Abstract
The rapid increase in the number of mitochondrial genomes in public databases provides opportunities for insect phylogenetic studies; but it also provides challenges because of gene rearrangements and variable substitution rates among both lineages and sites. Typically, phylogenetic studies use mitochondrial sequence data but exclude other features of the mitochondrial genome from analyses. Here, we undertook large-scale sequencing of mitochondrial genomes from a worldwide collection of specimens belonging to Braconidae, one of the largest families of Metazoa. The strand-asymmetry of base composition in the mitochondrial genomes of braconids is reversed, providing evidence for monophyly of the Braconidae. We have reconstructed a backbone phylogeny of the major lineages of Braconidae from gene order of the mitochondrial genomes. Standard phylogenetic analyses of DNA sequences provided strong support for both Cyclostomes and Noncyclostomes. Four subfamily complexes, that is, helconoid, euphoroid, sigalphoid, and microgastroid, within the Noncyclostomes were reconstructed robustly, the first three of which formed a monophyletic group sister to the last one. Aphidiinae was recovered as a lineage sister to other groups of Cyclostomes, while the Ichneutinae was recovered as paraphyletic. Separate analyses of the subdivided groups showed congruent relationships, employing different matrices and methods, for the internal nodes of the Cyclostomes and the microgastroid complex of subfamilies. This research, using multiple lines of evidence from mitochondrial genomes, illustrates multiple uses of mitochondrial genomes for phylogenetic inference in Braconidae.
Keywords: strand asymmetry, gene rearrangement, phylogeny, Hymenoptera, Braconidae
Introduction
Mitochondrial genomes are considered powerful markers of phylogenetic relationships, because of their maternal inheritance (Barr et al. 2005), rare recombination (Boore 1999), relatively high evolutionary rate, and conserved gene components (Curole and Kocher 1999). This organelle genome has been widely used, at both deep and shallow taxonomic scales, from sub-kingdom (Bernt, Bleidorn, et al. 2013) and class (Simon and Hadrys 2013) to population level studies (Ma et al. 2012). Other features of the mitochondrial genome, such as gene arrangement or base composition, have also been used to illuminate phylogenetic relationships (Boore et al. 1995; Boore and Brown 1998; Timmermans and Vogler 2012).
The use of mitochondrial genomes for phylogenetic reconstruction is problematic in some cases, particularly in resolving deep relationships. Many studies have shown that substitution saturation (Liu et al. 2014), among-lineage compositional heterogeneity (Lartillot et al. 2009; Cameron 2014) and codon-usage bias (Stenøien 2004), may all negatively affect the reconstruction of phylogenetic relationships (Maddison and Maddison 2007; Simon and Hadrys 2013). Although an accelerated rate of gene rearrangement has been found in many groups, and some derived patterns of gene arrangement are linked to specific lineages (Boore et al. 1998; Timmermans and Vogler 2012), gene arrangement patterns have been used rarely for phylogenetic reconstruction in insects (Dowton 1999; Timmermans and Vogler 2012).
Mitochondrial genomes in the order Hymenoptera have been found to exhibit several extreme features, such as exceptionally high A+T content (Wei et al. 2009), frequent gene rearrangement in the Apocrita (Dowton, Cameron, Austin, et al. 2009), large-scale rearrangement of protein-coding genes (Wei et al. 2014), and rapid substitution rates (Oliveira et al. 2008). Both among-lineage rate heterogeneity (Dowton, Cameron, Dowavic, et al. 2009) and independent evolution of gene rearrangement (Dowton, Cameron, Dowavic, et al. 2009; Wei, Shi, Sharkey, et al. 2010, 2014) challenge the use of mitochondrial genomes in higher-level phylogenetic reconstruction of the Hymenoptera. In contrast, these same genomic features might be useful for lower-level phylogenetic reconstruction in the Hymenoptera (Wei et al. 2014).
Braconidae is one of the most species-rich families of Hymenoptera and one of the largest families of Metazoa, with more than 1,040 genera and more than 19,000 species described (Yu et al. 2012). The species in this family parasitize insects from 120 families, and many benefit humans in biological and natural control of pests in agriculture and forestry (Overholt et al. 1994; Day 1996; Ribeiro et al. 2013). Parasitism by members of Braconidae provides a good model system for the study of host–parasitoid interactions (Pennacchio and Strand 2006; Yu et al. 2008; Shi et al. 2013; CHu et al. 2014) and the evolution of parasitism (Whitfield 1992; Belshaw and Quicke 1997; Belshaw et al. 1998). Most braconids exhibit either koinobiont endoparasitism or idiobiont ectoparasitism (Shaw and Huddleston 1991). The origin of endoparasitism has long been a controversial topic (Quicke and van Achterberg 1990; Whitfield 1992). Quicke and van Achterberg’s (1990) study proposed that all Noncyclostome endoparasitoids form a single monophyletic lineage, derived from a basal paraphyletic grade of ectoparasitoid braconids. Their research was based on morphological and life history characters, and was corroborated by analyses based on 28S D2 rDNA gene sequences (Belshaw and Quicke 1997; Belshaw et al. 1998). However, different evidence led to the viewpoint that there have been multiple independent transitions from ectoparasitism to endoparasitism, in analyses based on adult morphological, larval, and biological characters and 16S rDNA data (Whitfield 1992; Dowton et al. 1998; Quicke and Belshaw 1999; Zaldivar-Riverón, Shaw, et al. 2008). The incongruence among studies may be caused by convergence among morphological characters resulting from shared life history strategies (Quicke and Belshaw 1999), or incomplete taxon sampling and a lack of overlap of sampling across different studies.
Reconstruction of the phylogenetic relationships among the subfamilies of Braconidae is essential to understanding the evolutionary origins of parasitism in the group. Phylogenetic reconstruction of Braconidae was originally based on morphology (van Achterberg 1984; Quicke and van Achterberg 1990; Wharton et al. 1992), however, it is problematic to choose useful morphological characters because of the high level of convergent evolution (Sharanowski et al. 2011). Nuclear genes, mitochondrial genes, and combinations of the two (Belshaw et al. 1998; Dowton et al. 1998, 2002; Shi et al. 2005; Zaldivar-Riverón et al. 2006) and complete mitochondrial genomes (Wei, Shi, Sharkey, et al. 2010) have been used in phylogenetic analyses of Braconidae. Most studies were conducted either with limited gene markers and a broad spectrum of taxa (Dowton et al. 1998; Belshaw et al. 2000; Belshaw and Quicke 2002; Dowton, Belshaw, et al. 2002; Shi et al. 2005; Zaldivar-Riverón et al. 2006), or multiples genes and limited taxa sampling (Wei, Shi, Sharkey, et al. 2010). Increasing the sampling of both genetic markers and taxa improved phylogenetic resolution in Sharanowski et al.’s (2011) study.
