Abstract
Understanding microbial adaptation to environmental stressors is crucial for interpreting broader ecological patterns. In the most extreme hot and cold deserts, cryptic niche communities are thought to play key roles in ecosystem processes and represent excellent model systems for investigating microbial responses to environmental stressors. However, relatively little is known about the genetic diversity underlying such functional processes in climatically extreme desert systems. This study presents the first comparative metagenome analysis of cyanobacteria-dominated hypolithic communities in hot (Namib Desert, Namibia) and cold (Miers Valley, Antarctica) hyperarid deserts. The most abundant phyla in both hypolith metagenomes were Actinobacteria, Proteobacteria, Cyanobacteria and Bacteroidetes with Cyanobacteria dominating in Antarctic hypoliths. However, no significant differences between the two metagenomes were identified. The Antarctic hypolithic metagenome displayed a high number of sequences assigned to sigma factors, replication, recombination and repair, translation, ribosomal structure, and biogenesis. In contrast, the Namib Desert metagenome showed a high abundance of sequences assigned to carbohydrate transport and metabolism. Metagenome data analysis also revealed significant divergence in the genetic determinants of amino acid and nucleotide metabolism between these two metagenomes and those of soil from other polar deserts, hot deserts, and non-desert soils. Our results suggest extensive niche differentiation in hypolithic microbial communities from these two extreme environments and a high genetic capacity for survival under environmental extremes.
Keywords: stress response, deserts, comparative metagenomics, Antarctica, Namib Desert, hypoliths, soils, biomes
Introduction
Understanding the mechanisms driving ecological processes in extreme environments remains a major objective (Casanueva et al. 2010; Andrei et al. 2012; Makhalanyane et al. 2015), particularly since environmental stressors are often associated with diminished ecosystem capacity and functionality (Petchey et al. 1999; Schimel et al. 2007; Harrison et al. 2013; Ferrenberg et al. 2015). Factors which are characteristic of desert soil ecosystems, in particular low levels of bioavailable water, wide temperature fluctuations, oligotrophy, and high levels of ultraviolet radiation, all contribute to high levels of biotic stress (Cowan et al. 2014; Makhalanyane et al. 2015). In such depauperate environments, some of which are marked by the total absence of higher organisms, life is often at the ‘limit’ and frequently associated with refuge niches (Pointing and Belnap 2012). Colonization is often restricted to soil surfaces (i.e., Biological Soil Crusts), or the interior (i.e., endoliths), and subsurface (i.e., hypoliths) of translucent rocks (Gorbushina 2007; Chan et al. 2012). These niche communities are recognized as ‘ecosystem engineers’ due to the important role they play in soil biogeochemical processes in the desert environment (Jones et al. 1996; Elbert et al. 2009). Hypoliths, cyanobacteria-dominated assemblages adhering to the ventral surfaces of translucent rocks, are widely distributed in both hot and cold hyperarid deserts (Stomeo et al. 2013; Valverde et al. 2015). These communities are considered to be good models for understanding the link between community structure and function, especially under severe environmental conditions such as water stress (Stevenson et al. 2015).
Adaptations to the physicochemical conditions of deserts ecosystems have been well characterized for many plants and animals; however, the responses of microbial communities to such stressors in arid environments remain poorly understood (Valentine 2007; Manzoni et al. 2012). These responses are nevertheless critical, given the known association between biodiversity and ecosystem functioning in a range of habitats, particularly in prokaryote-dominated desert soils (Swift and Anderson 1994; Yachi and Loreau 1999; Loreau et al. 2001) and the projections of climate-change-related expansion of arid ecosystems (Pointing and Belnap 2012). It is currently unclear how changing environmental conditions in desert ecosystems will affect microbial functionality. For example, the extent to which climate change may change the ecological strategies of microbial guilds in desert soils and the manner in which these processes regulate key biogeochemical processes is currently unknown (Maestre et al. 2013; Evans and Wallenstein 2014). In addition, the variations in the genetic capacity and redundancy within these systems between climatically extreme areas are also unclear, although it has been shown that changes in environmental stressors may lead to alterations in the community composition, CO2 gas exchange rates and C-allocation of biological soil crusts in Antarctic deserts (Colesie et al. 2014).
The Namib Desert, located in the coastal southwest of the African continent, is one of the oldest dryland regions in the world (Eckardt et al. 2012). The desert zone is characterized by a longitudinal water gradient (Eckardt et al. 2012), with the western coastal zone receiving some water input via sea-fog (Eckardt et al. 2012) and the eastern inland zone receiving substantial seasonal rainfall. The western and central zones are generally classified as hyperarid (Eckardt et al. 2012). A series of recent studies have demonstrated that both surface soils and the extensive quartz hypolith fields across these zones support complex microbial assemblages (Makhalanyane, Valverde, Lacap, et al. 2013; Stomeo et al. 2013; Valverde et al. 2015).
The Miers Valley is one of the smaller maritime valleys of the East Antarctic McMurdo Dry Valleys, a region which represents the largest single ice-free area of the Antarctic continent (Cowan et al. 2014). This ice-fee coastal region of East Antarctica is characterized by very low levels of precipitation, all in the form of snow, episodic katabatic winds, high salt and low organic carbon content mineral soils and extremely low mean annual temperatures (Convey 2010; Cowan et al. 2014).
Both the hot Namib Desert and the cold Miers Valley zones are characterized by distinct desert pavements, where embedded quartz rocks and pebbles form a significant component of the pavement coverage (Cowan et al. 2010). Both regions show substantial hypolithic colonization (Chan et al. 2012; Makhalanyane et al. 2015). Previous studies have shown that niche (habitat) and deterministic selection by environmental factors may play a role in structuring the microbial communities in these locales (Caruso et al. 2011; Makhalanyane, Valverde, Birkeland, et al. 2013). Although a geochip-based metagenomic study of Antarctic Dry Valley soils reported the capacity for a range of basic microbial processes (autotrophic, heterotrophic, and diazotrophic metabolism: Chan et al 2013), little is actually known of the detailed metabolic functions in these communities.
