Abstract
Molecular chaperones, also known as heat-shock proteins, refold misfolded proteins and help other proteins reach their native conformation. Thanks to these abilities, some chaperones, such as the Hsp90 protein or the chaperonin GroEL, can buffer the deleterious phenotypic effects of mutations that alter protein structure and function. Hsp70 chaperones use a chaperoning mechanism different from that of Hsp90 and GroEL, and it is not known whether they can also buffer mutations. Here, we show that they can. To this end, we performed a mutation accumulation experiment in Escherichia coli, followed by whole-genome resequencing. Overexpression of the Hsp70 chaperone DnaK helps cells cope with mutational load and completely avoid the extinctions we observe in lineages evolving without chaperone overproduction. Additionally, our sequence data show that DnaK overexpression increases mutational robustness, the tolerance of its clients to nonsynonymous nucleotide substitutions. We also show that this elevated mutational buffering translates into differences in evolutionary rates on intermediate and long evolutionary time scales. Specifically, we studied the evolutionary rates of DnaK clients using the genomes of E. coli, Salmonella enterica, and 83 other gamma-proteobacteria. We find that clients that interact strongly with DnaK evolve faster than weakly interacting clients. Our results imply that all three major chaperone classes can buffer mutations and affect protein evolution. They illustrate how an individual protein like a chaperone can have a disproportionate effect on the evolution of a proteome.
Keywords: molecular chaperones, mutational robustness, experimental evolution, protein evolution, Escherichia coli, DnaK
Introduction
Robustness is one of the fundamental properties of living systems (de Visser et al. 2003; Wagner 2005; Masel and Siegal 2009; Fares 2015). This property describes the ability of a biological system to preserve its phenotype in a particular environment despite perturbations that it encounters. The robustness of a system against perturbations that are environmental (e.g., a change in temperature) is referred to as environmental robustness, whereas robustness against perturbations caused by genetic mutations receives the name of mutational or genetic robustness. Molecular chaperones (Ellis 1987) are one of the best-known sources of both types of robustness (Fares 2015). Chaperones, also called heat-shock proteins, assist proteins in reaching their native conformations, prevent protein aggregation, and refold misfolded proteins (Young et al. 2004; Hartl and Hayer-Hartl 2009; Hartl et al. 2011). Thanks to these roles, chaperones can restore the native conformation of proteins destabilized by environmental perturbations, thus providing environmental robustness to organisms coping with stressful conditions. Because some chaperones can buffer the deleterious effects of mutations that affect protein folding, they are also a source of mutational robustness. In the context of protein evolution, chaperones are able to increase a protein’s mutational robustness because they alter the mapping from protein genotypes into protein phenotypes, that is, into the structures that proteins form (Rutherford 2003). Specifically, they increase the number of amino acid sequences that fold into the same structure and that can perform the function associated with this structure.
There are three main chaperone systems, which are the Hsp90 system, the Hsp70 system, and the Hsp60 system (or chaperonins), of which the bacterial GroEL is a prominent member (Hartl et al. 2011). Overwhelming evidence shows that Hsp90 and GroEL can buffer mutations (Bogumil and Dagan 2012), but whether the same holds for any major chaperone from the Hsp70 system is to our knowledge unknown. A recent study has shown that RNA chaperones—they help RNA molecules to fold properly, and comprise a class of chaperones different from these three systems—can also buffer deleterious mutations in Escherichia coli (Rudan et al. 2015).
Pioneering work carried out by Rutherford and Lindquist (1998) showed that inhibition of the chaperone Hsp90 can unveil cryptic genetic variation—genotypic variation without phenotypic variation—in the fruit fly Drosophila melanogaster. Subsequently, similar observations have been made in the plant Arabidopsis thaliana (Queitsch et al. 2002), the yeast Saccharomyces cerevisae (Cowen and Lindquist 2005) and the fish Astyanax mexicanus (Rohner et al. 2013). Further support was provided by Burga et al. (2011), who found that high induction of Hsp90 during development of the nematode Caenorhabditis elegans reduced the penetrance of certain mutations. Additionally, Lachowiec et al. (2013) found that paralogs of duplicated kinase-coding genes that encode a substrate of Hsp90 (i.e., a Hsp90 “client”) in Saccharomyces cerevisiae often evolve faster than paralogs encoding nonclients. In general, the rate at which nonconservative substitutions—those that alter physicochemical properties of amino acids—accumulate is especially accelerated in Hsp90 clients (Pechmann and Frydman 2014).
Multiple studies also demonstrate mutational buffering mediated by the bacterial chaperonin GroEL. For example, Fares et al. (2002) showed that overexpressing GroEL considerably improved the fitness of E. coli strains with a high load of deleterious mutations, a pattern that was also observed later in Salmonella enterica (Maisnier-Patin et al. 2005). Moreover, GroEL overexpression in E. coli increases the ability of GroEL client proteins to tolerate mutations (Tokuriki and Tawfik 2009; Bershtein et al. 2013; Wyganowski et al. 2013; Sabater-Muñoz et al. 2015), as well as their ability to undergo adaptive evolution (Tokuriki and Tawfik 2009; Wyganowski et al. 2013). Buffering of destabilizing mutations accelerates the evolutionary rates of GroEL clients (Bogumil and Dagan 2010; Warnecke and Hurst 2010; Williams and Fares 2010; Pechmann and Frydman 2014).
While no Hsp70 chaperone has been directly implicated in mutational buffering, pertinent circumstantial evidence exists. For example, DnaK—the major bacterial Hsp70 chaperone—is overexpressed together with GroEL in S. enterica lineages with reduced fitness caused by the accumulation of deleterious mutations (Maisnier-Patin et al. 2005). In addition, D. melanogaster populations showing inbreeding depression, where increased homozygosity exposes recessive deleterious mutations, significantly up-regulate the expression of Hsp70 compared with outbred populations (Pedersen et al. 2005).
The chaperones from the Hsp70 system are very conserved from bacteria to humans (Powers and Balch 2013). They play a central role in proteome integrity, and are involved both in co- and post-translational folding (Hartl et al. 2011). In bacteria, the Hsp70 chaperone DnaK (together with GroEL and the Trigger Factor) is one of the main molecular chaperones, where it is the central hub in the chaperone network of the cytosol (Bukau and Walker 1989; Calloni et al. 2012). It interacts with at least ∼700 mostly cytosolic proteins (Calloni et al. 2012). The DnaK interactome was characterized by the isolation of DnaK interactors using immobilized metal affinity chromatography, followed by liquid chromatography mass spectrometry. These regular clients of DnaK are enriched for proteins with low intrinsic solubility, proteins that tend to be members of hetero-oligomeric complexes and/or proteins that show a high density of hydrophobic patches flanked by positive residues (Calloni et al. 2012). DnaK is highly expressed constitutively and essential at 42 °C (Bukau and Walker 1989; Calloni et al. 2012). During its ATP-dependent reaction cycle, DnaK interacts with the Hsp40 co-chaperone DnaJ, which determines the client binding specificity of DnaK (Straus et al. 1990; Hoffmann et al. 1992), and the nucleotide exchange factor GrpE (Hartl et al. 2011). The chaperone system formed by these three proteins can both fold nascent proteins and refold denatured proteins. It does so by binding to exposed hydrophobic patches in unfolded or partially folded protein substrates, thus preventing detrimental interactions with other polypeptides in the crowded cellular milieu. By successively binding and releasing a protein substrate in a cyclic process that consumes ATP, the chaperone system DnaK–DnaJ–GrpE allows the substrate to gradually explore its complex folding energy landscape (Hartl and Hayer-Hartl 2009; Hartl et al. 2011). For some proteins (∼20% of the total proteome), several of these bind-release cycles are enough to achieve the native conformation. However, other proteins (∼10% of the total proteome) still require the downstream chaperone system GroEL/ES (Hartl et al. 2011). The importance of the DnaK–DnaJ–GrpE system in the bacterial chaperone network is obvious from its strong conservation across bacteria, except for two species from the order Aquificales that have lost the entire system, and individual losses of dnaJ and grpE in obligate endosymbionts that have experienced considerable genome reductions (Warnecke 2012).
