Abstract
We developed a computational pipeline for homology based identification of the complete repertoire of olfactory receptor (OR) genes in the Asian honey bee species, Apis florea. Apis florea is phylogenetically the most basal honey bee species and also the most distant sister species to the Western honey bee Apis mellifera, for which all OR genes had been identified before. Using our pipeline, we identified 180 OR genes in A. florea, which is very similar to the number of ORs identified in A. mellifera (177 ORs). Many characteristics of the ORs including gene structure, synteny of tandemly repeated ORs and basic phylogenetic clustering are highly conserved. The composite phylogenetic tree of A. florea and A. mellifera ORs could be divided into 21 clades which are in harmony with the existing Hymenopteran tree. However, we found a few nonorthologous OR relationships between both species as well as independent pseudogenization of ORs suggesting separate evolutionary changes. Particularly, a subgroup of the OR gene clade XI, which had been hypothesized to code cuticular hydrocarbon receptors showed a high number of species-specific ORs. RNAseq analysis detected a total number of 145 OR transcripts in male and 162 in female antennae. Most of the OR genes were highly expressed on the female antennae. However, we detected five distinct male-biased OR genes, out of which three genes (AfOr11, AfOr18, AfOr170P) were shown to be male-biased in A. mellifera, too, thus corroborating a behavioral function in sex-pheromone communication.
Keywords: genome-wide survey for ORs, phylogeny of ORs, eusociality, Hymenoptera, transmembrane helix prediction, antennal transcriptome
Introduction
Honey bees are the most important pollinators for global food supply and commercial food production. Recent losses in commercially kept colonies of the Western honey bee, A. mellifera led to an extensive research on environmental and agricultural risks affecting honey bee health (Evans and Schwarz 2011; Berenbaum 2014). Unfortunately, basic and applied research mainly focused on A. mellifera, neglecting all the other honey bee species which mainly occur in the Asian tropics (Arias and Sheppard 2005; Oldroyd and Wongsiri 2006; Lo et al. 2009). Given the predicted human population growth and its impact on food supply in Asian countries, an intensification of research on Asian honey bee species is highly desirable.
Besides the economical importance, the A. mellifera has been a successful model system in the study of sensory and behavioral capabilities as well as communication and social organization in insects (Frisch 1965; Seeley 1995; Zayed and Robinson 2012; Giurfa 2015). Particularly, olfaction plays an important role in honey bee life, in both finding food sources and social communication (Frisch 1965; Bortolotti and Costa 2014). Given the behavioral importance of odors, A. mellifera exhibits a well-developed olfactory system comprising 177 olfactory receptor (ORs) genes and similar number of corresponding olfactory glomeruli in the first brain neuropiles, antennal lobes (Galizia et al. 1999; Robertson and Wanner 2006; Smith, Zimin, et al. 2011; Smith, Smith, et al. 2011; Brill et al. 2013; Kropf et al. 2014).
In addition, females and males (drones) show a pronounced sexual dimorphism in the olfactory system. Males exhibit an enlarged sex-pheromone sensitive olfactory system and reduced specific components of the system present in females (Esslen and Kaissling 1976; Arnold et al. 1985; Brockmann and Brückner 1998, 2001; Sandoz 2006; Kropf et al. 2014). Wanner, Nichols, et al. (2007) successfully used this sexual dimorphism to identify the first olfactory receptor gene (AmOr11) in honey bees. AmOr11 binds 9-oxo-2-decenoic acid (9-ODA), the so-called queen substance, which functions as a sex-pheromone in mating behavior and as a component of the queen signal in colony integration (Free 1987; Bortolotti and Costa 2014). More recently, two additional olfactory receptors (AmOr151, AmOr152) with higher expressions in females were identified to bind floral odorants. AmOr151 binds linalool, a major odorant component of many plants (Claudianos et al. 2014).
As all honey bee species are thought to be generalist flower-visitors (Oldroyd and Wongsiri 2006), the interesting question arises how similar or different their olfactory systems are? So far, there are no comparative studies on differences in olfactory perception of floral odorants, but there is emerging evidence that there are changes in pheromone communication. For example, sex-pheromone communication likely became more complex in A. mellifera compared with other honey bee species (Brockmann and Brückner 1998, 2001, 2003; Nagaraja and Brockmann 2009; Bastin et al. 2014). With genome sequences available for Apis florea, Apis cerana, and Apis dorsata, comparative studies on OR genes could be a good starting point for a detailed research on olfaction in Asian honey bee species.
Among Asian honey bee species, A. florea, is an interesting species to start the comparative research, because it is phylogenetically the most basal extant honey bee species (Alexander 1991). So far (by the end of 2014), automated annotation identified about 100 ORs for A. florea. In contrast, Pilot anatomical studies of the antennal lobe of A. florea suggested a similar number of glomeruli in A. florea and A. mellifera (Brockmann and Brückner 2001; Brockmann A, personal communication). Given the correlation between glomeruli number and OR genes, we hypothesize that A. florea should also have a similar number of ORs as A. mellifera.
Identification and annotation of insect ORs is complicated for several reasons. First, many insects and, particularly, Hymenopteran species (wasps, ants and bees) have a huge number of OR genes (Robertson and Wanner 2006; Kirkness et al. 2010; Robertson et al. 2010; Nishimura et al. 2012; Zhou et al. 2012; Gress et al. 2013; Yin et al. 2013; Engsontia et al. 2015). Second, different from vertebrate ORs, insect ORs contain hardly any of the characteristic GPCR motifs (Benton et al. 2006). Third, there is evidence that many OR families follow a birth and death model of evolution, so that there are many lineage specific expansions and deletions (Nei and Rooney 2005; Zhou et al. 2012, 2015; Engsontia et al. 2015). As a consequence, direct orthologous relationships are rare among closely related taxonomic families and any easy extrapolation from model systems like Drosophila ORs is not possible.
Here, we report the identification of ORs on the scaffolds of A. florea genome. Our biology-informed comprehensive annotation of the OR genes of A. florea suggests a total number of 180 ORs. They were extensively validated using known gene models, transmembrane helix prediction methods and domain analysis. RNA expression analysis confirmed good coverage of the ORs attained through this genome-wide survey. We further performed phylogenetic reconstruction of the ORs from A. mellifera and A. florea, to detect perfect orthologous copies or species-specific characteristics. Finally, differential expression of the ORs between female and male antennae supports putative pheromone function of specific receptors.
Materials and Methods
Genome-Wide Survey
Odorant/Olfactory/chemosensory receptor protein sequences were collected from National Center for Biotechnology Information (NCBI) RefSeq (Pruitt et al. 2012). Sequences with less than 100 or more than 600 amino acids were not considered. Redundant sequences at 95% or more identity were discarded using CD-HIT (Li and Godzik 2006). Of these representative sequences, ones with 7tm_6 (PF02949—7 transmembrane Drosophila like odorant receptors) Pfam family signature were retained (Finn et al. 2014). Remaining sequences were manually curated for odorant receptor gene ontology (GO) annotations in UniProt and NCBI and included into the previous data set (Pruitt et al. 2012; The UniProt Consortium 2014). Sequences with uncertain functions (such as the ones with 7tm_7 Pfam domain–chemosensory receptors comprising both olfactory and gustatory receptors) were removed. Finally, a curated insect OR protein data set of 2,382 sequences was prepared. In addition to these, 394 OR gene sequences from species belonging to genus Apis were retrieved from NCBI (Maglott et al. 2010). AmOr protein sequences were collected from the authors (Robertson and Wanner 2006; Smith, Zimin, et al. 2011; Smith, Smith, et al. 2011).
OR protein sequences were aligned to the A. florea genome sequence [Aflo_1.0 from NCBI GenBank with permission from The Honey Bee Genome Sequencing Consortium (HBGC)] using tblastn at the E-value cutoff of 10 −5 and nucleotide sequences for Apis OR genes were aligned using blastn with E-value cutoff of 10 −10 (Altschul et al. 1997; Gertz et al. 2006; Elsik et al. 2015). Information from both the resources was integrated to extract putative OR containing regions from the genome along with their approximate exonic regions. In cases of multiple queries aligning and overlapping the same region on the genome, maximal region obtained from all the alignments was chosen. Exonerate protein2genome module was used to generate alignments of 177 A. mellifera OR sequences (AmOr) on these selected genomic regions, allowing for maximum intron size to be 2,000 and 10,000 (Slater and Birney 2005; Robertson and Wanner 2006; Smith, Zimin, et al. 2011; Smith, Smith, et al. 2011). For every AmOr, best scoring alignments were chosen from the hits. If the proteins remained incompletely aligned due to the stringent criterion of 2,000 intron size, the alternate option of distant exons was chosen to complete the genic regions, wherever possible. If the exons were either too distant or if the gene models were still incomplete, association to distantly related ORs was exploited to finalize such gene models, so long as there was high coverage to the genomic region under question. The putative OR gene containing regions recognized from the BLAST hits but not through good Exonerate hits, were separately re-examined.
