Abstract
Campylobacter iguaniorum is most closely related to the species C. fetus, C. hyointestinalis, and C. lanienae. Reptiles, chelonians and lizards in particular, appear to be a primary reservoir of this Campylobacter species. Here we report the genome comparison of C. iguaniorum strain 1485E, isolated from a bearded dragon (Pogona vitticeps), and strain 2463D, isolated from a green iguana (Iguana iguana), with the genomes of closely related taxa, in particular with reptile-associated C. fetus subsp. testudinum. In contrast to C. fetus, C. iguaniorum is lacking an S-layer encoding region. Furthermore, a defined lipooligosaccharide biosynthesis locus, encoding multiple glycosyltransferases and bounded by waa genes, is absent from C. iguaniorum. Instead, multiple predicted glycosylation regions were identified in C. iguaniorum. One of these regions is > 50 kb with deviant G + C content, suggesting acquisition via lateral transfer. These similar, but non-homologous glycosylation regions were located at the same position on the genome in both strains. Multiple genes encoding respiratory enzymes not identified to date within the C. fetus clade were present. C. iguaniorum shared highest homology with C. hyointestinalis and C. fetus. As in reptile-associated C. fetus subsp. testudinum, a putative tricarballylate catabolism locus was identified. However, despite colonizing a shared host, no recent recombination between both taxa was detected. This genomic study provides a better understanding of host adaptation, virulence, phylogeny, and evolution of C. iguaniorum and related Campylobacter taxa.
Keywords: Campylobacter iguaniorum, reptile, comparative genomics, recombination, phylogeny, evolution
Introduction
The majority of the Campylobacter species are associated with endothermic mammalian and avian hosts. Recently, a novel Campylobacter species has been described, Campylobacter iguaniorum (Cig), which is predominantly isolated from ectothermic reptilian hosts (Gilbert et al. 2015). Chelonians and lizards in particular appear to be a primary reservoir of this Campylobacter species. Reported overall prevalence in reptiles based on culturing was 8.2%; 15.6% of the chelonians and 6.1% of the lizards, but none of the snakes examined, carried this Campylobacter species (Gilbert et al. 2014a). Recently, Cig has also been isolated from a non-reptilian host (Miller et al. 2016a).
Within the genus Campylobacter, this species forms a clearly separated phylogenetic clade, together with the closely related taxa C. fetus, C. hyointestinalis, and C. lanienae (collectively called the C. fetus clade). C. fetus currently comprises three subspecies: mammal-associated C. fetus subsp. fetus (Cff) and C. fetus subsp. venerealis (Cfv), and reptile-associated C. fetus subsp. testudinum (Cft) (Fitzgerald et al. 2014). As with Cig, Cft has been shown to be associated primarily with reptiles, and both taxa are the most frequently isolated Campylobacter in reptiles (Harvey and Greenwood 1985; Wang et al. 2013; Gilbert et al. 2014a). Also, C. hyointestinalis has been infrequently isolated from reptiles (Gilbert et al. 2014a). Interestingly, Campylobacter species commonly found in various avian and mammalian hosts, such as C. coli, C. jejuni, and C. lari, were not isolated from reptiles, despite culturing conditions suitable for these species. It was speculated that the host body temperature, which is on average lower and more fluctuating in reptiles than in mammals and birds, is associated with this remarkable species distribution (Gilbert et al. 2014a). Indeed, Cig, C. fetus, and to a lesser extent C. hyointestinalis, were found able to grow at lower temperatures than most other Campylobacter species (Gilbert et al. 2015).
The pathogenicity of Cig in reptiles is unknown; isolates have been recovered both from reptiles with and without clinical signs of disease (Benejat et al. 2014; Gilbert et al. 2014a). In contrast to reptile-associated Cft, which has been shown to cause infection in humans (Tu et al. 2004; Patrick et al. 2013), no human Cig infections have been reported to date.
The first closed and annotated Cig genome has been described previously (Gilbert et al. 2014b). Genome analysis and comparison provide valuable insights into host adaptation, virulence, phylogeny, and evolution of this reptile-associated Campylobacter species. Comparison with reptile-associated Cft could identify factors specific for adaptation to their shared reptilian host. Here we report the complete whole genome sequences of two Cig strains isolated from reptiles, and compare them to those of the closely related taxa Campylobacter fetus, C. hyointestinalis, and C. lanienae.
