et al. 2013). Another gene that is regulated by FGF4 in mouse embryo is the transcription factor Sox17 (Frankenberg et al. 2011), which in turn can act as an enhancer for Fn1 (Shirai et al. 2005). Both of these hypoblast markers were found to be upregulated in the hypoblast subpopulation in the present experiment. FN1 encodes fibronectin 1, a component of extracellular matrix. In the bovine, FN1 is primarily localized to ICM although a small fraction of immunoreactive FN1is associated with what could be hypoblast (Goossens et al. 2009).
Another growth factor important for hypoblast survival and expansion in the mouse is Pdgf (Artus et al. 2010, 2013). The receptor for this growth factor, PDGFRA, was overexpressed in the hypoblast subpopulation. Hypoblast cells also overexpressed two genes involved in gap junction formation - GJA1, and CDH2, which was also highly expressed in epiblast. Gap junctions play and important role in Hippo signaling in the ICM by regulating localization of Cdh1 which is a direct regulator of the Hippo signaling (Sasaki 2015). Perhaps these specialized junctional complexes are important for differentiation or maintenance of the hypoblast.
Although hypoblast cells represent a more differentiated cell type than epiblast, gene expression patterns suggest that this cell line retains some degree of an undifferentiated estate, at least at this stage of development. Thus, hypoblast cells overexpressed ALPL, which is highly expressed in embryonic stem cells in mouse (Tielens et al. 2006) and pluripotent stem cells derived from cloned bovine embryos (Wang et al. 2005). The hypoblast is also apparently undergoing epigenetic regulation as indicated by overexpression of HDAC1 and HDAC8. Hdac1 is a histone deacetylase that is embryonic lethal (Montgomery et al. 2007) and is involved in recruiting silencing complexes to maintain pluripotency of mouse embryonic stem cell (Dovey et al. 2010). In addition, Hdac1 cooperates with Brg1 to silence Nanog in the TE (Carey et al. 2015). Perhaps the same phenomenon occurs in hypoblast. Inhibition of Nanog expression is sufficient to induce hypoblast formation in mouse embryos (Frankenberg et al. 2011).
In the current experiment, a large number of genes involved in chemokine signaling were selected for analysis because of a recent finding that suggests CCL24 participates in positioning of hypoblast precursors to the outside of the ICM (Negrón-Pérez et al. 2017). This chemokine gene reaches maximal expression at Day 7 of development (Negrón-Pérez et al. 2017) and is more highly expressed in the ICM than in the TE at Day 7–9 (Ozawa et al. 2012; Brinkhof et al. 2015; Hosseini et al. 2015; Zhao et al. 2016). Moreover, inhibition of CCL24 signaling by addition of receptor antagonists or knockdown of CCL24 mRNA reduces the proportion of GATA6+ cells in the outer portion of the ICM (Negrón-Pérez et al. 2017). Based on these results, it was expected to observe higher expression of CCL24 in epiblast and of various chemokine receptor genes in hypoblast. In fact, however, CCL24 was overexpressed in one population of TE cells and there was no evidence for overexpression of chemokine receptors in hypoblast. One possibility is that the apparent expression of CCL24 in ICM reflects contamination with TE. This could be possible because, as discussed below, the TE4 population may represent polar TE that is adjacent to ICM. However, immunolocalization of CCL24 indicated that the protein was most commonly localized to the ICM (Negrón-Pérez et al. 2017). It may be more likely, therefore, that the site of CCL24 expression changes as the embryo advances in development so that by day 8.75 of development, the time examined here, CCL24 expression by cells of the ICM becomes inhibited and the TE begins expressing CCL24.
CDX2 is the transcription factor upregulated in TE as compared to epiblast or hypoblast that has been shown to be necessary for development of the trophoblast in the bovine. Zygotic deletion of CDX2 does not prevent blastocyst formation (Berg et al. 2011) but is important for upregulation of GATA3 (Sakurai et al. 2016a), and maintenance of tight junctions in the TE (Goissis & Cibelli 2014). Transgenic CDX2-deficent embryos were unable to undergo trophoblast elongation at Day 14 of gestation after transfer to females but expression of IFNT at this stage was not blocked (Berg et al. 2011). A recent paper from the mouse indicates that maternally-derived Cdx2 is important for blastocyst formation (Jedrusik et al. 2015) and it remains to be determined whether a similar phenomenon occurs in the bovine.