The split of braconids into Cyclostomes and Noncyclostomes according to their mouth morphology has been widely published (van Achterberg 1984; Quicke and van Achterberg 1990; Wharton et al. 1992). Phylogenetic relationships among and within many subfamilies, however, are particularly controversial. Some studies based on morphological analyses moved Aphidiinae between the Cyclostomes and Noncyclostomes (Quicke and van Achterberg 1990; van Achterberg and Quicke 1992). More recent studies support a sister-group relationship between Aphidiinae and Mesostoinae at the base of the Cyclostomes (Belshaw et al. 2000; Dowton, Belshaw, et al. 2002; Zaldivar-Riverón et al. 2006). Sharanowski et al. (2011) named a clade, the Aphidioid complex, consisting of (Mesostoinae (Aphidiinae + Maxfischeria)), that is sister to the cyclostome complex of subfamilies. Although the monophyly of Doryctinae was recovered in the morphological studies or combination with molecular analyses of several publications (van Achterberg 1984; Quicke and van Achterberg 1990; Dowton, Belshaw, et al. 2002), most molecular studies supported the paraphyly of Doryctinae as well as Ichneutinae and Rogadinae (Belshaw and Quicke 2002; Zaldivar-Riverón et al. 2006; Pitz et al. 2007; Zaldivar-Riverón, Belokobylskij, et al. 2008; Sharanowski et al. 2011). By using ∼4 kb of sequence data from both mitochondrial and nuclear genomes for 139 taxa, Sharanowski et al. (2011) recovered well-supported relationships among Noncyclostomes. However, relationships within the Cyclostome complex were poorly supported, probably due to the lower taxonomic sampling relative to Noncyclostomes.
In this study, we sequenced 21 mitochondrial genomes from a worldwide collection of braconids, and attempted to reconstruct the phylogenetic relationships among major lineages of the Braconidae using sequences as well as gene arrangement pattern of the mitochondrial genomes. Our research provides a robust phylogenetic hypothesis of relationships among subfamilies of Braconidae.
Materials and Methods
Sample and DNA Extraction
In total, 21 species from 18 subfamilies were used for mitochondrial genome sequencing (table 1). Specimens were stored in 100% ethanol at −80 °C. Genomic DNA was extracted from a single adult using a DNeasy tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. All voucher specimens are kept in the Evolutionary Biology Laboratory of Zhejiang University, China.
Table 1.
General Information of the Braconid Mitochondrial Genomes used in this Study
| Species | Length (bp) | Subfamily | GenBank Accession No. | Collection Location |
|---|---|---|---|---|
| Acanthormius sp. | 13,051 | Lysiterminae | KF385867 | Hainan, China |
| Afrocampsis griseosetosus | 10,104 | Acampsohelconinae | KJ412474 | Pool Department, Republic of Congo |
| Aphidius gifuensis | 11,996 | Aphidiinae | GU097658 | Hangzhou, China |
| Capitonius sp. | 13,078 | Cenocoeliinae | KF385869 | Kentucky, USA, |
| Cardiochiles fuscipennis | 14,390 | Cardiochilinae | KF385870 | Fuyang, China |
| Cotesia vestalis | 15,543 | Microgastrinae | FJ154897 | Hangzhou, China |
| Diachasmimorpha longicaudata | 13,850 | Opiinae | GU097655 | Guangzhou, China |
| Elasmosoma sp. | 13,326 | Euphorinae | KJ412470 | Kentucky, USA |
| Eumacrocentrus sp. | 14,080 | Helconinae | KF385872 | West Virginia, USA |
| Euurobracon breviterebrae | 12,957 | Braconinae | KF385871 | Hainan, China |
| Histeromerus sp. | 13,168 | Histeromerinae | KF418765 | West Virginia, USA |
| Homolobus sp. | 13,927 | Homolobinae | KF385873 | Ningxia, China |
| Ichneutes sp. | 13,092 | Ichneutinae | KF385874 | Florida, USA |
| Macrocentrus camphoraphilus | 15,801 | Macrocentrinae | GU097656 | Jiaxing, China |
| Meteorus pulchricornis | 10,186 | Euphorinae | GU097657 | Nanjing, China |
| Mirax sp. | 13,664 | Miracinae | KJ412471 | Kentucky, USA |
| Pambolus sp. | 13,175 | Pambolinae | KF385875 | Hainan, China |
| Paroligoneurus sp. | 13,413 | Ichneutinae | KJ412472 | Florida, USA |
| Phaenocarpa sp. | 9,981 | Alysiinae | KJ412475 | Ningxia, China |
| Phanerotoma flava | 10,171 | Cheloninae | GU097654 | Jiaxing, China |
| Proterops sp. | 12,883 | Ichneutinae | KJ412477 | Kentucky, USA |
| Pselaphanus sp. | 13,204 | Pselaphaninae | KF385876 | Guyana, French |
| Pseudognaptodon sp. | 13,190 | Gnamptodontinae | KJ412473 | Kentucky, USA |
| Sigalphus bicolor | 12,744 | Sigalphinae | KF385878 | West Virginia, USA |
| Spathius agrili | 15,425 | Doryctinae | FJ387020 | Tianjin, China |
| Therophlius festivus | 14,216 | Agathidinae | KF385868 | Beijing, China |
| Triraphis sp. | 13,162 | Rogadinae | KF385877 | Fuyang, China |
| Xiphozele sp. | 9,160 | Xiphozelinae | KJ412476 | Trang, Thailand |
Note.—Species name in bold indicates the sequence was published in Wei, Shi, Sharkey, et al. (2010).
PCR Amplification and Sequencing
Initially, a set of universal primers for animal mitochondrial genomes (Simon et al. 1994, 2006) were used for amplification and sequencing of a range of segments. Subsequently, species-specific primers were designed, according to the segments obtained, to fill in the missing areas of sequence (supplementary table S1, Supplementary Material online). PCRs were done with Takara LA Taq (Takara Biomedical, Japan) under the following conditions: initial denaturation for 3 min at 96 °C and then 95 °C 15 s, 45–57 °C 15 s, 60 °C 2–3 min for 40 cycles, 60 °C 10 min. PCR components were added following the Takara LA Taq protocols. The PCR products were directly sequenced by Sangon Biotech Company at Shanghai using a primer-walking strategy, from both strands.
Genome Annotation and Base Composition Analysis
Putative tRNA genes were identified using the tRNAscan-SE search server (Lowe and Eddy 1997). The COVE cut-off score was reduced to 5 when expected tRNA genes could not be found. The tRNAs that could not be identified by the tRNA-scan search server were determined by alignment with their homologs in related species. The gene boundaries of protein-coding and rRNA genes were determined based on the ends of neighbouring tRNAs and by alignment with their homologs using MEGA6 (Tamura et al. 2013). The AT and GC skews were calculated according to formula AT skew = (A%− T%)/(A% + T%) and GC skew = (G%−C%)/(G% + C%). Another 41 hymenopteran mitochondrial genomes downloaded from GenBank (supplementary table S2, Supplementary Material online) were added for analyses. We used 11 well-sequenced protein-coding genes (excluding nad1 and nad2) for calculation, to avoid the negative influence of missing genes in some species.