Campylobacter iguaniorum genome features and comparison
The genome size of strain 1485E is 1,684,608 bp with a 70,030 bp megaplasmid; strain 2364D is 1,809,624 bp with an estimated 54,764 bp megaplasmid. The general features of Cig strains 1485E and 2463D are summarized in supplementary table S1, Supplementary Material online.
Multiple genomic regions were specific for Cig, i.e. no orthologs were identified in all or most members of the C. fetus clade or the entire Campylobacter genus. In total, 59 genes were conserved in both Cig strains, which were absent from all other C. fetus clade members (supplementary table S2, Supplementary Material online). Multiple genes involved in sulfur metabolism were identified, indicating that this is important in Cig biology. Indeed, phenotypic testing showed that Cig and C. hyointestinalis are one of the few Campylobacter species that produce H2S on sulfate containing TSI agar (Gilbert et al. 2015). Notably, the gene coding for l-lactate permease (lctP) was absent from Cig and C. hyointestinalis subsp. hyointestinalis, but was well conserved in the other C. fetus clade members and most other Campylobacter species. In C. jejuni, transport of l-lactate occurs primarily via LctP, although at least one other transport route also exists (Thomas et al. 2011). This predicts that Cig relies less on exogenous l-lactate as carbon or energy source than other Campylobacter species encoding lctP.
Several genes were found to be shared specifically by both reptile-associated taxa Cig and Cft (supplementary table S2, Supplementary Material online). As in reptile-associated Cft, a putative tricarballylate catabolism locus tcuRABC (CIG1485E_0479-0482; CIG2463D_0480-0483) was identified in both Cig strains. This locus is partially present in C. hyointestinalis subsp. lawsonii, but as tcuC is a truncated pseudogene, it is likely not functional. Tricarballylate could potentially be used as a carbon and energy source by Cig, as this locus has been shown to function in the catabolism of tricarballylate (a citrate analog) (Lewis et al. 2004). Tricarballylate is considered to cause grass tetany in ruminants, a disease characterized by acute magnesium deficiency. In the ruminant rumen, trans-aconitate present in grass is rapidly reduced to tricarballylate, a toxic end product of ruminal fermentation (Russell 1985). Neither the ruminant nor the normal rumen flora can catabolize tricarballylate efficiently. However, it has been shown that Salmonella enterica serovar Typhimurium strain LT2 can use tricarballylate as a carbon and energy source; the end product of tricarballylate metabolism is cis-aconitate, which then enters into the citric acid cycle (Gutnick et al. 1969; Lewis et al. 2004). Conservation of the tcuRABC locus in the reptile-associated taxa suggests that tricarballylate is ubiquitous in the niche inhabited by Cig and Cft, i.e., the mucosa of the reptilian intestines. As the tcuRABC locus is present in all highly prevalent reptile-associated Campylobacter taxa, this locus could confer an advantage in colonization of the reptilian host.
In contrast to most Campylobacter species, both Cig strains lack a defined lipooligosaccharide (LOS) region, bounded by waa genes and containing multiple glycosyltransferases (supplementary table S2, Supplementary Material online). The lack of waaDEF, which are highly conserved in Campylobacter and Arcobacter, is especially remarkable. Instead, multiple separate predicted glycosylation regions were identified (fig. 1). A large glycosylation region (>50 kb) was present in both strains. Although syntopic in both strains, large parts of this region were not homologous and, in combination with the deviant G + C content (29.0–30.0%), suggests acquisition via lateral transfer. However, this region was highly homologous in strain 2463D and C. hyointestinalis subsp. hyointestinalis. In strains 1485E and 2463D, respectively 20.6% (7/34) and 22.0% (9/41) of the GC tracts were identified within this glycosylation region.
