Abstract
The root of Polygonum multiflorum Thunb (PM) has been used in China to treat a variety of diseases, such as constipation, early graying of the hair and hyperlipemia. Recent evidence shows that PM causes idiosyncratic drug-induced liver injury (IDILI) in humans. In this study, we investigated the molecular basis of PM-induced liver injury in a rat model of IDILI based on a non-hepatotoxic dose of LPS. SD rats were orally administered 3 potentially hepatotoxic compounds of PM: cis-stilbene glucoside (cis-SG, 50 mg/kg), trans-SG (50 mg/kg) or emodin (5 mg/kg), followed by injection of LPS (2.8 mg/kg, iv). Serum and liver histology were evaluated 7 h after LPS injection. Among the 3 compounds tested, cis-SG, but not emodin or trans-SG, induced severe liver injury in rats when combined with LPS. The levels of AST and ALT in plasma and inflammatory cytokines in both plasma and liver tissues were markedly elevated. The liver tissues showed increased injury, hepatocyte apoptosis, and macrophage infiltration, and decreased cell proliferation. Microarray analysis revealed a negative correlation between peroxisome proliferator-activated receptor-γ (PPAR-γ) and LPS/cis-SG-induced liver injury. Immunohistochemical staining and RT-PCR results further confirmed that cis-SG significantly inhibited activation of the PPAR-γ pathway in the liver tissues of LPS/cis-SG-treated rats. Pre-treatment with a PPAR-γ agonist pioglitazone (500 g/kg, ig) reversed LPS/cis-SG-induced liver injury, which was associated with inhibiting the nuclear factor kappa B (NF-κB) pathway. These data demonstrate that cis-stilbene glucoside induces immunological idiosyncratic hepatotoxicity through suppressing PPAR-γ in a rat model of IDILI.
Keywords: idiosyncratic drug-induced liver injury, Polygonum multiflorum Thunb, cis-stilbene glucoside, LPS, PPAR-γ, NF-κB, pioglitazone
Introduction
PM, the root of Polygonum multiflorum Thunb, has been used for several decades as an herbal treatment in East Asia and some parts of North America. PM extracts have been shown to exert different pharmacological effects and have been used to treat a wide variety of diseases, such as constipation, early graying of the hair and hyperlipemia1,2. Similarly to other traditional Chinese medicines, PM was initially considered to be non-toxic, but reports of hepatotoxicity have recently increased3.
Several cases of clinically apparent hepatic injuries caused by the administration of PM have been reported, particularly in Hong Kong, mainland China, Korea, Japan, Canada, Britain and Australia. Supervised usage of PM is recommended by drug regulatory agencies in Canada, Britain and Australia4.
High-performance liquid chromatography has been used to demonstrate that the concentrations of certain compounds differ among chemically processed PM extracts and raw unprocessed extract. In particular, the content of stilbene glycoside is decreased by 55.8% and that of emodin is increased by 34.0% in processed PM extracts5. The processing of Chinese herbal medicines is important because it has been shown to decrease toxicity and alter the therapeutic efficacy of the extracts5. PM-induced liver injury may therefore be related to the chemical components of the herbal plant, stilbene glycoside and emodin5. Structurally, stilbene glycoside consists of two benzene rings bonded with ethylene, whereas cis-SG is obtained by exposing a trans-SG aqueous solution to ultraviolet light or sunlight6. Although the structures are similar, there may be differences between their bioavailability and bio-toxicity, as has been observed for resveratrol, perfluoroalkyl substances, and bromopropane7,8,9,10. Therefore, it is important to study the bio-toxicity of cis-SG and trans-SG separately to determine their roles in inducing hepatotoxicity.
It has previously been demonstrated that high doses of emodin markedly inhibit the proliferation of L02 and HepG2 cells in vitro11, and high doses of emodin induce liver injury in animal models12. Although the exact mechanism of PM-induced liver injury is unknown, several studies have reported the occurrence of idiosyncratic reactions that are either related to genetic polymorphisms or immune-mediated hepatic injuries13. We have previously found that PM extracts combined with non-hepatotoxic doses of LPS, but not PM extracts alone, cause severe liver injury in an IDILI model. These findings suggest that the PM-induced liver injury in rats is indeed idiosyncratic14, but the molecular basis of LPS/PM-induced hepatotoxicity remains to be elucidated. Previous studies have successfully reproduced human idiosyncratic drug-induced liver injury (IDILI) in rats by co-exposure to ranitidine (RAN) and LPS15,16,17; therefore, we used RAN as a positive control in this study. The purpose of this study was to assess whether cis-SG, trans-SG and emodin induce liver injury in an idiosyncratic in vivo model.
Materials and methods
Preparation of methanol solutions of trans-SG and cis-SG
Cis-SG is not commercially available, owing to its instability in solid form and was therefore obtained from a 5 mg/mL methanol solution of trans-SG by exposure to 365-nm ultraviolet light at room temperature for 40 min. Subsequently, volatilized methanol was added for preservation, and the product was further purified by preparative liquid chromatography, in which the mobile phase consisted of a mixture of methanol, water and formic acid (36:64:0.1; v/v/v) with a flow rate of 60 mL/min. The sample volume was consistent (150 mg in 10 mL of 50% methanol/water), and the detector wavelengths were set at 330 nm and 280 nm. The fractions were collected and freeze-dried. Preparative liquid chromatography was conducted using a Hanbon preparative liquid chromatography system (Hanbon Science & Technology, Jiangsu, China) equipped with an NU3000 series UV detector (dual wavelength channel, 200–400 nm), an NP7000 series pump (0–100 mL/min, maximum pressure 10 mpa), and a Megres C18 column (50 mm×650 mm, 10 μm).
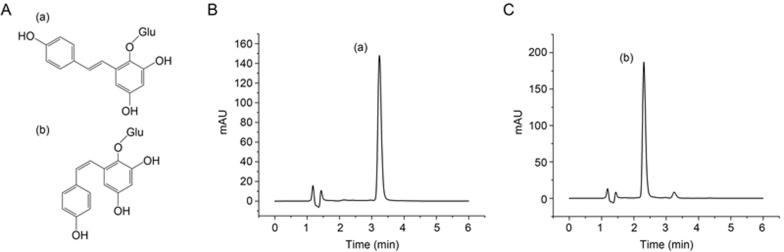
Pure cis-SG was detected with an Agilent 1290 series UHPLC system, and the separation was carried out on a ZORBAX RRHD 300 SB-C18 column (2.1 mm×100 mm, 1.8 μm) (Agilent, USA). The mobile phases used were solvent A (acetonitrile) and solvent B (water spiked with 0.1% formic acid) with isocratic elution: 20% A and 80% B. The total run time was 6 min, and the mobile phase was delivered at a flow rate of 0.2 mL/min. A trans-SG solution (1 μL) was injected into the column, which was preserved at 25 °C. The spectrophotometer was operated at two wavelengths—280 nm, at which the maximum UV absorbance of cis-SG occurs, and 320 nm, at which the maximum UV absorbance of trans-SG occurs. The chemical structures of cis-SG and trans-SG are shown in Figure 1A.
Figure 1.
UV-induced isomerization of stilbene glycosides (SG). (A) Chemical structures of (a) trans-SG and (b) cis-SG. HPLC chromatograms of SG exposed to ultraviolet radiation for 40 min (+ control) with UV detection at (B) 320 nm and (C) 280 nm.
Animal experiments
Male Sprague-Dawley (SD) rats, weighing 160–190 g, were purchased from the Laboratory Animal Center of the Academy of Military Medical Sciences (certification number SCXK-JUN 2007-004). The animals were given a standard chow diet and water. They were housed in a temperature- and humidity-controlled environment under a 12-h light/dark cycle. All experimental rats were maintained according to the institutional guidelines, and the in vivo procedures were approved by the Committee on the Ethics of Animal Experiments of the 302 Military Hospital (Approval ID: IACUC-2015-035).