The putative OR gene containing regions were further manually refined for the gene and intron–exon boundaries. Partial sequences were completed with the nearest START and/or STOP codons wherever possible. Genes found in the A. florea were named as AfOr followed by the number/s of the closest homologue from A. mellifera genome. Pseudogene-like sequences, identified using in-frame STOP codons or frameshifts, were suffixed with letter “P”. Gene names of the truncated sequences with only N-termini were suffixed with letter “N” and those with only C-termini were suffixed with letter “C”. Genes lacking both the termini and/or exons were suffixed with letter “F”. Probable amino acid sequences of the genes and pseudogenes were predicted from their intact codons in the predicted exonic regions, with STOP codons and frameshifts substituted by letter “X”. Sequence positions containing unknown amino acids were denoted with letter “Z”. Scaffolds with high representation of ORs from A. florea and their corresponding A. mellifera scaffolds were studied using Integrated Genomics Viewer (IGV) for their synteny (Robinson et al. 2011; Thorvaldsdóttir et al. 2013).
Validation of Predicted OR Sequences Using Sequence Similarity, Number of Exons, Transmembrane Helix Prediction, Motifs and Domains
AfOr protein sequences, obtained from GWS, were queried against NCBI-NR database to discover the closest non-A. florea homologues and their average identity with the closest orthologue was calculated. Similarly, the complete sequences between A. mellifera and A. florea were compared for their identities.
Ten highly specific motifs in the AfOrs were designed using default MEME parameters trained with negative set of A. mellifera gustatory receptor sequences. These were compared with the motifs from AmOrs (Bailey and Elkan 1994; Miller and Tu 2008; Bailey et al. 2009). Pfam-based protein family annotation was performed on AfOrs and compared with those of AmOrs. In cases of no Pfam family connections, Interpro-scan and CD-search were employed to further identify protein domains (Marchler-Bauer et al. 2011; Jones et al. 2014).
AfOrs and AmOrs were also subjected to transmembrane helix (TMH) prediction using TMHMM, HMMTOP and PolyPhobius (Sonnhammer et al. 1998; Tusnády and Simon 1998, 2001; Krogh et al. 2001; Käll et al. 2005, 2007). If any two of the three methods predicted an amino acid to be a part of a helix, it was reported as part of a helix using consensus method (Nagarathnam et al. 2014). Consensus transmembrane helix predictions for AfOrs were compared with those of AmOrs on the basis of the number of helices predicted. Perfect and complete 126 orthologous pairs of ORs from the two species were used to compute correlation of transmembrane helix predictions between the two species. Consensus TMH predictions were mapped onto a composite A. mellifera and A. florea OR protein alignment described later to predict the topology of the Apis ORs. The gene models were studied and compared in terms of the number of exons and their lengths with other known ORs, primarily from A. mellifera.
Multiple Sequence Alignment of OR Sequences
The OR sequences from the Apis genomes were aligned using MAFFT 7 algorithm (E-INS-i strategy) and JTT200 matrix to maximize the alignment of probable multiple conserved domains within transmembrane helices interspersed with long gaps (Katoh and Standley 2013). Gaps were reduced by aligning gappy regions (with large inserts) as much as possible. AmOrco (AmOr2) and AfOrco (AfOr2), OR co-receptors, which are known to be at the root of the OR evolution in insects, were added later using MAFFT “-add” method (Katoh and Frith 2012). This helped to minimize the gaps induced due to few distant sequences in the alignment. The alignments were visually evaluated and edited to re-align partial sequences at their respective positions and to remove low quality positions using Jalview version 1.6.0_27 (Waterhouse et al. 2009). Representative ant OR sequences from Indian Jumping ant, Harpegnathos saltator (HsOrs), were included to this alignment using MAFFT “-add” method and “-keeplength” option to avoid introduction of new gaps (Katoh and Frith 2012; Zhou et al. 2012). The alignments were carefully studied for the conservation patterns and TMH predictions. Both alignments, with and without HsOrs, were used for phylogenetic reconstruction described in detail in the next section.
Available OR sequences from few representative or neighboring Hymenopteran species were included into the above alignment to observe clustering and patterns of conservation in the clades. These include Apis cerana—AcOrs (obtained from authors of Park et al. 2015), Apis dorsata—AdOrs (NCBI), Bombus terrestris—BtOrs (Sadd et al. 2015), Lasioglossum albipes—LaOrs (obtained by CD-search for 7tm-6 domain on predicted proteome from the genome, Kocher et al. 2013), Megachile rotundata—MrOrs (NCBI), Nasonia vitripennis—NvOrs (Robertson et al. 2010), Cerapachys biroi—CbOrs (NCBI). MAFFT was used with “-add” method, JTT200 matrix, “Auto” strategy and “-keeplength” option to maintain the original length of the alignment and remove gap-inducing regions from the added sequences (Katoh and Frith 2012).
Phylogenetic Tree Reconstruction
Two phylogenetic trees (with and without HsOrs) were reconstructed for OR protein alignments described in previous section using maximum likelihood method in RAxML version 7.4.2 with PROTCATJTT matrix and 100 bootstraps (Stamatakis 2006). AfOrco, AmOrco and HsOrco were specified as outgroups. Tree were visualized using FigTree (Rambaut and Drummond 2009). Phylogenetic tree was categorized into 22 clades based roughly on their gene models and the available Hymenopteran tree (Zhou et al. 2012, 2015). Properties of each clade were closely studied with respect to their gene models, loci, previous annotations of co-clustering ORs, ligand information, RNAseq expression data, MEME Motifs and conserved residues.
Alignment of representative Hymenopteran sequences was used to study rough clustering of the other Hymenopteran sequences with the known Apis OR sequences and their clades using MAFFT UPGMA method with 100 bootstrap (Kuraku et al. 2013). Co-clustering sequences from other species along with the previously defined clade X group a, clade XI, clade XVIII and clade XXI sequences were studies for their alignment and conserved residues.
Analysis of Transcription within Male and Worker Antennae (RNAseq)
Apis florea males and workers were collected from a colony, maintained at NCBS, Bangalore and were snap-frozen in liquid nitrogen. All bees were collected at the same time point to avoid any variation due to daily internal rhythm. Antennae were dissected on dry ice and 30 antennae were pooled for each sample. Total RNA was extracted from two biological replicates for both, male and female, using TRIzol reagent method (Invitrogen Cat. No. 15596-026) and stored in −80 °C after nanodrop quantification.
RNA was shipped on dry ice to Genotypic Technology’s Genomics Facility, Bangalore for sequencing. Further quality check was done on Bioanalyzer (Agilent) and mRNA was extracted from 1 μg of total RNA by Poly A purification. Library preparation was done following the “TruSeq RNA Sample Preparation Guide” (Part # 15008136; Rev. A; Nov 2010). Transcriptome sequencing was performed on an Illumina NextSeq500 platform and 150-bp-long paired-end reads were obtained.
Transcriptome data from female and male A. florea were assembled separately using Trinity (Grabherr et al. 2011; Haas et al. 2013). Full-length transcript analysis was performed twice on these transcripts using analyze_blastPlus_topHit_coverage.pl utility and two databases—one of AfOrs and another which is a composite of all available ORs from A. florea, A. mellifera, A. dorsata and A. cerana. During each run, blastp was used with E-value cut-off of 10 −20 and only the best hits were collected for each query. As few sequences within the OR subset are highly similar to one or more ORs, high coverage of the query in the alignment is important. For each analysis, all the hits with >80% coverage and sequence identity between the OR and the transcript were collected and named as “highly significant” hits. Hits with >50%, but <80%, coverage were named as “moderately significant” hits. Hits with <50% coverage were called “lowly significant hits”.
For quantitative differential gene expression analysis between females and males, reads were mapped to A. florea reference genome, GCF_000184785.1 (available at NCBI), using STAR (Dobin et al. 2013). Gene annotations were obtained from NCBI and a bed file with all the A. florea gene boundaries, including our newly predicted AfOr genes, was generated. Number of raw reads falling within start and stop position of each mRNA genes were counted using bedtools (Quinlan and Hall 2010) and differential gene expression analysis was done using DESeq (Anders and Huber 2010) package in R. Volcano plot was generated after removing 28 AfOrs, which had less than 10 supporting reads in either condition. Genes, for which absolute log2-fold change was ≥1.6 and P value is <10 −22, were labeled.