Fig. 1.—
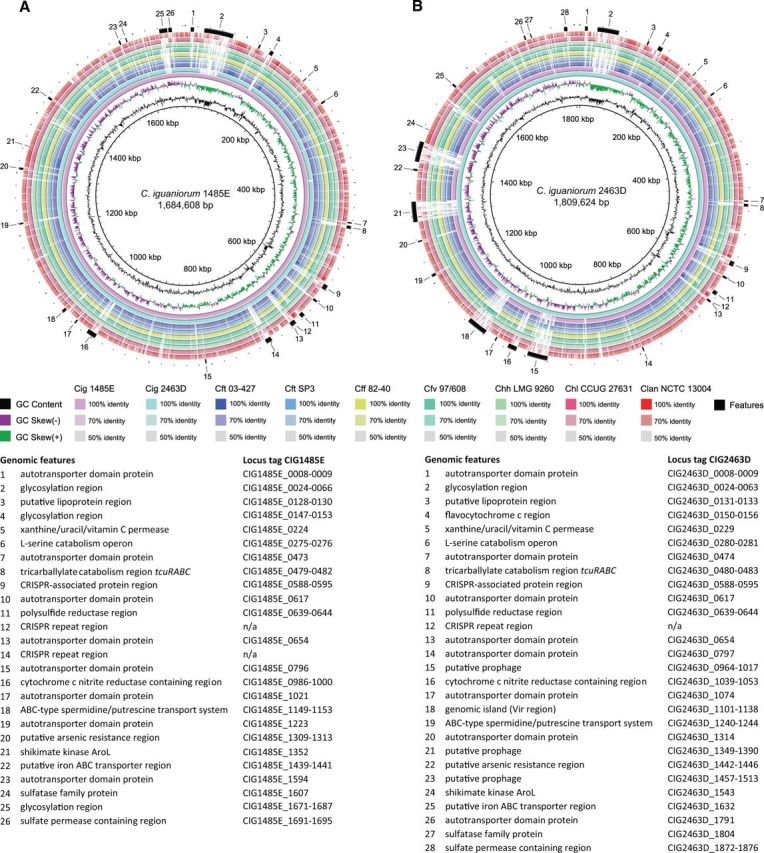
BRIG plots of C. iguaniorum and most closely related Campylobacter species. BLASTN-based genomic comparison of C. iguaniorum strains 1485E and 2463D, C. fetus subsp. testudinum (Cft) strains 03-427 and SP3, C. fetus subsp. fetus (Cff) strain 82-40, C. fetus subsp. venerealis (Cfv) strain 97/608, C. hyointestinalis subsp. hyointestinalis (Chh) strain LMG 9260, C. hyointestinalis subsp. lawsonii (Chl) strain CCUG 27631, and C. lanienae (Clan) strain NCTC 13004. Characteristic features of C. iguaniorum have been highlighted. Reference genome is C. iguaniorum strain 1485E (A); reference genome is C. iguaniorum strain 2463D (B).
As in other Campylobacter species many of the hypervariable GC tracts reside in surface structure-related genes showing phase variation. However, in strains 1485E and 2463D, respectively 23.5% (8/34) and 20.0% (8/41) of the GC tracts were located within autotransporter domain-containing genes, whose role in Cig biology remains to be determined. In total, seven autotransporter domain-containing genes were found conserved in both strains and showed no or low homology with other Campylobacter species. Noteworthy, 89% (8/9) of the autotransporter domain-containing genes in both strains contained a GC tract.
The cas genes present in Cig were homologous with those in C. hyointestinalis subsp. hyointestinalis, but not with those in C. fetus, indicating that at least two different CRISPR/Cas systems are present within the Campylobacter fetus clade (fig. 1).
One of the characteristics shared by Cig, C. fetus, and most Arcobacter species is the ability to grow at lower temperatures (≤25 °C) than most other Campylobacter species (On et al. 1996; Gilbert et al. 2015). Also, these species have been shown to occur together in reptilian hosts, which often show a broad body temperature range and on average lower body temperatures compared with most endothermic mammals and birds (Gilbert et al. 2014a). As in C. fetus, but in contrast to C. jejuni in which nuoE and nuoF are lacking (Kelly 2008), all NADH:quinone oxidoreductase complex I subunits (NuoA-N) are present in Cig, suggesting that NADH is an important electron donor in this species. Interestingly, NuoA-N present in Cig and all other C. fetus clade members showed higher homology to species from the Arcobacter genus (≤72%), than to other species from the Campylobacter genus (≤55%). The high homology observed in the NADH:quinone oxidoreductase complex I subunits might be related to low temperature adaptation. Indeed, in other organisms NADH:quinone oxidoreductase complex I is considered the most thermolabile protein complex of oxidative phosphorylation (Downs and Heckathorn 1998). Within the Campylobacter genus, the ability to exclusively proliferate at lower temperatures (18–37 °C) is unique for Cig. This feature may have helped Cig colonize hosts which have a low and variable body temperature.
