Abstract
γ-Secretase is an intramembrane aspartyl protease that cleaves the C99 fragment of amyloid precursor protein to generate extracellular Aβ peptides. These peptides can oligomerize and aggregate to form amyloid plaques, processes that are widely believed to be causal for Alzheimer's disease. In spite of this critical function, it remains unknown how γ-secretase recognizes C99 and its other substrates, including Notch. In this study we determined E22-K55 as the minimal C99 fragment that was sufficient and required for initial cleavage. Within this fragment, we identified four determinants: (i) a transferable extracellular determinant that differed between C99 and Notch, and which included negative charge in the case of C99, (ii) the amino acid sequence of the C-terminal half of the transmembrane helix, (iii) an invariant lysine or arginine at the intracellular membrane border, and (iv) a positive charge cluster that included the invariant lysine/arginine. We demonstrated that the charge clusters of C99 and Notch receptors could directly bind phosphatidylinositol 4,5-bisphosphate (PIP2). The PIP2-binding cluster was required for γ-secretase cleavage, and modulation of membrane PIP2 levels strongly affected γ-secretase cleavage levels and the Aβ40/Aβ42 ratio, providing support for the importance of the PIP2 interaction in cells. Together, these studies provide critically needed insight into substrate recognition by γ-secretase.
Keywords: Alzheimer's disease, amyloid beta (Aβ), amyloid precursor protein (APP), C99, γ-secretase, minimum substrate, Notch protein, recognition model
Introduction
Alzheimer's disease (AD) is a devastating neurodegenerative disease that is characterized by the progressive formation of proteinaceous deposits in the brain, including extracellular amyloid plaques and intracellular neurofibrillary tangles1,2. Extracellular amyloid plaques are mainly composed of amyloid peptides (Aβ), which are the product of the proteolytic processing of the amyloid precursor protein (APP)1,2,3. APP is a type I integral transmembrane protein that is known to be processed at three sites via non-amyloidogenic and amyloidogenic pathways in vivo3,4,5. Non-amyloidogenic processing involves initial APP cleavage by α-secretase between K687 and L688 in the middle of the Aβ luminal region, releasing a soluble extracellular fragment of APP (sAPPα) and a remnant membrane-bound 83-residue C-terminal fragment (C83), which can be further cleaved by γ-secretase, thereby precluding Aβ generation. Alternatively, in the amyloidogenic pathway, APP is first cleaved by β-secretase within the extracellular domain, producing a soluble extracellular fragment of APP (sAPPβ) and a membrane-bound 99-residue C-terminal fragment (C99) that undergoes subsequent cleavage by γ-secretase within the transmembrane region, resulting in the production of the 37–42 amino-acid Aβ peptides4,5,6,7,8. The proteolytic processing of APP is comparable to Notch signaling activation, which undergoes similar sequential proteolytic cleavages by α-secretase and γ-secretase9,10,11. Given the central role of the amyloidogenic pathway in AD, the substrate recognition of γ-secretase has been under intense investigation. A better understanding of structure, substrate recognition and potential substrate interaction mechanisms will likely provide new insights into AD pathogenesis and possible therapies.
γ-Secretase is an aspartyl protease that consists of four components: presenilin (PS), anterior pharynx-defective 1 (APH-1), nicastrin (NCT), and presenilin enhancer protein 2 (PEN2)12,13,14. PS forms the catalytic core15,16,17, while the other three accessory proteins are involved in the assembly of this protease18,19,20. Although the atomic structure of human γ-secretase has recently been reported21, the precise roles of these three cofactors are still ambiguous. While NCT has been proposed to be involved in substrate recognition22, γ-secretase lacking NCT remains catalytically active23. Recent molecular dynamic simulation suggest that the APP TMD can directly bind the catalytic PS1 subunit of γ-secretase24, and mutagenesis suggests that the interaction involves three distinct amino acid-binding pockets in the active site25. Use of genetically inserted photocrosslinkable amino acids confirmed the catalytic subunit as principal substrate binding site and identified its direct interaction with C99 TMD residues26.
To elucidate substrate recognition determinants in living cells, we utilized a γ-secretase Epsilon-Cleavage assay to comprehensively investigate the cleavage of C99 by γ-secretase in cells27,28. From a series of substrate fragments derived from C99, we identified E22-K55 as the shortest region of C99 that can still be efficiently cut by γ-secretase in the membrane of live cells. We then determined the contribution of each individual C99 residue by alanine scanning mutagenesis and of secondary structure elements by double proline mutagenesis. In combination with an analysis of charge requirements at the extra- and intracellular side of C99, we identified key substrate determinants, including an unanticipated interaction between a conserved cluster of positively charged residues at the intracellular membrane junction and PIP2 in the membrane.
Materials and methods
Cell culture
Adherent HTL wildtype (WT) or PS deletion cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen™ Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS) (Invitrogen™ Life Technologies) at 37 °C under humidified 5% CO2 atmosphere. HTL cells were split at 5000 cells per well in a 24-well plate one day before transfection.
Cloning
All mutants were generated by site direced mutagenesis and confirmed using DNA sequencing.
γ-Secretase Epsilon-Cleavage assay
We used the same cell-based γ-secretase Epsilon-Cleavage assay as before28. Briefly, we generated fusion constructs consisting of C99, T4 lysozyme (T4L), and the transcriptional activator rTA (C99-T4L-rTA). Endogenous γ-secretase was used in this assay, and 20 ng C99-T4L-rTA, 5 ng phRG-tk Renilla, 40 ng pBSK mock plasmid were co-transfected into HTL cells using X-tremeGENE 9 Reagent (1 μg DNA:3 μL reagent)27. After one day of growth, cells were harvested and lysed for luciferase detection. Relative activity was presented in arbitrary units using WT as 100. The red dashed line indicated the level of WT activity. Some activities were too low to show and marked as less than 5.
AlphaLISA assay
AlphaLISA assay was conducted as described previously28. HTL cells were transfected with 320 ng DNA using X-tremeGENE 9 Reagent (1 μg DNA:3 μL reagent). Media supernatants were collected the following day for Aβ42/Aβ40 detection. Briefly, 5 μL reaction samples were incubated at 23 °C with 5 μL AlphaLISA Aβ1–40/42 Acceptor beads and biotinylated anti-Aβ antibody for 1 h. After another 30-min incubation with 10 μL AlphaLISA Aβ1–40/42 donor beads in the dark at 23 °C, the samples were read in 384-well plates using an Envision-Alpha Reader (PerkinElmer).
Liposome preparation and AlphaScreen assay
Lipids were purchased from Avanti Polar Lipids, Inc. In preparation of liposome, lipids dissolved in chloroform were mixed at desired composition and ratio, and the mixture was dried under nitrogen to a thin film. For AlphaScreen use, the dried lipid film was resuspended in 10 mmol/L Tris pH 7.4 to give a final concentration of 20 mmol/L. After gently shaking for 1 h, the lipid suspension was sonicated in a bath sonicator for 5 min to form liposomes.