The rats were divided into 10 groups on the basis of the treatment: negative control, LPS, cis-SG, trans-SG, emodin, ranitidine, LPS/cis-SG, LPS/trans-SG, LPS/emodin, and LPS/ranitidine. We selected the doses of cis-SG, trans-SG and LPS on the basis of previous studies, and the dose of emodin was calculated according to the ratio of emodin and stilbene glycoside content in Polygonum multiflorum18. The rats in all the groups except the negative control and LPS-treated group were administered the following doses by gavage or injections: the cis-SG group and LPS/cis-SG group were gavaged with 50 mg/kg cis-SG dissolved in 20% sodium carboxymethyl cellulose (CMC-Na), the trans-SG group and LPS/trans-SG group were gavaged with 50 mg/kg trans-SG dissolved in 20% CMC-Na, the emodin group and LPS/emodin group were gavaged with 5 mg/kg emodin dissolved in CMC-Na, and the ranitidine group and LPS/ranitidine group were injected through the caudal vein with 30 mg/kg19 ranitidine dissolved in sterile PBS. After the respective treatments were administered, all the groups except the negative control and drug alone groups were injected through the caudal vein with 2.8 mg/kg LPS (E. coli055:B5, Sigma) dissolved in a sterile saline vehicle solution. Serum and liver histology were evaluated 7 h after LPS stimulation14.
To study the role of PPAR-γ in inducing liver injury, the rats were also treated with pioglitazone, a selective PPAR-γ agonist. The rats were divided into 6 groups: negative control, LPS, pioglitazone, LPS/cis-SG, LPS/pioglitazone, and LPS/cis-SG/pioglitazone. With the exception of the negative control, LPS and LPS/cis-SG groups, the rats were pretreated with pioglitazone (500 μg/kg, Hainan Honz Pharmaceutical, Haikou, China) by gavage for 2 days. On day 3, the pioglitazone, LPS/pioglitazone and LPS/cis-SG/pioglitazone groups were gavaged with 500 μg/kg pioglitazone dissolved in CMC-Na, whereas, the LPS/cis-SG and LPS/cis-SG/pioglitazone groups were gavaged with 50 mg/kg cis-SG dissolved in 20% CMC-Na. After oral administration, all the groups except the negative control and pioglitazone groups were injected through the caudal vein with 2.8 mg/kg LPS dissolved in sterile saline. Blood samples and liver tissues were collected for examination 7 h after LPS stimulation14.
Ten hours later, blood samples were collected into tubes with anticoagulant to harvest plasma and then centrifuged, and the plasma was stored at −20 °C for further analysis. ALT and aspartate transaminase (AST) were measured in the plasma after one freeze-thaw cycle by using the Reitman-Frankel assay (Nanjing Jianchneg Bioengineering Institute, China). The left lateral lobe of the liver was collected and fixed in 10% formalin for subsequent liver biochemical tests and histopathology. Liver tissues were collected in RNase-free tubes in liquid nitrogen. Frozen tissue samples were stored at −80 °C until processing for protein extraction.
Histopathology and immunohistochemistry
The rat liver tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned (thickness of 4 μm), and stained with H&E. The following antibodies were used to specifically study hepatocellular proliferation, macrophage infiltration, nuclear translocation of the p65 subunit of NF-κB, and PPAR-γ gene expression: anti-Ki67 rabbit polyclonal antibody (1:500) (Santa Cruz, USA); anti-CD68 rabbit polyclonal antibody (1:500) (Santa Cruz, USA); anti-p65 rabbit polyclonal antibody (1:200) (Cell Signaling Technology, USA); and anti-PPAR-gamma rabbit polyclonal antibody (1:50) (Abcam, UK), respectively. Slides were incubated with a biotin-conjugated anti-rabbit secondary antibody (DAKO, Denmark) followed by peroxidase-conjugated streptavidin (DAKO, Denmark). To visualize the immune complexes, liquid 3,3′-diaminobenzidine (DAB) was used. The sections were counterstained with hematoxylin. TUNEL staining was performed by using an in situ cell death detection kit POD (Roche, Switzerland) according to the manufacturer's instructions. The samples were visualized under a fluorescence microscope (Nikon, Japan) and photographed.
Immunoexpression analysis and quantitative real-time polymerase chain reaction (RT-qPCR) analysis
The serum levels of interferon (IFN)-γ, tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-1β, and induced nitric oxide synthase (iNOS) were determined by enzyme-linked immunosorbent assay (ELISA; IFN-γ SEA049Ra, IL-6 SEA079Ra, IL-1β SEA563Ra, TNF-α SEA133Ra, iNOS SEA837Ra; Cloud-Clone Corp, Houston, TX, USA) according to the manufacturer's protocol. The intensity of the final colorimetric reaction in proportion to the amount of bound protein was measured with a plate reader (ELx800, BioTek) at 450 nm. The results were calculated using a calibration curve constructed from standard solutions of known concentrations.
IL-6, TNF-α, IL-1β, IFN-γ, PPAR-γ and iNOS in the liver were analyzed using RT-qPCR. Relative quantification was performed with Power SYBR® Green PCR Master Mix (Applied Biosystems, USA) and a 7500 Real-Time PCR System (Applied Biosystems, USA) according to the manufacturer's instructions. PCR was performed using 100 ng of cDNA. The PCR conditions consisted of AmpliTaq Gold® Enzyme activation for 10 min at 95 °C, followed by 40 cycles of heating to 95 °C for 15 s and cooling to 60 °C for 1 min. The mRNA levels were normalized to those of GAPDH. Sequence-specific PCR primers were purchased from Invitrogen Corp (Shanghai, China). The following primers were used for PCR: GAPDH: sense, 5′-GGCAAGTTCAATGGCACAGT-3′ antisense, 5′-TGGTGAAGACGCCAGTAGA CTC-3′. IFN-γ: sense, 5′-AAAGACAACCAGGCCATCAG-3′ antisense, 5′-CTGGATCTGTGGGTTGTTCA-3′. IL-6: sense, 5′-ACCACCCACAACAGACC AGT-3′ antisense, 5′-ACAGTGCATCATTCGCTGTTC-3′. IL-1β: sense, 5′-TGACTCGTGGGATGATGACG-3′ antisense, 5′-CTGGAGACTGCCCATTCTC G-3′. TNF-α: sense, 5′-GCTCCCTCTCATCAGTTCC-3′ antisense, 5′-CTCCTCT GCTTGGTGGTTTG-3′. PPAR-γ: sense, 5′-TGTGGACCTCTCTGTGATGG-3′ antisense, 5′-CATTGGGTCAGCTCTTGTGA-3′ and iNOS: sense, 5′-AGCGGCTCCA TGACTCTCA-3′ antisense, 5′-TGCACCCAAACACCAAGGT-3′.