Campylobacter iguaniorum phylogeny and diversity
A phylogenomic reconstruction accounting for the effects of homologous recombination was performed, based on a 1,042,737 nucleotide gapless alignment of the whole genomes of Cig and most closely related species C. fetus, C. hyointestinalis, and C. lanienae (supplementary fig. S1, Supplementary Material online). Cig was clearly distinct from the other Campylobacter species. The first split occurred between C. lanienae and the other species. Excluding the more distantly related C. lanienae, the branch lengths indicated that Cig is less related to the last common ancestor than the other species. Interestingly, Cig, C. fetus, and C. hyointestinalis branch of the last common ancestor at the same point, suggesting that these species started diverging at the same time.
In order to examine the genomic relatedness in further detail, the average nucleotide identity (ANI) was determined for the whole genomes of both Cig strains and strains of the most closely related taxa C. fetus, C. hyointestinalis, and C. lanienae (supplementary table S3, Supplementary Material online). ANI values were highest for Cig and both C. hyointestinalis and C. fetus. Homology was higher between Cig and reptile-associated Cft than between Cig and mammal-associated C. fetus subspecies fetus and venerealis, indicating that both reptile-associated taxa Cig and Cft share genomic regions, which might be associated with adaptation to their shared reptilian hosts. Higher ANI values between Cig strain 2463D and Cft strain SP3, C. fetus subsp. venerealis, and C. hyointestinalis subsp. hyointestinalis can be explained by shared laterally transferred genomic regions, such as prophages, and shared glycosylation regions (fig. 1). In addition to this, the genes shared by Cig and other members of the C. fetus clade (≥50% identity) showed that most genes were shared between Cig and C. hyointestinalis subsp. hyointestinalis, followed by both Cft strains, suggesting that these taxa are most closely related (supplementary table S4, Supplementary Material online). Nevertheless, on a species level, differences based on shared genes and ANI are small and in support of the whole genome-based phylogeny showing similar divergence between Cig, C. fetus, and C. hyointestinalis.
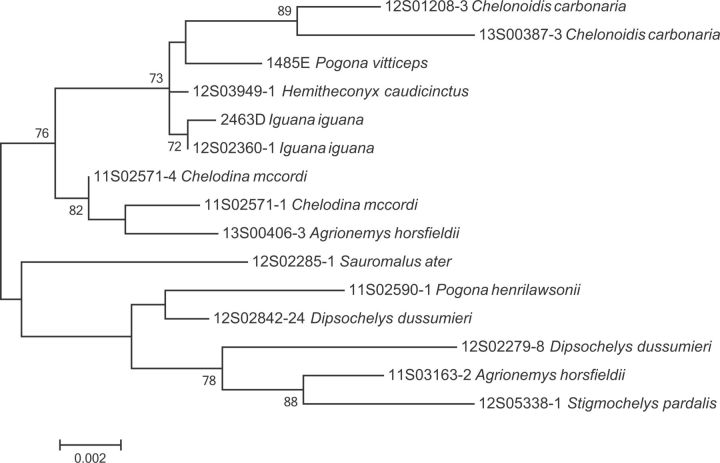
MLST analysis of 18 Cig strains showed two distinct lineages and a high intraspecies diversity without any identical sequence type (fig. 2). No clear association with host type was observed, as Cig isolates originating from lizards and chelonians were found in both lineages. Nevertheless, host association was observed at the species level to some degree, as Cig isolates originating from animals of the same species, but from different locations were mostly clustering together. This was most clear in Iguana iguana, in which two Cig isolates, which were obtained from different animals in 2003 and 2012, clustered closely together. In contrast, two isolates obtained from the same animal (11S02571-1 and 11S02571-4) showed a large genetic distance, indicating that Cig diversity within the same animal can be high.