Here, we use metagenomic sequence data from mDNA extracts of hot (Namib) and cold (Antarctic) desert hypolithic communities to gain insights into stress adaptation. We have also compared these metagenomes to those of soils from other biomes including polar, hot and non-deserts. These insights contribute to our understanding of how changing climatic regimes may affect microbial communities and stress responses in soil systems and in desert-based microbial communities.
Materials and Methods
Sample Collection, DNA Extraction, and Sequencing
Hypolithic samples were collected from the Namib Desert (S 23°32.031′, E 15°01.813′: April 2010) (Vikram et al. 2016) and in Antarctica (78°60′, 164°00′E: January 2012) as described elsewhere (Makhalanyane, Valverde, Birkeland, et al. 2013; Makhalanyane, Valverde, Lacap, et al. 2013). Samples were stored at −20°C for transport, and then at −80°C until required. DNA was extracted from 0.5 g aliquots of hypolithic biomass (n = 50 for both habitats) using the PowerSoil DNA Isolation Kit (MoBio, West Carlsbad, CA, USA) following the manufacturer’s instructions. DNA preparations from each habitat were combined before sequencing at GATC (Konstanz, Germany) using Illumina HiSeq-2000 paired-end technology (2 × 101 bases). An in-house python script was used to trim 5′ ambiguous bases (N) of reads and to filter bases with a quality score below 25. Assembly of high-quality reads into contigs was performed by Velvet v1.2.10 (Zerbino and Birney 2008) with a hash length of 51 (Vikram et al. 2016). The reads were then quantified against the assembled contigs (Li et al. 2009; Li and Durbin 2010; Quinlan and Hall 2010).
Taxonomic and Functional Analyses
Contigs (with depth of coverage ≥5×) were compared with the Non Redundant (NR) NCBI database by performing BlastX using sequences with E values <1e−5. The results were then imported into MEGAN (Huson et al. 2007), which uses the lowest common ancestor algorithm to assign sequences to the most specific taxonomic group. Open reading frames (ORFs) were identified on contigs using MetaGeneMark (Zhu et al. 2010). The ORFs of both hypolith metagenomes were used to perform BlastP (E values ≤1e−5) against the Cluster of Orthologous groups (COGs) (Tatusov et al. 2000) and an in-house stress response protein (SRPs) database, constructed from 45 ‘seed’ genes related to stress response obtained from the GeoChip database (Tu et al. 2014). Only genes related to stress response, including genes that belong to any one of five functional ‘stress’ categories: temperature, osmolarity, oxidative stress and oxygen limitation, nutrient limitation, and protein stress, were retrieved. These gene families were chosen because they are covered by more than 10,000 sequences and are also widely used to analyze microbial communities from various habitats (Tu et al. 2014).
The key word queries were first submitted to the GenBank Protein Database to recover all candidate amino acid sequences, which were then used to search against the NR NCBI database with an E value ≤1e−20 and a percent identity ≥99%. Gene families were obtained by assigning orthologous groups based on the OrthoMCL database (Li et al. 2003; Chen et al. 2006). Duplicate sequences were removed before further analysis. Ultimately, the SRP database contained 557 proteins, divided into 42 orthologous groups [six gene families, namely cspA, cspB (cold shock proteins); tnr, glnR (regulatory genes for nitrogen limitation), and katA, katE (peroxidase catalase) were assigned to three orthologous groups, reducing the number of orthogroups from 45 to 42].
Statistical Analysis
To identify biologically relevant differences between the two metagenomes, Statistical Analysis of Metagenomic Profiles (STAMP) software was used (Parks and Beiko 2010; Parks et al. 2014). The inputs were tables of the relative abundance of sequences, assigned to different taxa and functional systems (i.e., COGs and SRPs) for each metagenome. Statistically significant differences between functional systems and taxonomic groups of both metagenomes were established by Fisher’s exact test. The Newcombe–Wilson method was used for calculating confidence intervals (nominal coverage of 95%). A false-discovery-rate method (Benjamini-Hochberg method) was applied to indicate the percentages of false positives (reported q values). All features with a q-value of >0.05 were removed.
Comparison of Hypolith and Other Soil Metagenomes
A multiple comparative analysis of the two hypolith metagenomes and different hot, cold, and nondesert metagenomes was performed using the MG-RAST server. An additional 16 metagenomes were compared with both hypolith metagenomes, including three hot desert metagenomes (MD3, SF2, and SV1), six polar metagenomes (EB017, EB019, EB020, EB021, EB024, and EB026), and seven non-desert metagenomes (AR3, BZ1, CL1, DF1, KP1, PE6, and TL1) (Fierer et al. 2012). The contigs of both hypolith metagenomes were compared with protein-coding gene databases using Meta Genome Rapid Annotation with Subsystem Technology (MG-RAST) server version 3 (Meyer et al. 2008). MG-RAST applies BlastX to compare contigs with the COGs database. Only matches with an E value of ≤10−5 were included. The COG results of sixteen biomes were retrieved from the MG-RAST server and ORFs were predicted using the MetaGeneMark annotator. These ORFs were further compared with sequences in the SRPs database by using BlastP with a maximum E value of 1e−5. Tables of frequencies of best hits for each functional group (i.e., COG and SRP gene families) were generated for each metagenome and normalized by applying the “scale” function in R (version 3.1.1).