Most mutations affecting proteins are neutral or deleterious (Eyre-Walker and Keightley 2007), and functionally important mutations often destabilize proteins (Tokuriki et al. 2008; Wyganowski et al. 2013). If DnaK buffers destabilizing mutations, then the deleterious effects of mutations in highly interacting (strong) clients should be lower than in sporadic (weak) clients, where they should be lower than in nonclients. In other words, the more strongly a protein’s integrity depends on DnaK, the higher should be its tolerance to mutations, and the lower the signature of purifying selection that purges those mutations. With this reasoning in mind, we here use laboratory experiments to evaluate the effect of DnaK buffering on the evolution of its client proteome on short evolutionary time scales. We complement our experimental observations with sequence analyses to study the effect of DnaK on intermediate and long evolutionary time scales.
Results
Experimental Evolution of E. coli under DnaK Overexpression
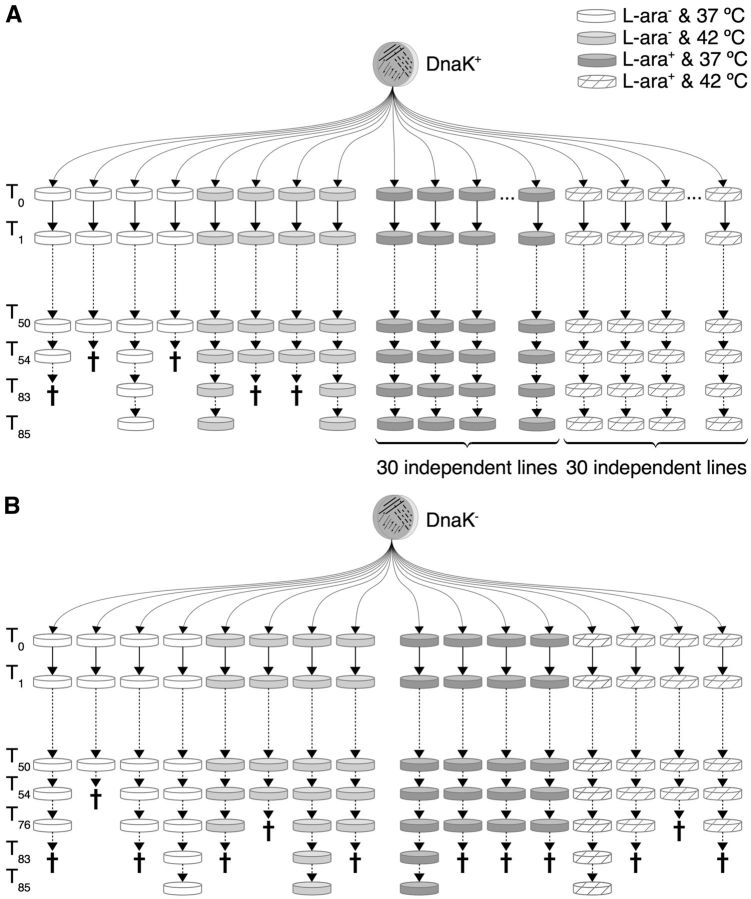
To study the effect of DnaK overexpression on protein evolution experimentally, we performed mutation accumulation experiments similar to those we reported recently for the chaperonin GroEL, but for DnaK overexpression (Sabater-Muñoz et al. 2015). Briefly, we initiated 68 parallel and independent clonal lines of evolution, all of which derived from the same hypermutable clone (E. coli K12 MG1655 ΔmutS) (Sabater-Muñoz et al. 2015) (fig. 1A). Cells of the 68 lines all harbored the plasmid pKJE7, which contains the operon dnaK–dnaJ–grpE under the control of the l-arabinose-inducible araB promoter PBAD (Nishihara et al. 1998). We refer to this strain as DnaK+. We evolved 60 of the 68 DnaK+ lines through repeated single-cell bottlenecks in the presence of the inducer, to ensure overexpression of DnaK, as well as of the cochaperone DnaJ and the nucleotide exchange factor GrpE. All evolving lineages were passaged after 24 hours of incubation. Because of the bottlenecks to which we exposed the populations, genetic drift was strong and the efficiency of selection was weak during the experiment, such that nonlethal mutations are free to accumulate (Barrick and Lenski 2013). We evolved 30 of the 60 clonal lines at 37 °C, and the other 30 at 42 °C. The higher temperature serves to increase the deleterious effect of destabilizing mutations in the bacterial proteome (Bukau and Walker 1989). Finally, the remaining 8 DnaK+ lines were evolved in the absence of inducer, and therefore without DnaK overexpression (4 lines at 37 °C and the other 4 at 42 °C).
Fig. 1.—
Mutation accumulation experiment. Evolutionary history of the populations evolved in this study from the first daily transfer or single-cell bottleneck (T0) until the end of the evolution experiment (T85). We constructed two strains derived from an ancestral Escherichia coli K-12 MG1655 strain lacking the mismatch repair gene mutS. The DnaK+ strain harbours the ∼15-copy plasmid pKJE7 that contains the DnaK/DnaJ/GrpE chaperone system under the control of the promoter PBAD inducible by L-arabinose (Nishihara et al. 1998). The DnaK− strain contains a control pKJE7-derived plasmid where the operon dnaK–dnaJ–grpE has been deleted. We evolved in parallel multiple independent populations of both strains through single-cell bottlenecks under the effect of strong genetic drift at two different temperatures (37 °C and 42 °C). At each temperature we evolved some populations in the presence of L-arabinose (L-ara+), and some in the absence of this expression inducer (L-ara–). (A) During the evolution of 68 DnaK+ populations, five out of eight lines evolving in the absence of inducer went extinct (indicated by a cross). None of the 60 lines evolving under DnaK overexpression experienced any extinction. (B) Of the 16 independent DnaK− populations, 12 populations went extinct. We finished the evolution experiment after 85 single-cell bottlenecks (T85), or ∼1,870 generations.