Luminescence proximity AlphaScreen (PerkinElmer) assays were performed for determining the polybasic region-PIP2 interaction with a hexahistidine detection kit. 100 nmol/L of the polybasic region-containing peptides and 15 μmol/L biotinylated liposome were incubated with 5 μg/mL Streptavidin-coated Donor beads and 5 μg/mL Nickel-chelate Acceptor beads in a buffer of 10 mmol/L Tris, pH 7.4, and 0.1 mg/mL bovine serum albumin for 1.5 h in the dark at room temperature. Thus, the donor and acceptor beads were brought into proximity by the interaction between PIP2 and polybasic peptide. When excited by a laser beam of 680 nm, the photosensitizer in the donor beads converts ambient oxygen into short-lived singlet oxygen that can transfer energy to the thioxene derivatives in the acceptor beads, which then release photons of 520–620 nm as the binding signal. For the competition assays, untagged polybasic peptides were added at increasing concentrations while His6-tagged Ni-Acceptor bead-bound peptides were kept at a constant concentration (100 nmol/L). The IC50 values were obtained from curve fitting to the competitive inhibitor model using GraphPad Prism.
PIP2 modulations and Aβ analyses
To modulate cellular PIP2 levels, DMSO (control), 10 μmol/L edelfosine (the PLC inhibitor) and 15 μmol/L m-3M3FBS (the PLC activator) were added to the culture medium during DNA transfection, respectively. After one day treatment, HTL cells or supernatant were harvested for γ-secretase Epsilon-Cleavage assay or AlphaLISA assay.
Protein isolation and Western blot analysis
HTL PS deleted cells were transfected with the same amount of DNA as for γ-secretase Epsilon-Cleavage assays, by X-tremeGENE 9 Reagent for substrate expression quantification. Cells were harvested and lysed in the following day. Western blot analysis was carried out using primary antibodies against FLAG tag (Sigma-Aldrich A8592), or β-actin (Abcam Ab6276). The β-actin level was used as the internal control.
Results
E22-K55 is the minimum region of C99 that is required for γ-secretase recognition and cleavage in living cells
C99 is the transmembrane C-terminal β-secretase cleavage product of APP, which undergoes subsequent cleavage by γ-secretase to release amyloid-forming Aβ peptides29,30. The latter cleavage event occurs within the transmembrane region and proceeds by successive removal of tripeptide units8. In spite of the importance of this cleavage event, it remains unclear how C99 is recognized by γ-secretase and which region of the C99 substrate is necessary and sufficient for γ-secretase recognition and initial cleavage in living cells. Since C83, the product of α-cleavage in the non-amyloidogenic pathway, is also recognized and cleaved by γ-secretase30, the N-terminal 16 residues of C99 do not seem to be required for substrate recognition.
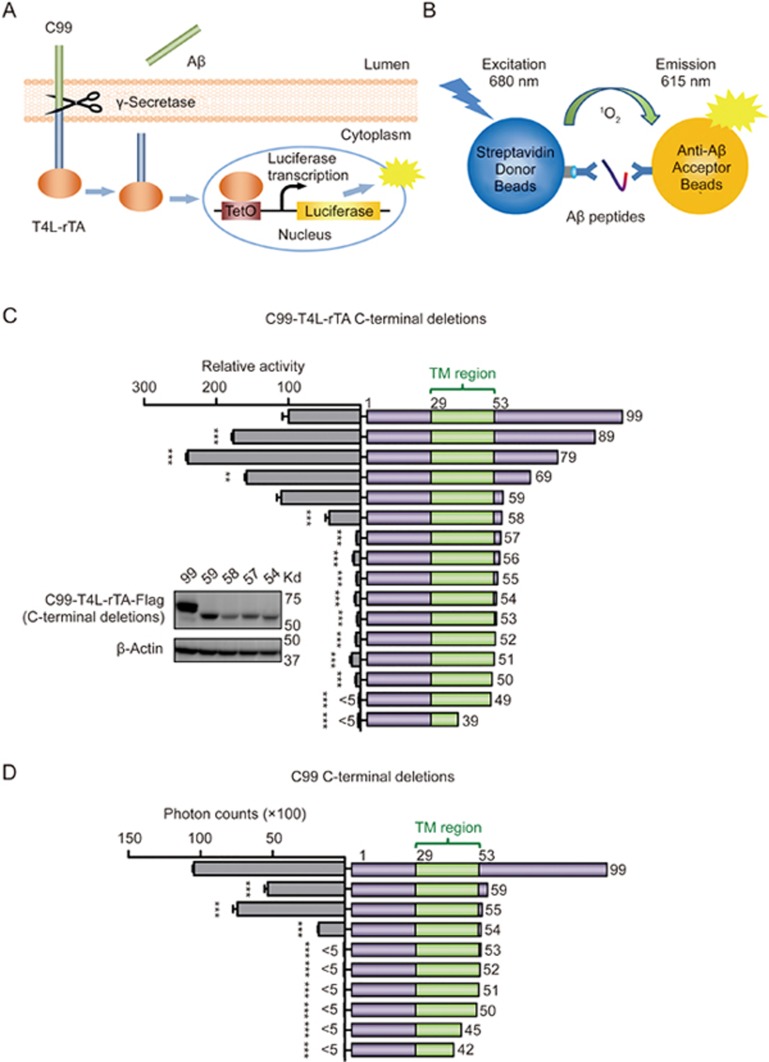
To define the minimum substrate of γ-secretase, we first utilized the highly selective γ-secretase Epsilon-Cleavage assay to investigate the in-cell cleavage of C99 by γ-secretase28. In this assay, the C-terminus of C99 was fused to the transcriptional transactivator rTA as well as T4 lysozyme (T4L), whose rigid structure increases the stability of the fusion protein. Cleavage by γ-secretase releases T4L-rTA from the cell membrane to allow its transfer into the nucleus where it activates a luciferase reporter gene (Figure 1A).
Figure 1.