In the mouse, transcription of Cdx2 depends upon formation of a complex of Yap1 and Tead4 after inactivation of Hippo signaling in the outer cells of the embryo [reviewed by (Lorthongpanich & Issaragrisil 2015)]. It is not known whether a similar signaling system is involved in the bovine. Expression of TEAD4 and YAP1 were not different between cell types of the bovine blastocyst and other components of Hippo signaling were increased in epiblast (SAV1), all four TE subpopulations (SFN), TE1 (AMOT, LATS1, or LATS2), or TE2 (MST1 or TAZ). Bovine blastocysts can form, appear to show no differences of ICM and TE and have normal CDX2, GATA6, IFNT and POU5F1 levels in the presence of reduced amounts of mRNA for TEAD4 (Sakurai et al. 2016b).
Use of single cell analysis of blastocyst gene expression revealed that the cells of the TE are heterogeneous. There are at least four subpopulations of TE cells at day 8.75 of development with TE1 forming one subclade and TE2, TE3 and TE4 forming a second subclade. The existence of so many populations of TE could reflect variation among individual blastocysts or the presence of more than one population of cells in an individual blastocyst. Blastocysts used for the experiment included a mixture of genetic types and a variety of stages (i.e. early, expanding, hatching and hatched blastocysts) and some of the heterogeneity in TE populations could reflect variation among blastocysts in genetic background or stage of development. Moreover, there are differences between male and female blastocysts in gene expression (Bermejo-Alvarez et al. 2010) and such differences could lead to differences in gene expression among TE cells. However, it is also likely that the TE is not functionally uniform within an individual blastocyst. Indeed, there is evidence that gene expression differs between mural and polar TE cells in the bovine (Nagatomo et al. 2013) and immunolocalization of IFNT in the TE is not uniform (Johnson et al. 2006).
Additional studies will be required to identify temporal, spatial and functional differences between the TE subpopulations. However, there are indications that TE1, which forms a separate subclade from TE2, TE3 and TE4, may represent immature TE cells. This population of cells had low expression of the maternal recognition of pregnancy signal IFNT as well as the lowest amounts of the transcription factor EOMES. Secretion of IFNT was higher for expanded blastocysts than for non-expanded blastocysts (Kubisch et al. 2001) and increased as blastocysts were cultured for 24 or 48 h (Kubisch et al. 2004). Eomes is a late TE marker in the mouse blastocyst and its transcription depends on actions of Tead4 and Cdx2 (Probst & Arnold 2017). TE1 cells also exhibited elevated expression of three molecules involved in Hippo signaling – AMOT, LATS1 and LATS2. Moreover, TE1 cells expressed high amounts of TET2 which is involved in inhibition of differentiation of mouse embryonic stem cells to trophoblast (Koh et al. 2011). The fact that TE1 cells highly express CCL26, a marker of polar TE in the bovine (Nagatomo et al. 2013), could mean that these cells are preferentially localized in this region of the TE. While trophoblast stem cells (TSC) are also located in polar TE, at least in the mouse (Probst & Arnold 2017), it is unlikely that TE1 cells represent TSC because of the low expression of EOMES, which is highly expressed in TSC of the mouse (Probst & Arnold 2017).
A better candidate for TSC are cells in the TE4 subpopulation. These cells highly express EOMES and two markers of polar TE (Nagatomo et al. 2013) – ASGR1 and (non-significantly) TMEM232. TE4 cells also express high amounts of SOX2, which has been implicated in maintenance of TSC in mouse (Adachi et al. 2013) as well as the transcription factor SOX30, a member of the SRY family whose function is not well understood.