Western blot analysis
Lysis buffer (RIPA, Beyotime, China) was used to isolate total proteins from the liver samples. Aliquots from each sample containing equal amounts of protein (100 mg) were boiled for 10 min, separated by 12% SDS-polyacrylamide gel electrophoresis and electro-transferred onto a nitrocellulose membrane. The membranes were then incubated in blocking buffer containing 5% nonfat milk in 0.1% Tris-buffered saline containing Tween 20 (TBST) for 2 h at room temperature. The following primary antibodies diluted in TBST were added, and the membranes were incubated at 4 °C overnight: β-actin (1:1000), PPAR-γ (1:400), and IκB-α (1:1000). The membranes were then washed three times (5 min each) in TBST and incubated with goat anti-rabbit HRP-conjugated secondary antibody (1:3000, Santa Cruz Biotechnology, USA) for 2 h at room temperature. The protein bands were visualized by the chemiluminescence detection method, and digital images were taken by an automatic digital gel image analysis system (Tanon-4200, Tanon, Shanghai, China). The protein bands were quantified by densitometry scanning using the AlphaEaseFC 4.0 software (Alpha Innotech Corp, San Leandro, CA, USA). The quantitative densitometry values of PPAR-γ and IκB-α were normalized to those of β-actin.
Microarray experiments
Gene expression profiling was conducted with samples from rats treated with LPS/cis-SG and LPS/trans-SG by using a Rat Gene Expression Microarray (Agilent Technologies). The core set of probe clusters was used in a Crystal Core® cRNA amplification tag kit (CapitalBio Corporation, Beijing, China). The dataset was imported into GeneSpring GX (Agilent Technologies), and one-way analysis of variance (ANOVA) and principal component analysis were then used to evaluate differences in gene expression between the LPS/cis-SG and LPS/trans-SG groups. Genes with P ≤ 0.05 and an absolute fold change of at least 2 were clustered and visualized using a cluster gram heat map. The list of differentially expressed genes was loaded into KEGG (http://www.genome.jp/kegg/) to conduct biological network and functional analyses.
Cells and cell culture
Human L02 hepatocytes were purchased from the China Center for Type Culture Collection (Wuhan, China) and were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) supplemented with penicillin (100 U/mL) and streptomycin (100 μg/mL). The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2. All cell culture reagents were obtained from GIBCO (MD, USA).
Cell proliferation assays
Cell proliferation assays were performed using a Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer's protocol. Briefly, cells were cultured in a 96-well plate at a concentration of 8×104 cells/mL for 24 h. Next, cis-SG (7.6875, 15.375, 30.75, 61.5, 123, and 246 μmol/L) or trans-SG (7.6875, 15.375, 30.75, 61.5, 123, and 246 μmol/L) diluted in cell culture medium was added to each well, and the plates were incubated at 37 °C for 24 h. After incubation, the wells were washed with PBS, CCK-8 (10%) diluted in cell culture medium was added to each well, and the plates were incubated for 40 min at 37 °C. Cell proliferation was determined by measuring the absorbance at 450 nm by using a Synergy H2 microplate reader (Biotek Instruments, USA).
Statistical analysis
All data are expressed as the mean±SEM. A paired Student's t-test and ANOVA were performed to evaluate differences between groups. The statistical analysis was performed using SPSS software (SPSS version 22.0, Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
UV-induced isomerization of stilbene
As described previously, we used ultraviolet radiation to convert trans-SG to cis-SG6. Additionally, cis-SG was purified by preparative liquid chromatography, and UHPLC was used to determine the relative proportions of trans-SG and cis-SG. UHPLC allowed for quick and efficient separation of trans-SG and cis-SG. The trans-SG eluted first, with a retention time of 3.3 min, followed by cis-SG at 2.3 min, thus indicating a noticeable difference in the hydrophobicity of these two compounds (Figure 1B, 1C).
Development of liver injury in rats co-treated with LPS and cis-SG
To directly evaluate the effects of cis-SG and trans-SG on hepatocytes, we measured their cytotoxicity by using CCK8 assays. The results showed that up to 24 h of exposure to cis-SG and trans-SG did not alter the viability of L02 cells, thus suggesting that these compounds do not induce hepatic parenchymal cell injury (Supplementary Table S1). We have previously reported that PM administered alone does not induce liver injury in vivo; however, PM extracts combined with non-hepatotoxic doses of LPS cause liver injury14. Therefore, in this study, we evaluated the effects of cis-SG and trans-SG on hepatoxicity in an in vivo LPS model.
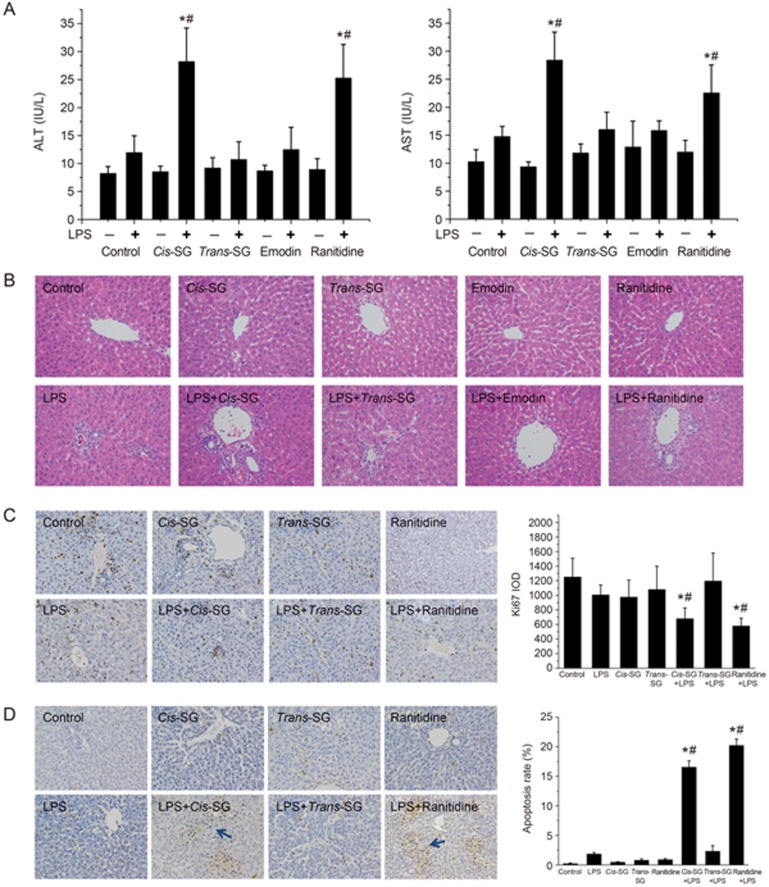
As shown in Figure 2, no significant differences were observed in the levels of plasma ALT and AST in rats administered a non-hepatotoxic dose of LPS compared with control rats. Co-treatment with LPS and cis-SG or ranitidine (RAN), but not trans-SG and emodin, resulted in significantly increased plasma ALT and AST levels. Moreover, there were no significant differences in the levels of AST and ALT in rats treated with cis-SG, trans-SG, emodin or ranitidine alone (Figure 2A and B).
Figure 2.
Effect of cis-SG co-treatment on LPS-induced hepatic injury. (A) Blood samples were collected from the post cava, and the levels of ALT and AST in the plasma were quantified by Reitman-Frankel assays. (B) Photomicrographs of H&E-stained sections of livers from rats treated with different conditions (200×). (C) Representative micrographs of positive Ki67 staining (indicative of cell proliferation) in liver tissues obtained from rats treated with different conditions (200×). The integral optical density (IOD) of Ki67 was calculated by Image-Pro Plus 6.0. (D) Representative micrographs of positive TUNEL staining (indicative of cell apoptosis) in liver tissues obtained from rats subjected to different treatment conditions. The cellular apoptosis rate was calculated by using Image-Pro Plus 6.0 and is represented as the apoptosis rate%. The results are represented as mean±SEM (n=8). *P<0.05 vs control. #P<0.05 vs LPS as determined by ANOVA.