Defining the C99 minimum boundary for γ-secretase cleavage. (A) An overall view of the γ-secretase Epsilon-Cleavage assay. Once C99-T4L-rTA hybrid protein is cleaved by endogenous γ-secretase, Aβ peptides are released into the culture medium, and the AICD-T4L-rTA hybrid protein is translocated into the nucleus, where it binds tetO and activates luc transcription to generate a luciferase signal as measurement of total C99 cleavage. (B) An overall view of the AlphaLISA assay. The biotinylated anti-Aβ antibody binds to Streptavidin-coated donor beads, and AlphaLISA acceptor beads are directly conjugated to the anti-Aβ antibody. The presence of Aβ peptides brings the two beads into close proximity to generate a light emission signal as measurement of secreted Aβ levels. (C and D) Defining the C-termini of the minimum substrate by γ-secretase Epsilon-Cleavage assay (C) and AlphaLISA assay (D). Cartoon illustration of the C-terminally truncated C99 fragments (right side) and their cleavage efficiencies (left side). The numbers in the cartoon illustration indicate the last residue in each construct. The inlet panel of C shows relative protein expression of some critical boundary constructs, with corresponding construct numbers marked on top of each lane. Aβ55, which contains first 55 amino acids of C99, is the shortest substrate among these C99 C-terminal truncations that retained similar activity as wild-type. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
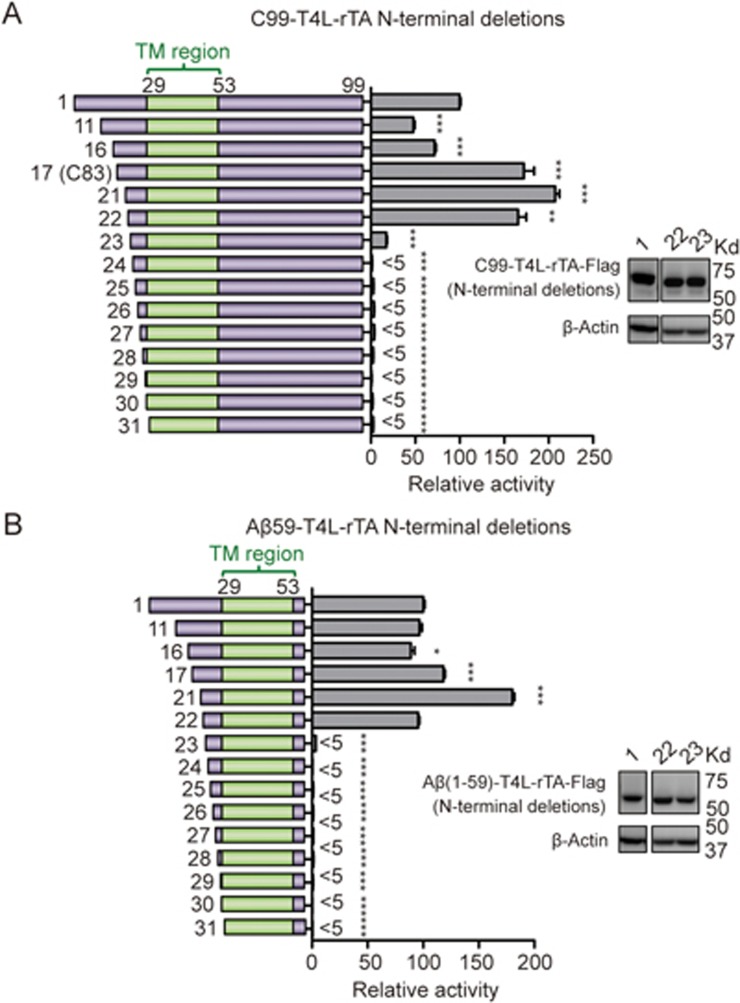
We generated a series of truncated C99 fusion constructs by sequentially removing residues from the N- and C-terminal flanking regions of the C99 transmembrane helix. Deletions from the C-terminal cytoplasmic region identified Aβ(1-59) as shortest N-terminal fragment that maintained a similar cleavage level as WT C99 (Figure 1C). Since the C-terminally fused T4L-rTA may affect γ-secretase's accessibility to C-terminally truncated C99 fragments, this fragment is sufficient for γ-secretase cleavage, but not all C-terminal residues of the 59 amino acid fragment may necessarily be required for cleavage by γ-secretase. To clarify the exact C-terminal boundary, we, therefore, determined the amount of the major extracellular C99 cleavage product, Aβ40, for each of the non-fused truncated C99 proteins. Aβ40 levels were quantified by a commercial AlphaLISA assay, in which simultaneous interaction of Aβ40 with two different antibodies generates a luminescence proximity-based binding signal (Figure 1B). This assay identified K55 as the C-terminal border of the minimum region required for efficient cleavage by γ-secretase (Figure 1D). We also validated the expression levels of some key truncations (Figure 1C, insert). In spite of some variation in expression, the cleavage efficiency did not correlate with protein expression, supporting that differences in cleavage activity primarily reflect differences in substrate recognition and preference.
Sequential deletions of the N-terminal extracellular region of either full-length C99 or the Aβ59 fragment identified in Figure 1C demonstrated that the first 21 amino acids are not required for maintaining a similar cleavage level as full-length C99 in the Epsilon-Cleavage assay (Figure 2). Deletion of certain parts of the N-terminus increased the cleavage efficiency, which is consistent with γ-secretase's preference for substrates with a short extracellular domain31 and a previous report that identified the L17-A21 region containing inhibiting activity of γ-secretase32. Since all truncated N-termini are freely accessible in this assay (only the C-terminus of C99 is fused to T4L-rTA), E22 defines the minimal N-terminal amino acid region required for efficient cleavage by γ-secretase (Figure 2). Note that the AlphaLISA assay cannot be used to define the minimal N-terminal region as this assay depends on antibody recognition of the V18-E22 epitope of Aβs. The dramatically decreased cleavage efficiency of fragments lacking the N-terminal 22 residues of C99 was not caused by changes in protein expression as seen in immunoblot of the D1, E22 and D23 constructs (Figure 2, insert). Together, these results identify E22-K55 as the shortest region of C99 that is necessary and sufficient for substrate recognition and γ-secretase cleavage in living cells.
Figure 2.
Defining the N-terminal minimum boundary for γ-secretase cleavage. Defining the N-termini of the minimum substrate by the γ-secretase Epsilon-Cleavage assay based on C99 (A) and Aβ59 (B) hybrid proteins. Cartoon illustration of N-terminal C99 truncations (left side) and their cleavage efficiencies (right side). The numbers in the cartoon illustration indicate the first residue in each construct. The inlet panels show relative protein expression of some critical boundary constructs, with corresponding construct numbers marked on top of each lane. Aβ peptide (E22-K55) is the minimum substrate of γ-secretase that retains a close to normal cleavage level. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
Identification of individual amino acids and secondary structure elements for γ-secretase cleavage
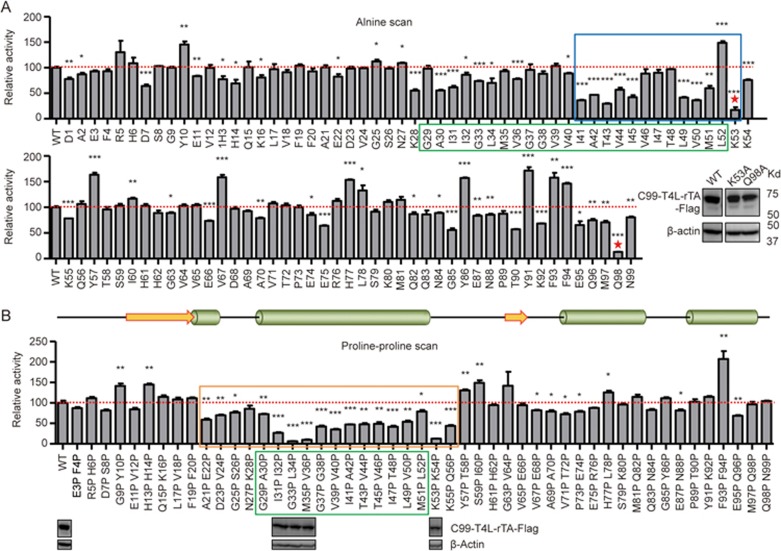
After determining the minimal C99 region that still functions as efficient γ-secretase substrate, we next performed a complete C99 alanine scan to identify individual residues that are important for cleavage. Consistent with our previous analysis of known familial AD-linked APP mutations28, identified residues were clustered in the C-terminal half of the C99 transmembrane (TM) domain (residues 41–53), among which K53 at the TM boundary was most critical (Figure 3A), consistent with a previous study28. In addition, mutation of Q98 also led to a strong decrease in cleavage, which, as in the case of C99 K53A, was not caused by changes in protein expression (Figure 3A insert). The Q98A mutation result was unexpected as deletion of the C-terminal cytoplasmic region including Q98 did not affect cleavage (Figure 1C and 1D). We therefore also tested different constructs with synonymous changes in the Q98A codon, yet all of them maintained the same low-level cleavage efficiency (data not shown). We do not understand the mechanism by which Q98A decreases cleavage efficiency, yet fragments lacking Q98 are cleaved with efficiency near or higher than WT C99.