Perhaps the most differentiated of the TE subpopulations is the TE2 subclade. This subpopulation had the highest number of genes that were upregulated in expression compared to other TE subpopulations. Among the 18 genes upregulated in TE2 are three implicated in placental function in other species: the Hippo signaling molecule MST1, the transcriptional regulator ID1 and the ephrin receptor EPHA4. Mst1 is important for placental differentiation in the mouse (Du et al. 2014), ID2, which is closely related to ID1, participates in regulation of basic helix-loop-helix transcription factors in human placenta (Liu et al. 2004), and interactions between ephrin A and ephrin receptors have been proposed to block premature binding of blastocysts to endometrium (Fujii et al. 2006). Another two genes that were upregulated were STAT3 and TAZ. STAT3 is an intracellular signal transduction molecule for various growth factors affecting embryonic development including, in the mouse, Lif and Egf (Cheng et al. 2016). TAZ is another Hippo signaling molecule and is directly related to YAP1 (Kanai et al. 2000). In mouse embryos, Taz works in conjunction with Yap1 to induce Cdx2 and an increase of Taz is enough to upregulate CDX2 (Nishioka et al. 2009). Another gene upregulated in TE2 was TET1. This gene, which encodes for an enzyme that converts 5-methylcytosine to 5-hydroxymethylcytosine, is not required for successful development in mice but knocking out the gene is associated with increased development of trophectoderm (Dawlaty et al. 2011).
Analysis of single cells indicated expression of some genes previously reported as not being expressed by the bovine blastocyst, most notably the chemokine receptors CCR3 and CCR5 (Negrón-Pérez et al. 2017) and EOMES (Berg et al. 2011). One possibility is that these genes are expressed at low levels by specific cell populations in the TE. Alternatively, it may be that the low levels of these transcripts detected in specific cell populations represent non-specific amplification or low amounts of DNA contamination.
We conclude that the bovine late-blastocyst cell population is heterogeneous and composed of epiblast, hypoblast and four TE cell types. Each cell population may be identified with specific markers previously described and with novel markers identified here. Further studies are needed to confirm whether subpopulations of TE cells represent different maturity stages in development of a committed TE phenotype.
Supplementary Material
SUPPLEMENTAL FILE 1. Information about genes selected for analysis of transcript abundance. Data shown are the gene symbol, forward and reverse primers used for RT-PCR, the reference sequence identification number, the full gene name, reason for selection/previously reported effects and references for previous reports. Not shown are data on KLF2, because primers did not meet validation criteria.
Acknowledgments
Research was supported by Agriculture and Food Research Initiative Competitive Grant No. 2011-67015-30688 from the USDA National Institute of Food and Agriculture and from funds from the L.E. “Red” Larson Endowment. Verónica Negrón-Pérez was supported by a McKnight Doctoral Fellowship from the Florida Education Fund, Inc. The authors thank owners and employees of Central Beef Packing Co. (Center Hill, FL), Adena Meat Products L.P. (Fort McCoy, FL), and Florida Beef Inc. (Zolfo Springs, FL) for providing ovaries; William Rembert and Eddie Cummings for ovary collection; Mei Zhang from the Gene Expression and Genotyping Core, Interdisciplinary Center for Biotechnology Research, University of Florida for assisting in the sample processing development; Maxine McClain and Pedja Sekaric from the Fluidigm Corporation for assisting in the troubleshooting process, and the Miami Center for AIDS Research (CFAR) at the University of Miami Miller School of Medicine, which is funded by a grant (P30AI073961) from the National Institutes of Health, and which performed qPCR.
Footnotes
CONFLICT OF INTEREST
PJH is a co-owner of Cooley Biotech LLC, the manufacturer of BBH7.