We applied agglomeration hierarchical clustering analysis to group metagenomic data into groups (clusters) according to similarities by using a Pearson correlation dissimilarity metric and an average clustering criterion. The inputs of the cluster analysis were the tables of frequencies of sequences, which were the best matches to COGs and SRPs databases, for each metagenome. All analyses were performed in R. Bootstrap analysis was performed by applying the “Pvclust” package (Suzuki and Shimodaira 2006) to the object data. Pvclust calculates probabilities values (P-values) for each cluster using bootstrap resampling techniques. Ten thousand bootstrap samples were generated by randomly sampling elements from the data, and bootstrap replicates of the dendogram were obtained through repeatedly applying cluster analysis (Efron 1979; Felsenstein 1981). The AU value was then used to determine whether the two hypolith metagenomes were significantly similar.
Nucleotide Accession Number
The high-quality paired end short reads for Namib and Antarctic hypoliths metagenome were submitted to the NCBI under the SRP IDs SRP061443 and SRP073918, respectively. The SRA accession number are SRR2124832 (Namib hypoliths) and SRR3471615 (Antarctic hypoliths).
Results and Discussion
Hypolith Communities in Hyperarid Environments
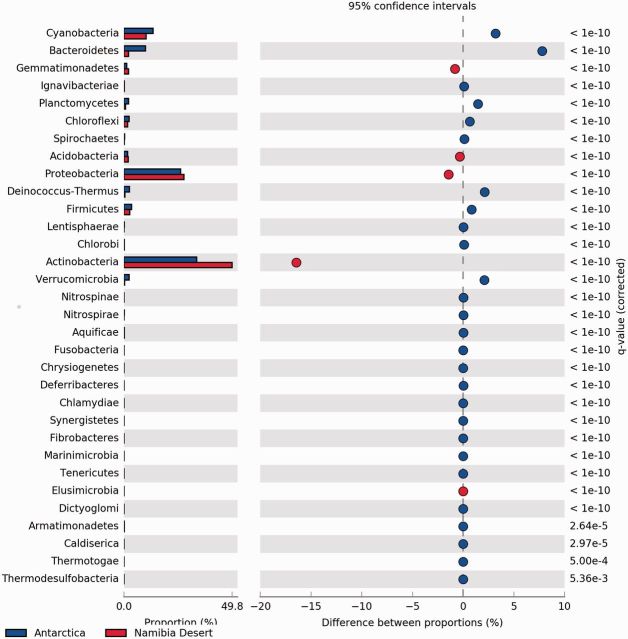
To better understand the relationship between biodiversity and microbial stress–response genes, we first compared the taxonomic composition of the two hypolithic metagenomes (Antarctic and the Namib Desert). Bacterial species abundance, as determined by the relative number of phylotypic assignments, was similar for the Antarctic and Namib Desert metagenomes, that is, 92% of reads. High bacterial abundance in desert niches is consistent with previous studies of both hot and cold desert soils (Pointing and Belnap 2012; Makhalanyane, Valverde, Birkeland, et al. 2013; Makhalanyane, Valverde, Lacap, et al. 2013). Our analysis indicated that three phyla, namely Actinobacteria, Proteobacteria, and Cyanobacteria, were the most abundant in both hypolithic metagenomes, albeit in different proportions (fig. 1).
Fig. 1.—
Statistical analysis of taxonomic profiles (at phylum level) of Antarctic and the Namib Desert metagenomes by BlastX of contigs against Non Redundant NCBI database. Phyla, which were in high relative abundance, have significant indicators.
Interestingly, Cyanobacteria, which are known to be functionally important as photoautotrophs in desert soils (Valverde et al. 2015), contributed a statistically lower percentage of Namib Desert sequences (10%) compared with Antarctic sequences (∼13%). Members of the Oscillatoriales, Chroococcales, and Nostocaceae lineages, were the dominant Cyanobacteria in both metagenomes. Two orders, the Oscillatoriophycideae and Nostocales, were more significant in the Namib Desert than in Antarctic samples. Within Nostocales, the Nostaceae family was more abundant in the Namib Desert (19%) than in Antarctic desert (7%). Within the Oscillatoriophycideae, Chroococcales were more abundant in the Antarctic metagenome (47%) than in the Namib Desert (18%) (supplementary table S1, Supplementary Material online).
Actinobacteria phylotypic sequences were more abundant in the Namib Desert (49%) metagenome than in the Antarctic metagenome (33%). Proteobacteria accounted for 26% of the Antarctic sequences and 27% of the Namib Desert sequences, while Bacteroidetes were 4-fold more abundant in the Antarctic than in the Namib Desert sequence dataset (fig. 1). Other phyla, such as Planctomycetes, Deinococcus-Thermus, Verrucomicrobiota, Firmicutes, and Chloroflexi, were generally more abundant in the Antarctic than in the Namib Desert metagenomes (fig. 1). Within Proteobacteria, there were also differences between the two hypolithons: Beta- and Gammaproteobacteria were more common in the Antarctica than in the Namib Desert dataset, while Alphaproteobacteria sequences showed the reverse trend (supplementary fig. S1, Supplementary Material online).
Comparisons of metagenomic sequences and from 16S rRNA gene amplicon sequences showed some notable differences (Makhalanyane, Valverde, Birkeland, et al. 2013; Makhalanyane, Valverde, Lacap, et al. 2013). The discrepancies in apparent diversity between the metagenome and amplicon sequence datasets might be attributable to several factors (Raes et al. 2007; Steven et al. 2012; Poretsky et al. 2014). First, more phyla were identified in the metagenomic analysis than in the amplicon analysis. Second, we found significant differences in the frequency of dominant phyla (Actinobacteria, Proteobacteria, Cyanobacteria, and Bacteroidetes). Third, it is known that species richness (number of species) and evenness (relative abundance of species) values can be affected by analytical factors such as amplicon length and primer pair choice (Poretsky et al. 2014). Fourth, the use of different and incomplete databases (Raes et al. 2007) to identify and bin environmental sequences into taxonomic units may also substantially influence diversity results and ecological inferences (Steven et al. 2012). Our results suggest that phylogenetic inference from shotgun metagenome may be a more valid method for taxonomic analysis of microbial communities.