Figure 3.
Validation of minimum substrate. (A) Scanning alanine mutagenesis experiments. The TM domain and a C-terminal portion of the TM domain plus the TM boundary residue (amino acids 41-53) are highlighted by green and blue boxes, respectively; K53 and Q98 are marked by red stars. The relative protein expression levels of C99 WT, K53A and Q98A are shown by western blot in the inlet panel. (B) Scanning double proline mutagenesis experiments. The minimum region (E22-K55) required for efficient γ-secretase cleavage was marked by an orange box. Top: Predicted secondary structure (predicted by PSIPRED Protein Sequence Analysis)40. The relative protein expression levels of C99 WT and some crucial mutations are shown at the corresponding positions. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
To further analyze the cleavage profile, we tested whether secondary structure elements of C99 affect γ-secretase cleavage. Specifically, we designed a complete “double proline scan mutagenesis”, in which consecutive pairs of C99 amino acids were replaced with prolines to introduce kinks, which destabilize α-helices and β-strands. As shown in Figure 3B, most of the proline-proline mutations within the identified minimal region of C99 (E22-K55) led to substantial decreases in cleavage. Notably, this region contains two predicted helices, the TM helix and a short N-terminal helix. In contrast, residues outside of the minimal C99 γ-secretase recognition fragment were relatively insensitive to double proline replacements (Figure 3B; immunoblots of critical double proline mutant constructs are shown underneath the respective activity bars). Together, these data validate E22-K55 as minimum γ-secretase substrate and indicate that secondary structure and individual C-terminal residues of this fragment are important γ-secretase substrate determinants.
N-terminal negative charge is important for C99 recognition by γ-secretase in living cells
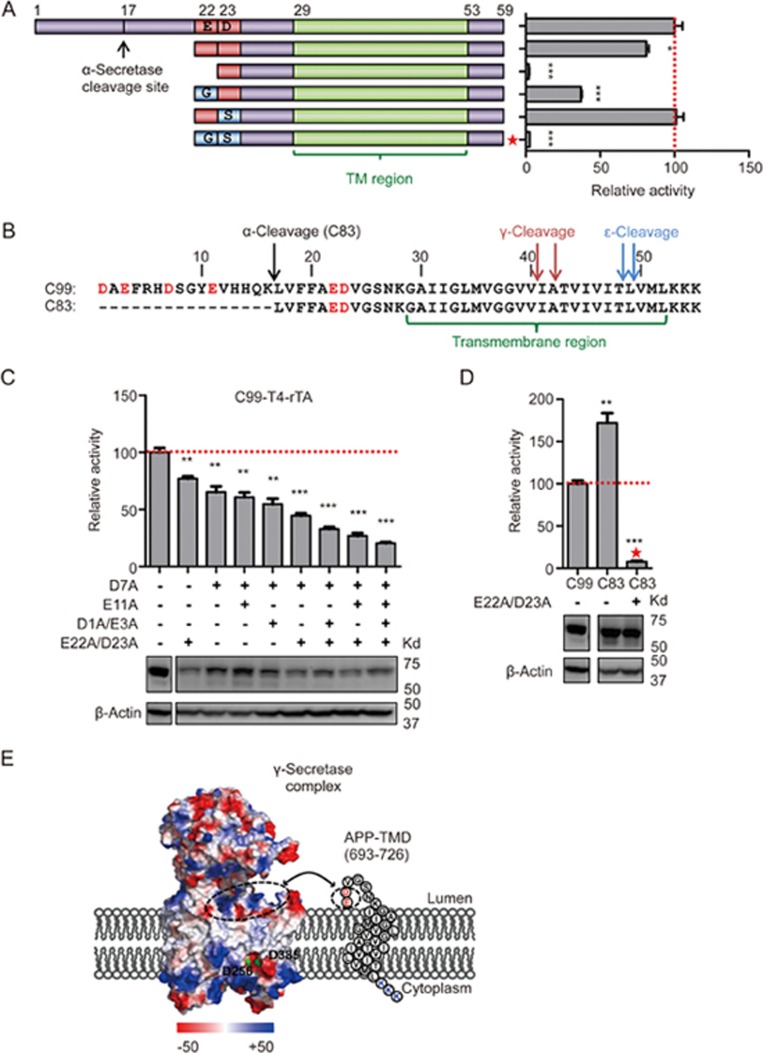
While individual C-terminal residues in the E22-K55 fragment of C99, especially the positively charged K53, are critical for γ-secretase cleavage, the continuous deletion of N-terminal residues also led to an elimination of C99 cleavage (Figure 2). To understand this requirement, we introduced random mutations into the N-terminal portion of the minimal region. Mutations that removed both negative charges at the N-terminus of the minimal fragment abolished its cleavage by γ-secretase (Figure 4A). This suggested that charge, rather than a specific amino acid, may be responsible for the requirement for cleavage, supporting the substrate diversity of γ-secretase. The extracellular region of C99 contains a total of six negatively charged residues including E22 and D23 (Figure 4B, marked in red). We speculate that if the charge would indeed be the key feature for the requirement of E22 and D23 in the minimal fragment for γ-secretase cleavage, then their requirement might be overcome by negative charges N-terminal to the minimum fragment. When we mutated E22 and D23 to alanine in the context of C99 (Figure 4C), cleavage efficiency was only subtly affected. However, when we mutated these two residues together with the other four negatively charged residues in the extracellular domain of C99 (D1, E3, D7, and E11), or in the context of C83 (Figure 4D), which lacks the N-terminal four negatively charged residues, cleavage activity was greatly reduced. Moreover, when we mutated these residues one by one, cleavage efficiency gradually decreased with decreased charge (Figure 4C). Together, these data strongly suggest that negative charge on the extracellular side of C99, rather than any specific amino acid position, is important for γ-secretase recognition. This observation further suggests that the negatively charged extracellular side of C99 may form electrostatic interactions with a positively charged surface of γ-secretase, such as the cluster of positively charged residues on the extracellular membrane side of γ-secretase identified in the cryo-EM structure of human γ-secretase21 (Figure 4E).
Figure 4.