Reconstruction of Phylogenetic Relationships from tRNA Rearrangements
We inferred phylogenetic relationships among the major lineages of Braconidae using tRNA rearrangement patterns. Two main approaches of treating gene order data were utilized as reviewed by Bernt, Braband, et al. (2013). The rearrangement-based approach assumes that certain well-defined elementary operations, that is, inversion, transposition, inverse transposition, tandem duplication, and random loss, are responsible for evolutionary changes in gene order, while the gene-cluster-based approach compares the properties of the gene orders shared among species (Bernt, Braband, et al. 2013). We used the Maximum Likelihood (ML) method based on gene-order data implemented in the MLGO web server to construct the phylogenetic tree (Hu et al. 2014). We also constructed phylogenetic trees based on the pairwise rearrangement distances using the neighbor-joining (NJ), unweighted pair group method with arithmetic mean (UPGMA) and Fitch–Margoliash (FM) methods in the web server ‘Sorting genomes and reconstructing phylogenetic trees by Reversals, generalized Transpositions and Translocations (SoRT2)’ (Huang et al. 2010). Pairwise distances from genome order data was calculated based on the double cut and join (DCJ) rearrangement model, that is, genes are cut in two places with a subsequent re-joining in a different order (Yancopoulos et al. 2005). The sorting of genomes was set to reversals, generalized transpositions, and translocations, as observed in our data. The jackknife analysis was performed with 100 replicates to evaluate statistical reliability of the constructed trees. We selected 16 tRNA genes sequentially located in the putative ancestral mitochondrial genome of insects in 14 species for analysis (two tRNA clusters near the boundaries of nad2 were not used, i.e., trnI-trnQ-trnM and trnW-trnC-trnY, which failed to sequence in most species). The type of the genomes was set to linear rather than circular due to the inclusion of the incomplete set of the 22 tRNA genes.
Multiple Sequence Alignment, Alignment Masking, and Data Partition
Protein-coding and RNA genes were aligned using the consistency-based algorithms implemented in MAFFT version 7.205 (Katoh and Standley 2013). G-INS-i and Q-INS-I algorithms in MAFFT (Golubchik et al. 2007) were used for protein-coding and RNA gene alignment, respectively. The alignment of nucleotide sequences was guided by the amino acid sequence alignment using the Perl script TranslatorX version 1.1 (Abascal et al. 2010).
Phylogenetic analyses can be impeded by random similarity of sequences (Misof and Misof 2009). Masking of blocks of sites in alignments can be employed systematically to reduce the influence of random similarity of sequences on the resultant phylogenetic tree (Kück et al. 2014). We used a sliding window approach implemented in Aliscore version 02.2 (Misof and Misof 2009) to identify blocks of sites with putative ambiguities or random similarity in individual genes, and masked these with Alicut version 2.3 (Kück 2009). Default settings were used in both analyses. The genes were concatenated into a matrix using the Perl script FASconCAT-G version 1.0 (Kück and Longo 2014).
To accommodate substitution heterogeneity among genes and codon positions, the PartitionFinder version 1.1.1 (Lanfear et al. 2012) was used to simultaneously choose partitioning schemes and substitution models for the matrix. The maximum partition scheme that could be entered into the PartitionFinder software was defined by codon position for nucleotide sequences of protein-coding genes and RNA genes, and by gene for amino acid sequences of protein-coding genes. The search models for DNA and amino acid sequences were set to be “mrbayes” and “all protein”, respectively. The greedy algorithm was used, with branch lengths estimated, to search for the best-fit partitioning scheme.
Although we reduced noise in the aligned genes by using a masking method, tree reconstructions could still be misled, because masking methods are relatively insensitive to strong sequence divergence in a single taxon (Kück et al. 2014). Thus, we used a method based on a sliding window and a Monte Carlo resampling approach to detect strongly divergent nucleotide sequences that could have the potential to bias tree reconstruction and nodal support; this was implemented in Aligroove version 1.05 (Kück et al. 2014). Each partition defined by gene and codon position was assessed by Aligroove employing default settings. There is no specific criterion on which to base the inclusion or exclusion of a partition in subsequent phylogenetic analyses. We used a stringent criterion that partitions with more than 10% negative pairwise similarity scores (44 out of 435 in our data with 30 taxa) were excluded in subsequent analyses. For the amino acid sequences, MARE (http://mare.zfmk.de, last accessed 8 Aug, 2016) (Meusemann et al. 2010), based on weighted geometry quartet mapping (Nieselt-Struwe and von Haeseler 2001), was used to calculate the relative quality of information of each single gene within the matrix. Genes with lower quality information were excluded for subsequent analyses. Finally, 11 amino acid genes were used for all phylogenetic analyses. In our phylogenetic analyses of the Braconidae, the Cyclostomes, the helconoid complex and the microgastroid complex of subfamilies, 15, 22, 18, and 21 partitions were retained in the nucleotide data matrix matrices, respectively (supplementary table S3, Supplementary Material online).
Phylogenetic Analyses
In the phylogenetic analyses, 21 mitochondrial genomes generated in this study and 7 from previous research (Wei, Shi, Sharkey, et al. 2010) were included, representing 25 subfamilies of Braconidae. The sister-group relationship between Ichneumonidae and Braconidae is now well accepted (Quicke and van Achterberg 1990; Sharkey and Wahl 1992; Belshaw et al. 1998; Quicke et al. 1999; Dowton, Belshaw, et al. 2002; Shi et al. 2005; Wei, Shi, Sharkey, et al. 2010; Sharanowski et al. 2011). Thus, we selected Diadegma semiclausum and Enicospilus sp. from Ichneumonidae as outgroups. In separate phylogenetic analyses of the Cyclostomes, Eumacrocentrus sp. and Homolobus sp. were set as outgroups, while in the phylogenetic analyses of the helconoid and microgastroid complexes, Histeromerus sp. and Triraphis sp. were set as outgroups, respectively, following the results of Sharanowski et al. (2011).
The protein-coding gene nad2 in 25 taxa and 6 transfer RNAs, that is, trnC, trnG, trnI, trnM, trnW, and trnY were missing in more than 20 taxa due to sequencing failure either in the presently sequenced or the previously published mitochondrial genomes. Although including missing data may not impede accuracy in phylogenetic analyses (Ho and Phillips 2009; Papadopoulou et al. 2010), we excluded these genes because of their absence in most taxa.