Fig. 2.—
Maximum likelihood dendrogram of C. iguaniorum based on concatenated MLST sequences, using 500 bootstraps. Isolate numbers are followed by the host species.
Both Cig and Cft colonize the same reptilian hosts (Gilbert et al. 2014a), facilitating potential lateral gene transfer between these two taxa. Indeed, both reptile-associated taxa share specific genomic regions which might confer certain competitive advantages to survive in the reptilian host. Furthermore, similar prophages were identified in the accessory genomes of both taxa, which can serve as a vehicle for novel genetic material and enable gene flow between both taxa. However, no recent recombination events between both taxa were detected in the core genomes. Despite the close genetic relationship and shared host type, the recombination rate between both reptile-associated taxa can be considered low compared with certain other Campylobacter species colonizing a shared host (Sheppard et al. 2008). This could be explained by a recent introduction in the same host or by barriers such as an intrinsic resistance to interspecific recombination or separated niches within the host, as has been shown for C. jejuni (Sheppard et al. 2014). Instead, multiple regions showed higher than expected homology in C. iguaniorum, C. hyointestinalis, and members of the C. concisus clade, suggesting gene flow between these distantly related Campylobacter taxa.
Material and Methods
Strains
Characteristics of all strains used in this study are summarized in table 1. Cig strain 1485E (= CCUG 66346 = LMG 28143) was isolated in 2003 from a bearded dragon (Pogona vitticeps) with a hypertrophic and perforated colon. Cig strain 2463D (= CCUG 66347) was isolated in 2003 from a green iguana (Iguana iguana) with chronic interstitial nephritis. All strains were grown on Columbia agar with 5% sheep blood (Oxoid, the Netherlands) in a microaerobic atmosphere (83.3 N2, 7.1% CO2, 3.6% H2, and 6% O2) at 37 °C for 48 h.
Table 1.
Features of the Campylobacter Strains Used in this Study
| Species | Strain | Source organism | Source type | Location | Sequence data | Sequence method | Reference | Accession number |
|---|---|---|---|---|---|---|---|---|
| Cig | 1485E | Lizard (Pogona vitticeps) | Feces | NL | WGS | 454, Illumina, PacBio | Gilbert et al. 2014b | CP009043-CP009044 |
| Cig | 2463D | Lizard (Iguana iguana) | Feces | NL | WGS | 454, Illumina, PacBio | Gilbert et al. 2015 | CP010995 |
| Cig | 11S02571-1 | Chelonian (Chelodina mccordi) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697811, KU697824, KU697837, KU697850, KU697863, KU697876, KU697889 |
| Cig | 11S02571-4 | Chelonian (Chelodina mccordi) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697812, KU697825, KU697838, KU697851, KU697864, KU697877, KU697890 |
| Cig | 11S02590-1 | Lizard (Pogona henrilawsonii) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697813, KU697826, KU697839, KU697852, KU697865, KU697878, KU697891 |
| Cig | 11S03163-2 | Chelonian (Agrionemys horsfieldii) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697814, KU697827, KU697840, KU697853, KU697866, KU697879, KU697892 |
| Cig | 12S01208-3 | Chelonian (Chelonoidis carbonaria) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697815, KU697828, KU697841, KU697854, KU697867, KU697880, KU697893 |
| Cig | 12S02279-8 | Chelonian (Aldabrachelys gigantea) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697816, KU697829, KU697842, KU697855, KU697868, KU697881, KU697894 |
| Cig | 12S02285-1 | Lizard (Sauromalus ater) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697817, KU697830, KU697843, KU697856, KU697869, KU697882, KU697895 |
| Cig | 12S02360-1 | Lizard (Iguana iguana) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697818, KU697831, KU697844, KU697857, KU697870, KU697883, KU697896 |