At each of the two temperatures, we additionally evolved 8 control clonal lines founded from the same parental strain, but carrying a pKJE7-derived plasmid where the operon dnaK–dnaJ–grpE is deleted (fig. 1B). Cells of all 16 control lines therefore cannot overexpress DnaK, even though their growth medium contains l-arabinose (DnaK− lines). At each temperature, half of the lines evolved in the presence of l-arabinose, whereas the other half evolved in medium devoid of this expression inducer. In total, we evolved 86 bacterial populations: 68 DnaK+ lines and 16 DnaK− lines. We stopped the evolution experiment after 85 single-cell bottlenecks, or ∼1,870 generations (assuming conservatively ∼22 generations per daily growth cycle).
Evolving Lineages Tend to Go Extinct in the Absence of DnaK Overexpression
One of the first indications that DnaK overexpression could be buffering deleterious mutations accumulated during the evolution experiment is the observed pattern of extinctions (fig. 1). Some evolving lines went extinct, presumably due to high levels of mutational load, and remarkably, all extinctions occurred in lines that were not overexpressing DnaK. They were either DnaK− lines or DnaK+ evolved in the absence of the inducer. More specifically, 75% of the DnaK− lines (12 of 16) went extinct before the end of the evolution experiment (fig. 1B). Among the DnaK+ lines, 62.5% of the lines (5 of 8) evolving in the absence of the inducer went extinct, whereas none of the 60 lines evolving in the presence of the inducer experienced any extinction (fig. 1A). This observation strongly suggests that overexpressing the chaperone DnaK has increased the robustness of the cells to the accumulation of deleterious mutations, helping them cope with mutational load.
Overexpressing DnaK Increases the Robustness to Nonsynonymous Mutations of DnaK Clients
In order to study the effect of DnaK buffering on genome evolution, we sequenced the genomes of some lines at the end of the evolution experiment, after 85 passages, and compared them to the ancestral genome, which we had sequenced in a previous study (Sabater-Muñoz et al. 2015). Among the clonal lines evolved in the presence of the inducer l-arabinose, we randomly selected for sequencing 3 DnaK+ lines evolved at 37 °C, and another 3 at 42 °C. We also sequenced the only two surviving control DnaK− lines evolved with l-arabinose in the medium, each at a different temperature. Although all sequenced lines evolved in the presence of the inducer, only DnaK+ lines are able to overexpress the chaperone.
In order to evaluate if a significant difference existed in the mutation rate (or generation time) between the sequenced DnaK+ and DnaK− lines, we compared the number of accumulated synonymous mutations between them (supplementary table S1, Supplementary Material online). We observed an average number of 78 synonymous mutations per DnaK+ line and 66 synonymous substitutions per DnaK− line, which is not significantly different (binomial test, P = 0.359; supplementary table S1, Supplementary Material online). We also did not observe any significant difference in the number of accumulated nonsynonymous mutations (binomial test, P = 0.646; table 1), the number of indels (binomial test, P = 0.332; supplementary table S1, Supplementary Material online), or the ratio of transitions to transversions (χ2 test, P = 0.273; supplementary table S1, Supplementary Material online).
Table 1.
Distribution of Nonsynonymous Nucleotide Substitutions among DnaK Client and Nonclient Proteins After ∼1,870 Generations of Evolution in Mutation Accumulation Experiments Conducted at 37 °C and 42 °C
| Temperature | Line a |
Number of mutations
|
|||
|---|---|---|---|---|---|
|
Clients
|
Nonclients | ||||
| Strong b | Weak c | Total | |||
| 37 °C | DnaK+ #1 | 3 | 0 | 12 | 50 |
| DnaK+ #2 | 7 | 2 | 19 | 91 | |
| DnaK+ #3 | 11 | 2 | 25 | 91 | |
| DnaK− | 12 | 1 | 17 | 108 | |
| 42 °C | DnaK+ #1 | 11 | 1 | 22 | 95 |
| DnaK+ #2 | 13 | 3 | 27 | 103 | |
| DnaK+ #3 | 9 | 2 | 22 | 115 | |
| DnaK− | 11 | 1 | 15 | 99 | |
aExperimental evolution lines sequenced in this study. For each temperature, we sequenced three lines overexpressing the DnaK–DnaJ–GrpE chaperone system (DnaK+ lines) and a control line where this system is expressed at wild type levels (DnaK− line).
bStrong clients are those with a high relative enrichment factor on DnaK within the third quartile of the distribution.
cWeak clients are those with a low relative enrichment factor on DnaK within the first quartile of the distribution.
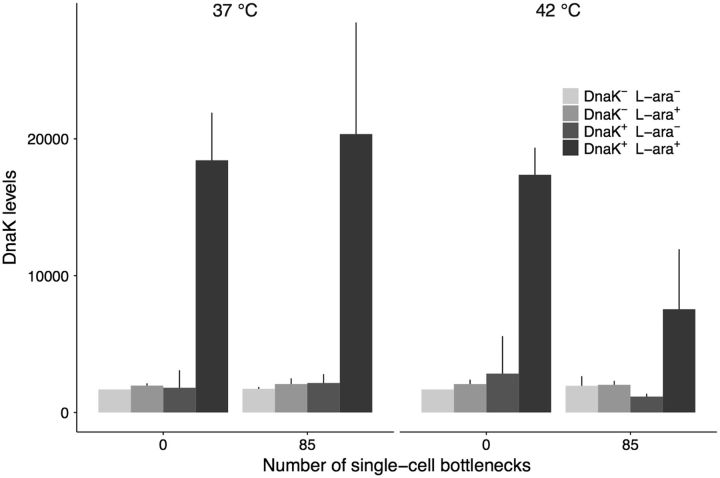
We also verified that DnaK was still overexpressed at the end of the experiment in the 8 sequenced lines. The overexpression of DnaK may be energetically costly, just as is the case for the chaperonin GroEL (Fares et al. 2002; Sabater-Muñoz et al. 2015). In principle, this cost could favor the accumulation of mutations that lead to a decrease in the expression of DnaK during the evolution experiment, especially if the energetic cost of overproducing the chaperone is greater than the benefits derived from mutational buffering (Sabater-Muñoz et al. 2015). However, we observed that for the sequenced lines the overexpression of DnaK was maintained through the mutation accumulation experiment at both 37 °C and 42 °C (fig. 2; supplementary fig. S1, Supplementary Material online). In the presence of the inducer l-arabinose, all DnaK+ lines overexpressed DnaK not only at the start of the evolution experiment, but also at the end, except for one of the DnaK+ lines evolved at 42 °C. However, this loss of overexpression occurred towards the end of the experiment and even then DnaK was still overexpressed for most of the daily growth cycle of this line (supplementary fig. S2, Supplementary Material online). In no line did we observe overexpression in the absence of the inducer. The control DnaK− lines always exhibited wild-type expression levels of DnaK.