We analyzed four different data matrices, with the Bayesian inference method (BI) implemented in Mrbayes v3.2.2 (Ronquist et al. 2012) and PhyloBayes v3.3 (Lartillot et al. 2009), and the ML method implemented in RAxML v7.9.6 (Stamatakis 2006) to reconstruct the phylogenetic relationships of Braconidae for each set of taxa. Data matrices of the nucleotide and amino acid sequences were used in separate analyses. In Bayesian analyses, substitution models were applied for each partition chosen by the software Partitionfinder (supplementary table S4, Supplementary Material online). Four independent Markov chains for 50 million MCMC generations were run with tree sampling every 5,000 generations and a burn-in of 2,500 trees. After 50 million generations, all runs reached stationarity, as determined by the program Tracer v1.4.0 (Effective sample sizes >200) (http://tree.bio.ed.ac.uk/software/tracer/, last accessed 8 Aug, 2016). In the PhyloBayes analyses, the CAT-GTR model was used with a run of 20,000 generations. In ML analyses, the GTRGAMMA and MtArtF models were used for nucleotide and amino acid partitions, respectively. For each ML analysis 200 runs were conducted to find the highest-likelihood tree, followed by analysis of 1,000 bootstrap replicates.
Results
Sequencing of the Mitochondrial Genomes from Braconidae
We sequenced 21 braconid mitochondrial genomes from 18 subfamilies, bringing the total of known braconid mitochondrial genomes to 28, representing 25 subfamilies (table 1). All of the sequenced mitochondrial genomes are incomplete, which is common in most previously studied species of Hymenoptera (Cameron et al. 2008; Wei, Shi, Sharkey, et al. 2010; Mao et al. 2012; Tang et al. 2014). Nonetheless, the complete set of protein-coding genes and most tRNA genes were successfully sequenced for most species, providing sufficient data for further comparative and phylogenetic analyses. The regions that failed to sequence were usually around the A+T-rich region (control region), and near nad2 with high A+T content and frequent repeat units, which may have influenced the amplification and sequencing of the region (Mao et al. 2012).
Reversal of Strand Asymmetry within Braconidae
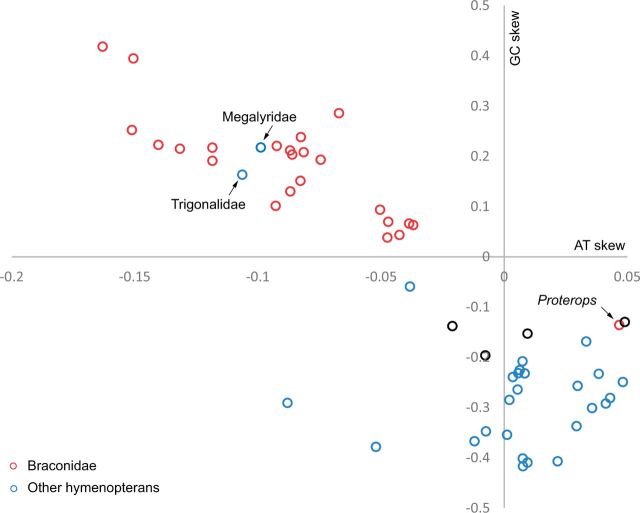
We evaluated the strand asymmetry of base composition by calculating the AT and GC skew values for 28 braconid mitochondrial genomes as well as 41 from other Hymenoptera. In most insect mitochondrial genomes the AT skew is positive, while the GC skew is negative. Reversal of strand asymmetry was found in three groups of insects, including Braconidae: that is, more Ts than As and more Cs than Gs on the majority strand (Wei, Shi, Chen, et al. 2010). Our analysis confirmed the reversal of base composition strand asymmetry in Braconidae as a synapomorphy, although exceptions were found in Proterops (fig. 1, supplementary fig. S1, Supplementary Material online). The Proterops has a normal strand asymmetry pattern on the sequenced region of majority strand and protein-coding genes. Detailed analysis on each of the protein-coding genes of Proterops indicated that 10 of 11 genes showed normal GC skew values, of which, seven of eight genes coded on the minority strand showed positive AT skew values and four genes coded on the majority strand showed negative AT skew values (supplementary tables S2 and S5, Supplementary Material online). All analyses indicated a reversal of strand asymmetry in Proterops within Braconidae, that is, a normal strand asymmetry pattern for other Hymenoptera (Wei, Shi, Chen, et al. 2010). In addition, we found that two nonichneumonoid species (Megalyridae and Trigonalidae) also showed reversal of strand asymmetry (fig. 1, supplementary fig. S1, Supplementary Material online).
Fig. 1.—
Scatterplots of AT and GC skew values calculated for sequenced majority strand of 65 species of Hymenoptera. Reversal of both AT and GC skew are found in most species of Braconidae except for Proterops sp. (0.0468, −0.1360) and two species from family Megalyridae and Trigonalidae. Other species in Hymenoptera all have normal GC-skew values.
Phylogenetic Relationships within Braconidae Revealed by tRNA Rearrangement
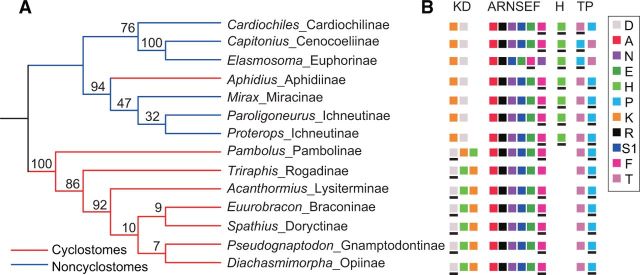
Frequent gene rearrangements were found in Braconidae. Most of the rearranged genes were tRNA genes. All protein-coding genes in all species were arranged in the ancestral pattern of the putative ancestral insect mitochondrial genome, except for the Cotesia vestalis (Wei, Shi, Sharkey, et al. 2010) indicating within-lineage heterogeneity of gene rearrangement rate in Braconidae. Three tRNA gene-rearrangement hotspots were found in Braconidae, that is, trnK- trnD, trnA-trnR-trnN- trnS1-trnE-trnF, and trnT-trnP, two of which have previously been reported in studies using partial regions of mitochondrial genomes (Dowton 1999; Dowton and Austin 1999). The rearranged genes in the Cyclostomes are usually located in the tRNA cluster trnK -trnD, while those in the Noncyclostomes are in tRNA clusters trnA–trnR–trnN–trnS1–trnE–trnF and trnT–trnP (fig. 2). The ML, NJ, UPGMA, and FM trees inferred from the arrangement pattern of 16 tRNA genes all recovered the two major lineages of Cyclostomes and Noncyclostomes (fig. 2, supplementary fig. S2, Supplementary Material online), as accepted by previous analyses (Wei, Shi, Sharkey, et al. 2010; Sharanowski et al. 2011) except for the position of Aphidiinae, which was recovered within the Noncyclostomes. Capitonius and Elasmosoma (the euphoroid complex) were always recovered as sisters in the NJ, UPGMA, and FM trees (supplementary fig. S2, Supplementary Material online). However, the relationships within Cyclostomes and Noncyclostomes are mostly unacceptable.