It has been previously reported that co-treatment of the positive control drug ranitidine with LPS causes inflammation and necrosis in the liver20. Histopathological analysis of the liver tissues from rats treated with LPS/cis-SG and LPS/RAN revealed similar results (increased inflammatory lesions). In contrast, rats treated with LPS/trans-SG, LPS/emodin, LPS, cis-SG, trans-SG, emodin, or ranitidine alone exhibited liver histology similar to that of the control group (Figure 2B).
LPS is known to stimulate the immune system, thus eventually leading to increased secretion of inflammatory mediators involved in cell proliferation and apoptosis. Here, we evaluated both apoptosis and cell proliferation by immunocytochemistry. Hepatocellular proliferation was quantitatively estimated after Ki67 staining, and we observed significantly decreased cell proliferation in the livers of rats treated with LPS/cis-SG or LPS/RAN compared with control rats. In addition, no obvious changes in hepatocellular proliferation were observed in the control, LPS, LPS/trans-SG, cis-SG, or trans-SG groups (Figure 2C). Liver cell apoptosis was determined by TUNEL staining. Apoptotic cells were occasionally found in the livers of rats from most of the treatment groups, whereas the apoptosis rate was significantly higher in the livers of rats treated with LPS/cis-SG and LPS/RAN (Figure 2D).
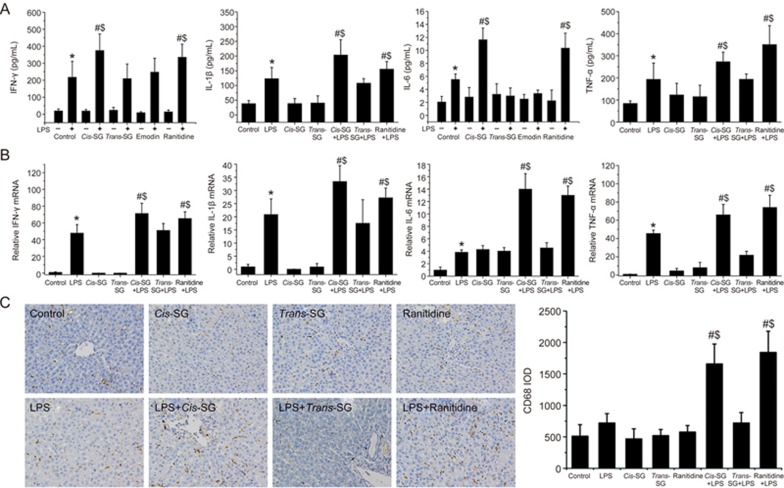
LPS/cis-SG modulates the expression of inflammatory cytokines in an LPS/cis-SG-induced liver injury model
LPS binds to Toll-like receptor (TLR) 4 on the surface of macrophages and promotes the activation of the NF-κB signaling pathway. Sustained activation of the NF-κB signaling pathway induces overexpression of various inflammatory cytokines and chemokines, thus ultimately leading to inflammation and liver parenchymal cell injury21,22. To elucidate the inflammatory responses induced by the PM extracts, we quantified the levels of IL-1β, IL-6, TNF-α and IFN-γ in plasma and liver tissues by both ELISA and RT-qPCR. As shown in Figure 3, LPS stimulation significantly increased the levels of IL-1β, IL-6, TNF-α and IFN-γ mRNA and protein compared with those in the control group. Interestingly, the expression of these cytokines was increased significantly at both the mRNA and protein levels in the LPS/RAN or LPS/cis-SG groups compared with the LPS group, whereas the expression levels in all other groups were comparable (Figure 3A, 3B).
Figure 3.
Effect of LPS/cis-SG-induced liver injury on inflammatory cytokines. (A) IFN-γ, IL-6, IL-1β and TNF-α secretion in rats treated with different conditions was measured by ELISA. (B) The mRNA levels of IFN-γ, IL-6, IL-1β and TNF-α in rats subjected to different treatment conditions were determined by qRT-PCR, and the expression levels were calculated relative to the levels of GAPDH. (C) Representative micrographs of CD68 staining in the liver tissues obtained from rats t subjected to different treatment conditions (200×). The results are represented as mean±SEM (n=8). *P<0.05 vs control. #P<0.05 vs LPS. $P<0.05 vs LPS/trans-SG as determined by ANOVA.
Macrophages are the main effector cells involved in hepatic injury after LPS stimulation and are well characterized as a target of LPS in the liver23. Therefore, we investigated the recruitment of macrophages to the site of liver injury by staining the tissue with the macrophage-specific marker CD68. The liver tissues from rats in the control, LPS/trans-SG, cis-SG and trans-SG groups exhibited mild staining for CD68 (+) macrophages. However, the number of CD68 (+) macrophages increased dramatically in the livers of rats co-treated with LPS and cis-SG or RAN, compared with rats in the LPS group, thus indicating significantly up-regulated macrophage infiltration (Figure 3C).
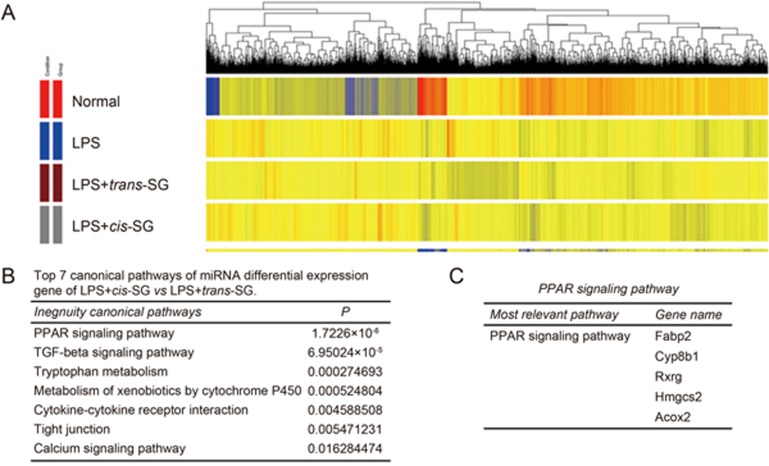
The PPAR-γ signaling pathway is down-regulated in LPS/cis-SG-induced liver injury
To understand the possible signaling pathway involved in LPS/cis-SG-induced liver injury, we analyzed liver gene expression patterns by using a 13 K cDNA microarray and cluster analysis of the significantly regulated genes (P<0.05) (Figure 4A). A total of 5342 genes had significant differences that were over two-fold. One hundred twenty-six genes were down-regulated, and 57 genes were up-regulated in the LPS/cis-SG-treated group compared with the LPS/trans-SG-treated group (Supplementary Table S2, S3). All the differentially expressed genes were entered into the KEGG database for pathway analysis, and 138 pathways were matched (Supplementary Table S4). Of these pathways, 30 were differentially regulated in the LPS/cis-SG-treated group compared with the LPS/trans-SG-treated group (Supplementary Table S5), and those that were significantly down-regulated are shown in Figure 4B. Notably, the Fabp2, Cyp8b1, Rxrg, Hmgcs2 and Acox2 genes, which are involved in PPAR-γ signaling, showed significantly altered expression (Figure 4C).
Figure 4.
Hierarchical clustering of LPS+cis-SG-induced gene expression. (A) Gene cluster analysis of microarray results (P<0.05; fold change>2.0) for the control, LPS, LPS+cis-SG and LPS+trans-SG groups. (B) The top 7 canonical pathways with differential gene expression in the LPS+cis-SG group vs the LPS+trans-SG group. (C) Expression of genes in the PPAR-γ signaling pathway.