Fig. 2.—
DnaK abundance at the beginning and the end of the mutation accumulation experiment. We measured the abundance of the chaperone DnaK for the 8 sequenced lines evolved trough 85 single-cell bottlenecks (∼1,870 generations) at 37 °C or 42 °C. For comparison, we also measured the abundance of the chaperone in the ancestral DnaK+ and DnaK− strains at both temperatures. We determined DnaK levels in the presence and absence of the inducer L-arabinose (L-ara+ and L-ara–, respectively), as described in Materials and Methods (“Verification of DnaK overexpression”), via the intensity of the DnaK band in a Western blot. The evolved lines did not lose the ability to overexpress DnaK in the presence of the inducer L-arabinose except for a DnaK+ line evolved at 42 °C (line #2), which explains the decrease in the average DnaK abundance at the end of the evolution experiment. However, this loss of overexpression occurred late in the evolution experiment, and it is not even complete for most of the daily growth cycle of this line (supplementary fig. S2, Supplementary Material online). The height of the bars indicates mean DnaK abundance across two experimental replicates per strain and condition. Error bars represent 1 SD of the mean.
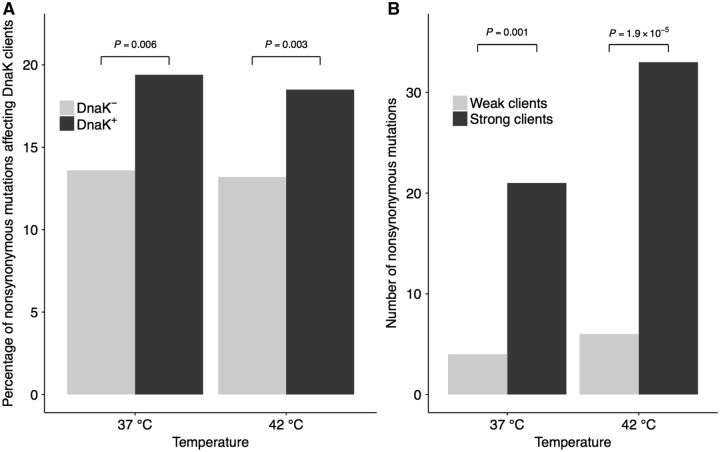
In the genomes of the evolved DnaK+ lines, we first studied the incidence of nonsynonymous nucleotide substitutions among DnaK clients and nonclients (table 1). In this analysis, we considered as nonclients all proteins from the E. coli proteome that are not part of a set of 674 DnaK clients determined by Calloni et al. (2012), and analyzed the lines evolved at 37 °C and 42 °C independently. To improve statistical power, we combined mutations across DnaK+ lines evolved at the same temperature after classifying them according to whether they affect DnaK clients or nonclients. If DnaK is buffering deleterious mutations, we would expect a higher proportion of mutations affecting clients in the lines evolved under DnaK overexpression.
In the DnaK− line evolved at 37 °C, ∼14% of nonsynonymous mutations (17 out of 125) affected DnaK clients. Compared with this proportion when DnaK is not overexpressed, the proportion of mutations in clients in the DnaK+ lines was significantly higher (56 out of the total 288 mutations, ∼19%; binomial test: P = 0.006; fig. 3A). Similarly, compared with the DnaK− line evolved at 42 °C, where ∼13% of all mutations affected DnaK clients (15 out of 114 mutations), the DnaK+ lines showed significantly more mutations in clients (71 out of 384 total mutations, ∼18%; binomial test: P = 0.003; fig. 3A). These results suggest that overexpressing DnaK does indeed increase the robustness of its clients to amino acid replacements. Temperature itself had no significant effect on the fraction of all mutations affecting DnaK clients in DnaK+ lines (Fisher’s exact test: odds ratio F = 1.06, P = 0.77) and DnaK− lines (Fisher’s exact test: odds ratio F = 1.04, P = 0.999).
Fig. 3.—
Nonsynonymous mutations accumulated in DnaK clients. (A) The proportion of nonsynonymous mutations that affect DnaK clients is significantly higher in DnaK+ lines that overexpress DnaK than in the control DnaK− lines that do not express the chaperone at such high levels. This is observed both for lines evolved at 37 °C and 42 °C. We combined mutations across DnaK+ lines evolved at the same temperature. The significance of the difference in the proportions was evaluated using a binomial test. (B) At both temperatures, strong clients have accumulated significantly more nonsynonymous substitutions than weak clients in DnaK+ lines. Strong clients include those clients with the highest DnaK dependency, whereas weak clients include clients with the lowest chaperone dependency. Statistical significance was evaluated using Fisher’s test.
Strong DnaK Clients Accumulate More Nonsynonymous Mutations than Weak Clients
Next, we studied if strongly interacting DnaK clients are more robust to mutations than weakly interacting clients, as evidenced by the pattern of mutations fixed in the mutation accumulation experiment under DnaK overexpression. To assess how strongly a protein depends on DnaK for folding, we used recent experimental proteomic data which determined how strongly 668 DnaK-interacting proteins interact with DnaK by measuring the fraction of cellular protein bound to DnaK at 37 °C, a property that correlates with chaperone dependency for folding and maintenance and residence time of the protein on DnaK (Calloni et al. 2012). In a ΔdnaK E. coli strain, strong clients are more prone to form aggregates than weak clients, indicating that the relative enrichment of a protein on DnaK is a good proxy for the dependence upon DnaK for folding (Calloni et al. 2012). We consider as strong clients those with a relative enrichment factor on DnaK within the third quartile of the distribution of DnaK dependency (N = 167), and weak clients those within the first quartile (N = 167). Therefore, strong clients include those clients with the highest DnaK dependency, whereas weak clients include clients with the lowest chaperone dependency.
In the DnaK+ lines evolved at 37 °C, we found more nonsynonymous substitutions in strong clients (21 mutations) than in weak clients (4 mutations) (table 1). Considering the number of nonsynonymous sites in strong clients (43,731 sites) and weak clients (41,238 sites), this difference was significant (Fisher’s exact test: F = 4.95, P = 0.001; fig. 3B). At 42 °C, the results were similar, with 33 mutations in strong clients and 6 in weak clients (Fisher’s exact test: F = 5.12, P = 1.9 × 10 − 5; fig. 3B; table 1). In conclusion, at both temperatures, client proteins that are more dependent upon DnaK for folding accumulate significantly more mutations than less dependent clients.