References
- Adachi K, Nikaido I, Ohta H, Ohtsuka S, Ura H, Kadota M, Wakayama T, Ueda HR, Niwa H. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Molecular Cell. 2013;52:380–392. doi: 10.1016/j.molcel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nature Reviews Molecular Cell Biology. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Artus J, Panthier J-J, Hadjantonakis A-K. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 2010;137:3361–3372. doi: 10.1242/dev.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Kang M, Cohen-Tannoudji M, Hadjantonakis AK. PDGF signaling is required for primitive endoderm cell survival in the inner cell mass of the mouse blastocyst. Stem Cells. 2013;31:1932–1941. doi: 10.1002/stem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Molecular Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. Trophectoderm lineage determination in cattle. Developmental Cell. 2011;20:244–255. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3394–3399. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley P, Fogarty NME, Del Valle I, Wamaitha SE, Hu TX, Elder K, Snell P, Christie L, Robson P, Niakan KK. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A, Bertone P. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Developmental Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhof B, van Tol HT, Groot Koerkamp MJ, Riemers FM, IJzer SG, Mashayekhi K, Haagsman HP, Roelen BA. A mRNA landscape of bovine embryos after standard and MAPK-inhibited culture conditions: a comparative analysis. BMC Genomics. 2015;16:277. doi: 10.1186/s12864-015-1448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey TS, Cao Z, Choi I, Ganguly A, Wilson Ca, Paul S, Knott JG. BRG1 governs Nanog transcription in early mouse embryos and embryonic stem cells via antagonism of histone H3 lysine 9/14 acetylation. Molecular and Cellular Biology. 2015;35:4158–4169. doi: 10.1128/MCB.00546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Developmental Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Cheng E-H, Liu J-Y, Lee T-H, Huang C-C, Chen C-I, Huang L-S, Lee M-S. Requirement of leukemia inhibitory factor or epidermal growth factor for pre-implantation embryogenesis via JAK/STAT3 signaling pathways. Plos One. 2016;11:e0153086. doi: 10.1371/journal.pone.0153086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–75. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denicol AC, Block J, Kelley DE, Pohler KG, Dobbs KB, Mortensen CJ, Ortega MS, Hansen PJ. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB Journal. 2014;28:3975–3986. doi: 10.1096/fj.14-253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs K, Khan F, Sakatani M. Regulation of pluripotency of inner cell mass and growth and differentiation of trophectoderm of the bovine embryo by colony stimulating factor 2. Biology of Reproduction. 2013;89:141. doi: 10.1095/biolreprod.113.113183. [DOI] [PubMed] [Google Scholar]
- Dominguez MH, Chattopadhyay PK, Ma S, Lamoreaux L, McDavid A, Finak G, Gottardo R, Koup RA, Roederer M. Highly multiplexed quantitation of gene expression on single cells. Journal of Immunological Methods. 2013;391:133–145. doi: 10.1016/j.jim.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Dong Y, Shi H, Li J, Kong S, Shi D, Sun LV, Xu T, Deng K, Tao W. Mst1 and Mst2 are essential regulators of trophoblast differentiation and placenta morphogenesis. PLoS ONE. 2014;9:e90701. doi: 10.1371/journal.pone.0090701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducibella T, Anderson E. Cell shape and membrane changes in the eight-cell mouse embryo: prerequisites for morphogenesis of the blastocyst. Developmental Biology. 1975;47:45–58. doi: 10.1016/0012-1606(75)90262-6. [DOI] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JRE, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Current Biology. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Developmental Cell. 2011;21:1005–1013. doi: 10.1016/j.devcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Fujii H, Tatsumi K, Kosaka K, Yoshioka S, Fujiwara H, Fujii S. Eph-ephrin A system regulates murine blastocyst attachment and spreading. Developmental Dynamics. 2006;235:3250–3258. doi: 10.1002/dvdy.20977. [DOI] [PubMed] [Google Scholar]
- Goissis MD, Cibelli JB. Functional characterization of CDX2 during bovine preimplantation development in vitro. Molecular Reproduction and Development. 2014;81:962–970. doi: 10.1002/mrd.22415. [DOI] [PubMed] [Google Scholar]