N-terminal positive charge recognition model. (A) Cartoon illustration of the truncated C99 N-terminal mutations (left panel) and cleavage measurement by Epsilon-Cleavage assay of these substrates (right panel). Mutations that completely remove N-terminal negative charge by replacement with Gly or Ser are indicated by a red star. (B) N-terminal sequences of C99 and C83 with secretase cleavage sites and transmembrane domain are highlighted. Red letters indicate the N-terminal negatively charged residues. (C) Stepwise reduction of N-terminal negative charges by alanine mutagenesis. Cleavage was measured by the Epsilon-Cleavage assay. The relative protein expression is shown under each bar. (D) N-terminal negative charge study on C83. The red star indictes the construct in which N-terminal negative charge residues were replaced by GS amino acids. The relative protein expression is shown underneath. (E) Charge distribution of human γ-secretase (PDB code: 5A63) and cartoon illustration of a possible orientation of the APP minimum substrate. Negatively and positively charged residues are labeled with red and blue, respectively (see charge potential color code at bottom). PS1 catalytic aspartates are presented with green dots. Potential charge interaction areas are encircled with dashed lines. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
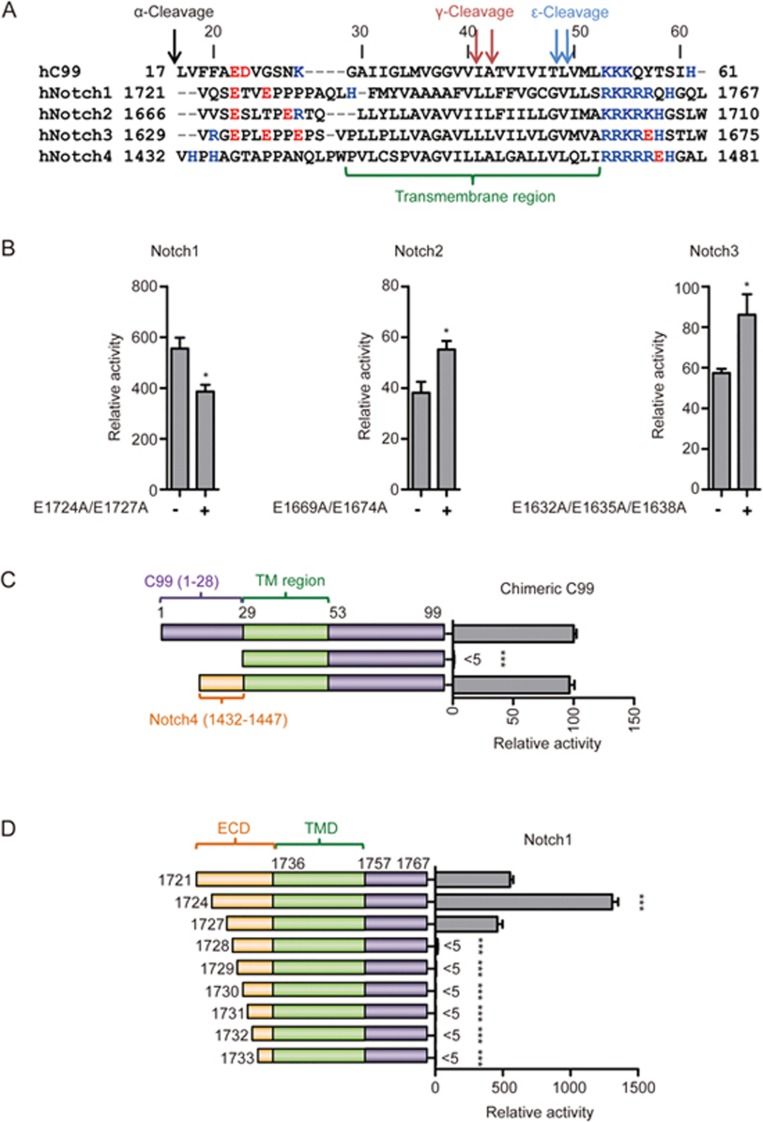
The extracellular domains of Notch and C99 are differently recognized by γ-secretase
The N-terminal negatively charged residues in C99 are not conserved across γ-secretase substrates, such as the family of Notch receptors (Figure 5A). We, therefore, investigated the general application of the N-terminal charge model by analyzing the cleavage efficiency of Notch fragments consisting of the TM domain and flanking regions (Notch1 [1721-1767], Notch2 [1666-1710], Notch3 [1629-1675], and Notch4 [1432-1481]; Figure 5A). While the Notch1, Notch2 and Notch3 fragments all have negatively charged residues in their N-terminal, extracellular regions, replacement of these residues with non-charged alanine had no obvious impact on Notch cleavage by γ-secretase (Figure 5B). Additionally, Notch4 completely lacks negatively charged residues in the N-terminal fragment (Figure 5A), yet in a domain swap experiment, its extracellular N-terminal sequence can functionally replace the C99 extracellular domain, which otherwise is absolutely required for γ-secretase cleavage (Figure 5C). These results, together with the fact that the C99 extracellular domain can be replaced with that from Notch1 without losing activity31, indicates that negative charge in the extracellular domain of Notch is not required for γ-secretase cleavage, but that Notch contains a transferable, alternative extracellular γ-secretase recognition motif. N-terminal truncation analysis of Notch1 [1721-1767] indicates that the first 6 residues are dispensable for this function (Figure 5D). Together, these data support that negative charge in the extracellular domain is not universally required for γ-secretase substrates, but plays a significant role in the recognition of the C99 fragment of APP.
Figure 5.
The N-terminal negative charge is not required for Notch cleavage by γ-secretase. (A) Sequence alignment of a subset of γ-secretase substrates (human C99 and human Notch family). Shown are TM regions together with flanking N-terminal luminal and C-terminal cytoplasmic regions (C99 [17-61], Notch1 [1721-1767], Notch2 [1666-1710], Notch3 [1629-1675], Notch4 [1432-1481]). Negatively and positively charged residues are highlighted by red and blue letters, respectively. (B) Abolishment of Notch1/2/3 N-terminal negative charge by alanine mutagenesis. (C) Cartoon illustration of C99, C99ΔN, and the C99 hybrid protein, in which the N-terminus is replaced by that of Notch4 [1432-1447] (left). Cleavage efficiencies of these constructs measured by Epsilon-Cleavage assay are shown on the right. (D) Defining the minimum substrate N-terminus for Notch1 by γ-secretase Epsilon-Cleavage assay in living cells. Cartoon illustration of the Notch1 N-terminal truncations (left) and the corresponding cleavage efficiencies (right). Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
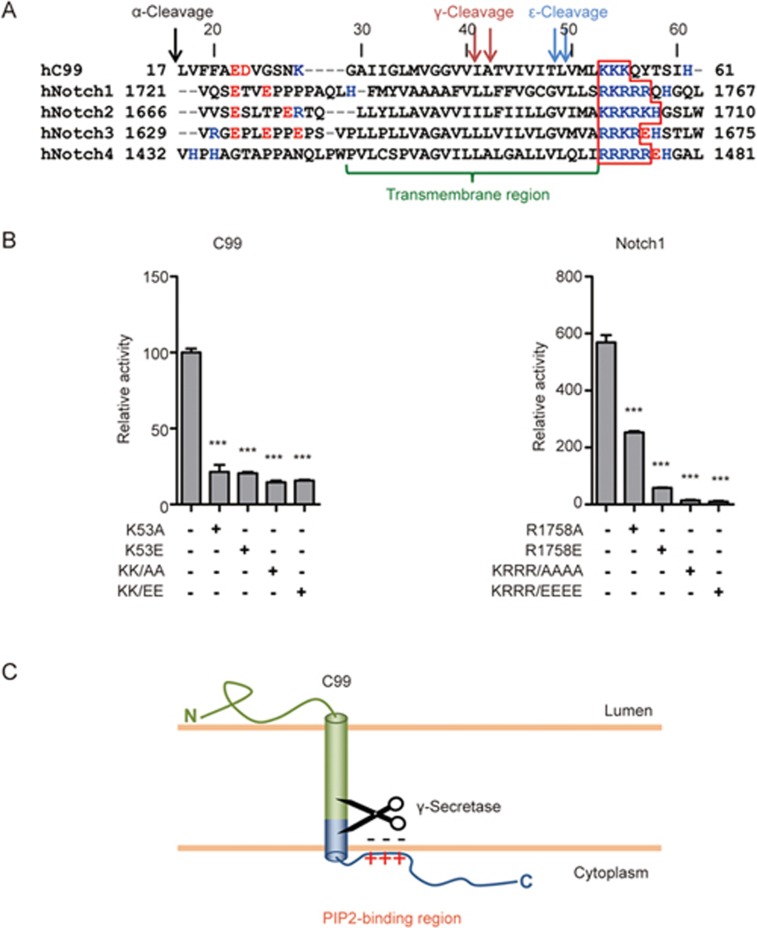
C99 and Notch1 polybasic regions bind PIP2 and are important for recognition by γ-secretase