Results
Discovery of 180 ORs
A total of 180 OR loci were identified in A. florea genome (supplementary files S1 and S2, Supplementary Material online). Fifty-three of these are completely new predictions and were not detected in gene prediction in the A. florea automated annotation available on March 2014 in NCBI genome, which contained about 100 OR genes. Sixty-two of the total ORs found were modified from their previous gene annotations in terms of their gene boundaries or splice sites. A good set of the modified ORs were resolved and separated from their fused gene predictions. Few cases underwent exon additions/deletions, shortening/elongation based on their closest homologue from A. mellifera.
Twenty-one partial genes, including seven pseudogenes, were observed. The lengths of the near-complete ORs normally vary within the range 370–420 amino acids (similar to AmOrs, with few exceptions like AfOrco with length 477 residues). ORs are known to be diverse across insect orders, but the OR co-receptor (Orco) is essential for signaling process and is highly conserved across insects (Benton et al. 2006; Missbach et al. 2014). AfOrco is highly conserved and was found to be 99% identical to the AmOrco and A. cerana and A. dorsata Orco sequences.
Annotations and Orthologue Search of AfOrs
As discussed before, the AfOr names are based on their orthology with AmOrs. AfOr6/7, AfOr8/9, AfOr36/37/38, AfOr41/42 are clear cases, where two OR genes from A. mellifera show identity to only one OR from A. florea. AfOr47/48, AfOr74/86like_1, AfOr154/155P are similar to two ORs from A. mellifera, in addition to the perfectly orthologous copies found in A. florea. AfOr64_1, AfOr64_2, AfOr91_1, AfOr91_2, AfOr101_1C, AfOr101_2C, AfOr112_1, AfOr112_2, AfOr151_1, AfOr163_1, AfOr163_2, AfOr166_1, AfOr166_2PC are the genes, where A. florea has two copies for a single OR in A. mellifera. There are peculiar cases, particularly for AmOr122 to AmOr138, for which complex orthologous relationships emerge—showing different parts of a single AmOr with better identity to different AfOrs. Hence, such AfOrs acquired complex names. Orthologous sequences for AmOr33, AmOr63, AmOr78, AmOr81, AmOr92, AmOr93, AmOr109, AmOr145 could not be found. AfOr178 and AfOr179 are few additional sequences.
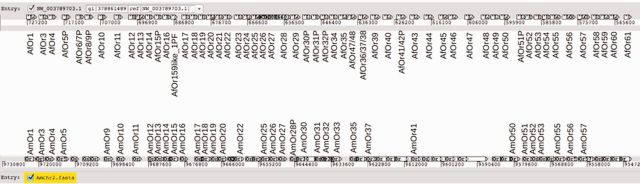
Most of the AfOrs were found to be present in tandem arrays, concentrated on only 30 out of the total 7,946 scaffolds. Homologous to chromosome 2 of A. mellifera, scaffold NW_003789703.1 of A. florea houses the maximum number of ORs, amounting to total 57. Comparison of order of these 57 with available OR gene information for chromosome 2 of A. mellifera confirms synteny (fig. 1). Next big cluster of ORs is present on NW_003791127.1 with 27 members which are orthologous to ORs present on chromosome 12, chromosome 15 and scaffold NW_003378215.1 of A. mellifera.
Fig. 1.—
Scaffold with 57 Apis florea OR genes and their synteny with known A. mellifera OR genes. Upper horizontal line represents A. florea scaffold NW_003789703.1 with 57 AfOrs. Horizontal line below represents chromosome 2 of A. mellifera with known AmOr genes mapped. Conserved synteny between the two species can be observed.
Apis florea has total 31 pseudogenous ORs (supplementary file S2, Supplementary Material online). Out of these, five show multiple stop codons and frameshift mutations. The remaining 26 AfOrs show only one or two frameshift mutations. Most of these have been edited at the mRNA level and reported as normal genes in the recent automated A. florea annotation report by NCBI (Annotation release 101), reasons behind which remain unknown. The comparison of the complete pseudogenes across the two species shows that only Or97 and Or173 are pseudogenized in both the species. Out of the orthologues of the remaining six A. mellifera OR pseudogenes, AfOr139like_1PF and AfOr159like_1PF1 are partial and may or may not be pseudogenes, based on their missing fragments. AfOr82P, AfOr119P, AfOr151_2PN, AfOr159like_1PF, AfOr176P and AfOr179P show more than one element of pseudogenization, but none of their orthologues in the A. mellifera genome are pseudogenes. From A. mellifera AmOr92PSE, AmOr93PSE and AmOr139PSE show extreme pseudogenization, but they do not find high identity single orthologues in the A. florea genome. Multiple exons of the AmOr92PSE and AmOr93PSE find orthology to different exons of AfOr91_1, AfOr91_2 and some other neighboring genes. Similarly, AmOr139PSE retains multiple exons which share similarity to different genes like AfOr131like_2 and AfOr136like_1.
Computational Validation of the OR Genes
Similarity to Other Known ORs
The average identity of AfOrs with the best non-A. florea homologues was ∼84% and the median was 90%. The hits were mainly from Hymenoptera and specifically more abundant in Apis. As many ORs are yet to be named according to their function, best hits for around 60 AfOrs were found to be named as uncharacterized proteins. When complete AfOr sequences were compared with the curated set of only A. mellifera ORs, average identity was 88% and median was 91%, demonstrating the close homology of the AfOr gene models.
The number and length of exons in the gene models holds evolutionary information and have been reflected in the phylogenetic clustering of ORs, e.g., most ORs from clade XI contain nine exons (supplementary file S2, Supplementary Material online).
TMH Prediction
Consensus transmembrane helix prediction shows similar trend in both A. mellifera and A. florea homologues (supplementary files S3 and S4, Supplementary Material online) with more than 60 ORs predicted to have seven transmembrane helices. The next highly populated category is of sequences with six transmembrane helices; largely due to the missing last transmembrane helix as seen in the comparison with transmembrane helix predictions for AmOrco and AfOrco sequences (supplementary files S4 and S5, Supplementary Material online).
General topology of the Apis ORs predicted from the analysis is: N-terminus intracellular region of around 25–30 amino acids with very few exceptions with longer lengths—TMH1—1st extracellular short loop of around 10 amino acids—TMH2—1st intracellular loop of around 35 residues—TMH3—2nd extracellular slightly variable loop of around 25–35 residues—TMH4—2nd intracellular variable loop of about 40–65 residues (long loop of 120 residues in Orco)—TMH5—3rd extracellular short loop of about 8–15 residues—TMH6—3rd intracellular loop of 40–50 residues (sometimes interrupted by additional TMH region, e.g., in few clade XI ORs)—TMH7—extracellular region. Some clade-specific differences observed in the TMH predictions are discussed later.
Motif and Domain Comparison
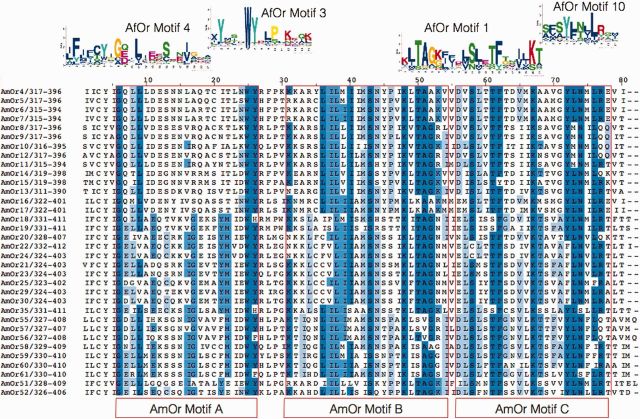
The AfOr motifs obtained by MEME analysis show great similarity to the AmOr motifs in the literature (fig. 2 and supplementary file S6, Supplementary Material online) (Miller and Tu 2008). All the 10 motifs show good E-value, but their distribution across 180 ORs is variable. Usual order of motifs from N terminus to C-terminus is 5-7-9-2-6-8-4-3-1-10. Motifs are clustered more towards C-terminus. Among the motifs, C-terminal ones are more conserved within OR sequences, whereas the N-terminus motifs tend to be more clade-specific (supplementary file S6, Supplementary Material online). Motif 4 and 3 are two consecutive motifs present towards C-terminus on Apis ORs which are present at 158 and 157 sites/ORs, respectively. Motif 3 is also characterized by the most conserved WY motif predicted to be part of third intracellular loop. The next most conserved motif is motif 2 which is located towards the end of fourth transmembrane helix and spanning the start of second intracellular loop. Motif 1 with the maximum confidence (2.9e-941) is present at the end of third intracellular loop and start of the seventh helix and is located at 120 sites. Detailed information on the 10 motifs and their distribution across AfOr sequences can be obtained from Supplementary material (supplementary file S6, Supplementary Material online).