To assess the validity of the phylogenetic data derived from the metagenome sequences, we used various methods of binning reads to SSU or universal marker genes, including Metaxa, MetaPhlAn, and mOTU (supplementary fig. S2, Supplementary Material online). All analysis showed that the frequencies of members of the four major phyla (i.e., Actinobacteria, Proteobacteria, Cyanobacteria, and Bacteroidetes) were relatively similar.
Comparatively little is known about lower eukaryote diversity in hyperarid soil systems (but see for example Rao et al. 2011; Gokul et al. 2013). We found a low proportion of Antarctic (0.2%) and Namib Desert (∼3%) sequences that matched known eukaryotic taxa, with fungal phylotypes being 4-fold more abundant in the Namib Desert metagenome than in the Antarctic metagenome. The majority of fungal phylotypes in the Namib Desert metagenome were assigned to Ascomyceta, in which Eurotiomycetes, Sordariomyceta, and Dothideomyceta were most highly represented (supplementary table S2, Supplementary Material online). Although a number of phylogenetic studies have focused on hypolithic microbial communities’ composition, few have included lower eukaryote diversity analyses (Pointing et al. 2009; Rao et al. 2011; Gokul et al. 2013), and those that exist are almost exclusively from cold desert habitats. A recent study reported a number of uncommon fungal phylotypes, such as Sclerotinia homeocarpa in Namib Desert soil fungal communities (Ramond et al. 2014). Ascomycete phylotypes have previously been identified in Antarctic hypolithic fungal communities based on 18S RNA gene marker analysis (Rao et al. 2011). Most of these phylotypes were related to the genus Acremonium, while some sequences were affiliated to Stromatonectria and Verrucaria. Other studies have found that Ascomycetes and Basidiomycetes were patchily distributed in Antarctic mineral soils (Fell et al. 2006; Pointing et al. 2009), whereas Ascomycetes had been reported as the only fungal taxa present in hypolithic communities (Khan et al. 2011). Free-living fungi are thought to be more susceptible to environmental extremes than bacteria (Cowan et al. 2014), However, the discovery of similar fungal ITS sequences from the Antarctic Ross Sea “expedition huts” (wood-degrading fungal isolates) and from geographically remote soils suggested that these fungal taxa were indigenous to Antarctic soils but responsive to organic matter brought into the region by the early Antarctic expeditions (Arenz et al. 2006). These findings are in agreement with the view that the degree of severity of an environment and the fungal diversity are inversely correlated (Rao et al. 2011). In addition, the lower fungal diversity of Antarctic hypoliths, compared with Namib Desert hypoliths, suggests that the cold Antarctic Dry Valley desert soils represent the more ‘extreme’ of the two environments (i.e., has lower water bioavailability and more extreme oligotrophy).
We found archaeal phylotypic sequences in even lower proportions. Only 0.43% Antarctic sequences and 0.38% of Namib Desert sequences matched with known archaeal phylotypes. Our analysis indicated that Halobacteria and Methanomicrobia (both belonging to the phylum Euryarchaeota) were predominant in both hypolith metagenomes. Nitrososphaerales (belonging to the phylum Thaumarchaeota) were more numerous in the Namib Desert metagenome (supplementary table S3, Supplementary Material online). Nevertheless, our results further expand the limited knowledge of archaea in these habitats. Archaea were thought to be largely absent from both hot and cold desert hypolithic communities, as early attempts to detect archaeal rRNA genes using molecular methods were unsuccessful (Smith et al. 2006; Warren-Rhodes et al. 2007; Pointing et al. 2009). Archaeal 16S rRNA gene signals have only recently been detected in Antarctic soils (Ayton et al. 2010; Bates et al. 2011). By comparison, archaeal sequences (i.e., Euryarchaeota) made up 8–40% of the community in the salt marsh located in the Thar Desert in the Kutch District of Gujarat (India), presumably due to the halophilic nature of this desert habitat (Pandit et al. 2014). The low occurrence of archaea may be linked to their inability to tolerate the xeric environmental stress conditions (Pointing et al. 2009), and these taxa are not thought to play a significant role in the development or function of niche habitats such as desert hypolithons (Khan et al. 2011).
Inferring Functional Capacity from Antarctic and Namib Desert Hypolith Metagenomes
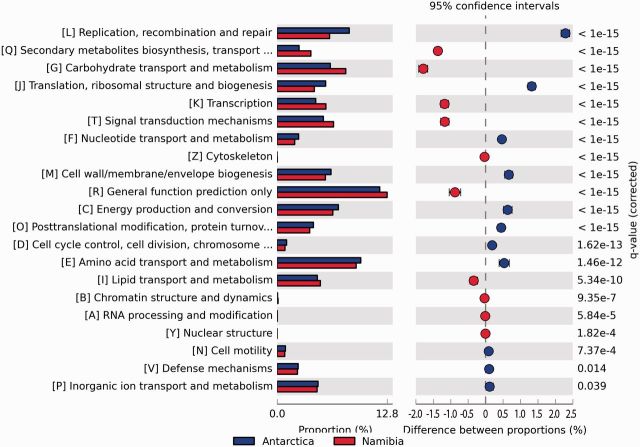
From a total of 867,382 ORFs in the Antarctic metagenome and 396,495 ORFs in the Namib Desert metagenome, only 47% and 56% ORFs, respectively, could be assigned to functional groups in the COGs database. To identify where the hot and cold desert hypolith metagenomes differed in their apparent functional capacity, a quantitative comparison of 23 COG functional categories was performed.