Fig. 2.—
Phylogenetic relationships among the major lineages of the Braconidae reconstructed from the arrangement patterns of tRNA genes. (A) The ML tree based on gene-order data inferred from the MLGO web server. (B) The arrangement of tRNA genes in rearrangement hotspots in Braconidae. The tRNA genes are denoted by one letter symbol according to the IPUC-IUB single-letter amino acid codes. The symbol in (B) with a bold black line in the bottom indicates that the correspondence gene is coded on the minority strand of the mitochondrial genome.
Phylogenetic Relationships within Braconidae Reconstructed from Gene Sequences
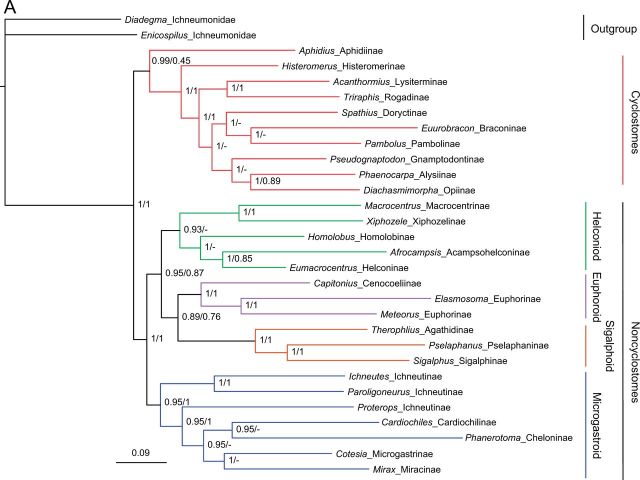
Phylogenetic relationships among 25 subfamilies of Braconidae were reconstructed using mitochondrial genome sequences. All analyses robustly support the division of Braconidae into Cyclostomes and Noncyclostomes as commonly accepted (fig. 3) (Quicke and van Achterberg 1990; Wei, Shi, Sharkey, et al. 2010; Sharanowski et al. 2011). Within each of the two major lineages, topologies varied among analyses based on different data matrices and analytical methods when all species from Braconidae were included. Among the different results, those generated from amino acid sequences of protein-coding genes using Bayesian and PhyloBayes methods recovered the traditional relationships (fig. 3). In order to validate the relationships within Cyclostomes and Noncyclostomes, we conducted separate analyses for each lineage, which improved the congruence of topologies among analyses in the Cyclostomes. Both the ML and BI methods, using nucleotide sequences, supported a congruent topology for the Cyclostomes and the microgastroid complex of subfamilies (supplementary fig. S3A and D, Supplementary Material online).
Fig. 3.—
Phylogenetic relationships among subfamilies of the Braconidae inferred from amino acid sequences of protein-coding genes using Bayesian and PhyloBayes methods. Bayesian posterior probabilities from Bayesian and PhyloBayes analyses are shown sequentially, separated by “/” near respective nodes.
Within the Cyclostomes, Aphidiinae was recovered as sister to the remaining Cyclostomes in all analyses followed by Histeromerinae and then Lysiterminae + Rogadinae. In analyses of amino acid sequences, Gnamptodontinae was sister to Opiinae + Alysiinae (fig. 3), corroborating the Alysioid subcomplex of Sharanowski et al. (2011), and the Braconinae formed a monophyletic lineage with Doryctinae and Pambolinae. However, in analyses of Cyclostomes based on nucleotide sequences, Braconinae were the sister-group to Opiinae + Alysiinae, which together were sister to Gnamptodontinae (supplementary fig. S3A, Supplementary Material online).
Within the Noncyclostomes, there were three well-supported lineages (fig. 3), that is, ((Xiphozelinae + Macrocentrinae) + (Homolobinae + (Acampsohelconinae + Helconinae))), (Agathidinae + (Sigalphinae + Pselaphaninae)) +(Cenocoeliinae + Euphorinae), corresponding to the helconoid, sigalphoid + euphoroid subfamily complexes (Sharanowski et al. 2011). These three subfamily complexes formed a monophyletic group, sister to the microgastroid complex of subfamilies. However, the relationships among these three lineages varied among analyses (fig. 3, supplementary fig. S3B and C, Supplementary Material online).
Within the microgastroid complex of subfamilies, the relationships of the ichneutine taxa, that is, Ichneutes sp., Paroligoneurus sp., and Proterops sp., suggest that this subfamily is paraphyletic (fig. 3), which has also been reported in other studies based on molecular markers (Belshaw and Quicke 2002; Pitz et al. 2007; Sharanowski et al. 2011). In our study the Protoperini ichneutines are sister to the other members of the microgastroid complex, similar to results found in previous studies (Belshaw et al. 1998, 2000; Pitz et al. 2007; Sharanowski et al. 2011). A lineage composed of Cardiochilinae, Cheloninae, Microgastrinae, and Miracinae was sister to Ichneutinae (fig. 3).
Discussion
Strand Asymmetry of Base Composition in Braconidae
The adding of mitochondrial genomes further confirmed the reversal of strand asymmetry as ancestral feature for Braconidae (Wei, Shi, Chen, et al. 2010). In our analyses, we did not find any relationship between the reversal of strand asymmetry and gene rearrangement rate in Braconidae. In Mirax and Proterops, the gene arrangement patterns are identical, but the former shows reversal of strand asymmetry while Proterops shows reversal of strand asymmetry again in Braconidae. This phenomenon is present in other hymenopteran groups, such as Nasonia species, which show accelerated gene rearrangements and normal strand asymmetry (Oliveira et al. 2008). The lack of correlation between strand asymmetry of base composition and the rate of gene rearrangement is consistent with the hypothesis that strand asymmetry reversal was caused by the inversion of the replication origin in the A + T-rich region (Wei, Shi, Chen, et al. 2010).
Gene Rearrangement in Braconidae
Gene rearrangement of mitochondrial genomes is considered to be a phylogenetic character of great potential in invertebrates (Dowton, Castro, et al. 2002). For example, the shared translocation of trnL (UUR) to the position between cox1 and cox2 in the Pancrustacea has linked the insects and crustaceans (Boore et al. 1998). However, with more mitochondrial genomes sequenced, it is clear that no gene rearrangements are shared between higher-level taxa of insects. The focus has shifted to taxa below the ordinal level, in examining gene rearrangements (Shao et al. 2001; Dowton, Cameron, Dowavic, et al. 2009; Wei et al. 2014). In this study, we used gene arrangement patterns as characters and thus corroborated the placement of the two major lineages within Braconidae except for the position of Aphidiinae (fig. 2). Recently, methods automatizing the comparative analysis of gene order, and publicly available software have been developed (Bernt, Braband, et al. 2013).