Fig. 2.—
AfOr motifs with similarity to AmOr motifs. C-terminus alignment of few AmOrs is highlighted with the motifs predicted in literature. AfOr predicted motifs matching AmOr motifs are placed at the top.
Analysis of Composite Apis mellifera and Apis florea OR Alignment and Phylogeny
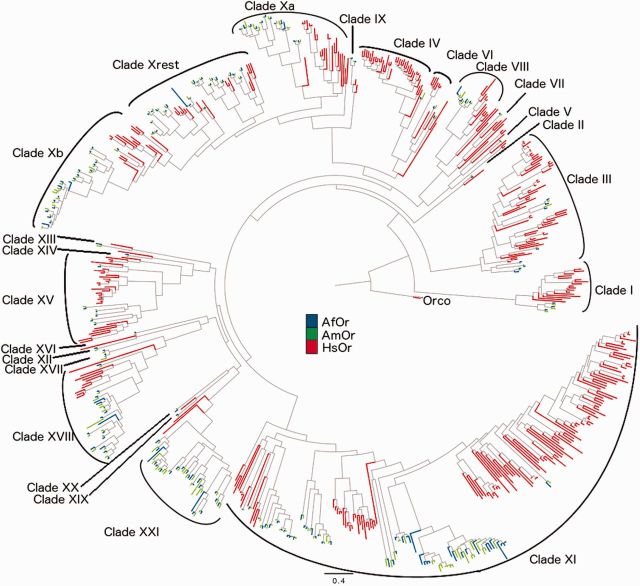
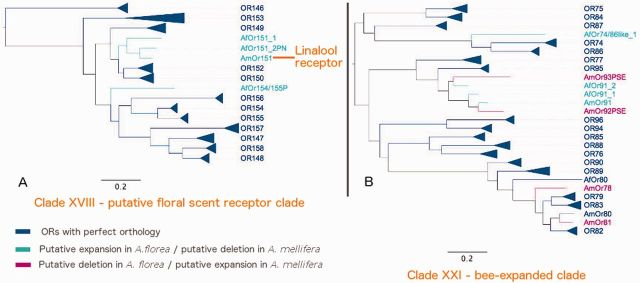
The alignment of the AmOrs and AfOrs reveals that only one position is thoroughly conserved in all the sequences, which is W in WY motif near the C-terminus (supplementary file S7, Supplementary Material online). Overall, C-terminus shows more conservation than the N-terminus. The phylogenetic tree has been divided into total 22 clades depending on their clustering and gene structure (fig. 3, table 1 and supplementary file S8, Supplementary Material online). It is compared with the Hymenopteran OR tree containing members from one bee (A. mellifera—AmOrs), one wasp (N. vitripennis—NvOrs) and four ants (L. humile—LhOrs, H. saltator—HsOrs, P. barbatus—PbOrs, C. floridanus—CfOrs), which was shown to have 24 well-supported subfamilies (Zhou et al. 2012). As many of these subfamilies (especially the basal members) are not expanded in Apis, as opposed to the other ant species, they contain very few members and also show less bootstrap support. These were still classified as separate clades for the purpose of maintaining uniformity and ease of comparison. Few of these new orphan clades were comparable to new subfamilies found in the recent analysis with 30 clades and have been thus included in the table 1 (Zhou et al. 2015). Results associated with N. vitripennis should be taken cautiously as the Hymenopteran tree does not cover/support about 80 out of 301 NvOrs. Results from recent phylogenetic co-clustering of ORs from Cephus cinctus (CcinOrs), B. terrestris (BtOrs), M. mediator (MmedOrs), A. cerana (AcOrs) with A. mellifera were also compared and included in the analysis (Gress et al. 2013; Sadd et al. 2015; Wang et al. 2015). Observations for few clusters are recorded in the discussions. Some interesting clusters include Clade X with putative pheromone receptors (fig. 4A), Clade XI with species-specific putative CHC receptors (fig. 4B), Clade XVIII with putative floral scent receptors (fig. 5A) and Clade XXI with unique bee-expanded set of ORs (fig. 5B).
Fig. 3.—
Composite phylogenetic tree of ORs from A. florea, A. mellifera and Harpegnathos saltator.
Table 1.
List of 22 clades corresponding to the phylogeny in Figure 3
| Clade name | Hymenopteran subfamilya | No. of ORs from A. florea | Apis orthologs belonging to the clade | No. of exons in AfOr genes | Expression support for females and males in A. florea |
|---|---|---|---|---|---|
| Orco | Orco | 1 | Or2 | 8 | Highly expressed in both |
| I | A | 3 | Or168–170 | 5 | AfOr170P—Females > Males, others in both |
| II | I | 1 | Or161 | 7 | Both |
| III | V | 8 | Or163–167 and Or118 | 6 | Slightly more enrichment in females than in males, except for AfOr164P |
| IV | U | 1 | Or121 | 7 | Both |
| V | Q | 1 | Or160 | 6 | Both |
| VI | T | 2 | Or114–115 | 6 | Both |
| VII | M | 1 | Or62 | 4 | Both |
| VIII | P | 5 | Or63–67 | Mostly 4 | Females>Males |
| IX | K | 2 | Or1 and Or3 | 6 | Both |
| X group a | L—putative pheromone receptors | 12 | Or4–17 | 5 | AfOr11—Males>Females. AfOr12—Females>Males. Most ORs show Males>Females |
| X group b | L | 19 | Or26–28, Or31–34 and Or36–50 | 4–5 | Both |
| X group rest | L | 23 | Or18–25, Or51–61, Or29,30 and Or35 | 5–6 | Mixed |
| XI | 9 exon—putative CHC receptors | 50 | AmOr97–113, AmOr122–138, AmOr140, AmOr159, AmOr162, AmOr172–177 and related A. florea homologs. | Mostly 9. | Few AfOrs expressed with good support only in females—e.g., Apis florea homologs similar to AmOr122–138. All AfOrs except AfOr162 show Females>Males. |
| XII | F | 1 | Or171 | 5 | Both |
| XIII | B | 1 | Or119 | 7 | Both |
| XIV | C | 1 | Or116 | 7 | Both |
| XV | E | 6 | Or68–73 | 5 | Both |
| XVI | Z | 1 | Or141 | 5 | Both |
| XVII | G | 2 | Or143–145 | 4–5 | AfOr143—Males>Females |
| XVIII | H—putative floral scent receptors | 16 | Or142 and Or146–158. | Mostly 6 | AfOR158 and AfOr154—Females>Males. Exception of AfOr155, highly abundant in males than in females. |
| XIX | W | 1 | Or120 | 5 | Males>Females |
| XX | Orphan | 1 | Or117 | 9 | Females>Males |
| XXI | J—bee expanded clade | 21 | Or74–96 | Mostly 6 | Most ORs show Females>Males except AfOr91_1. |
aFrom Zhou et al. 2012, 2015.
> indicates comparison of expression levels between two sexes for ORs in the clade.
“Both” denotes that the corresponding ORs are expressed in both sexes at similar transcription levels.
“Mixed” denotes that the pattern of expression across males and females varies for different ORs within the same clade.
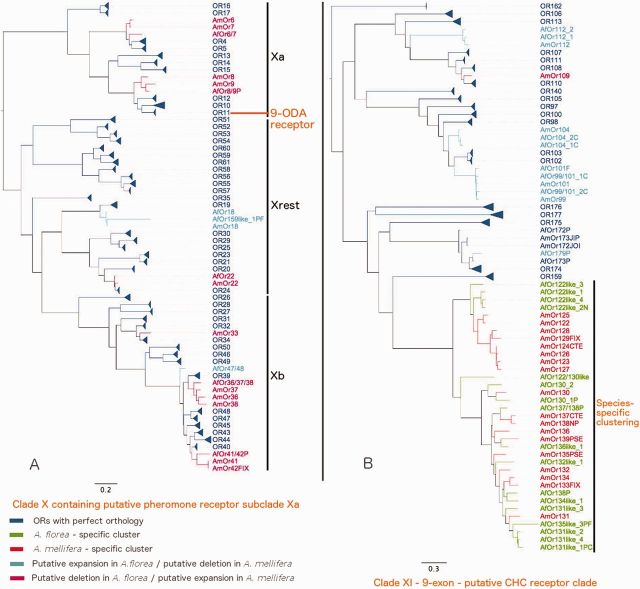
Fig. 4.—
(A) Clade X (putative pheromone receptor clade) and (B) Clade XI (9-exon—putative CHC receptor clade) from composite phylogenetic tree of AfOrs and AmOrs.