DnaK Accelerates Protein Evolution On Intermediate and Long Evolutionary Time Scales
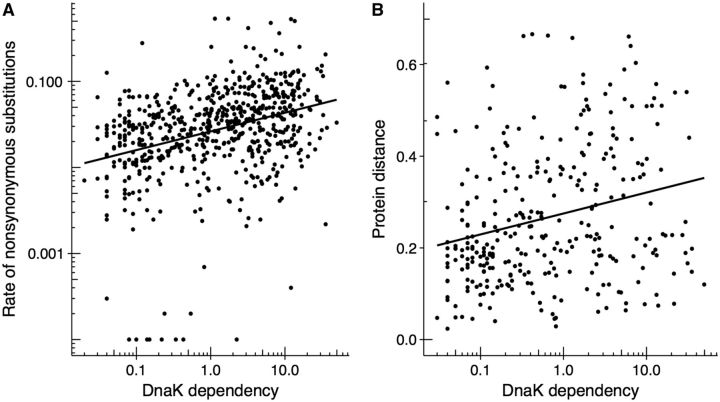
We wanted to find out if the DnaK-mediated mutational buffering we observed on the short time scales of laboratory evolution has also left signatures on longer evolutionary time scales. To this end, we determined two measures of evolutionary rates for protein-coding genes from gamma-proteobacteria. The first, nonsynonymous divergence among one-to-one orthologs of E. coli and S. enterica, is relevant for intermediate evolutionary time scales. The second, protein (amino acid) distance among orthologous proteins found in 85 gamma-proteobacterial genomes (including E. coli and S. enterica), is relevant for long time scales. We employ protein distance instead of nonsynonymous distance because amino acid replacements are less sensitive than nucleotide substitutions to the expected loss of phylogenetic signal between sequences of distantly related taxa. To assess how strongly a protein depends on DnaK for folding, we use the relative enrichment of the protein on DnaK as a proxy for the dependence of the protein upon DnaK for folding (Calloni et al. 2012). We note that this interaction strength is more likely to have remained unchanged during the divergence of E. coli and S. enterica, than during the divergence of all the other 83 gamma-proteobacterial species we analyzed.
We find a strong and highly significant positive association between DnaK dependency and the rate of nonsynonymous substitutions for S. enterica and E. coli (Spearman’s rank correlation coefficient, ρ = 0.367, N = 627, P < 2.2 × 10 − 16; fig. 4A). This indicates that the stronger the interaction of a protein with DnaK, the faster the protein evolves. The same pattern is obtained at the larger time scales of protein distances for 85 gamma-proteobacterial genomes (ρ = 0.257, N = 311, P = 4.4 × 10 − 6; fig. 4B). Gene expression level, which is the most important determinant of protein evolutionary rates, at least in unicellular organisms (Pál et al. 2001; Drummond et al. 2005; Zhang and Yang 2015), is a possible confounding factor in this analysis. For example, using codon usage bias (CUB) as a proxy for gene expression, we observe that genes with higher CUB show lower nonsynonymous divergence (ρ = −0.558, N = 1014, P < 2.2 × 10 − 16), protein distance (ρ = −0.255, N = 3159, P < 2.2 × 10 − 16) and DnaK dependency (ρ = −0.262, N = 627, P = 2.5 × 10 − 11). However, the association between DnaK dependency and evolutionary rate cannot be solely explained by this confounding factor: A partial correlation analysis shows that the association still holds after controlling for CUB, both on intermediate time scales (ρ = 0.295, N = 627, P = 1.2 × 10 − 14) and long time scales (ρ = 0.229, N = 311, P = 3.8 × 10 − 5). We use CUB instead of gene expression data here for two main reasons. First, we can compute CUB for all 631 DnaK clients in our data set, whereas expression data is only available for 457 clients. Second, gene expression data has been measured in just one environment and one strain of E. coli, whereas CUB is the result of selective pressures imposed by many different environments over long periods of time. Nonetheless, the association between evolutionary rate and DnaK dependency still holds after correcting for gene expression directly (supplementary information section 1.1, Supplementary Material online). Together, these results indicate that the chaperone DnaK affects protein evolution in accordance with the mutational buffering hypothesis. Importantly, this effect is not only independent of CUB and gene expression, but also of other biological factors, such as essentiality and number of protein–protein interactions (supplementary information section 1.2 and table S2, Supplementary Material online).
Fig. 4.—
DnaK accelerates protein evolution on intermediate and long evolutionary time scales. Scatter-plots showing the relationship between DnaK dependency (calculated as a relative enrichment factor that indicates the fraction of cellular protein bound to DnaK at 37 °C, horizontal axis) and the degree of divergence over (A) intermediate time scales, measured as nonsynonymous divergence (Spearman rank correlation coefficient, ρ = 0.367, N = 627, P < 2.2 × 10 − 16), and (B) long time scales, measured as protein (amino acid) distance (ρ = 0.257, N = 311, P = 4.4 × 10 − 6) (vertical axes). Solid lines represent the best fit to the points. Note the logarithmic scale on both axes.
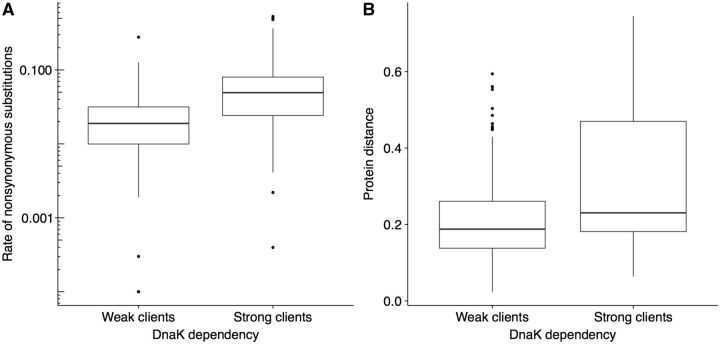
In a subsequent analysis, we find that clients evolve more slowly than nonclients (supplementary fig. S3 and supplementary information section 1.3, Supplementary Material online). This last difference cannot be explained by the number of protein–protein interactions, by essentiality, or by CUB as confounding factors (supplementary information section 1.3 and supplementary tables S3 and S4, Supplementary Material online). The reason for this observation could be that clients are intrinsically less robust to mutations than nonclients due to some general physicochemical difference. For example, Calloni et al. (2012) found that DnaK clients have generally low solubility, often belong to heterooligomeric complexes, and are prone to misfolding. However, in accordance with the mutational buffering hypothesis we observe that strong clients evolve faster than weak clients (fig. 5; supplementary information section 1.3, Supplementary Material online). The accelerated evolution of strong clients compared with weak clients exactly mirrors the greater accumulation of nonsynonymous mutations in strong clients during the evolution experiment (fig. 4B).
Fig. 5.—
Strong clients evolve faster than weak clients. (A) We find that strong clients evolve faster than weak clients on intermediate evolutionary time scales, measured as the rate of nonsynonymous substitutions (Wilcoxon rank-sum test, P < 2.2 × 10 − 16). (B) On long evolutionary time scales, we also find that strong clients evolve faster than weak clients (Wilcoxon rank-sum test, P = 2.3 × 10 − 3). The thick horizontal line in the middle of each box represents the median of the data, whereas the bottom and top of each box represent the 25th and 75th percentiles, respectively. Note the logarithmic scale on the y-axis in (A).