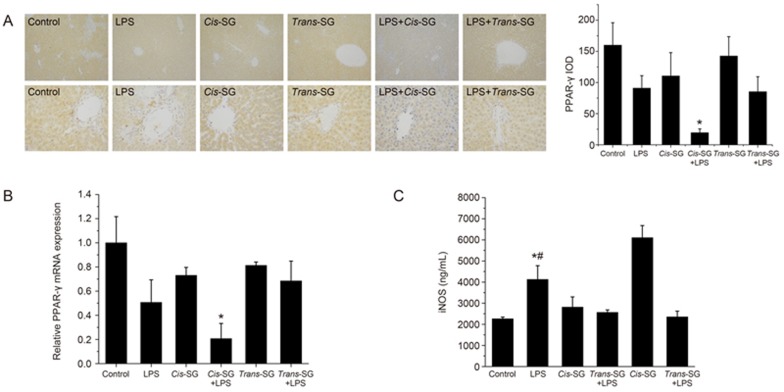
To validate the microarray data, we analyzed the expression of PPAR-γ in LPS/cis-SG-induced liver injury by immunohistochemical staining and RT-qPCR. The expression of PPAR-γ was significantly decreased in rats treated with LPS/cis-SG compared with that rats treated with LPS or cis-SG alone (Figure 5A, 5B). LPS induces the expression of iNOS in macrophages, and PPAR-γ agonists have been shown to negatively regulate iNOS expression24. In line with the results from these previous reports, we detected significantly up-regulated levels of iNOS protein in LPS/cis-SG-treated rats compared with cis-SG-treated rats (Figure 5C).
Figure 5.
Effect of LPS/cis-SG-induced liver injury on PPAR-γ expression. (A) Representative micrographs of PPAR-γ staining in the liver tissues obtained from rats subjected to different treatment conditions (100× and 400×). The integral optical density (IOD) of PPAR-γ (400×) was calculated by using Image-Pro Plus 6.0. (B) PPAR-γ mRNA expression in rats subjected to different treatment conditions was determined by qRT-PCR, and the expression levels were calculated relative to the levels of GAPDH. (C) iNOS secretion in rats subjected to different treatment conditions was measured by ELISA. The results are represented as mean±SEM (n=8). *P<0.05 vs control. #P<0.05 vs LPS as determined by ANOVA.
Effect of pioglitazone on LPS/cis-SG-induced hepatic injury
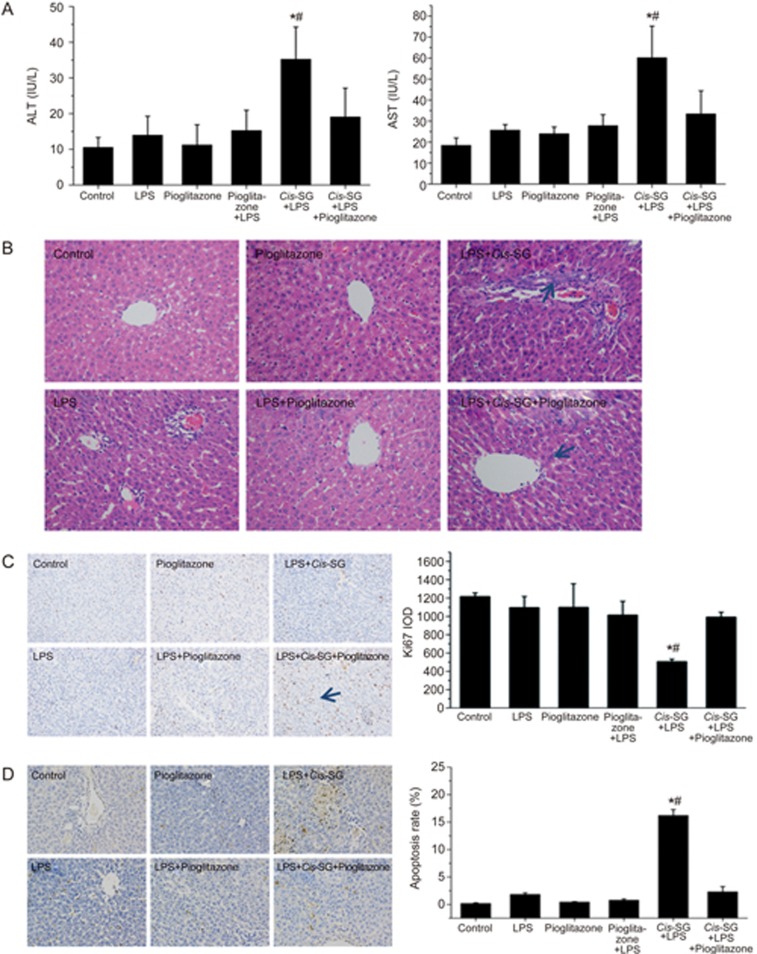
It has been previously reported that pioglitazone, a PPAR-γ agonist, antagonizes inflammatory responses through the transrepression of NF-κB target genes and prevents alcoholic liver disease through the abrogation of Kupffer cell sensitization to LPS25. To further validate the role of PPAR-γ in the LPS/cis-SG model, we pretreated rats with pioglitazone for 3 days and then established an LPS/cis-SG-induced liver injury model. Plasma analysis revealed no differences in ALT and AST activities among the control, LPS, pioglitazone and LPS/pioglitazone groups. Significantly increased ALT and AST activities were detected in the LPS/cis-SG group, but this increase was almost completely blocked by the administration of pioglitazone (Figure 6A). Animals given pioglitazone and LPS/pioglitazone showed normal liver histology. However, the livers of rats in the LPS/cis-SG group showed pronounced focal necrosis and neutrophil infiltration. In contrast, overt necrosis was absent in the LPS/cis-SG/pioglitazone group, which displayed only minor infiltration of inflammatory cells (Figure 6B).
Figure 6.
Effect of pioglitazone co-treatment on LPS/cis-SG-induced liver injury. (A) Blood samples were collected from the post cava, and the levels of ALT and AST in the plasma were quantified with Reitman-Frankel assays. (B) Photomicrographs of H&E-stained sections of livers from rats subjected to different treatment conditions (200×). (C) Representative micrographs of positive Ki67 staining (indicative of cell proliferation) in liver tissues obtained from rats subjected to different treatment conditions (200×). The integral optical density (IOD) of Ki67 was calculated by using Image-Pro Plus 6.0. (D) Representative micrographs of positive TUNEL staining (indicative of cell apoptosis) in liver tissues obtained from rats subjected to different treatment conditions. The cellular apoptosis rate was calculated by Image-Pro Plus 6.0 and represented as apoptosis rate. The results are represented as mean±SEM. n=8. *P<0.05 vs LPS. #P<0.05 vs LPS+cis-SG+pioglitazone as determined by ANOVA.
We further evaluated the effects of pioglitazone on the proliferation and apoptosis of hepatic parenchymal cells in injured liver tissues. We observed significant inhibitory effects of LPS/cis-SG on the expression of Ki67 in hepatic parenchymal cells, and this inhibitory effect on cell proliferation was markedly reversed when pioglitazone was co-administered with LPS/cis-SG (Figure 6C). Furthermore, using TUNEL staining, we observed significantly increased cell apoptosis in liver sections from rats treated with LPS/cis-SG. However, co-administration of pioglitazone with LPS/cis-SG reversed this effect on cell apoptosis. Apoptotic cells were occasionally found in the livers of rats in the control, LPS, pioglitazone and LPS/pioglitazone groups (Figure 6D).
Pioglitazone decreases the production of inflammatory cytokines in the LPS/cis-SG-induced hepatic injury model
It has been reported that the activation of PPAR-γ decreases liver injury by enhancing PPAR-γ DNA-binding activity and decreasing NF-κB p65 transcriptional activity26. Therefore, we evaluated the expression of PPAR-γ, p65 and IκB-α through immunohistochemical staining and Western blotting and quantified the inflammatory cytokines IL-6, IFN-γ, IL-1β and TNF-α by both ELISA and RT-qPCR.