- Goossens K, Van Soom A, Van Zeveren A, Favoreel H, Peelman LJ. Quantification of fibronectin 1 (FN1) splice variants, including two novel ones, and analysis of integrins as candidate FN1 receptors in bovine preimplantation embryos. BMC Developmental Biology. 2009;9:1. doi: 10.1186/1471-213X-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Developmental Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hermitte S, Chazaud C. Primitive endoderm differentiation: from specification to epithelium formation. Philosophical Transactions of the Royal Society of London Series B. 2014;369:20130537. doi: 10.1098/rstb.2013.0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue KI, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al. Polarity-dependent distribution of angiomotin localizes hippo signaling in preimplantation embryos. Current Biology. 2013;23:1181–1194. doi: 10.1016/j.cub.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Hosseini SM, Dufort I, Caballero J, Moulavi F, Ghanaei HR, Sirard MA. Transcriptome profiling of bovine inner cell mass and trophectoderm derived from in vivo generated blastocysts. BMC Developmental Biology. 2015;15:49. doi: 10.1186/s12861-015-0096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A, Cox A, Wicher K, Glover DM, Zernicka-Goetz M. Maternal-zygotic knockout reveals a critical role of Cdx2 in the morula to blastocyst transition. Developmental Biology. 2015;398:147–152. doi: 10.1016/j.ydbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Alvarez X, Borkhsenious ON, Kubisch HM. Nuclear and cytoplasmic localization of interferon-τ in in vitro-produced bovine blastocysts. Reproduction, Nutrition, Development. 2006;46:97–104. doi: 10.1051/rnd:2005060. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: A novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO Journal. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis A-K. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2013;140:267–279. doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhof N, Carnwath J, Anastassiadis K, Scholer H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. 2000;63:1698–1705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisch H, Larson M, Kiesling D. Control of interferon-τ secretion by in vitro-derived bovine blastocysts during extended culture and outgrowth formation. Molecular Reproduction and Development. 2001;58:390–397. doi: 10.1002/1098-2795(20010401)58:4<390::AID-MRD6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Kubisch HM, Sirisathien S, Bosch P, Hernandez-Fonseca HJ, Clements G, Liukkonen JR, Brackett BG. Effects of developmental stage, embryonic interferon-τ secretion and recipient synchrony on pregnancy rate after transfer of in vitro produced bovine blastocysts. Reproduction in Domestic Animals. 2004;39:120–124. doi: 10.1111/j.1439-0531.2004.00491.x. [DOI] [PubMed] [Google Scholar]
- Kuijk EW, Du Puy L, Van Tol HTA, Oei CHY, Haagsman HP, Colenbrander B, Roelen BAJ. Differences in early lineage segregation between mammals. Developmental Dynamics. 2008;237:918–927. doi: 10.1002/dvdy.21480. [DOI] [PubMed] [Google Scholar]
- Kuijk EW, van Tol LTA, Van de Velde H, Wubbolts R, Welling M, Geijsen N, Roelen BAJ. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139:871–882. doi: 10.1242/dev.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Kim T-S, Yang T-H, Koo B-K, Oh S-P, Lee K-P, Oh H-J, Lee S-H, Kong Y-Y, Kim J-M, et al. A crucial role of WW45 in developing epithelial tissues in the mouse. The EMBO Journal. 2008;27:1231–1242. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nature Communications. 2013;4:2251. doi: 10.1038/ncomms3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JB, Johnson MH, Goodall H, Maro B. The timing of compaction: control of a major developmental transition in mouse early embryogenesis. Journal of Embryology and Experimental Morphology. 1986;95:213–237. [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes and Development. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Burleigh D, Durning M, Hudson L, Chiu IM, Golos TG. Id2 is a primary partner for the E2-2 basic helix-loop-helix transcription factor in the human placenta. Mol Cell Endocrinol. 2004;222:83–91. doi: 10.1016/j.mce.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Lo CW, Gilula NB. Gap junctional communication in the preimplantation mouse embryo. Cell. 1979;18:399–409. doi: 10.1016/0092-8674(79)90059-x. [DOI] [PubMed] [Google Scholar]
- Lorthongpanich C, Issaragrisil S. Emerging role of the Hippo signaling pathway in positional-sensing and lineage-specification in mammalian preimplantation embryos. Biology of Reproduction. 2015;92:1–10. doi: 10.1095/biolreprod.114.127803. [DOI] [PubMed] [Google Scholar]