The single most important residue for C99 γ-secretase substrate recognition is K5328 (Figure 3A). Replacement of K53 by non-charged or negatively charged residues but not against arginine greatly decreased C99 cleavage28 (Figure 6B). Importantly, a lysine or arginine residue at the position corresponding to C99 K53 (ie, directly following the last amino acid of the TM domain), is strictly conserved in all known 69 γ-secretase substrates, and has been shown to be important for substrate recognition of both C99 and each of the four human Notch receptors28. A positively charged residue at the TM junction, therefore, is likely a universal requirement for γ-secretase substrates, and this residue has been proposed to interact with the negatively charged catalytic center of the presenilin subunit of γ-secretase28. We noticed that this basic residue is followed by one or several positively charged residues in γ-secretase substrates, including human APP and Notch receptors, to form a polybasic region of unknown function that is located close to the cytoplasmic layer of the membrane (Figure 6A). Our alanine scanning mutagenesis demonstrated a more than four-fold cleavage decrease for C99 K53A, but only subtle changes for alanine replacements of the other two residues of the polybasic region, K54 and K55 (Figure 3A). However, double-replacement of C99 K54/K55 with either alanine or glutamate resulted in greater cleavage defects than K53A (Figure 6B), consistent with a functional role of the C-terminal charge cluster in γ-secretase recognition. Similarly, replacement of the positively charged cluster C-terminal to the critical R1758 of Notch1 abolished cleavage by γ-secretase (Figure 6B, right panel). Together, these data suggest that substrate recognition of γ-secretase probably involves the whole C-terminal polybasic region, but unlike the critical first basic residue, the other basic residues in this region likely contribute indirectly to promoting substrate binding.
Figure 6.
The C-terminal polybasic regions of γ-secretase substrates are important for cleavage. (A) Sequence alignment of a subset of γ-secretase substrates (human C99 and human Notch family). C-terminal polybasic regions are marked by the red outline. (B) Mutational analysis of the positively charged conserved membrane junction sites (K53 of C99 or R1758 of Notch1) and the polybasic regions (K54-K55 of C99 and K1759-R1762 of Notch1) C-terminal to the junction site. (C) Schematic diagram of a PIP2-binding model of the substrate C-terminal polybasic regions. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
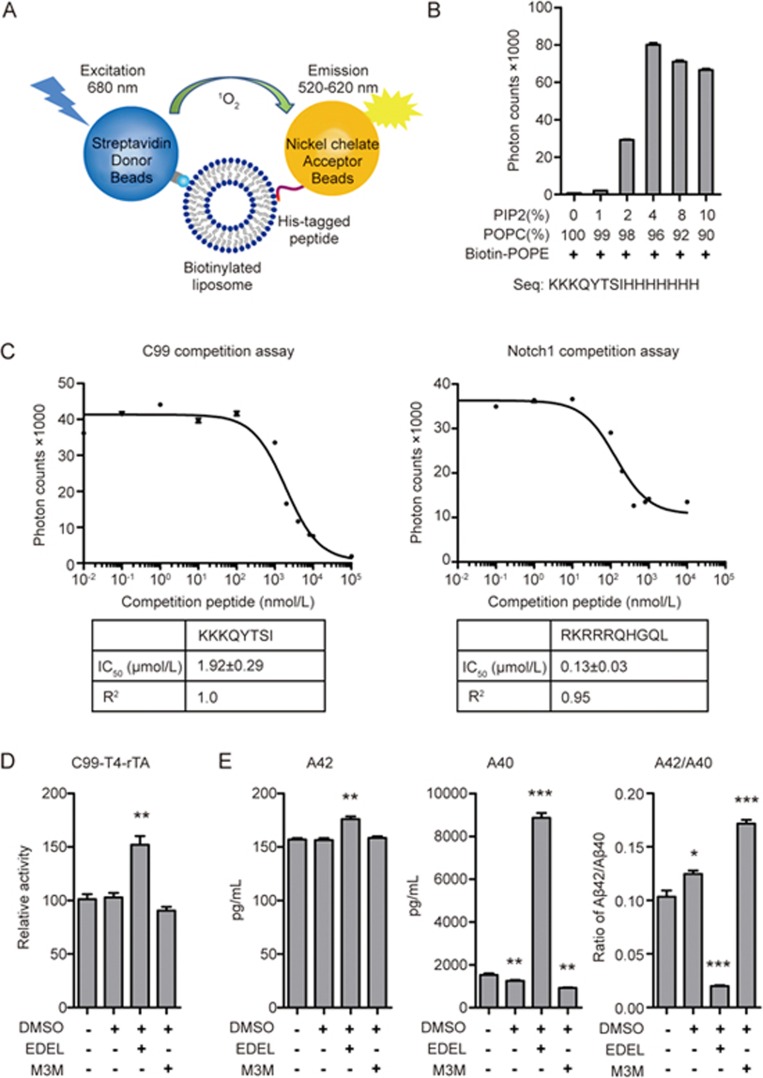
Given the position of the polybasic region at the cytoplasmic layer of the membrane, we speculated that it might anchor to components of the plasma membrane to correctly orientate C99 for cleavage (see the model in Figure 6C). Since PIP2 is the most abundant plasma membrane lipid that carries multiple negatively charged phosphates33, we developed an AlphaScreen assay to test whether the polybasic regions of C99 and Notch1 can interact selectively with PIP2-containing liposomes. Specifically, we prepared liposomes with different ratios of PIP2 and the single phosphate group-containing phospholipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) spiked with 1% biotin-labeled 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) to enable liposomes to bind streptavidin-coated AlphaScreen donor beads (Figure 7A). C99 (KKKQYTSIH) and Notch1 (RKRRRQHGQL) peptides containing the polybasic regions were synthesized with C-terminal His6 tags for immobilization on AlphaScreen nickel-chelated acceptor beads. The interaction between peptides and liposomes induces proximity between donor and acceptor beads that leads to a singlet oxygen-mediated energy transfer that can be measured as light emission at 680 nm (Figure 7A). Liposomes that lacked PIP2 did not show any interaction with the C99 and Notch1 polybasic peptides while increasing concentrations of PIP2 up to 4% PIP2 yielded increasingly stronger binding signals (Figure 7B), which is in the close range to typical plasma membrane PIP2 levels of 1%–3% of total lipid34. To estimate the strength of this interaction, we performed homologous competition assays with non-tagged C99 and Notch1 peptides to determine IC50 values of ∼1.9 μmol/L (C99) and ∼130 nmol/L (Notch1) for the interaction with liposomes spiked with 4% PIP2 (Figure 7C).