Significant differences between the two metagenomes were observed in many categories (fig. 2), indicating distinct genetic capacities between the two systems. The Antarctic metagenome contained more genes assigned to categories L, J, F, M, C, and O, whereas the Namib Desert metagenome had more genes assigned to categories Q, G, K, T, R, and I. The specific COG categories that were more abundant in the Namib Desert metagenome included carbohydrate metabolism and transport (G), secondary metabolite biosynthesis, transport, and catabolism (Q) (fig. 2). This was mainly due to a high number of proteins associated with permeases of the major facilitator super-family (COG0477), and dehydrogenases (COG1028) (supplementary fig. S3, Supplementary Material online). The predominance of genes related to carbohydrate metabolism and associated processes in the Namib Desert metagenome is probably related to seasonal biomass production in the Namib Desert (Southgate et al. 1996), which provides an intermittent but regular source of polymeric lignocellulosic substrates for heterotrophic metabolism. In contrast, the extreme conditions of the Antarctic Dry Valleys desert preclude the existence of any higher plants. Nevertheless, the presence in the Antarctic soil metagenome of degradative genes for polymeric substrates (such as lignins: Chan et al. 2013) suggests alternative sources of polymeric aromatic substrates, such as moss or algal tissues.
Fig. 2.—
Statistical analysis of functional gene analysis based on the COGs database.
The predominance of the genes assigned to the processes of replication, recombination and repair (L) in the Antarctic metagenome may be due to a higher number of proteins associated with bacterial transposases (COG2801) (supplementary fig. S3, Supplementary Material online). The high number of transposase genes allows us to speculate that microorganisms surviving in the more extreme Antarctic desert soils acquire a competitive advantage from transposase-associated evolutionary and adaptive processes (e.g., gene rearrangements, lateral gene transfer, etc.).
Genes Involved in Stress Response
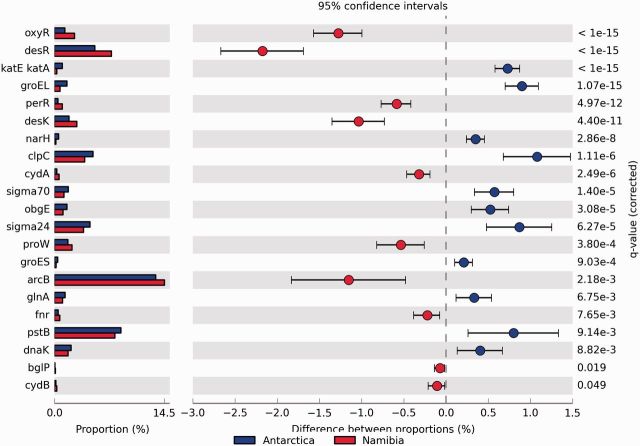
To decipher the role of environmental stressors in hot and cold hyperarid environments, we compared stress response genes in the two metagenomes. Our analysis showed that about 3.5% ORFs in the Antarctic and 3.9% in the Namib Desert metagenomes could be assigned to genes implicated on stress resistance using the SRP database (Blast P, E-value ≤1e−3) but that only a low number of gene families were statistically significantly different between the two datasets (fig. 3).
Fig. 3.—
Statistical analysis of functional gene analysis based on SRP database.
Two histidine kinases (desR and desK) known to be involved in temperature regulation (Mascher et al. 2006) were significantly enriched in the Namib Desert metagenome compared with Antarctic metagenome. Six genes families (oxyR, perR, cydA, cydB, fnr, and arcB) involved in oxidative stress and oxygen limitation (Lüthi et al. 1986; Melville and Gunsalus 1996; Kana et al. 2001; Chen et al. 2008) were also enriched in the Namib Desert metagenome compared with the Antarctic metagenome. In contrast, genes that were more abundant in the Antarctic metagenome included pstB, glnA, two sigma genes (sigma 24 and sigma 70), and opuE [encoding a transport system for osmoprotective proline uptake (Spiegelhalter and Bremer 1998)]. The gene pstB is one of the four genes (pstS, pstC, pstA, and pstB) encoding proteins required for phosphate transport via the Pst system (Qi et al. 1997), while glnA (glutamine synthase) is subject to transcriptional regulation in response to changes in nitrogen availability (Reyes et al. 1997). One of two genes belonging to the bglPH operon (bglH), which is implicated in glucose utilization, was present in both hypolith metagenomes. However, no Namib Desert sequences could be assigned to the bglP gene (fig. 4).
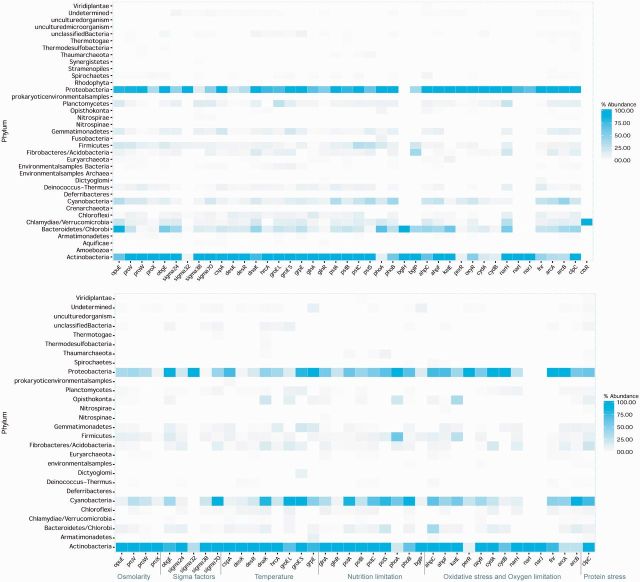
Fig. 4.—
Heat-map of genes assigned to SRPs, with the proportion of sequences corresponding to taxonomic binning. (a) The Antarctic metagenome, (b) the Namib Desert metagenome.