Phylogenetic Relationships within Braconidae
Our analyses, based on multiple lines of evidence from mitochondrial genomes, recovered the well-accepted major lineages within the family, such as the Cyclostomes and Noncyclostomes within Braconidae. Phylogenetic analyses based on sequence data supported the major lineages of helconoid, sigalphoid, euphoroid, microgastroid subfamily complexes and (helconoid + (sigalphoid + euphoroid)) within Noncyclostomes. Our study also robustly recovered several subfamily subcomplexes, such as the (Gnamptodontinae + (Opiinae + Alysiinae)) in analyses based on amino acid sequences, the Alysioid subcomplex of Sharanowski et al. (2011), and the Macrocentrinae acXiphozelinae, the Macrocentroid subcomplex of Sharanowski et al. (2011). Our study recovered Homolobinae + (Acampsohelconinae + Helconinae) within helconoid complex, whereas Sharanowski et al. (2011) recovered Homolobinae in the Macrocentroid subcomplex. Denser sampling from within the helconoid complex is needed to corroborate or refute our result, since several subfamilies were not included in our analysis, such as Orgilinae, Charmontinae, Amicrocentrinae and Microtypinae.
One of the most studied groups of Braconidae is the subfamily Aphidiinae, which was long considered as a separate family, Aphidiidae. As in all other recent studies our analyses confirmed placement of Aphidiinae in Braconidae. On the basis of earlier morphological analyses Aphidiinae was placed as a sister group of Noncyclostomes (Quicke and van Achterberg 1990; van Achterberg and Quicke 1992; Wharton et al. 1992). Recent molecular studies placed Aphidiinae + Mesostoinae as sister to the remaining Cyclostomes (Zaldivar-Riverón et al. 2006; Sharanowski et al. 2011). This is also where our analyses, based on both amino acid (fig. 3) and nucleotide sequences (supplementary fig. S3, Supplementary Material online), place the subfamily, though our analyses did not include representatives of the Mesostoinae or Maxifischeriinae.
Within Noncyclostomes, the clade consisting of Sigalphinae and Agathidinae (the sigalphoid complex) was recovered as in several previous analyses (Belshaw and Quicke 2002; Sharanowski et al. 2011). In our analyses, (Agathidinae + (Sigalphinae + Pselaphaninae)) was strongly supported. This agrees with Sharkey’s (1997) synonymy of Pselaphaninae with Sigalphinae that was also corroborated by Quicke et al. (2008). This complex was placed, with strong support, as the sister group to the helconoid and euphoroid subfamily complexes in nucleotide sequence analyses. Contrastingly, the sigalphoid complex was recovered as sister to the microgastroid complex in Sharanowski et al. (2011).
The phylogenetic placement of Acampsohelconinae was unstable in previous molecular and morphological analyses. It was placed either as a sister group to other members of the helconoid complex or as sister to Meteorideinae, based on CAD54 and 28S genes, respectively (Sharanowski et al. 2011). van Achterberg (2002) suggested that Acampsohelconinae was not closely related to Helconinae or Blacinae. Contrary to that study we recovered (Homolobinae + (Acampsohelconinae + Helconinae)), which was supported and congruent among the different analyses, although a denser sampling is needed in further studies.
Factors Influencing Phylogenetic Analyses
Phylogenetic analyses can be influenced by taxon sampling, choice of sequences, inference methods and coding of characters (Hassanin 2006; Song et al. 2010; Duchêne et al. 2011). In this study, we addressed taxon sampling by using a worldwide sampling of 25 representative subfamilies from Braconidae. We addressed issues of the evolution of our chosen sequences by using a data masking process to reduce noise and recoding the nucleotide sequences into amino acid sequences for protein-coding genes. We addressed potential methodological artifacts by using BI and ML methods, as well as separately analyzing subdivided groups to avoid among-lineage rate heterogeneity.
Both nucleotide and amino acid sequences of protein-coding genes were used in our phylogenetic analyses. Amino acid sequences are thought to be the best data source for analyzing higher-level insect phylogeny, using mitochondrial genomes (Rota-Stabelli et al. 2010; Talavera and Vila 2011), whereas for lower-level phylogenetic analysis, reducing nucleotide sequences to amino acid sequences may eliminate valuable phylogenetic signal (Cameron et al. 2006; Cameron 2014). Our analyses based on amino acid sequences inferred well-accepted phylogenetic relationships among major lineages of Braconidae, such as the Cyclostomes, Noncyclostomes and Noncyclostome subfamily complexes, but generated varied topologies for the lower-level relationships of the subfamilies when different methods were used (fig. 3, supplementary fig. S3, Supplementary Material online). Using nucleotide sequences, the Agathidinae, Pselaphaninae, and Sigalphinae were placed in the microgastroid complex of subfamilies, conflicting with analyses based on amino-acid sequences. In subsequent analyses of each group within Braconidae, nucleotide sequences performed better than amino acid sequences, corroborating the idea that amino acid sequences are more suitable for higher-level analyses, whereas the nucleotide sequences are better suited for lower-level analyses within Braconidae.
Supplementary Material
Acknowledgments
We thank Prof Kees van Achterberg for the valuable comments on the phylogeny of Braconidae. We also thank Dr Dennis Lavrov and two anonymous reviewers for their important comments and suggestions.
Funding
This study was supported by the State Key Program of the National Natural Science Foundation of China (Grant number 31230068), the National Natural Science Foundation of China (Grant numbers 31401996, 31472025, 31101661), the NSFC Innovative Research Groups (Grant number 31021003), and the 973 Program (Grant number 2013CB127600).