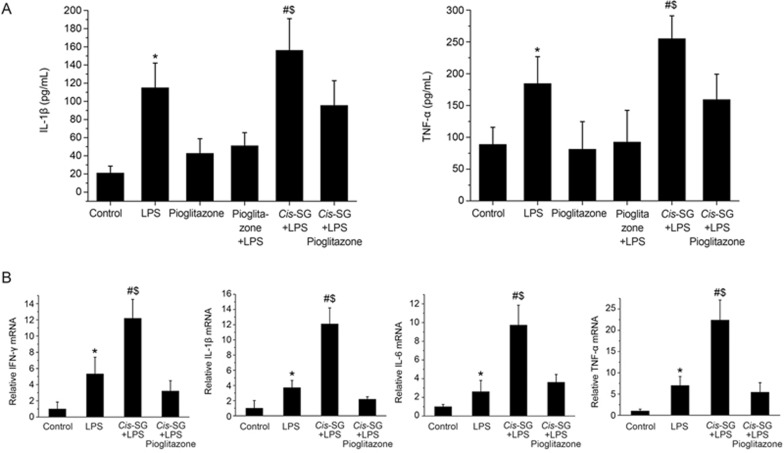
We found greatly increased levels of TNF-α and IL-1β mRNA and protein in the livers and plasma of rats treated with LPS and LPS/cis-SG; however, the levels were significantly lower in the LPS/cis-SG/pioglitazone group than in the LPS/cis-SG group. The levels of TNF-α and IL-1β protein in the livers of control, LPS, and LPS/pioglitazone rats were not significantly different (Figure 7A). Additionally, increased levels of IFN-γ and IL-6 mRNA were detected in the livers of rats treated with LPS compared with control rats, whereas the significantly increased levels in the LPS/cis-SG group were inhibited by co-administration of pioglitazone (Figure 7B).
Figure 7A, 7B.
Effect of LPS/cis-SG and pioglitazone co-treatment on inflammatory cytokines and PPAR-γ expression in the liver. (A) IL-1β and TNF-α secretion in rats subjected to different treatment conditions was measured by ELISA. (B) The mRNA levels of IFN-γ, IL-6, IL-1β and TNF-α in rats subjected to different treatment conditions were determined by qRT-PCR, and the expression levels were calculated relative to those of GAPDH. The results are represented as mean±SEM. n=8. *P<0.05 vs control. #P<0.05 vs LPS. $P<0.05 vs LPS/cis-SG/pioglitazone; as determined by ANOVA.
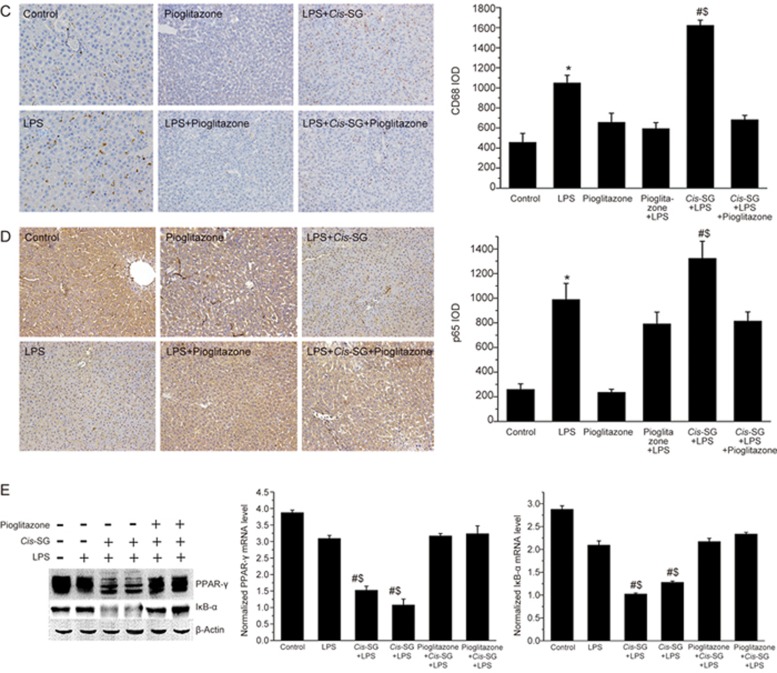
CD68 staining of the liver tissue showed increased macrophage infiltration in rats treated with LPS and LPS/cis-SG, and the co-administration of pioglitazone with LPS/cis-SG attenuated this effect on macrophage infiltration (Figure 7C). To study NF-κB activation in LPS/cis-SG-induced hepatic injury, we analyzed the levels of p65 in the nuclei of liver cells by immunohistochemistry. We found significantly increased p65 levels in the livers of rats treated with LPS, LPS/pioglitazone, LPS/cis-SG and LPS/cis-SG/pioglitazone compared with control rats. The levels of p65 were dramatically increased compared with those in the LPS group; however, the LPS/cis-SG/pioglitazone group appeared to have lower levels (Figure 7D).
Figure 7C–E.
Representative micrographs of CD68 (C) and p65 (D) staining in the liver tissues obtained from rats subjected to different treatment conditions (×200). The CD68 integral optical density (IOD) and p65 IOD were calculated by using Image-Pro Plus 6.0. (E) The protein levels of PPAR-γ and IκB-α in the liver tissues of rats subjected to different treatment conditions were detected by Western blotting. Densitometry analysis was used to calculate the protein levels of PPAR-γ and IκB-α, which were normalized to those of β-actin. The results are represented as mean±SEM. n=8. *P<0.05 vs control. #P<0.05 vs LPS. $P<0.05 vs LPS/cis-SG/pioglitazone; as determined by ANOVA.
Western blot analysis revealed constitutive expression of both PPAR-γ and IκB-α in the liver tissues of control and LPS-treated rats. The protein levels of IκB-α and PPAR-γ were dramatically lower in the LPS/cis-SG-treated group compared with the LPS-treated group. Pioglitazone co-administration increased the protein levels of PPAR-γ and IκB-α, which were inhibited by LPS/cis-SG (Figure 7E). Together, the data indicated that pioglitazone not only inhibits the nuclear translocation of NF-κB p65 but also inhibits the degradation of its upstream inhibitory protein IκB-α. Pioglitazone also increased the activation of PPAR-γ and the nuclear translocation of NF-κB p65 in the LPS/cis-SG-treated group.
Discussion
IDILI is a type of idiosyncratic drug reaction that occurs in a small fraction of people taking a drug and has an unclear association with dose or duration of therapy15. IDILI is mainly thought to be correlated with polymorphisms affecting drug metabolism genes, inflammatory stress or episodes of modest inflammation occurring during drug therapy27. A low dose of LPS co-administered with certain drugs has been shown to evoke mild concurrent inflammation in animals that mimic human IDILI28. Several drugs, including trovafloxacin, chlorpromazine, diclofenac, and RAN, that cause human IDILI, have been shown to induce hepatotoxicity in an idiosyncratic LPS-induced in vivo model29,30.
In the present study, we found that cis-SG combined with LPS not only induced the levels of plasma ALT and AST but also caused liver injury similar to that observed with LPS/RAN, whereas trans-SG and emodin did not induce liver injury. LPS activates macrophages, causing them to produce various inflammatory mediators such as IL-6 and TNF-α, which in turn directly or indirectly damage surrounding cells and tissues31. Our results showed decreased Ki67 staining (marker for cell proliferation) and increased TUNEL staining (marker for apoptosis) in rats treated with LPS/cis-SG or LPS/RAN, thus indicating increased cell death caused by LPS/cis-SG. Similar results were obtained when we evaluated inflammatory mediators (IFN-γ, IL-1β, IL-6 and TNF-α) and CD68+ macrophage expression. Our data suggested that a modest inflammatory response triggered by a non-hepatotoxic dose of LPS may decrease the hepatotoxicity threshold for cis-SG, thus inducing idiosyncratic liver injury and resulting in increased inflammatory cell infiltration, liver cell apoptosis, and inflammation.