In general, stress response genes were attributed to common phyla in both hypolith metagenomes, including Proteobacteria, Bacteroidetes, Actinobacteria, and Cyanobacteria (fig. 4). However, gene families implicated in oxidative stress and oxygen limitation were more abundant in the Namib Desert metagenome than in the Antarctic metagenome; conversely, sigma factor gene families were more abundant in the Antarctic metagenome (fig. 3). No significant differences were observed between two hypolith metagenomes in terms of other functional gene family categories: osmolarity, nutrition limitation, and protein stress.
Moisture Stress Response
Water is the primary determinant of life in desert ecosystems. In the Namib Desert, both rainfall and fog contribute to the annual precipitation budget (Henschel and Seely 2008; Eckardt et al. 2012), and both have been shown to be important for hot desert hypolithic communities (Pointing et al. 2007; Warren-Rhodes et al. 2007; Azúa-Bustos et al. 2011). In Antarctic Dry Valley desert soils, where precipitation is solely in the form of snow, liquid water is derived from both snowmelt and the seasonally-melted permafrost layer (Cowan and Ah Tow 2004). In comparing the water budgets of the two environments, it is evident that xeric stress is much greater in Antarctica deserts than in Namib Desert soils.
Our analysis showed a higher proportion of genes affiliated to replication, recombination and repair processes in the Antarctic metagenome (fig. 2), which may be a consequence of the higher desiccation levels (Hawes et al. 1992). Conversely, our analysis showed a comparatively higher proportion of genes linked to the metabolism and biosynthesis COG categories in the Namib Desert, suggesting that this community possesses a much higher metabolic capacity (and therefore resilience) than Antarctic edaphic systems (Cowan and Ah Tow 2004).
Comparison of Hyperarid Hypolith Metagenomes and Other Biomes
Hypolithic communities present valuable models for exploring niche differentiation in extreme environments. To further explore both the nature of hypolithic microbial community structures and the concept of niche differentiation in these habitats, we compared the hot and cold hyperarid desert hypolithic metagenomes to metagenomes from other edaphic biomes including both hot and cold deserts, and nondesert soils.
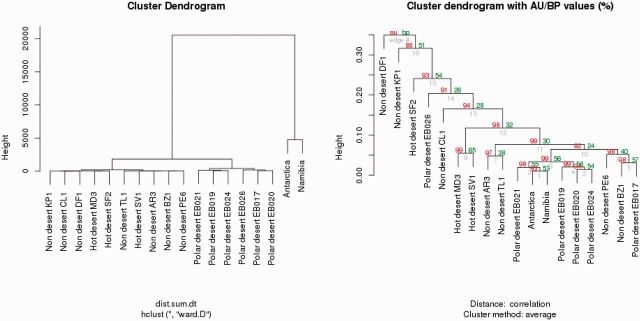
The two hyperarid hypolith metagenomes clearly contained a higher proportion of genes assigned to SRPs than other biomes (fig. 5a). In addition, both hypolith metagenomes formed a statistically significant cluster with an AU value of 90% when we performed a cluster analysis of all biomes based on the SRPs database (fig. 5b).
Fig. 5.—
Cluster analysis of the two hyper arid hypolith metagenomes against other biomes based on the SRP database.
These results suggest that stress response elements in microorganisms inhabiting both hypolith niches, despite their widely different thermal characteristics, are more similar to each other than to those from organisms inhabiting open soil niches. Our analysis therefore highlights the unique physicochemical characteristics of the hypolithic habitat relative to other systems. Exposure to high levels of desiccation provides a valid explanation for the higher abundance of genes associated with stress proteins (Chan et al. 2013).
When we analyzed all metagenomic datasets for genes associated with the COG categories amino acid transport and metabolism, we found that the two hypolithic metagenomes formed a unique cluster with very strong support (AU value of 99%). Similar differential groupings of nucleotide metabolism, transport elements (AU value of 89%), and lipid transport and metabolism elements (AU value of 97%) were also observed, supporting the view that hypolith communities may harbor distinct physiological functions (supplementary fig. S4, Supplementary Material online) compared with open soil communities. The higher abundance in the two hyperarid bacterial communities of the genes associated with amino acid and nucleotides metabolism, which are commonly associated with osmoregulation in bacteria (Harris 1981; Chan et al. 2013), may indicate that hypolithic microbial communities have a higher capacity to respond to osmotic stress than open soil communities.
The most striking observation from the cluster analysis of hypoliths, cold, hot, and nondesert soil metagenomes was found in the category for defense mechanisms, which included genes involved in biosynthesis of antibiotic resistance compounds (such as beta lactamases, streptomycin, and chloramphenicol) (supplementary fig. S4, Supplementary Material online). Both hypolith metagenomes showed more similarity to desert soil metagenomes (AU value of 96%) than to nondesert soil metagenomes. This finding is consistent with the observation that desert and nondesert soil microbial communities show differences in abundance in antibiotic resistance genes and that genes associated with antibiotic resistance were far less abundant in desert soils than in nondesert soils (Chan et al. 2013).