Figure 7.
Modulations of PIP2 levels correlate with C99 cleavage levels. (A) Schematic of the AlphaScreen Assay. Biotinylated liposome binds to Streptavidin-coated donor beads, and the nickel chelated acceptor beads bind to His-tagged peptide. The interaction between His-tagged peptides and biotinylated liposome brings the two beads into close proximity to generate a light emission signal. (B) Validation of PIP2 binding by C99 polybasic peptide. The PIP2 lipid content varies from 0 to 10%. Underneath is the sequence of His-tagged C99 polybasic peptide. (C) Competition assay of C99/Notch1 polybasic peptides in presence of 4% PIP2. The IC50 values were obtained from curve fitting using the GraphPad Prism competitive inhibitor model. (D) Modulations of PIP2 levels by the PLC pathway inhibitor edelfosine (EDEL) and the PLC activator m-3M3FBS (M3M) led to changes in total C99 cleavage. Cleavage was measured by the Epsilon-Cleavage assay. (E) Modulations of PIP2 levels affect Aβ40 levels and Aβ40/Aβ42 cleavage ratios as determined by AlphaLISA assay. Absolute concentrations were calculated using a previous calibration graph. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.
While the above experiments demonstrated that the polybasic regions of C99 and Notch can bind PIP2 with high affinity and are required for cleavage by γ-secretase, it is not clear whether PIP2-binding is important for γ-secretase recognition in cells. We therefore modulated cellular PIP2 levels in HTL cells treated with either DMSO (control), the PLC pathway inhibitor edelfosine (EDEL), which increases PIP2 levels35, or the PLC pathway activator m-3M3FBS (M3M), which decreases cellular PIP2 levels35. PIP2 upregulation by edelfosine treatment resulted in increased C99 cleavage, while activation of PLC by m-3M3FBS treatment led to a slightly lower level of C99 cleavage (Figure 7D). Using AlphaLISA assays, we further tested the levels of the cleavage products Aβ40 and Aβ42. While edelfosine treatment had only a small effect on the generation of Aβ42, it dramatically increased Aβ40 levels, leading to a strikingly reduced Aβ42/Aβ40 ratio. Conversely, the M3M treatment caused an about 30% increase of the Aβ42/Aβ40 ratio (Figure 7E). Together, these data provide strong support for an interaction of γ-secretase substrates with plasma membrane PIP2 and an important role of this interaction for substrate recognition and cleavage site specificity.
Discussion
In this study, we used the well-established γ-secretase Epsilon-Cleavage assay to investigate the initial cleavage of C99 by γ-secretase in living cells. Together with an AlphaLISA assay, we defined E22-K55 as the minimal C99-derived fragment that is required for γ-secretase initial cleavage in living cells. By further investigating the luminal and cytoplasmic regions flanking the TM domain of the minimum substrate, we obtained insight into the function of the N-terminal negative charge-rich region and the C-terminal polybasic region, which play important roles in substrate recognition. The minimum APP substrate we identified in living cells is different from the minimum APP fragments that can be cleaved by γ-secretase in cell-free conditions. It was shown that the APP transmembrane domain itself is recognized and efficiently cleaved by γ-secretase using a synthetic peptide incubated with crude CHAPSO extracted γ-secretase36. In addition, Aβ peptides such as Aβ48, Aβ49, and even Aβ42 can also be cleaved by γ-secretase in vitro37,38,39. In contrast, our results show that in cell cleavage of APP by γ-secretase requires the N-terminal domain negatively charged residues and the C-terminal positively charged domain embedded in the APP fragment of E22-K55.
Given the keen interest in developing drugs for the treatment of Alzheimer's disease, the mechanism of APP substrate recognition is under intense investigation. Although the near-atomic structure of human γ-secretase has recently been revealed21, the mechanism of substrate recognition has remained elusive. While NCT was believed to function as a substrate receptor that recognizes the N-terminus of C9922, our results demonstrate that the N-terminal 21 amino acids of C99 are not required for γ-secretase cleavage23. In contrast, we have found that the redundant negatively charged residues residing in the N-terminal luminal region of C99 are important for substrate recognition. While these are not conserved within the Notch family, they are clearly important in facilitating interactions between C99 and γ-secretase. Our previous work has established the basic residue K53 at the TM junction as a key determinant involved in substrate recognition, possibly by directly interacting with the catalytic pocket of γ-secretase28. In addition, we have demonstrated in this study that not only the basic junction residue but also the whole C-terminal polybasic region including K54 and K55 critically contributes to substrate recognition mediated by electrostatic PIP2 binding. Thus the negative charges in the extracellular side and the positive charges in the intracellular side of C99 are both required for γ-secretase cleavage.
The asymmetric charge distribution that is required for efficient C99 cleavage most likely aids in correctly orientating the membrane-bound substrate and enzyme through complementary electrostatic interactions. We speculate that correct substrate orientation would facilitate subsequent interaction between the K53 residue of C99, which resides just C-terminal to the initial (ε) γ-secretase cleavage site, and the catalytic pocket of γ-secretase, which is enriched with negatively charged residues. Several lines of evidence are in support of this model. First, the N-terminal residues 1-21 and the C-terminal residues 56-99 are not required for substrate recognition as truncated C99 lacking these two portions can still be recognized and cleaved by γ-secretase. Second, negatively charged residues in the N-terminus of both the minimum substrate and C83 are sensitive to mutations that change their charge properties (Figure 4A), yet they can be functionally replaced with negatively charged amino acids N-terminal to C83. Third, stepwise removal of the six negative charges in the N-terminus of full-length C99 causes corresponding stepwise decreases in C99 cleavage. Fourth, polybasic regions C-terminal to their TM helix are globally found in γ-secretase substrates28 and are conserved in both C99 and the members of the Notch family. While the basic residue closest to the TM helix is a key determinant of substrate cleavage, the other residues of the polybasic region have a subtle sensitivity to individual alanine mutations, but are highly sensitive to mutations that remove their positive charges. Fifth, peptides encompassing the C-terminal polybasic region specifically interact with liposomes that contain high physiological levels of PIP2, and, strikingly, pharmacological modulation of cellular PIP2 can affect both C99 cleavage efficiency as well as the cleavage selectivity. While Aβ42 levels were either subtly affected in our study or moderately decreased in a previous study35, Aβ40 levels and the Aβ40/Aβ42 ratio dramatically increased upon treatment with EDEL, in support of the importance of the PIP2 interaction for C99 recognition and cleavage by γ-secretase in cells.