Fig. 5.—
(A) Clade XVIII (putative floral scent receptor clade) and (B) Clade XXI (bee-expanded clade) from composite phylogenetic tree of AfOrs and AmOrs.
RNA Expression Analysis in Support of OR Expression
The total number of ORs showing expression (ranging from <10% coverage to >90% coverage) changes from males to females with 145 and 162 ORs, respectively. The “highly” significant (>80% coverage and identity), “moderately” significant (>50% coverage) and “lowly” significant (>10% coverage) categories, found through transcriptome assembly, can also roughly correlate with the expression levels of each OR. As the lowly significant ORs might lead to mis-annotation, they were considered as absent. About 64 and 72 AfOrs show high expression support and 32 and 45 show moderate expression support in males and females, respectively. Combined together, 94 genes have high expression support and additional 36 genes show moderate support in either or both cases, leading to total 130 AfOrs with moderate to high expression support. About 31 of the 53 new genes show high expression support by either male or worker assembly, which again validates the GWS analysis. In addition to these, three transcripts show high support of expression with ORs from other species—A. mellifera and A. dorsata. These might be completely new genes, not covered or found in the assembly, or they might be different isoforms of the existing ones. For example, one of these three transcripts shows high similarity to AmOr101, orthologue of which is partially covered in the existing scaffold NW_003796907.1 of A. florea and hence named as AfOr101F. Out of the other two transcripts, one was closely related to XP_006615191.1_OR_13a-l_iX4_Ad in turn to AfOr31 and the other was closely related to XP_006625152.1_OR_9a-l_iX1_Ad and in turn to AfOr160. All these are likely to be isoforms of the already defined ORs, as they have very high similarity to the other ORs. Hence these were not included as separate ORs in the subsequent analysis.
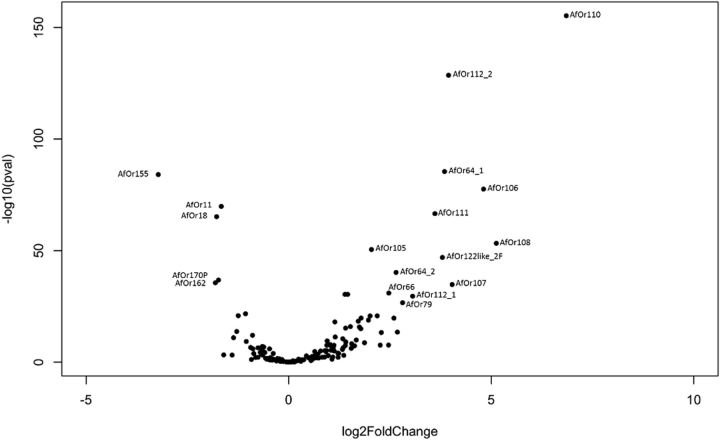
In the differential expression analysis, overall there are more number of genes which are highly expressed in females than in males (fig. 6). AfOr155, 162, 18, 170P, 11, 143, 164P, 120, 91_1, 32P are highly expressed in males and AfOr110, 138P, 172P, 131like_4, 131like_2, 132like_1, 134like_1, 131like_1PF, 175, 108, 106, 159C, 107, 112_2, 64_1, 122like_2F, 111, 112_1, 79, 158, 64_2, 98C, 66, 131like_3, 61F, 83, 65, 105, 40, 74, 97P, 80, 167, 90, 99/101_2F, 154, 117, 113, 101F, 166_1, 104_1C, 166_2PC, 163_2, 82P, 104_2C, 74/86_like1, 89, 69, 165, 12, 41/42P, 100, 67, 51P, 84, 47, 95, 43 are highly expressed in females (log2-fold > 1 and P value < 0.00001).
Fig. 6.—
Differential expression of ORs across female versus male antennae. Only ORs with highly significant differential expression are labeled.
Discussion
Identification of the Complete OR Repertoire of A. florea Using a Computational Pipeline
The principle significance of our study is that it demonstrates that automated annotations of OR repertoires miss to identify a huge number of candidate OR genes. For A. florea, we identified almost double the number of ORs (=180) compared with the first-pass automated annotation published in NCBI (=100). Our study emphasizes the importance of using more specialized genome-wide survey techniques for the resolution of gene models. Furthermore, dedicated search for tandemly duplicated proteins, accompanied with manual intervention is needed to resolve the gene and intron–exon boundaries. This procedure is especially useful for those proteins, which have recently undergone species-specific duplications.
We detected few partial genes, some of those might be ascribed to gaps in the scaffold level assembly and the full gene sequence would be resolved only through subsequent genome assembly revisions. Other partial genes might be remnants of the ancestral genes or might contribute to form alternative isoforms with the help of other exons from neighboring OR genes. Transcriptome data provides moderate to high expression support (>50% coverage of the gene) for 130 of 180 AfOrs. This analysis confirms that the OR set found through the analysis of genome is nearly complete. On the other hand, it demonstrates that transcriptomic data alone is not sufficient for complete coverage of entire OR gene repertoire.
The Total Numbers of ORs in Apis florea and Apis mellifera Are Similar
The number of ORs identified in A. florea via our genome-wide survey (180) is similar to the number of A. mellifera ORs (177) (supplementary file S2, Supplementary Material online). Furthermore, number of putative gene duplications and deletions in each OR clade matched, so that the numbers of OR genes in all clades are similar (maintained) in both species (supplementary file S8, Supplementary Material online).
The majority of OR genes of both species show a high sequence identity, a conservation of number of predicted TMHs and conserved motifs, which suggests that the OR-proteins likely bind similar odorants. Evolution of species-specific ORs occurred in only 1 (clade XI) of the 22 clades (fig. 4B). The sequences of the genes ascribed to clade XI show the least degree of identity between the two species. This finding nicely fits with the idea that these genes code for ORs binding cuticular hydrocarbons, which function as species-specific recognition signals in solitary and social insects (Howard and Blomquist 2005; Zhou et al. 2012; Sharma et al. 2015). The sequences of these genes were difficult to detect and refine, which in turn made it difficult to assign clear orthology to AmOr genes. As consequence, we decided to address some of the genes with complex names (e.g., AfOr122like_1, AfOr122like_2F and AfOr122/130like_1). Although perfect orthologous relationships were not found for these AfOrs, again the total number of AfOrs in this clade is similar to that in A. mellifera. Thus, one of the most surprising results of our study is the consistency of the total number of ORs and the number of ORs within the different gene-clades independent of evolutionary gains and losses of single ORs and profound sequence changes.
Our finding suggests that the number of olfactory genes in closely related species is similar, not because the same genes are conserved, but because their number might be constrained. If this is true, the question arises which organismal mechanisms may constrain the number of olfactory genes in the genome. In this respect, it is interesting that preliminary anatomical studies suggest a similar number of olfactory glomeruli in the antennal lobes, the first olfactory brain neuropils, of both species (Brockmann and Brückner 2001; Brockmann A, personal communication).
Synteny of ORs is Conserved Across A. florea and A. mellifera
Most of the tandemly duplicated ORs show similar synteny as those of A. mellifera (fig. 1). However, ORs on scaffold NW_003791127.1 of A. florea show orthology to ORs spread on distinct chromosomes and an unplaced scaffold of A. mellifera. Considering that A. florea traits more likely resemble those of ancestral Apis species, our findings suggest that chromosomal split might have occurred during evolution of A. mellifera.
Independent Pseudogenization of ORs is Observed in A. florea and A. mellifera
We identified a large number of 31 pseudogenous ORs in A. florea compared with only 8 pseudogenes in A. mellifera (AmOr 28, 92, 93, 97, 139, 153, 159, 173). Only Or97 and Or173 are pseudogenized in both the species and might have arisen before the origin of the two species. However, both of these ORs show only one frameshift in A. florea and do not show severe degradation characteristic of evolutionary old pseudogenes. Hence the time of origin of these pseudogenizing elements could still be debated and the possibility of alternative splicing events or RNA editing circumventing the pseudogenous elements cannot be ruled out at this stage.
Pseudogenes from the two genomes are dispersed throughout the phylogenetic tree, including the species-specific clusters which originated recently as compared with the other clades. Hence, evolution of pseudogenes in the two genomes seems to be independent of each other for most of the ORs and these events could be very recent. This is in agreement with the rapid pseudogenization and gain and loss of genes usually seen in the OR protein families from both mammals and insects and fits the birth-and-death model of evolution (Guo and Kim 2007; Niimura and Nei 2007; Sánchez-Gracia et al. 2009; Zhou et al. 2012; Engsontia et al. 2014).