Our microarray data revealed that the PPAR-γ pathway was negatively correlated with LPS/cis-SG-induced liver injury. PPAR-γ is an isotype of the peroxisome proliferator-activated receptors (PPARs), members of the type II nuclear receptor superfamily of transcription factors22. PPAR-γ is mainly involved in lipid and glucose metabolism, cell proliferation and apoptosis, regulation of food intake and body weight10. In addition to these roles, PPAR-γ has also been shown to regulate inflammation via diverse mechanisms28. Non-steroidal anti-inflammatory drugs (NSAIDs), 15-deoxy-Δ12,14-prostaglandin J232, and thiazolidinediones (e.g., rosiglitazone, pioglitazone, troglitazone), members of a new class of oral antidiabetic agents, are known PPAR-γ agonists. It has previously been reported that activation of PPAR-γ by its agonists results in an anti-inflammatory response in animal models of endotoxemia and sepsis33. In addition, pioglitazone, through PPAR-γ activation, can block TNF-α and abrogate Kupffer cell sensitization to LPS, thereby inhibiting inflammation in alcoholic liver disease25. Accordingly, we investigated whether pioglitazone could inhibit inflammation in LPS/cis-SG-induced liver injury by evaluating the liver function and inflammatory cytokines. The results revealed that co-treatment with pioglitazone and LPS/cis-SG attenuated cell apoptosis and hepatic injury and decreased the levels of plasma AST/ALT and inflammatory mediators, which were induced by LPS/cis-SG.
PPAR-γ has been shown to exert anti-inflammatory effects by decreasing the DNA-binding activity of NF-κB and directly targeting IκB-α and inhibiting NF-κB nuclear translocation, thereby decreasing cytokine production and tissue injury34,35,36. Moreover, the expression of PPAR-γ is down-regulated in macrophages after LPS stimulation37. Analyses of PPAR-γ, IκB-α and p65 expression by Western blotting and immunocytochemistry demonstrated increased levels of p65 and decreased IκB-α and PPAR-γ expression in LPS/cis-SG-treated rats; however, pioglitazone pretreatment reversed the effects of LPS/cis-SG. The data suggested that LPS/cis-SG induced liver injury via PPAR-γ down-regulation, thus further leading to activation of the NF-κB signaling pathway. These events together aggravate the inflammatory response and damage hepatocytes and eventually cause liver injury.
On the basis of previous studies and the data presented here, we suggest that cis-SG is the major factor responsible for PM-induced liver injury, but it is not the only component. We hypothesize that when the body is in an inflammatory state, the compounds in PM may further promote the immune response and cell apoptosis in the liver. Although the underlying mechanisms are not fully validated, our results reveal the immune mechanism of PM-induced hepatic injury and provide a foundation for future research on similar traditional Chinese medicines.
In summary, our results demonstrate that modest inflammatory responses triggered by a non-hepatotoxic dose of LPS potentiate cis-SG-induced liver injury in rats by down-regulating PPAR-γ. Furthermore, pioglitazone inhibits the occurrence of LPS/cis-SG-induced liver injury, thus suggesting that PPAR-γ agonists may be used as preventive drugs to decrease the incidence of PM-induced hepatotoxicity in the clinic. These results may also promote the development of integrated Chinese-Western therapy to prevent or limit the risks associated with cis-SG-containing herbs.
Author contribution
Ya-kun MENG, Zhao-fang BAI, Rong SUN, He-rong CUI, Ming NIU, Jia-bo WANG, and Xiao-he XIAO participated in research design; Ya-kun MENG, Lan-zhi HE, Ping YIN, Xiu-xiu SANG, and Ya WANG conducted the experiments; Ya-kun MENG, Cong-en ZHANG, Ming NIU, Ya-ming ZHANG, and Yu-ming GUO contributed new reagents or analytic tools; Ya-kun MENG, Chun-yu LI, and Rui-yu LI analyzed the data; and Ya-kun MENG, He-rong CUI, Cong-en ZHANG, Peng-yan LI, and Zhao-fang BAI wrote or contributed to the writing of the manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No 81503350 and 81630100), the "Major Drug Discovery" Science and Technology major projects (No 2015ZX09501-004-001-008), the National Public Welfare Industry Subject (No 201507004-04), the Beijing Natural Science Foundation (No 7152142), and the Beijing Nova Program (No xx2016098).
Footnotes
Supplementary information are available on the website of Acta Pharmacologica Sinica.
Supplementary Information
Supplementary data
the differential toxicity of Cis-SG, Trans-SG on the parenchymal hepatic cells
Down-expressed genes of Cis-SG vs Trans-SG
up- expressed genes of Cis-SG vs Trans-SG
The pathway of mRNA differential expression gene
The pathway of mRNA differential expression gene of Cis-SG vs Trans-SG
References
- Dong Q, Li N, Li Q, Zhang CE, Feng WW, Li GQ, et al. Screening for biomarkers of liver injury induced by Polygonum multiflorum: a targeted metabolomic study. Front Pharmacol 2015; 6: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Zhang N, Park DK. Topical application of Polygonum multiflorum extract induces hair growth of resting hair follicles through upregulating Shh and beta-catenin expression in C57BL/6 mice. J Ethnopharmacol 2011; 135: 369–75. [DOI] [PubMed] [Google Scholar]
- Yu J, Xie J, Mao XJ, Wang MJ, Li N, Wang J, et al. Hepatoxicity of major constituents and extractions of Radix Polygoni Multiflori and Radix Polygoni Multiflori Praeparata. J Ethnopharmacol 2011; 137: 1291–9. [DOI] [PubMed] [Google Scholar]
- Dong H, Slain D, Cheng J, Ma W, Liang W. Eighteen cases of liver injury following ingestion of Polygonum multiflorum. Complement Ther Med 2014; 22: 70–4. [DOI] [PubMed] [Google Scholar]
- Wu X, Chen X, Huang Q, Fang D, Li G, Zhang G. Toxicity of raw and processed roots of Polygonum multiflorum. Fitoterapia 2012; 83: 469–75. [DOI] [PubMed] [Google Scholar]
- Sun J, Huang X, Wu H, Huang F. Determination of content and light stability of cis-and trans-2, 3, 5, 4′-tetrahydroxystilbene-2-O-β-D-glucoside in Radix Polygoni multiflori by RRLC/DAD/MS. Chin Pharm J 2009; 44: 541–4. [Google Scholar]
- Camont L, Cottart CH, Rhayem Y, Nivet-Antoine V, Djelidi R, Collin F, et al. Simple spectrophotometric assessment of the trans-/cis-resveratrol ratio in aqueous solutions. Anal Chim Acta 2009; 634: 121–8. [DOI] [PubMed] [Google Scholar]
- Unsalan O, Kus N, Jarmelo S, Fausto R. Trans- and cis-stilbene isolated in cryogenic argon and xenon matrices. Spectrochim Acta A Mol Biomol Spectrosc 2015; 136: 81–94. [DOI] [PubMed] [Google Scholar]