We argue that the clustering of the metagenomic data is clear evidence of niche differentiation. Niche differentiation, the tendency for coexisting populations to occupy different niches or environmental requirements, has been demonstrated previously in microbial mats (Wong et al. 2015). However, this is a relatively poorly defined concept for microbial communities (compared to plants). Stress response is a valid niche dimension for microbial communities in these conditions, and our results have shown marked differences between hypoliths in hyperarid deserts versus other edaphic biomes.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the following organizations: The University of Pretoria Genomics Research Institute (PTL, TPM, DAC, and YVdP), South African National Antarctic Program (SANAP) and the National Research Foundation (NRF) for funding (LG, SV). We also thank Antarctica New Zealand and the Desert Research and Training Centre (Namibia) for logistics support. YVdP also acknowledges support from Ghent University [Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks”].
Literature Cited
- Andrei A-Ş, Banciu HL, Oren A. 2012. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol Lett. 330:1–9. [DOI] [PubMed] [Google Scholar]
- Ayton J, Aislabie J, Barker GM, Saul D, Turner S. 2010. Crenarchaeota affiliated with group 1.1b are prevalent in coastal mineral soils of the Ross Sea region of Antarctica. Environ Microbiol. 12:689–703. [DOI] [PubMed] [Google Scholar]
- Azúa-Bustos A, et al. 2011. Hypolithic cyanobacteria supported mainly by fog in the coastal range of the Atacama Desert. Microb Ecol. 61:568–581. [DOI] [PubMed] [Google Scholar]
- Arenz BE, et al. 2006. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol Biochem. 38(10):3057–3064. [Google Scholar]
- Bates ST, et al. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso T, et al. 2011. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 5:1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva A, Tuffin M, Cary C, Cowan DA. 2010. Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol. 18:374–381. [DOI] [PubMed] [Google Scholar]
- Chan Y, et al. 2012. Hypolithic microbial communities: between a rock and a hard place. Environ Microbiol. 14:2272–2282. [DOI] [PubMed] [Google Scholar]
- Chan Y, Van Nostrand JD, Zhou J, Pointing SB, Farrell RL. 2013. Functional ecology of an Antarctic Dry Valley. Proc Natl Acad Sci. 110:8990–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34:D363–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, et al. 2008. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One 3:e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colesie C, Green TGA, Haferkamp I, Büdel B. 2014. Habitat stress initiates changes in composition, CO2 gas exchange and C-allocation as life traits in biological soil crusts. ISME J. 8:2104–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convey P. 2010. Terrestrial biodiversity in Antarctica—recent advances and future challenges. Polar Sci. 4:135–147. [Google Scholar]
- Cowan DA, Ah Tow L. 2004. Endangered antarctic environments. Annu Rev Microbiol. 58:649–690. [DOI] [PubMed] [Google Scholar]
- Cowan DA, Khan N, Pointing SB, Cary SC. 2010. Diverse hypolithic refuge communities in the McMurdo Dry Valleys. Antarct Sci. 22:714–720. [Google Scholar]
- Cowan DA, Makhalanyane TP, Dennis PG, Hopkins DW. 2014. Microbial ecology and biogeochemistry of continental Antarctic soils. Front Microbiol. 5:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt F, et al. 2012. The nature of moisture at Gobabeb, in the central Namib Desert. J Arid Environ. 93:7–19. [Google Scholar]
- Efron B. 1979. Bootstrap methods: another look at the jackknife. Ann Stat. 7:1–26. [Google Scholar]
- Elbert W, Weber B, Büdel B, Andreae M, Pöschl U. 2009. Microbiotic crusts on soil, rock and plants: neglected major players in the global cycles of carbon and nitrogen? Biogeosci Discuss. 6:6983–7015. [Google Scholar]
- Evans SE, Wallenstein MD. 2014. Climate change alters ecological strategies of soil bacteria. Ecol Lett. 17:155–164. [DOI] [PubMed] [Google Scholar]
- Fell JW, Scorzetti G, Connell L, Craig S. 2006. Biodiversity of micro-eukaryotes in Antarctic Dry Valley soils with<5% soil moisture. Soil Biol Biochem. 38:3107–3119. [Google Scholar]
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17:368–376. [DOI] [PubMed] [Google Scholar]
- Ferrenberg S, Reed SC, Belnap J. 2015. Climate change and physical disturbance cause similar community shifts in biological soil crusts. Proc Natl Acad Sci U S A. 112:12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, et al. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A. 109:21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokul JK, Valverde A, Tuffin M, Cary SC, Cowan DA. 2013. Micro-eukaryotic diversity in hypolithons from miers valley, Antarctica. Biology 2:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbushina AA. 2007. Life on the rocks. Environ Microbiol. 9:1613–1631. [DOI] [PubMed] [Google Scholar]
- Harris R. 1981. Effect of water potential on microbial growth and activity In: Parr J, Gardner W, Elliott L, editors. Madison, WI: Soil Science Society of America; 23–95. [Google Scholar]
- Harrison JP, Gheeraert N, Tsigelnitskiy D, Cockell CS. 2013. The limits for life under multiple extremes. Trends Microbiol. 21:204–212. [DOI] [PubMed] [Google Scholar]
- Hawes I, Howard-Williams C, Vincent WF. 1992. Desiccation and recovery of Antarctic cyanobacterial mats. Polar Biol. 12:587–594. [Google Scholar]
- Henschel JR, Seely MK. 2008. Ecophysiology of atmospheric moisture in the Namib Desert. Atmos Res. 87:362–368. [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CG, Lawton JH, Shachak M. 1996. Organisms as ecosystem engineers. Ecosyst Manage. 69:373–386. [Google Scholar]
- Kana BD, et al. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J Bacteriol. 183:7076–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, et al. 2011. Hypolithic microbial communities of quartz rocks from Miers Valley, McMurdo Dry Valleys, Antarctica. Polar Biology, Springer.