- Lorthongpanich C, Doris TPY, Limviphuvadh V, Knowles BB, Solter D. Developmental fate and lineage commitment of singled mouse blastomeres. Development. 2012;139:3722–3731. doi: 10.1242/dev.086454. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes and Development. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Santanach A, Di Croce L. Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Molecular and Cellular Biology. 2015;35:2716–2728. doi: 10.1128/MCB.00266-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Teo RTY, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Graham SJL, Jedrusik A, Zernicka-Goetz M. The differential response to Fgf signalling in cells internalized at different times influences lineage segregation in preimplantation mouse embryos. Open Biology. 2013;3:130104. doi: 10.1098/rsob.130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatomo H, Kagawa S, Kishi Y, Takuma T, Sada A, Yamanaka K-I, Abe Y, Wada Y, Takahashi M, Kono T, et al. Transcriptional wiring for establishing cell lineage specification at the blastocyst stage in cattle. Biology of Reproduction. 2013;88:158. doi: 10.1095/biolreprod.113.108993. [DOI] [PubMed] [Google Scholar]
- Negrón-Pérez V, Vargas-Franco D, Hansen P. Role of CC cytokines in spatial arrangement of the inner cell mass of the bovine embryo. Biology of Reproduction. 2017;96:945–959. doi: 10.1093/biolre/iox037. [DOI] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nature Cell Biology. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Developmental Biology. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nikas G, Ao A, Winston RM, Handyside AH. Compaction and surface polarity in the human embryo in vitro. Biology of Reproduction. 1996;55:32–37. doi: 10.1095/biolreprod55.1.32. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mechanisms of Development. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue KI, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Developmental Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Ortega MS, Wohlgemuth S, Tribulo P, Siqueira LGB, Null DJ, Cole JB, Da Silva MV, Hansen PJ. A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biology of Reproduction. 2017;96:652–663. doi: 10.1093/biolre/iox004. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Sakatani M, Yao J, Shanker S, Yu F, Yamashita R, Wakabayashi S, Nakai K, Dobbs KB, Sudano MJ, et al. Global gene expression of the inner cell mass and trophectoderm of the bovine blastocyst. BMC Developmental Biology. 2012;12:33. doi: 10.1186/1471-213X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam M, Sarkeshik A, Yates JR, Fernandes MJG, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Molecular Biology of the Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearton DJ, Broadhurst R, Donnison M, Pfeffer PL. Elf5 regulation in the trophectoderm. Developmental Biology. 2011;360:343–350. doi: 10.1016/j.ydbio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Probst S, Arnold S. T-Box Genes in Development and Disease. 1st. Ed Z Kruze; London: 2017. Eomesodermin - At dawn of cell fate decisions during early embryogenesis; pp. 93–111. [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Ross PJ, Rodriguez RM, Iager AE, Beyhan Z, Wang K, Ragina NP, Yoon SY, Fissore RA, Cibelli JB. Activation of bovine somatic cell nuclear transfer embryos by PLCZ cRNA injection. Reproduction. 2009;137:427–437. doi: 10.1530/REP-08-0419. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Saha A, Pandian GN, Sato S, Taniguchi J, Hashiya K, Bando T, Sugiyama H. Synthesis and biological evaluation of a targeted DNA-binding transcriptional activator with HDAC8 inhibitory activity. Bioorganic and Medicinal Chemistry. 2013;21:4201–4209. doi: 10.1016/j.bmc.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Current Biology. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Sakurai N, Takahashi K, Fujii T, Hirayama H, Kageyama S, Hashizume T, Sawai K. The necessity of OCT-4 and CDX2 for early development and gene expression involved in differentiation of inner cell mass and trophectoderm lineages in bovine embryos. Cell Reprogram. 2016;18:309–318. doi: 10.1089/cell.2015.0081. [DOI] [PubMed] [Google Scholar]