Literature Cited
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168:39–50. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Dowton M, Quicke DL, Austin AD. 2000. Estimating ancestral geographical distributions: a Gondwanan origin for aphid parasitoids? Proc R Soc Lond Ser B: Biol Sci. 267:491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Fitton M, Herniou E, Gimeno C, Quicke DLJ. 1998. A phylogenetic reconstruction of the Ichneumonoidea (Hymenoptera) based on the D2 variable region of 28S ribosomal RNA. Syst Entomol. 23:109–123. [Google Scholar]
- Belshaw R, Quicke DLJ. 1997. A molecular phylogeny of the aphidiinae (Hymenoptera: Braconidae). Mol Phylogenet Evol. 7:281–293. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Quicke DLJ. 2002. Robustness of ancestral state estimates: evolution of life history strategy in ichneumonoid parasitoids. Syst Biol. 51:450–477. [DOI] [PubMed] [Google Scholar]
- Bernt M, Bleidorn C, et al. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69:352–364. [DOI] [PubMed] [Google Scholar]
- Bernt M, Braband A, et al. 2013. Bioinformatics methods for the comparative analysis of metazoan mitochondrial genome sequences. Mol Phylogenet Evol. 69:320–327. [DOI] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 8:668–674. [DOI] [PubMed] [Google Scholar]
- Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM. 1995. Deducing the pattern of arthropod phylogeny from mitochondrial-DNA rearrangements. Nature 376:163–165. [DOI] [PubMed] [Google Scholar]
- Boore JL, Lavrov DV, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature 392:667–668. [DOI] [PubMed] [Google Scholar]
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. [DOI] [PubMed] [Google Scholar]
- Cameron SL, Barker SC, Whiting MF. 2006. Mitochondrial genomics and the new insect order Mantophasmatodea. Mol Phylogenet Evol. 38:274–279. [DOI] [PubMed] [Google Scholar]
- Cameron SL, et al. 2008. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 51:800–808. [DOI] [PubMed] [Google Scholar]
- Chu Y, et al. 2014. Performance of Microplitis tuberculifer (Hymenoptera: Braconidae) parasitizing Mythimna separata (Lepidoptera: Noctuidae) in different larval instars. Biol Control 69:18–23. [Google Scholar]
- Curole JP, Kocher TD. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. 14:394–398. [DOI] [PubMed] [Google Scholar]
- Day WH. 1996. Evaluation of biological control of the tarnished plant bug (Hemiptera: Miridae) in alfalfa by the introduced parasite Peristenus digoneutis (Hymenoptera: Braconidae). Environ Entomol. 25:512–518. [Google Scholar]
- Dowton M. 1999. Relationships among the cyclostome braconid (Hymenoptera: Braconidae) subfamilies inferred from a mitochondrial tRNA gene rearrangement. Mol Phylogenet Evol. 11:283–287. [DOI] [PubMed] [Google Scholar]
- Dowton M, Austin AD. 1999. Evolutionary dynamics of a mitochondrial rearrangement “hot spot” in the Hymenoptera. Mol Biol Evol. 16:298–309. [DOI] [PubMed] [Google Scholar]
- Dowton M, Austin AD, Antolin MF. 1998. Evolutionary relationships among the Braconidae (Hymenoptera: Ichneumonoidea) inferred from partial 16S rDNA gene sequences. Insect Mol Biol. 7:129–150. [DOI] [PubMed] [Google Scholar]
- Dowton M, Belshaw R, et al. 2002. Simultaneous molecular and morphological analysis of Braconid relationships (Insecta: Hymenoptera: Braconidae) indicates independent mt-tRNA gene inversions within a single wasp family. J Mol Evol. 54:210–226. [DOI] [PubMed] [Google Scholar]
- Dowton M, Castro LR, Austin AD. 2002. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr Syst. 16:345–356. [Google Scholar]
- Dowton M, Cameron SL, Austin AD, et al. 2009. Phylogenetic approaches for the analysis of mitochondrial genome sequence data in the Hymenoptera—a lineage with both rapidly and slowly evolving mitochondrial genomes. Mol Phylogenet Evol. 52:512–519. [DOI] [PubMed] [Google Scholar]
- Dowton M, Cameron SL, Dowavic JI, et al. 2009. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol Biol Evol. 26:1607–1617. [DOI] [PubMed] [Google Scholar]
- Duchêne S, Archer FI, Vilstrup J, Caballero S, Morin PA. 2011. Mitogenome phylogenetics: the impact of using single regions and partitioning schemes on topology, substitution rate and divergence time estimation. PLoS One 6:e27138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubchik T, Wise MJ, Easteal S, Jermiin LS. 2007. Mind the gaps: evidence of bias in estimates of multiple sequence alignments. Mol Biol Evol. 24:2433–2442. [DOI] [PubMed] [Google Scholar]
- Hassanin A. 2006. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol. 38:100–116. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 58:367–380. [DOI] [PubMed] [Google Scholar]
- Hu F, Lin Y, Tang J. 2014. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics 15:354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Huang CC, Tang CY, Lu CL. 2010. SoRT2: a tool for sorting genomes and reconstructing phylogenetic trees by reversals, generalized transpositions and translocations. Nucleic Acids Res. 38: W221–W227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück P. 2009. ALICUT: a Perlscript which cuts ALISCORE identified RSS. Version 2.3. Department of Bioinformatics, Zoologisches Forschungsmuseum A. Koenig (ZFMK; ), Bonn, Germany. [Google Scholar]
- Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11:81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück P, Meid SA, Gross C, Wagele JW, Misof B. 2014. AliGROOVE—visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinformatics 15:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cox CJ, Wang W, Goffinet B. 2014. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst Biol. 63:862–878. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, et al. 2012. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol Ecol. 21:4344–4358. [DOI] [PubMed] [Google Scholar]
- Maddison W, Maddison D. 2007. Mesquite: a modular system for evolutionary analysis. Version 2.01. In: http://mesquiteproject.wikispaces.com/?responseToken=07f8a546fc1bdae4e17d8e55a3221496c, last accessed 8th Aug, 2016.