DnaK-Mediated Acceleration of Protein Evolution Is Independent of GroEL Buffering
The ability of DnaK to facilitate the accumulation of nonsynonymous mutations in DnaK clients resembles the well-studied mutational buffering by the chaperonin GroEL (Fares et al. 2002; Tokuriki and Tawfik 2009; Bershtein et al. 2013; Wyganowski et al. 2013; Sabater-Muñoz et al. 2015). Additionally, the observed correlation between DnaK dependency and protein evolutionary rates is similar to the previously reported acceleration of protein evolution by GroEL (Bogumil and Dagan 2010; Williams and Fares 2010). We therefore removed known GroEL clients from our data set to investigate if our observations are independent of the effect of GroEL on mutation accumulation and evolutionary rates. We defined the GroEL interactome in E. coli as the union of two previously reported sets of DnaK interactors (Kerner et al. 2005; Fujiwara et al. 2010). Of the 253 GroEL clients that comprise the GroEL interactome, there are 122 proteins that are also clients of DnaK.
The observation that DnaK overexpression increases the proportion of nonsynonymous substitutions affecting DnaK clients is still significant after removing GroEL clients. Combining mutations from the DnaK+ lines evolved at 37 °C and 42 °C we find that ∼16% of mutations (97 out of 621) affected DnaK clients, which is significantly higher than what we find in the DnaK− lines (28 out of 230 mutations, ∼12%; binomial test: P = 0.01). Similarly, considering the number of nonsynonymous sites in strong clients (36,026 sites) and weak clients (33,047 sites) after removing GroEL clients, we still find that strong DnaK clients accumulate more nonsynonymous substitutions than weak clients (Fisher’s exact test: odds ratio F = 4.7, P = 2 × 10 − 5). Finally, the positive association between DnaK dependency and evolutionary rates still holds after removing GroEL clients and controlling for CUB in a partial correlation analysis, both on intermediate time scales (ρ = 0.318, N = 511, P = 3.9 × 10 − 14) and long time scales (ρ = 0.226, N = 240, P = 3.6 × 10 − 4).
Discussion
We show how the overexpression of the DnaK–DnaJ–GrpE chaperone system over the course of a mutation accumulation experiment increases the proportion of nonsynonymous substitutions affecting DnaK clients. In addition, strong clients accumulate more nonsynonymous mutations than weak clients. Additional evidence of mutational buffering by DnaK is provided by the observation that evolving lines overproducing this chaperone avoid extinction after experiencing 85 single-cell bottlenecks. Recently, we obtained similar results in hypermutable E. coli cells evolving in identical conditions but overproducing the GroEL-GroES chaperonin system (Sabater-Muñoz et al. 2015). There, we observed that lines evolving with high levels of GroEL were not only less prone to extinction under strong genetic drift than control lines, but also that they were accumulating significantly more indels and replacements between amino acids belonging to different physicochemical categories.
We also find that DnaK-mediated mutational buffering has left a trace in DnaK clients during the divergence of 85 different gamma-proteobacterial species over much longer evolutionary time scales than those explored in our laboratory evolution experiment. We find that clients that depend more on DnaK for folding tend to evolve faster than less interacting clients. Similar chaperone-mediated accelerations of protein evolution have been observed in GroEL clients (Bogumil and Dagan 2010; Williams and Fares 2010) and Hsp90 clients (Lachowiec et al. 2013; Pechmann and Frydman 2014). However, we notice that DnaK clients evolve slower than proteins not known to be DnaK interactors (Calloni et al. 2012). This is likely the result of important physicochemical differences between clients and nonclients. For example, clients are prone to aggregation and misfolding (Calloni et al. 2012), which may make them intrinsically less robust to destabilizing mutations.
Despite the great differences in the mechanism of chaperone action between the three major chaperone families—chaperonins, Hsp90 chaperones and Hsp70 chaperones—(Young et al. 2004; Hartl et al. 2011; Bogumil and Dagan 2012; Kim et al. 2013), at least some of their members seem to have qualitatively comparable effects on protein evolution. But protein chaperones are not the only chaperones that can increase the mutational robustness of their substrates: A recent study has found that some RNA chaperones can buffer deleterious mutations in E. coli and therefore affect RNA evolution (Rudan et al. 2015). These chaperones, which are completely unrelated to protein chaperones, are RNA-binding proteins that facilitate the proper folding of RNA molecules. Elucidating to what extent the buffering mechanisms of all these chaperones differ is an important future direction of enquiry.
Thanks to their fostering of mutational robustness, chaperones can facilitate evolutionary innovations (Rutherford 2003), even though we do not study such innovations here. The increase in the mutational robustness of a protein caused by chaperone interactions reduces the efficiency of purifying selection in purging mutations in the protein. Thanks to chaperone-mediated buffering, many such mutations are neutral and can persist in a population. Importantly, these cryptic genetic variants may include preadaptive mutations that can generate evolutionary innovations in new environments (Tokuriki and Tawfik 2009; Wyganowski et al. 2013). To illuminate if and how DnaK can increase the ability to evolve functional innovations of its client proteome will also be an interesting subject for future work.
In summary, we analyzed the evolution of proteins that are subject to DnaK-assisted folding on short, intermediate, and long evolutionary time scales through a combination of experimental and comparative approaches. Most of our evidence indicates that the bacterial chaperone DnaK can buffer mutations in its client proteins, and that these proteins therefore evolve faster than in the absence of DnaK-mediated folding. This is, to our knowledge, the first demonstration that a member of the Hsp70 family can buffer the effect of mutations, with long-term consequences on protein evolution (Bogumil and Dagan 2012). Through its role in protein folding, an individual chaperone such as DnaK can have a disproportionate effect on proteome evolution, and thus on genome evolution.
Materials and Methods
Bacterial Strains and Plasmids
We obtained E. coli K-12 substr. MG1655 ΔmutS::FRT from Ivan Matic (Université Paris Descartes, INSERM U1001, Paris, France) through Jesús Blázquez (Centro Nacional de Biotecnología, CSIC, Madrid, Spain) (Sabater-Muñoz et al. 2015). In this E. coli strain, the gene encoding the protein MutS has been deleted. This protein is a component of the mismatch repair system that recognizes and binds mispaired nucleotides so that the mispairing can be corrected by two further repair proteins, MutL and MutH. The strain MG1655 ΔmutS has a predicted mutation rate that is 1000-fold higher than the wild type (Turrientes et al. 2013), which ensures that a sufficient number of mutations occur during the mutation accumulation experiment. We transformed this strain with the plasmid pKJE7 (Takara, Cat. #3340), which contains an operon encoding DnaK, and its co-chaperones DnaJ and GrpE under the regulation of a single promoter inducible by l-arabinose (Nishihara et al. 1998). We generated a control strain by transforming the same ΔmutS strain with a plasmid that lacks the operon dnaK–dnaJ–grpE but is otherwise identical to pKJE7. We refer to this plasmid as pKJE7-DEL(dnaK–dnaJ–grpE). This control plasmid was derived from the plasmid pKJE7 by removal of the operon dnaK–dnaJ–grpE with a restriction digest using BamHI and SpeI, followed by religation, after obtaining permission for plasmid modification from Takara.