- O'Brien JM, Austin AJ, Williams A, Yauk CL, Crump D, Kennedy SW. Technical-grade perfluorooctane sulfonate alters the expression of more transcripts in cultured chicken embryonic hepatocytes than linear perfluorooctane sulfonate. Environ Toxicol Chem 2011; 30: 2846–59. [DOI] [PubMed] [Google Scholar]
- Ichihara G, Li W, Shibata E, Ding X, Wang H, Liang Y, et al. Neurologic abnormalities in workers of a 1-bromopropane factory. Environ Health Perspect 2004; 112: 1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Sun Y, Li H, Sun W. Influence of main component of Heshouwu such as emodin, rhein and toluylene glycoside on hepatic cells and hepatoma carcinoma cells. Modern J Integr Tradit Chin Western Med 2010; 19: 1315–17. [Google Scholar]
- Lei X, Chen G, Chen K, Li J. Acute toxicity studies of emodin in mice. Pharmacol Clin Chin Mat Med 2008; 24: 29. [Google Scholar]
- Empey PE. Genetic predisposition to adverse drug reactions in the intensive care unit. Crit Care Med 2010; 38: S106–16. [DOI] [PubMed] [Google Scholar]
- Li C, Li X, Tu C, Li N, Ma Z, Pang J, et al. The idiosyncratic hepatotoxicity of Polygonum multiflorum based on endotoxin model. Yao Xue Xue Bao 2015; 50: 28–33. [PubMed] [Google Scholar]
- Luyendyk JP, Maddox JF, Cosma GN, Ganey PE, Cockerell GL, Roth RA. Ranitidine treatment during a modest inflammatory response precipitates idiosyncrasy-like liver injury in rats. J Pharmacol Exp Ther 2003; 307: 9–16. [DOI] [PubMed] [Google Scholar]
- Roth RA, Luyendyk JP, Maddox JF, Ganey PE. Inflammation and drug idiosyncrasy—is there a connection? J Pharmacol Exp Ther 2003; 307: 1–8. [DOI] [PubMed] [Google Scholar]
- Roth RA, Harkema JR, Pestka JP, Ganey PE. Is exposure to bacterial endotoxin a determinant of susceptibilityto intoxication from xenobiotic agents? Toxicol Appl Pharmacol 1997; 147: 300–11. [DOI] [PubMed] [Google Scholar]
- Li CY, Niu M, Bai ZF, Zhang CN, Zhao YL, Li RY, et al. Screening for main components associated with the idiosyncratic hepatotoxicity of a tonic herb, Polygonum multiflorum. Front Med 2017; 11: 253–65. doi: 10.1007/s11684-017-0508-9. [DOI] [PubMed] [Google Scholar]
- Li SF, Zheng CZ, Zhang L, Yao WF, Lan CW, He DX, et al. Effect of harvest seasons on main bioactive constituents in Polygoni Multiflori Radix. Modern Chin Med 2015; 17: 1177–83. [Google Scholar]
- Luyendyk JP, Lehman-McKeeman LD, Nelson DM, Bhaskaran VM, Reilly TP, Car BD, et al. Coagulation-dependent gene expression and liver injury in rats given lipopolysaccharide with ranitidine but not with famotidine. J Pharmacol Exp Ther 2006; 317: 635–43. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Yoneda M, Tomeno W, Imajo K, Shinohara Y, Fujita K, et al. Peroxisome proliferator-activated receptor gamma exacerbates concanavalin a-induced liver injury via suppressing the translocation of NF-kappaB into the nucleus. PPAR Res 2012; 2012: 940384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RB. The role of PPARs in inflammation and immunity. J Leukoc Biol 2002; 71: 388–400. [PubMed] [Google Scholar]
- Faure E, Equils O, Sieling PA, Thomas L, Zhang FX, Kirschning CJ, et al. Bacterial lipopolysaccharide activates NF-kappaB through toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J Biol Chem 2000; 275: 11058–63. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 2005; 437: 759–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N, Takei Y, Hirose M, Konno A, Shibuya T, Matsuyama S, et al. Prevention of ethanol-induced liver injury in rats by an agonist of peroxisome proliferator-activated receptor-gamma, pioglitazone. J Pharmacol Exp Ther 2003; 306: 846–54. [DOI] [PubMed] [Google Scholar]
- Han Z, Zhu T, Liu X, Li C, Yue S, Liu X, et al. 15-Deoxy-Delta12,14 -prostaglandin J2 reduces recruitment of bone marrow-derived monocyte/macrophages in chronic liver injury in mice. Hepatology 2012; 56: 350–60. [DOI] [PubMed] [Google Scholar]
- Ganey PE, Roth RA. Concurrent inflammation as a determinant of susceptibility to toxicity from xenobiotic agents. Toxicology 2001; 169: 195–208. [DOI] [PubMed] [Google Scholar]
- Roth RA, Luyendyk JP, Maddox JF, Ganey PE. Inflammation and drug idiosyncrasy–is there a connection? J Pharmacol Exp Ther 2003; 307: 1–8. [DOI] [PubMed] [Google Scholar]
- Waring JF, Liguori MJ, Luyendyk JP, Maddox JF, Ganey PE, Stachlewitz RF, et al. Microarray analysis of lipopolysaccharide potentiation of trovafloxacin-induced liver injury in rats suggests a role for proinflammatory chemokines and neutrophils. J Pharmacol Exp Ther 2006; 316: 1080–7. [DOI] [PubMed] [Google Scholar]
- Buchweitz JP, Ganey PE, Bursian SJ, Roth RA. Underlying endotoxemia augments toxic responses to chlorpromazine: is there a relationship to drug idiosyncrasy? J Pharmacol Exp Ther 2002; 300: 460–7. [DOI] [PubMed] [Google Scholar]
- Xaus J, Comalada M, Valledor AF, Lloberas J, Lopez-Soriano F, Argiles JM, et al. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-alpha. Blood 2000; 95: 3823–31. [PubMed] [Google Scholar]
- Scher JU, Pillinger MH. 15d-PGJ2: the anti-inflammatory prostaglandin? Clin Immunol 2005; 114: 100–9. [DOI] [PubMed] [Google Scholar]
- Zingarelli B, Sheehan M, Hake PW, O'Connor M, Denenberg A, Cook JA. Peroxisome proliferator activator receptor-gamma ligands, 15-deoxy-Delta(12,14)-prostaglandin J2 and ciglitazone, reduce systemic inflammation in polymicrobial sepsis by modulation of signal transduction pathways. J Immunol 2003; 171: 6827–37. [DOI] [PubMed] [Google Scholar]
- Standiford TJ, Keshamouni VG, Reddy RC. Peroxisome proliferator-activated receptor-{gamma} as a regulator of lung inflammation and repair. Proc Am Thorac Soc 2005; 2: 226–31. [DOI] [PubMed] [Google Scholar]
- Liu D, Zeng BX, Shang Y. Decreased expression of peroxisome proliferator-activated receptor gamma in endotoxin-induced acute lung injury. Physiol Res 2006; 55: 291–9. [DOI] [PubMed] [Google Scholar]
- Li CC, Yang HT, Hou YC, Chiu YS, Chiu WC. Dietary fish oil reduces systemic inflammation and ameliorates sepsis-induced liver injury by up-regulating the peroxisome proliferator-activated receptor gamma-mediated pathway in septic mice. J Nutr Biochem 2014; 25: 19–25. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wu R, Dong W, Jacob A, Wang P. Endotoxin downregulates peroxisome proliferator-activated receptor-gamma via the increase in TNF-alpha release. Am J Physiol Regul Integr Comp Physiol 2008; 294: R84–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
the differential toxicity of Cis-SG, Trans-SG on the parenchymal hepatic cells
Down-expressed genes of Cis-SG vs Trans-SG
up- expressed genes of Cis-SG vs Trans-SG
The pathway of mRNA differential expression gene
The pathway of mRNA differential expression gene of Cis-SG vs Trans-SG