- Mao M, Valerio A, Austin AD, Dowton M, Johnson NF. 2012. The first mitochondrial genome for the wasp superfamily Platygastroidea: the egg parasitoid Trissolcus basalis. Genome 55:194–204. [DOI] [PubMed] [Google Scholar]
- Meusemann K, et al. 2010. A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol. 27:2451–2464. [DOI] [PubMed] [Google Scholar]
- Misof B, Misof K. 2009. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Syst Biol. 58:21–34. [DOI] [PubMed] [Google Scholar]
- Nieselt-Struwe K, von Haeseler A. 2001. Quartet-mapping, a generalization of the likelihood-mapping procedure. Mol Biol Evol. 18:1204–1219. [DOI] [PubMed] [Google Scholar]
- Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH. 2008. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol Biol Evol. 25:2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholt W, et al. 1994. Biological Control ecological considerations of the introduction of Cotesia flavipes Cameron (Hymenoptera: Braconidae) for biological control of Chilo partellus (Swinhoe) (Lepidoptera: Pyralidae) in Africa. Biocontrol News Inf. 15:19N–24N. [Google Scholar]
- Papadopoulou A, Anastasiou I, Vogler AP. 2010. Revisiting the insect mitochondrial molecular clock: the mid-Aegean trench calibration. Mol Biol Evol. 27:1659–1672. [DOI] [PubMed] [Google Scholar]
- Pennacchio F, Strand MR. 2006. Evolution of developmental strategies in parasitic Hymenoptera. Annu Rev Entomol. 51:233–258. [DOI] [PubMed] [Google Scholar]
- Pitz KM, et al. 2007. Phylogenetic relationships among the Braconidae (Hymenoptera: Ichneumonoidea): a reassessment of Shi et al. (2005). Mol Phylogenet Evol. 43:338–343. [DOI] [PubMed] [Google Scholar]
- Quicke DL, Belshaw R. 1999. Incongruence between morphological data sets: an example from the evolution of endoparasitism among parasitic wasps (Hymenoptera: Braconidae). Syst Biol. 48:436–454. [Google Scholar]
- Quicke DLJ, Basibuyuk HH, Fitton MB, Rasnitsyn AP. 1999. Morphological, palaeontological and molecular aspects of ichneumonoid phylogeny (Hymenoptera, Insecta). Zoologica Scripta 28:175–202. [Google Scholar]
- Quicke DLJ, Sharkey MJ, Laurenne NM, Dowling A. 2008. A preliminary molecular phylogeny of the Sigalphinae (Hymenoptera: Braconidae), including Pselaphanus Szepligeti, based on 28S rDNA, with descriptions of new Afrotropical and Madagascan Minanga and Malasigalphus species. J Nat Hist. 42:2703–2719. [Google Scholar]
- Quicke DLJ, van Achterberg C. 1990. Phylogeny of the subfamilies of the family Braconidae (Hymenoptera: Ichneumonoidea). Zoologische Verhandelingen Leiden 258:1–180. [Google Scholar]
- Ribeiro RC, et al. 2013. Trichospilus diatraeae (Hymenoptera: Eulophidae): a potential biological control agent of Lepidopteran pests of oil palm in the Brazilian Amazon. Flor Entomol. 96:676–678. [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O, et al. 2010. Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda. Genome Biol Evol. 2:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao RF, Campbell NJH, Schmidt ER, Barker SC. 2001. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol Biol Evol. 18:1828–1832. [DOI] [PubMed] [Google Scholar]
- Sharanowski BJ, Dowling APG, Sharkey MJ. 2011. Molecular phylogenetics of Braconidae (Hymenoptera: Ichneumonoidea), based on multiple nuclear genes, and implications for classification. Syst Entomol. 36:549–572. [Google Scholar]
- Sharkey MJ. 1997. Sigalphinae. In: Wharton RA Marsh PM Sharkey MJ. eds. Manual of the New World Genera of the Family Braconidae (Hymenoptera). Special Publication of the International Society of Hymenoptera 1, 1–439. [Google Scholar]
- Sharkey MJ, Wahl DB. 1992. Cladistics of the Ichneumonoidea (Hymenoptera). J Hymenoptera Res. 1:15–24. [DOI] [PubMed] [Google Scholar]
- Shaw M, Huddleston T. 1991. Classification and biology of braconid wasps. Handbk Ident Br Insects 7:1–126. [Google Scholar]
- Shi M, Chen XX, van Achterberg C. 2005. Phylogenetic relationships among the Braconidae (Hymenoptera: Ichneumonoidea) inferred from partial 16S rDNA, 28S rDNA D2, 18S rDNA gene sequences and morphological characters. Mol Phylogenet Evol. 37:104–116. [DOI] [PubMed] [Google Scholar]
- Shi M, Wang YN, Zhu N, Chen XX. 2013. Four heat shock protein genes of the endoparasitoid wasp, Cotesia vestalis, and their transcriptional profiles in relation to developmental stages and temperature. PLoS One 8:e59721.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Syst. 37:545–579. [Google Scholar]
- Simon C, et al. 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 87:651–701. [Google Scholar]
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69:393–403. [DOI] [PubMed] [Google Scholar]
- Song H, Sheffield NC, Cameron SL, Miller KB, Whiting MF. 2010. When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol. 35:429–448. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Stenøien HK. 2004. Adaptive basis of codon usage in the haploid moss Physcomitrella patens. Heredity 94:87–93. [DOI] [PubMed] [Google Scholar]
- Talavera G, Vila R. 2011. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol. 11:315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 302725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166-e166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJTN, Vogler AP. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic Elateriform beetles (Dryopoidea). Mol Phylogenet Evol. 63:299–304. [DOI] [PubMed] [Google Scholar]
- van Achterberg C. 1984. Essay on the phylogeny of Braconidae (Hymenoptera: Ichneumonoidea). Entomologisk Tidskrift 105:41–58. [Google Scholar]
- van Achterberg C. 2002. Revision of the genus Canalicephalus Gibson and the recognition of the Acampsohelconinae (Hymenoptera: Braconidae) as extant. Zoologische Mededelingen, Leiden 76:347–370. [Google Scholar]
- van Achterberg K, Quicke DLJ. 1992. Phylogeny of the subfamilies of the family Braconidae: a reassessment assessed. Cladistics 8:237–264. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Li Q, van Achterberg K, Chen XX. 2014. Two mitochondrial genomes from the families Bethylidae and Mutillidae: independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol Phylogenet Evol. 77:1–10. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Shi M, Chen XX, et al. 2010. New views on strand asymmetry in insect mitochondrial genomes. PLoS One 5:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Shi M, He JH, Sharkey MJ, Chen XX. 2009. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome 52:308–319. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX. 2010. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics 11:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RA, et al. 1992. Phylogeny of the subfamilies of the family Braconidae (Hymenoptera, Ichneumonoidea)—a reassessment. Cladistics 8:199–235. [DOI] [PubMed] [Google Scholar]
- Whitfield JB. 1992. The polyphyletic origin of endoparasitism in the cyclostome lineages of Braconidae (Hymenoptera). Syst Entomol. 17:273–286. [Google Scholar]
- Yancopoulos S, Attie O, Friedberg R. 2005. Efficient sorting of genomic permutations by translocation, inversion and block interchange. Bioinformatics 21:3340–3346. [DOI] [PubMed] [Google Scholar]
- Yu DS, Horstmann K, van Achterberg C. 2012. World Ichneumonoidea 2012: taxonomy, biology, morphology and distribution. CD. Taxapad, Vancouver. [Google Scholar]
- Yu RX, Shi M, Huang F, Chen XX. 2008. Immature development of Cotesia vestalis (Hymenoptera: Braconidae), an endoparasitoid of Plutella xylostella (Lepidoptera: Plutellidae). Ann Entomol Soc Am. 101:189–196. [Google Scholar]
- Zaldivar-Riverón A, Belokobylskij SA, Leon-Regagnon V, Briceno-G R, Quicke DLJ. 2008. Molecular phylogeny and historical biogeography of the cosmopolitan parasitic wasp subfamily Doryctinae (Hymenoptera: Braconidae). Invertebr Syst. 22:345–363. [Google Scholar]
- Zaldivar-Riverón A, Mori M, Quicke DL. 2006. Systematics of the cyclostome subfamilies of braconid parasitic wasps (Hymenoptera: Ichneumonoidea): a simultaneous molecular and morphological Bayesian approach. Mol Phylogenet Evol. 38:130–145. [DOI] [PubMed] [Google Scholar]
- Zaldivar-Riverón A, Shaw MR, et al. 2008. Evolution of the parasitic wasp subfamily Rogadinae (Braconidae): phylogeny and evolution of lepidopteran host ranges and mummy characteristics. BMC Evol Biol. 8:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.