Evolution Experiment
We evolved 68 clonal lines of the hypermutable E. coli ΔmutS strain containing pKJE7 (DnaK+ lines) and 16 lines containing the control plasmid pKJE7-DEL(dnaK–dnaJ–grpE) (DnaK− lines) by daily passaging them through single-cell bottlenecks on solid LB medium (agar plates; Pronadisa #1551 and #1800) supplemented with 20 μg/ml of chloramphenicol (Sigma-Aldrich #C0378) (fig. 1). Except for 8 DnaK+ lines and 8 DnaK− lines, all the remaining lines were evolved in the presence of 0.2% (w/v) of l-arabinose (Sigma-Aldrich #A3256), which induces the expression of DnaK/DnaJ/GrpE from the plasmid pKJE7 but not from the control plasmid pKJE7-DEL(dnaK–dnaJ–grpE). We passaged both the DnaK− and DnaK+ lines during 85 days or ∼1,870 generations (conservatively assuming ∼22 generations per daily growth cycle), except for those lines that went extinct before reaching the end of the experiment. We evolved half of the DnaK+ and DnaK− lines under mild heat-stress (42 °C) whereas the other half remained at 37 °C.
Verification of DnaK Overexpression
We grew the ancestral and evolved strains (DnaK+ and DnaK–, at 37 °C and 42 °C) from glycerol stocks in liquid LB medium supplemented with 20 μg/ml of chloramphenicol in the presence or absence of the inducer l-arabinose (0.2%). After 24 h of growth, we pelleted cells by centrifugation at 12,000 rpm. We resuspended the pelleted cells in 100 μl lysis buffer (containing 200 mM Tris–HCl pH 6.8, 10 mM DTT, 5% SDS, 50% glycerol). To prepare a crude extract, we first boiled resuspended cells at 95 °C for 15 min. After the removal of cell debris by centrifugation, we quantified soluble proteins using the Bradford method (Bradford 1976). We loaded 1 μg of total protein for each sample in SDS–PAGE gels (12.5% resolving gel). In addition, we loaded onto all gels samples from the ancestral DnaK− and DnaK+ strains grown in the presence of inducer at 37 °C, as controls to facilitate inter-gel comparisons. We detected DnaK protein by Western blotting using as primary antibody a mouse monoclonal antibody specific to E. coli DnaK (Abcam #ab69617) at a 1:10,000 dilution, and as secondary antibody a goat polyclonal (alkaline phosphatase-conjugated) antibody specific to mouse IgG1 (Abcam #ab97237). We scanned membranes after colorimetric detection of conjugated antibodies with the BCIP®/NBT-Purple liquid substrate system (Sigma-Aldrich #BP3679), and used ImageJ to quantify the intensity of DnaK bands on the Western blots (Schneider et al. 2012). We used the control samples to normalize abundances, which allow the comparison of DnaK levels across experiments.
We examined the change in DnaK levels along a daily cycle of growth for a DnaK+ line evolved at 42 °C (line #2, supplementary fig. S2, Supplementary Material online) that showed a lowed DnaK level after ∼1,870 generations of mutation accumulation. After 24 h of exponential growth at 42 °C in liquid LB medium supplemented with chloramphenicol, we diluted the culture to OD ∼0.3, and induced DnaK expression by adding 10 mM of l-arabinose. We allowed the culture to grow for another 24 h in the presence of this expression inducer. Each hour, 1 ml of culture was removed and the DnaK level following the protocol described earlier was measured.
Whole-Genome Resequencing
We sequenced the genomes of 2 DnaK− and 6 DnaK+ lines after 85 single-cell bottlenecks. All of these lines evolved in the presence of l-arabinose in the medium, although only DnaK+ cells are able to overexpress DnaK. Half of the sequenced DnaK− and DnaK+ lines evolved at 37 °C, whereas the other lines evolved at 42 °C. We used the genome sequence of the ancestral ΔmutS strain from which both the DnaK+ and DnaK− lines were derived from our previous study (Sabater-Muñoz et al. 2015).
Specifically, for the evolved lines we performed paired-end Illumina whole-genome sequencing. For DNA extraction, we used the QIAmp DNA mini kit (Qiagen, Venlo [Pays Bas], Germany) in a QiaCube automatic DNA extractor using bacterial pellets obtained from ∼10 ml cultures. We constructed multiplexed DNAseq libraries from each clonal evolution line using the TrueSeq DNA polymerase chain reaction-free HT sample preparation kit (Illumina). We performed paired-end sequencing on an Illumina HiSeq2000 platform, using a 2 × 100 cycles configuration.
We converted sequencing reads from Illumina quality scores into Sanger quality scores. Subsequently, we used the breseq v 0.24rc4 (version 4) pipeline (Deatherage and Barrick 2014) for aligning the Illumina reads to our E. coli parental genome and for identifying single nucleotide polymorphisms and indels using bowtie2 (Langmead and Salzberg 2012). We performed individual runs of breseq, with junction prediction disabled but otherwise default parameters for each of the evolved lines. We deposited the data from this project at the NCBI Sequence Read Archive under the accession SRP074414.
Sequence Data
We obtained the complete genomes of E. coli K-12 MG1655 (NC_000913) and S. enterica serovar Typhimurium LT2 (NC_003197) from GenBank Genomes (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). We also used a data set from Williams and Fares (2010) that consists of 1092 multiple sequence alignments of conserved orthologous proteins from 85 gamma-proteobacterial genomes.
DnaK Dependency
We obtained information about DnaK clients from Calloni et al. (2012). This study used quantitative proteomics to identify 674 DnaK interactors or client proteins. For 668 of these proteins, the investigators calculated a relative enrichment factor that indicates the fraction of cellular protein bound to DnaK at 37 °C. We used this measure as a proxy for DnaK dependency. We excluded from our analyses the transposases InsC, InsH and InsL of the insertion sequences IS2, IS5 and IS186, respectively. In the genome of E. coli K-12 MG1655 there are 6 copies of insC, 11 of insH and 3 of insL.
GroEL Dependency
We obtained information about 253 GroEL clients from Kerner et al. (2005) and Fujiwara et al. (2010). Our set of GroEL clients is the union of the slightly different GroEL interactomes characterized in these two studies. We excluded from our analyses the transposase insH of the insertion sequences IS5 and 3 clients reported by Kerner et al. (2005), which are encoded on plasmids (SwissProt Accession Numbers: P00810, P29368 and Q9339).
Orthology
We identified 3159 one-to-one orthologs in E. coli and S. enterica genomes as reciprocal best hits (Tatusov et al. 1997) using the Basic Local Alignment Search Tool (BLAST, i.e., BLASTP with an E-value cut-off of 10 − 10). We identified 631 and 242 S. enterica orthologs to DnaK and GroEL clients, respectively. We aligned each pair of orthologous proteins with the Needleman–Wunsch dynamic programming algorithm, using the Needle program from the EMBOSS package (Rice et al. 2000). We translated the resulting alignments into codon-based nucleotide alignments with PAL2NAL (Suyama et al. 2006).