In summary, our study reported here describes a framework for substrate recognition by γ-secretase. For C99, we have identified a 34 amino acid region consisting of the 24 amino acid TM helix, seven N-terminal and three C-terminal residues, as being required and sufficient for recognition. Both the negatively charged residues N-terminal to the TMD and the positively charged cluster C-terminal to the TMD allow C99 to fit into the γ-secretase catalytic site by electrostatic interactions, which may include clustering in PIP2-rich subdomains of the membrane through direct PIP2 binding of the polybasic region (K53-K55) of C99. This positioning through long-range electrostatic interactions provides an environment for a subsequent residue-specific interaction of C99 K53 that likely involves the negatively charged catalytic pocket given the proximity of K53 to the initial γ-secretase cleavage site.
Author contribution
Yan YAN and Ting-Hai XU designed and conducted the experiments and wrote the paper; H Eric XU and Karsten MELCHER designed the experiments. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
This work was supported by the Van Andel Research Institute, the National Natural Science Foundation of China (31300607, 31300245 and 91217311), Ministry of Science and Technology grants of China (2012ZX09301001, 2012CB910403, 2013CB910600, XDB08020303, and 2013ZX09507001), Shanghai Science and Technology Committee (13ZR1447600), Shanghai Rising-Star Program (14QA1404300) and the National Institute of Health grants DK071662 (H Eric XU), GM102545 and GM104212 (Karsten MELCHER). We thank members of the Xu/Melcher laboratories for discussion.
References
- Zheng H, Koo EH. Biology and pathophysiology of the amyloid precursor protein. Mol Neurodegener 2011; 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci 1997; 20: 154–9. [DOI] [PubMed] [Google Scholar]
- Nunan J, Small DH. Regulation of APP cleavage by alpha-, beta- and gamma-secretases. FEBS Lett 2000; 483: 6–10. [DOI] [PubMed] [Google Scholar]
- Jiang S, Li Y, Zhang X, Bu G, Xu H, Zhang YW. Trafficking regulation of proteins in Alzheimer's disease. Mol Neurodegener 2014; 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang M, Cai F, Song W. Biological function of Presenilin and its role in AD pathogenesis. Transl Neurodegener 2013; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Ihara Y. Alzheimer's disease: beta-Amyloid protein and tau. J Neurosci Res 2002; 70: 392–401. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer's disease. Mol Brain 2011; 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami M, Nagashima Y, Sano Y, Ishihara S, Morishima-Kawashima M, Funamoto S, et al. gamma-Secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment. J Neurosci 2009; 29: 13042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih IeM, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res 2007; 67: 1879–82. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci 2009; 66: 1631–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol 2007; 194: 459–74. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol 2003; 5: 486–8. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron 2003; 38: 9–12. [DOI] [PubMed] [Google Scholar]
- Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, et al. The role of presenilin cofactors in the gamma-secretase complex. Nature 2003; 422: 438–41. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Vassar R, Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol 2010; 6: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Iwatsubo T, Wolfe MS. Presenilins and gamma-secretase: structure, function, and role in Alzheimer disease. Cold Spring Harb Perspect Med 2012; 2: a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, et al. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature 2006; 440: 1208–12. [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Fraering PC, Ostaszewski BL, Ye W, Kimberly WT, Wolfe MS, et al. Assembly of the gamma-secretase complex involves early formation of an intermediate subcomplex of Aph-1 and nicastrin. J Biol Chem 2003; 278: 37213–22. [DOI] [PubMed] [Google Scholar]
- Lee SF, Shah S, Yu C, Wigley WC, Li H, Lim M, et al. A conserved GXXXG motif in APH-1 is critical for assembly and activity of the gamma-secretase complex. J Biol Chem 2004; 279: 4144–52. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Tomita T, Sato C, Kitamura T, Morohashi Y, Iwatsubo T. Pen-2 is incorporated into the gamma-secretase complex through binding to transmembrane domain 4 of presenilin 1. J Biol Chem 2005; 280: 41967–75. [DOI] [PubMed] [Google Scholar]
- Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, et al. An atomic structure of human gamma-secretase. Nature 2015; 525: 212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell 2005; 122: 435–47. [DOI] [PubMed] [Google Scholar]
- Zhao G, Liu Z, Ilagan MX, Kopan R. Gamma-secretase composed of PS1/Pen2/Aph1a can cleave notch and amyloid precursor protein in the absence of nicastrin. J Neurosci 2010; 30: 1648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang W, Han W. Initial substrate binding of gamma-secretase: the role of substrate flexibility. ACS Chem Neurosci 2017. doi: 10.1021/acschemneuro.6b 00425. [DOI] [PubMed]
- Bolduc DM, Montagna DR, Seghers MC, Wolfe MS, Selkoe DJ. The amyloid-beta forming tripeptide cleavage mechanism of gamma-secretase. Elife 2016; 5. pii: e17578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori A, Steiner H. Substrate recruitment of gamma-secretase and mechanism of clinical presenilin mutations revealed by photoaffinity mapping. EMBO J 2016; 35: 1628–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, et al. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A 2008; 105: 64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu TH, Yan Y, Kang Y, Jiang Y, Melcher K, Xu HE. Alzheimer's disease-associated mutations increase amyloid precursor protein resistance to gamma-secretase cleavage and the Abeta42/Abeta40 ratio. Cell Discov 2016; 2: 16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer's disease. Nat Rev Neurosci 2007; 8: 499–509. [DOI] [PubMed] [Google Scholar]
- Haapasalo A, Kovacs DM. The many substrates of presenilin/gamma-secretase. J Alzheimers Dis 2011; 25: 3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S, Sasaki T, Ishihara S, Nobuhara M, Nakano M, Watanabe-Takahashi M, et al. Substrate ectodomain is critical for substrate preference and inhibition of gamma-secretase. Nat Commun 2013; 4: 2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Bassit B, Chau D, Li YM. An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat Struct Mol Biol 2010; 17: 151–8. [DOI] [PubMed] [Google Scholar]
- Suh BC, Hille B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu Rev Biophys 2008; 37: 175–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP2 and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct 2002; 31: 151–75. [DOI] [PubMed] [Google Scholar]
- Landman N, Jeong SY, Shin SY, Voronov SV, Serban G, Kang MS, et al. Presenilin mutations linked to familial Alzheimer's disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc Natl Acad Sci U S A 2006; 103: 19524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YI, Bassit B, Zhu L, Yang X, Wang C, Li YM. gamma-Secretase substrate concentration modulates the Abeta42/Abeta40 ratio: implications for Alzheimer disease. J Biol Chem 2007; 282: 23639–44. [DOI] [PubMed] [Google Scholar]
- Okochi M, Tagami S, Yanagida K, Takami M, Kodama TS, Mori K, et al. gamma-Secretase modulators and presenilin 1 mutants act differently on presenilin/gamma-secretase function to cleave Abeta42and Abeta43. Cell Rep 2013; 3: 42–51. [DOI] [PubMed] [Google Scholar]
- Fernandez MA, Klutkowski JA, Freret T, Wolfe MS. Alzheimer presenilin-1 mutations dramatically reduce trimming of long amyloid beta-peptides (Abeta) by gamma-secretase to increase 42-to-40-residue Abeta. J Biol Chem 2014; 289: 31043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Hung AY, Schlossmacher MG, Teplow DB, Selkoe DJ. beta-Amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J Biol Chem 1993; 268: 3021–4. [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 2013; 41: W349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]