Phylogenetic Clustering Helps to Identify Characteristics of Each Clade
In the following, we summarize the important clade-wise observations of our study (table 1):
Orco
The olfactory receptor coreceptors (Orco) have been shown to be the origin of the other tuning ORs and are highly conserved across all insects (Krieger et al. 2003; Larsson et al. 2004; Benton et al. 2006; Missbach et al. 2014). Hence, the OR-coreceptors in A. florea and A. mellifera, AfOrco and AmOrco, respectively, were used as the outgroup for phylogenetic reconstruction. They form a very distant cluster from the other tuning ORs. AfOrco is the highest expressed OR in all the A. florea samples. They both also lack the Motif 10 (SYFT type) which is closest to the C-terminus.
Clade I
This clade contains Or168–170 present on chromosome 7 in A. mellifera. The sequences of these proteins are very different from the other ORs. AfOr168 and AfOr169 possess only three to four motifs, and AfOr170P has only motif 4. AfOr170 was predicted to be a pseudogene with a frameshift mutation but showed a high confidence transcription in both sexes according to the transcriptome assembly. Similar to its A. mellifera orthologue AmOr170 it was highly expressed in male than in female antennae (fig. 6 and supplementary file S2, Supplementary Material online; Wanner, Nichols, et al. 2007). In general, Or170 seems to be a very ancestral OR gene with homologous genes in ants. In ants, however, the expression of this gene is higher in worker than male antennae (Zhou et al. 2015).
Clade III
This clade contains Or163–167 and Or118. AmOr163 and AmOr166 find two very similar orthologues each in A. florea. In both cases, one out of the two copies in A. florea seems to have evolved earlier and clusters separately from the other A. mellifera and A. florea pair. AfOr166_2PC is one such putative ancestral gene, which is partial and a pseudogene. This OR might actually be isoform of the AfOr166_1 itself, which has recently undergone pseudogenization. These ancestral ORs might have been lost completely from the A. mellifera species. All of them show expression in females as well as males with usually slightly more enrichment in females than in males, except for AfOr164P (supplementary file S2, Supplementary Material online). This clade is highly expanded in ants with more than 40 members.
Clade IV
Clade IV, which corresponds to the rapidly expanding OR subfamily U in hymenopterans, comprises only one basal 7 exon gene (Or121) in both the bee species. The gene is expressed in antenna of both sexes.
Clade VI
This clade is also not as highly expanded in honey bees as in other Hymenopteran species, especially N. vitripennis. Both Or114 and Or115 show support for expression in both females and males.
Clade X
Clade X comprises OR genes Or4–Or61 (fig. 4A). In A. mellifera all these genes are lined up on chromosome 2, which houses altogether 60 ORs (AmOr1 to AmOr61 except AmOrco). Most of these genes belong to subfamily L in the Hymenopteran phylogenetic tree (Zhou et al. 2012, 2015). In A. florea, 57 orthologues of these ORs are arranged in the similar syntenic order on scaffold NW_003789703.1 (fig. 1).
We subdivided Clade X OR genes in three subgroups: Clade X group a, Clade X group b, Clade X “rest”.
Clade X group a comprises the genes AmOr4–17 and their A. florea orthologues. They have a 5 exon gene structure. AmOr11 is highly expressed in male antennae and was shown to bind the decenoic acid 9-ODA, the major component of the queen sex-pheromone, synthesized in the queen mandibular glands (Plettner et al. 1997; Brockmann et al. 2006; Wanner, Nichols, et al. 2007). Similar to A. mellifera, we also found a male-biased expression of AfOr11 in A. florea antennae (fig. 6). In addition, all other genes in this group (except AfOr5 and AfOr12) are also expressed at higher levels in male antennae, but with weak statistical support (fig. 6 and supplementary file S2,Supplementary Material online). Together, our results suggest that Or4–17 from honey bees are candidate receptors for a group of similar components of the queen mandibular glands, most likely the different 8- and 10-carbon functionalized fatty acids (Plettner et al. 1996). If this is true, all ORs in this group may be potential fatty acid receptors and might function as “pheromone receptors” in Hymenopteran or other insect species that have evolved fatty acids as communication signals (Blomquist and Howard 2003; Blomquist et al. 2011).
This subgroup includes 12 A. florea, 14 A. mellifera, 13 A. cerana, 12 A. dorsata, 6 B. terrestris, 21 H. saltator, 8 C. biroi ORs. Though the numbers of ORs are comparable for Apis species, the bumblebee (primitively eusocial) ORs are almost half in number. There was no representation of ORs from the two closest bees L. albipes (mainly primitively eusocial), M. rotundata (solitary) and the wasp N. vitripennis (solitary) in this subgroup according to their current OR protein data set. Patterns of conservation for this clade can be accessed from supplementary material (supplementary file S9, Supplementary Material online).
Clade X group b contains the Apis Or26–28, Or31–34 and Or36–50 which generally have 4–5 exon gene structure. In Hymenopteran tree this group includes HsOr55 (H. saltator), that was shown to respond in dose-dependent manner to 4-methoxyphenylacetone found in anise essential oil without any differential expression across sexes (Zhou et al. 2012). AfOrs in this group also show heterogeneous expression pattern (supplementary file S2, Supplementary Material online).
Remaining honeybee ORs—AfOr18–25, AfOr51–61, AfOr29 and AfOr35 do not form a single bootstrap-confident clade, but are kept together in Clade X group rest and show 5–6 exon gene structure.
Clade XI
This clade contains AmOr97–113, AmOr122–138, AmOr140, AmOr159, AmOr162, AmOr172–177 and related A. florea homologs. In the Hymenopteran tree, genes in this clade are characterized by nine exon gene structure shared by most of its members (Zhou et al. 2012). The clade is highly expanded in all the Hymenopteran ORs available, but especially in ants (e.g., 126 HsOrs, 276 CbOrs vs. 43 AmOrs, 41 AfOrs). The huge expansion of this family in ants together with a mostly worker-enriched expression led to the hypothesis that the OR genes code receptors for cuticular hydrocarbons which play a major role in ant social communication, e.g., nest mate recognition (Zhou et al. 2012).
Many of the ORs in this clade are species-specific, which likely leads to complication in automated computational annotation. For example, automated annotations for A. dorsata and A. cerana identified only 15 and 21 ORs, respectively. In contrast, we expect that the correct number of ORs in this clade in both species will be similar to those in A. mellifera and A. florea.
In honey bees, most of the genes of this clade are widely distributed over the genome. In A. florea the genes are found on scaffolds NW_003790919.1, NW_003789177.1, NW_003789977.1 and NW_003791794.1 and in A. mellifera on chromosome 4, chromosome 11, as well as other chromosomes.
The results of our phylogenetic analysis slightly differed from previous studies (Zhou et al. 2012). First, we added the genes Or172–177 which mostly show 9 exon gene structure to this clade, although with low bootstrap support. In addition, we newly included Or162 with 9-exon gene structure to this clade. It forms a distant outgroup (99% bootstrap) to most of the genes in this clade in the phylogenetic tree of only AfOrs and AmOrs (fig. 4B and supplementary file S8, Supplementary Material online).
On the basis of our phylogenetic analysis, one can distinguish two major gene groups in this cluster: a subgroup with perfect orthologous ORs in A. mellifera and A. florea, and a second subgroup which consists of many species-specific gene groups that do not have any direct orthologous ORs in the other species. The first group of genes seems to have evolved much earlier than the others and does not show any signs of rapid evolution, whereas some of the genes in the second group show rapid evolution (see, e.g., AmOr122–138 and the homologous AfOrs; fig. 4B). For ant ORs of this clade, particularly predicted transmembrane helix 3 and 6 appeared to show signs of positive selection and were suggested to be involved in evolving ligand specificity (Engsontia et al. 2015).
Our RNAseq analysis showed that most of the genes in this clade are highly expressed in female (worker antennae) (fig. 6 and supplementary file S2, Supplementary Material online). The top 15 genes with worker-biased expression all belong to this clade. Furthermore, many AfOrs of this clade, particularly those similar to AmOrs 122–138, appear to be not expressed at all in male antennae. This result corresponds with the reduction of specific sensilla on male antennae (Esslen and Kaissling 1976) and the reduction of regular-sized glomeruli in the antennal lobe (Arnold and Masson 1985; Brockmann and Brückner 2001).
Conservation pattern mapped onto alignment of this clade can be obtained from supplementary material (supplementary file S10, Supplementary Material online).