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinforma Oxf Engl. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreau M, et al. 2001. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294:804–808. [DOI] [PubMed] [Google Scholar]
- Lüthi E, Mercenier A, Haas D. 1986. The arcABC operon required for fermentative growth of Pseudomonas aeruginosa on arginine: Tn5-751-assisted cloning and localization of structural genes. J Gen Microbiol. 132:2667–2675. [DOI] [PubMed] [Google Scholar]
- Maestre FT, et al. 2013. Changes in biocrust cover drive carbon cycle responses to climate change in drylands. Glob Change Biol. 19:3835–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhalanyane TP, Valverde A, Birkeland N-K, et al. 2013. Evidence for successional development in Antarctic hypolithic bacterial communities. ISME J. 7:2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhalanyane TP, Valverde A, Lacap DC, et al. 2013. Evidence of species recruitment and development of hot desert hypolithic communities. Environ Microbiol Rep. 5:219–224. [DOI] [PubMed] [Google Scholar]
- Makhalanyane TP, et al. 2015. Microbial ecology of hot desert edaphic systems. FEMS Microbiol Rev. 39:203–221. [DOI] [PubMed] [Google Scholar]
- Manzoni S, Schimel JP, Porporato A. 2012. Responses of soil microbial communities to water stress: results from a meta-analysis. Ecology 93:930–938. [DOI] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev MMBR 70:910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville SB, Gunsalus RP. 1996. Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc Natl Acad Sci U S A. 93:1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, et al. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit AS, et al. 2014. Metagenomes from the saline desert of kutch. Genome Announc. 2:e00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. 2010. Identifying biologically relevant differences between metagenomic communities. Bioinforma Oxf Engl. 26:715–721. [DOI] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinforma Oxf Engl. 30:3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petchey OL, McPhearson PT, Casey TM, Morin PJ. 1999. Environmental warming alters food-web structure and ecosystem function. Nature 402:69–72. [Google Scholar]
- Pointing SB, Warren-Rhodes KA, Lacap DC, Rhodes KL, McKay CP. 2007. Hypolithic community shifts occur as a result of liquid water availability along environmental gradients in China’s hot and cold hyperarid deserts. Environ Microbiol. 9:414–424. [DOI] [PubMed] [Google Scholar]
- Pointing SB, et al. 2009. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci U S A. 106:19964–19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointing SB, Belnap J. 2012. Microbial colonization and controls in dryland systems. Nat Rev Microbiol. 10:551–562. [DOI] [PubMed] [Google Scholar]
- Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D, Konstantinidis KT. 2014. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Kobayashi Y, Hulett FM. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J Bacteriol. 179:2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma Oxf Engl. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Foerstner KU, Bork P. 2007. Get the most out of your metagenome: computational analysis of environmental sequence data. Curr Opin Microbiol. 10:490–498. [DOI] [PubMed] [Google Scholar]
- Ramond J-B, Pienaar A, Armstrong A, Seely M, Cowan DA. 2014. Niche-partitioning of edaphic microbial communities in the Namib Desert gravel plain Fairy Circles. PLoS One 9:e109539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, et al. 2011. Low-diversity fungal assemblage in an Antarctic Dry Valleys soil. Polar Biol. 35:567. [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ. 1997. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol. 179:2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimel J, Balser TC, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Tow LA, Stafford W, Cary C, Cowan DA. 2006. Bacterial diversity in three different Antarctic Cold Desert mineral soils. Microb Ecol. 51:413–421. [DOI] [PubMed] [Google Scholar]
- Southgate R, Masters P, Seekly M. 1996. Precipitation and biomass changes in the Namib Desert dune ecosystem. 33(3):267–280. [Google Scholar]
- Spiegelhalter F, Bremer E. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol Microbiol. 29:285–296. [DOI] [PubMed] [Google Scholar]
- Steven B, Gallegos-Graves LV, Starkenburg SR, Chain PS, Kuske CR. 2012. Targeted and shotgun metagenomic approaches provide different descriptions of dryland soil microbial communities in a manipulated field study. Environ Microbiol Rep. 4:248–256. [DOI] [PubMed] [Google Scholar]
- Stevenson A, et al. 2015. Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. Environ Microbiol. 17:257–277. [DOI] [PubMed] [Google Scholar]
- Stomeo F, et al. 2013. Hypolithic and soil microbial community assembly along an aridity gradient in the Namib Desert. Extrem Life Extreme Cond. 17:329–337. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinforma Oxf Engl. 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- Swift M, Anderson J. 1994. Biodiversity and ecosystem function in agricultural systems. Biodiversity and ecosystem function. Heidelberg: Springer, pp. 15–41. [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, et al. 2014. GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour. 14:914–928. [DOI] [PubMed] [Google Scholar]
- Valentine DL. 2007. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 5:316–323. [DOI] [PubMed] [Google Scholar]
- Valverde A, Makhalanyane TP, Seely M, Cowan DA. 2015. Cyanobacteria drive community composition and functionality in rock-soil interface communities. Mol Ecol. 24:812–821. [DOI] [PubMed] [Google Scholar]
- Vikram S, et al. 2016. Metagenomic analysis provides insights into functional capacity in a hyperarid desert soil niche community. Environ Microbiol. 18(6):1875–1888. [DOI] [PubMed] [Google Scholar]
- Warren-Rhodes KA, et al. 2007. Cyanobacterial ecology across environmental gradients and spatial scales in China’s hot and cold deserts. FEMS Microbiol Ecol. 61:470–482. [DOI] [PubMed] [Google Scholar]
- Wong HL, Smith D-L, Visscher PT, Burns BP. 2015. Niche differentiation of bacterial communities at a millimeter scale in Shark Bay microbial mats. Sci Rep. 5:15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi S, Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proc Natl Acad Sci U S A. 96(4):1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.