Evolutionary Rates
We estimated the rate of nonsynonymous substitutions (dN) using the program codeml from the package PAML 4.7 (one-ratio model M0) (Yang 2007). We calculated protein distances for the gamma-proteobacterial alignments from Williams and Fares (2010), using PROTDIST from the PHYLIP package (Felsenstein 2005) and the Jones, Taylor and Thornton (JTT) substitution matrix (Jones et al. 1992). We calculated an average distance for each cluster of orthologous proteins as the mean of all pairwise distances.
Codon Usage Bias
We computed the Codon Adaptation Index (CAI) using the program CAI from the EMBOSS package (Rice et al. 2000). We calculated Codon Usage Bias (CUB) for each pair of E. coli—S. enterica orthologs as the mean of the CAI values for each pair of orthologs. We used CUB as a proxy for gene expression.
Protein–Protein Interactions
We obtained the number of protein–protein interactions (PPI) for each E. coli K-12 protein from Rajagopala et al. (2014). The binary interactions considered for this study are a combination of the following: (1) literature curated interactions supported by multiple studies or methods and (2) interactions identified by yeast two-hybrid (Y2H) screening. We removed interactions involving DnaK or DnaJ.
Essentiality
We obtained data about gene dispensability for E. coli K-12 in rich media from Baba et al. (2006).
Gene Expression
We obtained gene expression data for E. coli K-12 MG1655 grown in rich media (LB) at 37 °C from Chen and Zhang (2013), where gene expression levels are measured as number of RNA-seq reads per gene length.
Statistical Tests
We carried out all statistical analyses and plotted data with R (R Development Core Team 2016) using the packages “base”, “pcor”, “ggplot2”, “dplyr” and “gridExtra”.
Supplementary Material
Acknowledgments
The authors thank Xiaoshu Chen and Jianzhi Zhang for kindly providing us with the gene expression data. This work was supported by the Forschungskredit program of the University of Zurich (grant FK-14-076 to J.A.), the Swiss National Science Foundation (grant 31003A_146137 to A.W.), the University Priority Research Program in Evolutionary Biology at the University of Zurich (to A.W.), the Science Foundation Ireland (grant 12/IP/1673 to M.A.F.), and the Spanish Ministerio de Economía y Competitividad (grant BFU2012-36346 to M.A.F.). We posted an earlier version of this paper in bioRxiv (doi: http://dx.doi.org/10.1101/040600) on 22 February 2016.
Literature Cited
- Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. 2013. Genome dynamics during experimental evolution. Nat Rev Genet. 14:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Serohijos AWR, Zhou J, Shakhnovich EI. 2013. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell 49:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogumil D, Dagan T. 2010. Chaperonin-dependent accelerated substitution rates in prokaryotes. Genome Biol Evol. 2:602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogumil D, Dagan T. 2012. Cumulative impact of chaperone-mediated folding on genome evolution. Biochemistry 51:9941–9953. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- Bukau B, Walker GC. 1989. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 171:2337–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga A, Casanueva MO, Lehner B. 2011. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature 480:250–253. [DOI] [PubMed] [Google Scholar]
- Calloni G, et al. 2012. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1:251–264. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J. 2013. No gene-specific optimization of mutation rate in Escherichia coli. Mol Biol Evol. 30:1559–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189. [DOI] [PubMed] [Google Scholar]
- de Visser JAGM, et al. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57:1959–1972. [DOI] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 1151:165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. 1987. Proteins as molecular chaperones. Nature 328:378–379. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet. 8:610–618. [DOI] [PubMed] [Google Scholar]
- Fares MA. 2015. The origins of mutational robustness. Trends Genet. 31:373–381. [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E. 2002. Endosymbiotic bacteria: GroEL buffers against deleterious mutations. Nature 417:398.. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle. [Google Scholar]
- Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. 2010. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29:1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475:324–332. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 16:574–581. [DOI] [PubMed] [Google Scholar]
- Hoffmann HJ, Lyman SK, Lu C, Petit MA, Echols H. 1992. Activity of the Hsp70 chaperone complex–DnaK, DnaJ, and GrpE–in initiating phage lambda DNA replication by sequestering and releasing lambda P protein. Proc Natl Acad Sci U S A. 89:12108–12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8:275–282. [DOI] [PubMed] [Google Scholar]
- Kerner MJ, et al. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122:209–220. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 82:323–355. [DOI] [PubMed] [Google Scholar]
- Lachowiec J, et al. 2013. The protein chaperone HSP90 can facilitate the divergence of gene duplicates. Genetics 193:1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S, et al. 2005. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat Genet. 37: 1376–1379. [DOI] [PubMed] [Google Scholar]
- Masel J, Siegal ML. 2009. Robustness: mechanisms and consequences. Trends Genet. 25:395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol. 64:1694–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158:927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. 2014. Interplay between chaperones and protein disorder promotes the evolution of protein networks. PLoS Comput Biol. 10:e1003674.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KS, Kristensen TN, Loeschcke V. 2005. Effects of inbreeding and rate of inbreeding in Drosophila melanogaster – Hsp70 expression and fitness. J Evol Biol. 18:756–762. [DOI] [PubMed] [Google Scholar]
- Powers ET, Balch WE. 2013. Diversity in the origins of proteostasis networks – a driver for protein function in evolution. Nat Rev Mol Cell Biol. 14:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster T. a, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2016. R: a language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing.
- Rajagopala SV, et al. 2014. The binary protein-protein interaction landscape of Escherichia coli. Nat Biotechnol. 32:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the european molecular biology open software suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Rohner N, et al. 2013. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342: 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudan M, Schneider D, Warnecke T, Krisko A. 2015. RNA chaperones buffer deleterious mutations in E. coli. Elife 4:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL. 2003. Between genotype and phenotype: protein chaperones and evolvability. Nat Rev Genet. 4:263–274. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396:336–342. [DOI] [PubMed] [Google Scholar]
- Sabater-Muñoz B, et al. 2015. Fitness trade-offs determine the role of the molecular chaperonin GroEL in buffering mutations. Mol Biol Evol. 32:2681–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. a, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D, Walter W, Gross C., a 1990. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev. 4:2202–2209. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin E, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Serrano L, Tawfik DS. 2008. How protein stability and new functions trade off. PLoS Comput Biol. 4:e1000002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. 2009. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459:668–673. [DOI] [PubMed] [Google Scholar]
- Turrientes M-C, et al. 2013. Normal mutation rate variants arise in a mutator (Mut S) Escherichia coli population. PLoS One 8:e72963.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. 2005. Robustness and evolvability in living systems. Princeton (NJ): Princeton University Press. [Google Scholar]
- Warnecke T. 2012. Loss of the DnaK-DnaJ-GrpE chaperone system among the Aquificales. Mol Biol Evol. 29:3485–3495. [DOI] [PubMed] [Google Scholar]
- Warnecke T, Hurst LD. 2010. GroEL dependency affects codon usage–support for a critical role of misfolding in gene evolution. Mol Syst Biol. 6:340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Fares MA. 2010. The effect of chaperonin buffering on protein evolution. Genome Biol Evol. 2:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyganowski KT, Kaltenbach M, Tokuriki N. 2013. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol. 425:3403–3414. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 5:781–791. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang J-R. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16:409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.