Clade XV
Clade XV comprises Apis Ors 68–73. The clade corresponds to the subfamily E of the Hymenopteran tree. All the genes show 5 exons and are present in single orthologous copies in both A. mellifera (chromosome 13) and A. florea (NW_003789264.1). Earlier studies concluded that this clade might have been reduced only in A. mellifera (Zhou et al. 2012), but our results now suggest that this reduction likely happened in all honey bees.
Clade XVIII
This clade contains Apis Or142 and Or146–158 and corresponds to subfamily H in the Hymenopteran tree (fig. 5A). All AfOrs in this clade, except AfOr148CP, show 6 exons. AfOr148CP is a partial pseudogene with multiple stop codons and frameshifts and possesses 8 exons.
AmOr151 and AmOr152 were reported to be recently duplicated and differentially spliced protein paralogs (Robertson and Wanner 2006). In A. florea, we found an additional paralog for AmOr151. We have named the two genes as AfOr151_1 and the pseudogene AfOr151_2PN. Similarly, an additional pseudogene with one frameshift mutation and similar to both AmOr154 and AmOr155 was identified and named as AfOr154/155P. Conservation pattern for this clade can be accessed from supplementary material (supplementary file S11, Supplementary Material online).
Physiological studies demonstrated that AmOr151 and AmOr152 respond to a variety of floral scents, e.g., linalool and nerol (AmOr151) and neral, myrcene and 6-methyl-5-heptene-2-one (AmOr152) (Claudianos et al. 2014). Based on these results and the trend that these ORs are generally highly expressed in workers compared with males, it was hypothesized that all the ORs of this clade might bind floral odor components (Zhou et al. 2015).
The RNAseq transcription analysis showed for one of genes of this clade, AfOr155, a strong male-biased expression. This strong male-biased expression suggests a behavioral function in mating behavior.
Clade XXI
This clade contains Or74–96 from A. florea and A. mellifera and corresponds to subfamily J in Hymenopteran tree (fig. 5B). Most of the AfOrs are six exon genes, respectively. All four honey bee species with the genome sequenced have similar numbers of OR genes in this clade (22 AmOrs, 21 AfOrs, 18 AdOrs, 17 AcOrs). Furthermore, numbers of genes in this clade are also similar in other bee species (17 BtOrs, 15 MrOrs, 12 LaOrs) whereas ants only have two or three genes in this clade and wasps (N. vitripennis) none. Thus, this clade seems to have expanded exclusively in bees (Zhou et al. 2015). The current data suggests that the common ancestor of bees has acquired multiple genes belonging to this clade through gene duplications. There also seems to be weak support for gradual increase in the OR numbers in further divergence from this common ancestor into various primitively eusocial to eusocial forms.
All AfOrs of this clade, except AfOr91_1, AfOr91_2 and AfOr86, displayed strong worker-enriched expression and half of them with statistical significance (P value < 0.00001). Alignment of ORs from this clade with highlighted conserved residues can be obtained from supplementary material (supplementary file S12, Supplementary Material online).
Detailed comparison of motif distribution and functionally important residues from four important clades (Xa, XI, XVIII and XXI) are discussed in supplementary material (supplementary file S13, Supplementary Material online).
Female (Worker) and Male Antennae Show Characteristic Differences in Olfactory Gene Expression
The major result of our RNAseq study is that worker antennae expressed a higher number of ORs and that in most of the cases the ORs in the female antenna showed higher abundance than their counterparts on the male antenna. This expression pattern nicely corresponds to the finding that in honey bees the male olfactory system is specialized for sex-pheromone detection and this specialization includes a reduction in the olfactory sub-systems involved in detecting odorants involved in worker activities (Esslen and Kaissling 1976; Brockmann and Brückner 2001; Bortolotti and Costa 2014).
Regarding the OR clades, we found that almost all OR genes of the putative CHC receptor clade showed higher expression in the female antenna (fig. 6 and supplementary file S2, Supplementary Material online) corroborating the idea that these OR genes play a major role in worker behavior and communication (Ozaki et al. 2012; Van Oystaeyen et al. 2014; Sharma et al. 2015).
Five OR genes, AfOr11, AfOr18, AfOr170P, AfOr155, and AfOr162, showed a distinct higher expression in male antennae (fig. 6 and supplementary file S2, Supplementary Material online). The A. mellifera orthologues of AfOr11, AfOr18, and AfOr170P also showed a higher expression in A. mellifera male antennae and AmOr11 was demonstrated to bind 9-ODA, the major component of the sex-pheromone. This conservation of male-biased expression supports studies indicating that all honey bee species use the same group of sex-pheromone components (Free 1987; Plettner et al. 1997; Brockmann et al. 2006; Nagaraja and Brockmann 2009).
Somewhat surprisingly AfOr155 (Clade XVIII) showed the highest expression difference between male and worker antennae. AfOr155 is closely related to AfOr151 and AfOr152 (orthologues of AmOr151 and AmOr152), which show a worker-biased expression in A. florea and A. mellifera and were shown to detect floral odors in A. mellifera (Claudianos et al. 2014). Unfortunately, there is no expression data on AmOr155 that would verify whether our results are specific for A. florea; nor are there any studies suggesting that A. florea males might be attracted to floral odors or that there is a sex-pheromone component that is also a component of flower scents (Free 1987; Oldroyd and Wongsiri 2006).
AfOr162 belongs to the XI-CHC clade that comprises putative cuticular hydrocarbon binding ORs. This finding is interesting as there is some evidence in A. mellifera that components of the queen’s tergal glands, which synthesize hydrocarbons, play a role in close range attraction and copulation activity of males (Renner and Vierling 1977; Smith et al. 1993).
In summary, we identified full repertoire of 180 ORs in the A. florea genome and established their orthologous relationships with the A. mellifera ORs. Our study is the first to compare OR genes in two closely related non-Drosophila insect species. As expected the total number of OR genes as well as the numbers in each OR clade are very similar between the two species. However, the conservation of gene number is not a result of simple orthologous relationships at all branches. Our findings raise the question whether OR gene numbers in closely related species might be constrained by some organismal mechanisms. Given our knowledge about honey bee behavior and the small number of species in this genus, honeybees might be a promising system to study OR evolution in a group of closely related species.
Supplementary Material
Acknowledgments
The authors acknowledge the Honey Bee Genome Consortium (HBGC) and the Baylor College of Medicine Human Genome Sequencing Center for making the A. florea genome and its automated annotations available before publication. The authors thank Hugh M. Robertson and Hyung Wook Kwon for sharing the OR sequences of A. mellifera and A. cerana, respectively. S.D.K. was supported by the Shyama Prasad Mukheerjee (SPM) fellowship funded by Council of Scientific and Industrial Research (CSIR) and R.J. by Indian Council of Medical Research (ICMR) fellowship. We also thank National Centre for Biological Sciences (NCBS) and Tata Institute of Fundamental Research (TIFR) for infrastructural facilities.
Literature Cited
- Alexander BA. 1991. Phylogenetic analysis of the genus Apis (Hymenoptera: Apidae). Ann Entomol Soc Am. 84:137–149. [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias MC, Sheppard WS. 2005. Phylogenetic relationships of honey bees (Hymenoptera:Apinae:Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol Phylogenet Evol. 37:25–35. [DOI] [PubMed] [Google Scholar]
- Arnold G, Masson C, Budharugsa S. 1985. Comparative study of the antennal lobes and their afferent pathway in the worker bee and the drone (Apis mellifera). Cell Tissue Res. 242:593–605. [Google Scholar]
- Bailey TL, et al. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37:202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36. [PubMed] [Google Scholar]
- Bastin F, Brandstaetter AS, Koeniger G, Koeniger N, Sandoz JC. 2014. Neuroethological study of pheromonal sex communication in honeybee drones. IUSSI 2014 Cairns Australia. Conference Abstract Book OR273.