| Cig | 12S02842-24 | Chelonian (Aldabrachelys gigantea) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697819, KU697832, KU697845, KU697858, KU697871, KU697884, KU697897 |
| Cig | 12S03949-1 | Lizard (Hemitheconyx caudicinctus) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697820, KU697833, KU697846, KU697859, KU697872, KU697885, KU697898 |
| Cig | 12S05338-1 | Chelonian (Stigmochelys pardalis) | Feces | FR | MLST | Sanger | Benejat et al. 2014 | KU697821, KU697834, KU697847, KU697860, KU697873, KU697886, KU697899 |
| Cig | 13S00387-3 | Chelonian (Chelonoidis carbonaria) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697822, KU697835, KU697848, KU697861, KU697874, KU697887, KU697900 |
| Cig | 13S00406-3 | Chelonian (Agrionemys horsfieldii) | Feces | NL | MLST | Sanger | Gilbert et al. 2014a | KU697823, KU697836, KU697849, KU697862, KU697875, KU697888, KU697901 |
| Cff | 82-40 | Human | Blood | US | WGS | Sanger | Perez et al. 1985 | CP000487 |
| Cft | 03-427 | Human | Blood | US | WGS | 454, Illumina, PacBio | Gilbert et al. 2013 | CP006833 |
| Cft | SP3 | Snake (Heterodon nasicus) | Feces | UK | WGS | 454, Illumina, PacBio | Gilbert et al. 2016 | CP010953 |
| Cfv | 97/608 | Bovine | Placenta | AR | WGS | 454, Illumina, PacBio | van der Graaf-van Bloois et al. 2014 | CP008810-CP008812 |
| Chh | LMG 9260 | Human | Feces | BE | WGS | 454, Illumina | Miller et al. 2016b | CP015575 |
| Chl | CCUG 27631 | Porcine | Gastric biopsy | SE | WGS | 454, Illumina | Miller et al. 2016b | CP015576 |
| Clan | NCTC 13004 | Human | Feces | SE | WGS | 454 | Logan et al. 2000 | CP015578 |
Cig, C. iguaniorum; Cff, C. fetus subsp. fetus; Cft, C. fetus subsp. testudinum; Cfv, C. fetus subsp. venerealis; Chh, C. hyointestinalis subsp. hyointestinalis; Chl, C. hyointestinalis subsp. lawsonii; Clan, C. lanienae.
AR, Argentina; BE, Belgium; FR, France; NL, Netherlands; SE, Sweden; UK, United Kingdom; US, United States. MLST, multilocus sequence typing; WGS, whole genome sequencing.
Whole Genome Sequencing
The sequencing of Cig strains 1485E and 2463D was performed using shotgun and paired-end reads obtained on a Roche 454 FLX Genome Sequencer. A total of 292,658 (1485E) or 318,374 (2463D) 454 reads were assembled using the Newbler assembler (v2.6) into single chromosomal scaffolds of 12 (1485E) or 11 (2463D) unique contigs and single megaplasmid scaffolds, providing draft genome sequences with coverages of 68× (1485E) or 65× (2463D). All 454 base calls were validated using 1,570,644 (1485E) or 2,300,370 (2463D) Illumina MiSeq reads, providing an additional 178× (1485E) or 263× (2463D) coverage. Scaffold gaps were filled as described (Merga et al. 2013). Sequences across the contig junctions were confirmed with Sanger sequencing. Assembly was confirmed using PacBio long reads for strain Cig 2463D. PacBio RS reads were assembled into contigs using Quiver (Pacific Bioscience, Menlo Park, CA, USA). Homopolymeric GC tracts were characterized using the high-depth MiSeq reads.
Genome Analysis
Protein-, rRNA-, and tRNA-encoding genes were identified as described (Merga et al. 2013). The 1485E genome was annotated based on Cft strain 03-427T (accession number CP006833) (Gilbert et al. 2013; Fitzgerald et al. 2014) and the annotation of the 2463D genome was based on that of 1485E, with further annotation using Artemis (Rutherford et al. 2000), the identification of Pfam domains (v.27.0) (Finn et al. 2014), and BLASTP comparisons to proteins in the NCBI non-redundant (nr) database. CRISPR regions were identified using CRISPRFinder (Grissa et al. 2007). The complete annotated genome sequence of Cig strain 1485E has been deposited in GenBank under accession numbers CP009043 (chromosome) and CP009044 (megaplasmid) (Gilbert et al. 2014b). The complete annotated genome sequence of the Cig strain 2463D chromosome has been deposited in GenBank under accession number CP010995. Accession numbers of all genomes used in this study can be found in table 1.