- Sakurai N, Takahashi K, Emura N, Hashizume T, Sawai K. Effects of downregulating TEAD4 transcripts by RNA interference on early development of bovine embryos. Journal of Reproduction and Development. 2017;18:309–310. doi: 10.1262/jrd.2016-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H. Position- and polarity-dependent Hippo signaling regulates cell fates in preimplantation mouse embryos. Seminars in Cell & Developmental Biology. 2015;48:1–8. doi: 10.1016/j.semcdb.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Schiffmacher AT, Keefer CL. CDX2 regulates multiple trophoblast genes in bovine trophectoderm CT-1 cells. Molecular Reproduction and Development. 2013;80:826–839. doi: 10.1002/mrd.22212. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sugar R, Dimond A, Javierre B-M, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nature Genetics. 2015;47:1179–1186. doi: 10.1038/ng.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T, Miyagi S, Horiuchi D, Okuda-Katayanagi T, Nishimoto M, Muramatsu M, Sakamoto Y, Nagata M, Hagiwara K, Okuda A. Identification of an enhancer that controls up-regulation of fibronectin during differentiation of embryonic stem cells into extraembryonic endoderm. Journal of Biological Chemistry. 2005;280:7244–7252. doi: 10.1074/jbc.M410731200. [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Sindhu C, Meissner A. Molecular features of cellular reprogramming and development. Nature Reviews Molecular Cell Biology. 2016;17:139–154. doi: 10.1038/nrm.2016.6. [DOI] [PubMed] [Google Scholar]
- Van Soom A, Boerjan M, Bols P, Vanroose G, Lein A, Coryn M, de Kruif A. Timing of compaction and inner cell allocation in bovine embryos produced in vivo after superovulation. Biology of Reproduction. 1997;57:1041–1049. doi: 10.1095/biolreprod57.5.1041. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tielens S, Verhasselt B, Liu J, Dhont M, Van Der Elst J, Cornelissen M. Generation of embryonic stem cell lines from mouse blastocysts developed in vivo and in vitro: relation to Oct-4 expression. Reproduction. 2006;132:59–66. doi: 10.1530/rep.1.00887. [DOI] [PubMed] [Google Scholar]
- Togashi K, Kumagai J, Sato E, Shirasawa H, Shimoda Y, Makino K, Sato W, Kumazawa Y, Omori Y, Terada Y. Dysfunction in gap junction intercellular communication induces aberrant behavior of the inner cell mass and frequent collapses of expanded blastocysts in mouse embryos. Journal of Assisted Reproduction and Genetics. 2015;32:969–976. doi: 10.1007/s10815-015-0479-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribulo P, Leão BCS, Lehloenya KC, Mingoti GZ, Hansen PJ. Consequences of endogenous and exogenous WNT signaling for development in the preimplantation bovine embryo. Biology of Reproduction. 2017;96:1129–1141. doi: 10.1093/biolre/iox048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twaroski K, Mallanna SK, Jing R, Difurio F, Urick A, Duncan SA. FGF2 mediates hepatic progenitor cell formation during human pluripotent stem cell differentiation by inducing the WNT antagonist NKD1. Genes and Development. 2015;29:2463–2474. doi: 10.1101/gad.268961.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese S, Waghmare I, Kwon H, Hanes K, Kango-Singh M. Scribble acts in the Drosophila Fat-Hippo pathway to regulate Warts activity. PLoS ONE. 2012;7:1–10. doi: 10.1371/journal.pone.0047173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Duan E, Sung L, Jeong B-S, Yang X, Tian XC. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biology of Reproduction. 2005;73:149–155. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- Xie D, Chen C, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20:804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature Structural & Molecular Biology. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yang QE, Fields SD, Zhang K, Ozawa M, Johnson SE, Ealy AD. Fibroblast growth factor 2 promotes primitive endoderm development in bovine blastocyst outgrowths. Biology of Reproduction. 2011;85:946–953. doi: 10.1095/biolreprod.111.093203. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes and Development. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zhao X-M, Cui L-S, Hao H-S, Wang H-Y, Zhao S-J, Du W-H, Wang D, Liu Y, Zhu H-B. Transcriptome analyses of inner cell mass and trophectoderm cells isolated by magnetic-activated cell sorting from bovine blastocysts using single cell RNA-seq. Reproduction in Domestic Animals. 2016;51:726–735. doi: 10.1111/rda.12737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL FILE 1. Information about genes selected for analysis of transcript abundance. Data shown are the gene symbol, forward and reverse primers used for RT-PCR, the reference sequence identification number, the full gene name, reason for selection/previously reported effects and references for previous reports. Not shown are data on KLF2, because primers did not meet validation criteria.