- Benton R, Sachse S, Michnick SW, Vosshall LB. 2006. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum MR. 2014. Bees in crisis: colony collapse, honey laundering, and other problems bee-setting American apiculture. Proc Am Philos Soc. 158:229–247. [PubMed] [Google Scholar]
- Blomquist GJ, Howard RW. 2003. 11 – Pheromone biosynthesis in social insects In: Blomquist GJ, Vogt RG, editors. Insect Pheromone Biochemistry and Molecular Biology. Elsevier. pp.323–340. doi: 10.1016/B978-012107151-6/50013-X. [Google Scholar]
- Blomquist GJ, Jurenka R, Schal C, Tittiger C. 2011. Pheromone production: biochemistry and molecular biology. In: Gilbert LI, editor. Insect endocrinology. Amsterdam: Elsevier; Chapter 12, p. 523–572. [Google Scholar]
- Bortolotti L, Costa C. Chemical Communication in the Honey Bee Society. In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 5. Available from: http://www.ncbi.nlm.nih.gov/books/NBK200983/ [Last accessed 2016 Aug 24]. [PubMed]
- Brill MF, et al. 2013. Parallel processing via a dual olfactory pathway in the honeybee. J Neurosci. 33:2443–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann A, Brückner D. 1998. The EAG response spectra of workers and drones to queen honeybee mandibular gland components: the evolution of a social signal. Naturwissenschaften 85:283–285. [Google Scholar]
- Brockmann A, Brückner D. 2001. Structural differences in the drone olfactory system of two phylogenetically distant Apis species, A. florea and A. mellifera. Naturwissenschaften 88:78–81. [DOI] [PubMed] [Google Scholar]
- Brockmann A, Brückner D. 2003. Drone antennae and the evolution of sex-pheromone communication in honeybees. Indian Bee J. 65:131–138. [Google Scholar]
- Brockmann A, Dietz D, Spaethe J, Tautz J. 2006. Beyond 9-ODA: sex pheromone communication in the European honey bee Apis mellifera L. J Chem Ecol. 32:657–667. [DOI] [PubMed] [Google Scholar]
- Claudianos C, et al. 2014. Odor memories regulate olfactory receptor expression in the sensory periphery. Eur J Neurosci. 39:1642–1654. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, et al. 2015. Hymenoptera Genome Database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 44(D1):D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Chotigeat W, Satasook C. 2014. Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: implications for their adaptation and speciation. J Mol Evol. 79:21–39. [DOI] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Robertson HM, Satasook C. 2015. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res Notes 8:380.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslen J, Kaissling KE. 1976. Zahl und Verteilung antennaler Sensillen bei der Honigbiene (Apis mellifera L.). Zoomorphologie 83:227–251. [Google Scholar]
- Evans JD, Schwarz RS. 2011. Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol. 19:614–620. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free JB. 1987. Pheromones of social bees. Ithaca (NY: ): Comstock. [Google Scholar]
- Frisch K. 1965. Tanzsprache und Orientierung der Bienen. Berlin, Heidelberg, New York: Springer Verlag. [Google Scholar]
- Galizia CG, McIlwrath SL, Menzel R. 1999. A digital three-dimensional atlas of the honeybee antennal lobe based on optical sections acquired by confocal microscopy. Cell Tissue Res. 295:383–394. [DOI] [PubMed] [Google Scholar]
- Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4(41):1741.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurfa M. 2015. Learning and cognition in insects. Wiley Interdiscip Rev Cogn Sci. 6:383–395. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gress JC, Robertson HM, Weaver DK, Dlakić M, Wanner KW. 2013. Odorant receptors of a primitive hymenopteran pest, the wheat stem sawfly. Insect Mol Biol. 22:659–667. [DOI] [PubMed] [Google Scholar]
- Guo S, Kim J. 2007. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol. 24:1198–1207. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard RW, Blomquist GJ. 2005. Ecological, behavioral, and biochemical aspects of insect hydrocarbons. Annu Rev Entomol. 50:371–393. 2005. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. 2005. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl 1):i251–i257. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. 2007. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 35:W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Frith MC. 2012. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28:3144–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, et al. 2010. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 107:12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, et al. 2013. The draft genome of a socially polymorphic halictid bee, Lasioglossum albipes. Genome Biol. 14:R142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J, Klink O, Mohl C, Raming K, Breer H. 2003. A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 189:519–526. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Kropf J, Kelber C, Bieringer K, Rossler W. 2014. Olfactory subsystems in the honeybee: sensory supply and sex specificity. Cell Tissue Res. 357:583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41:W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, et al. 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43:703–714. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Lo N, Gloag RS, Anderson DL, Oldroyd BP. 2009. A molecular phylogeny of the genus Apis suggests that the Giant Honey Bee of the Philippines, A. breviligula Maa, and the Plains Honey Bee of southern India, A. indica Fabricius, are valid species. Syst Entomol. 35:226–233. [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. 2010. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 39:D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Tu Z. 2008. Odorant receptor C-terminal motifs in divergent insect species. J Insect Sci. 8:1–10. [Google Scholar]
- Missbach C, et al. 2014. Evolution of insect olfactory receptors. Elife 3:e02115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja N, Brockmann A. 2009. males of the dwarf honey bee Apis florea are attracted to (2E)-9-oxodecenoic acid and (2E)-10-hydroxydecenoic acid. J Chem Ecol. 35:653–655. [DOI] [PubMed] [Google Scholar]
- Nagarathnam B, et al. 2014. Bioinformatics and biology insights DOR – a database of olfactory receptors – integrated repository for sequence and secondary structural information of olfactory receptors in selected eukaryotic genomes. Bioinform Biol Insights 8:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 39:121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One 2:e708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura O, Brillada C, Yazawa S, Maffei ME, Arimura G. 2012. Transcriptome pyrosequencing of the parasitoid wasp Cotesia vestalis: genes involved in the antennal odorant-sensory system. PLoS One 7:e50664.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd BP, Wongsiri S. 2006. Asian honey bees. Biology, conservation and human interactions. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- Ozaki M, Kidokoro-Kobayashi M, Hiraguchi T. 2012. Cuticular hydrocarbon sensillum for nestmate recognition in ants In Barth FG, Humphrey JAC, Srinivasan MV, editors. Frontiers in sensing. From biology to engineering. Wien: Springer Verlag; Chapter 10, p. 145–157. [Google Scholar]
- Park D, et al. 2015. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics 16:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plettner E, et al. 1997. Species- and caste-determined mandibular gland signals in honeybees (Apis). J Chem Ecol. 23(2):363–377. [Google Scholar]
- Plettner E, Slessor KN, Winston ML, Oliver JE. 1996. Caste-selective pheromone biosynthesis in honeybees. Science 271(5257):1851–1853. [Google Scholar]
- Pruitt K, Brown G, Tatusova T, et al. The Reference Sequence (RefSeq) Database. 2002. Oct 9 [Updated 2012 Apr 6]. In: McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-. Chapter 18. Available from: http://www.ncbi.nlm.nih.gov/books/NBK21091/ [Last accessed 2016 Aug 24].
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. 2009. FigTree version 1.3.1 [computer program] http://tree.bio.ed.ac.uk
- Renner M, Vierling G. 1977. Die Rolle des Taschendrüsenpheromons beim Hochzeitsflug der Bienenkönigin. The secretion of the tergal glands and the attractiveness of the queen honey bee to drones in the mating flight. Behav Ecol Sociobiol. 2:329–338. [Google Scholar]
- Robertson HM, Gadau J, Wanner KW. 2010. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol. 19(Suppl 1):121–136. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. 2006. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16:1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd BM, et al. 2015. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Rozas J. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103:208–216. [DOI] [PubMed] [Google Scholar]
- Sandoz J-C. 2006. Odour-evoked responses to queen pheromone components and to plant odours using optical imaging in the antennal lobe of the honey bee drone Apis mellifera L. J Exp Biol. 209:3587–3598. [DOI] [PubMed] [Google Scholar]
- Seeley TD. 1995. Wisdom of the hive. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- Sharma KR. et al. 2015. Cuticular Hydrocarbon Pheromones for Social Behavior and Their Coding in the Ant Antenna. Cell Rep. 12:1261–1271. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CDR, Zimin A, et al. 2011. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci U S A. 108:5673–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Smith CD, et al. 2011. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci U S A. 108: 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RK, Spivak M, Taylor OR, Bennent C, Smith ML. 1993. Maturation of tergal gland alkene profiles in European. J Chem Ecol. 19:133–142. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 6:175–182. [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. 2014. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády GE, Simon I., 1998Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 283:489–506. [DOI] [PubMed] [Google Scholar]
- Tusnády GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. [DOI] [PubMed] [Google Scholar]
- Van Oystaeyen A, et al. 2014. Conserved class of queen pheromones stops social insect workers from reproducing. Science 343(6168): 287–290. [DOI] [PubMed] [Google Scholar]
- Wang S-N, et al. 2015. Identification and expression analysis of putative chemosensory receptor genes in Microplitis mediator by antennal transcriptome screening. Int J Biol Sci. 11:737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner KW, Nichols AS, et al. 2007. A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc Natl Acad Sci U S A. 104:14383–14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X-W, et al. 2013. Odorant-binding proteins and olfactory coding in the solitary bee Osmia cornuta. Cell Mol Life Sci. 70:3029–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A, Robinson GE. 2012. Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu Rev Genet. 46:591–615. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. 2012. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8:e1002930.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2015. Chemoreceptor Evolution in Hymenoptera and Its Implications for the Evolution of Eusociali. Genome Biol Evol. 7:2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.