The C. iguaniorum strain 1485E genome was compared with the genome of strain 2463D and the completed genomes of the closely related taxa C. fetus subsp. testudinum (strains 03-427 and SP3), C. fetus subsp. fetus (strain 82-40), C. fetus subsp. venerealis (strain 97/608), C. hyointestinalis subsp. hyointestinalis (strain LMG 9260), C. hyointestinalis subsp. lawsonii (strain CCUG 27631), and C. lanienae (NCTC 13004). For Campylobacter species outside the C. fetus clade, BLASTP comparisons to proteins in the NCBI non-redundant (nr) database were performed. A local BLAST was performed based on the predicted proteomes of all genomes and the results were screened for features specific for one or both Cig strains and the other members of the C. fetus clade. Using JSpecies v.1.2.1 (Richter and Rosselló-Móra 2009) and BLAST v.2.2.26, average nucleotide identity (ANI) values based on the whole genome sequences were calculated for these strains as a measure of genetic relatedness. The BLAST parameters were: X = 150, q = −1, F = F, e = 1e−15, and a = 2. To visualize genomic regions specific for Cig, the BLAST ring image generator (BRIG) (Alikhan et al. 2011) was used, based on BLAST v.2.2.26.
Orthologous Grouping and Phylogenomic Reconstruction
An all versus all BLAST was performed for all predicted proteins of the whole genomes (table 1) at an E-value cutoff of 1E−6. To determine the orthologous relationships of all proteins, the BLAST output was parsed by Orthagogue (Ekseth et al. 2014). Proteins were considered for orthology clustering if the proteins had at least 50% identity and at least 50% overlap. To determine the orthologous groups, Markov clustering (MCL) was performed using MCL-edge (Enright et al. 2002). Genes encoding the proteins were aligned with each other within their respective orthologous groups using MUSCLE (Edgar 2004). A super alignment was created by concatenating the aligned genes according to their position in Cig strain 1485E if they were present in all isolates. Gaps were removed using Gblocks (Castresana 2000). Based on this super alignment phylogenomic reconstruction and prediction of recombination events was performed using Gubbins (Croucher et al. 2014) with the default settings. Phylogenetic dendrograms were created using Fasttree (Price et al. 2009). A BLAST search of the predicted recombination regions of both Cig strains against the genes of Cft was performed to search for particular recombination between these reptile-associated taxa.
MLST
Multilocus sequence typing (MLST) was performed for all Cig strains listed in table 1. The MLST loci were extracted from the genomes of 1485E and 2463D and sequenced as described previously for the other Cig isolates (Miller et al. 2012). For HFglt, the annealing temperature was lowered to 46 °C and 40 instead of 35 cycles were used. Sequences were trimmed and concatenated. In MEGA v.6.06, the concatenated sequences were aligned using MUSCLE and a Maximum likelihood dendrogram was created using 500 bootstraps. Accession numbers of the MLST sequence data can be found in table 1.
Supplementary Material
Acknowledgments
We thank Linda van der Graaf-van Bloois for valuable advice and Arjen Timmerman for technical support. We thank Mary Chapman and Nathaniel Simon for the generation of Illumina MiSeq reads and assistance in the final assembly and genome closure. This work was partially supported by USDA-ARS CRIS project 5325-42000-047-00D.
Literature Cited
- Alikhan N, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benejat L, et al. 2014. Characterization of a Campylobacter fetus‐like strain isolated from the faeces of a sick leopard tortoise (Stigmochelys pardalis) using matrix‐assisted laser desorption/ionization time of flight as an alternative to bacterial 16S rDNA phylogeny. Lett. Appl. Microbiol. 58:338–343. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, et al. 2014. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs CA, Heckathorn SA. 1998. The mitochondrial small heat-shock protein protects NADH: ubiquinone oxidoreductase of the electron transport chain during heat stress in plants. FEBS Lett. 430:246–250. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekseth OK, Kuiper M, Mironov V. 2014. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 30:734–736. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald C, et al. 2014. Description of Campylobacter fetus subsp. testudinum subsp. nov., isolated from humans and reptiles. Int. J. Syst. Evol. Microbiol. 64:2944–2948. [DOI] [PubMed] [Google Scholar]
- Gilbert MJ, et al. 2013. Complete genome sequence of Campylobacter fetus subsp. testudinum strain 03-427T. Genome Announc. 1:e01002–e01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, et al. 2014a. Occurrence, diversity, and host association of intestinal Campylobacter, Arcobacter, and Helicobacter in reptiles. PLoS One 9:e101599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, et al. 2014b. Complete genome sequence of Campylobacter iguaniorum strain 1485ET, isolated from a bearded dragon (Pogona vitticeps). Genome Announc. 2:e00844–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, Kik M, Miller WG, Duim B, Wagenaar JA. 2015. Campylobacter iguaniorum sp. nov., isolated from reptiles. Int. J. Syst. Evol. Microbiol. 65:975–982. [DOI] [PubMed] [Google Scholar]
- Gilbert MJ, et al. 2016. Comparative genomics of Campylobacter fetus from reptiles and mammals reveals divergent evolution in host-associated lineages. Genome Biol. Evol. 8:2006–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaf-van Bloois L, et al. 2014. Inconsistency of phenotypic and genomic characteristics of Campylobacter fetus subspecies requires re-evaluation of current diagnostics. J. Clin. Microbiol. 52:4183–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick D, Calvo JM, Klopotowski T, Ames BN. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S, Greenwood JR. 1985. Isolation of Campylobacter fetus from a pet turtle. J. Clin. Microbiol. 21:260–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DJ. 2008. Complexity and versatility in the physiology and metabolism of Campylobacter jejuni In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington D.C: ASM Press; p. 41–61. [Google Scholar]
- Lewis JA, Horswill AR, Schwem BE, Escalante-Semerena JC. 2004. The tricarballylate utilization (tcuRABC) genes of Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 186:1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan JM, Burnens A, Linton D, Lawson AJ, Stanley J. 2000. Campylobacter lanienae sp. nov., a new species isolated from workers in an abattoir. Int. J. Syst. Evol. Microbiol. 50:865–872. [DOI] [PubMed] [Google Scholar]
- Merga JY, Winstanley C, Williams NJ, Yee E, Miller WG. 2013. Complete genome sequence of the Arcobacter butzleri cattle isolate 7h1h. Genome Announc. 1:e00655–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, et al. 2012. Multilocus sequence typing methods for the emerging Campylobacter species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus. Front. Cell Infect. Microbiol. 2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Yee E, Huynh S, Chapman MH, Parker CT. 2016a. Complete genome sequence of the Campylobacter iguaniorum strain RM11343, isolated from an alpaca. Genome Announc. 4:e00646–e00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Yee E, Chapman MH. 2016b. Complete genome sequences of Campylobacter hyointestinalis subsp. hyointestinalis strain LMG 9260 and Campylobacter hyointestinalis subsp. lawsonii strain LMG 15993. Genome Announc. 4:e00665–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- On SL, Holmes B, Sackin MJ. 1996. A probability matrix for the identification of campylobacters, helicobacters and allied taxa. J. Appl. Bacteriol. 81:425–432. [DOI] [PubMed] [Google Scholar]
- Patrick ME, et al. 2013. Human infections with new subspecies of Campylobacter fetus. Emerg. Infect. Dis. 19:1678–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez GI, Hopkins JA, Blaser MJ. 1985. Antigenic heterogeneity of lipopolysaccharides from Campylobacter jejuni and Campylobacter fetus. Infect. Immun. 48:528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U S A. 106:19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB. 1985. Enrichment and isolation of rumen bacteria that reduce trans-aconitic acid to tricarballylic acid. Appl. Environ. Microbiol. 49:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. [DOI] [PubMed] [Google Scholar]
- Sheppard SK, et al. 2014. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol. Ecol. 23:2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MC. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237–239. [DOI] [PubMed] [Google Scholar]
- Thomas MT, et al. 2011. Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L‐lactate. Environ. Microbiol. 13:48–61. [DOI] [PubMed] [Google Scholar]
- Tu ZC, et al. 2004. Campylobacter fetus of reptile origin as a human pathogen. J. Clin. Microbiol. 42:4405–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CM, Shia WY, Jhou YJ, Shyu CL. 2013. Occurrence and molecular characterization of reptilian Campylobacter fetus strains isolated in Taiwan. Vet. Microbiol. 164:67–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.