Abstract
We report a Danio rerio transposon named DrTRT, for D. rerio Transposon Related to Tc1. The complete sequence of the DrTRT transposon is 1,563 base pairs (bp) in length, and its transposase putatively encodes a 338-amino acid protein that harbors a DD37E motif in its catalytic domain. We present evidence based on searches of publicly available genomes that TRT elements commonly occur in vertebrates and protozoa. Phylogenetic and functional domain comparisons confirm that TRT constitutes a new subfamily within the Tc1 family. Hallmark features of having no premature termination codons within the transposase, the presence of all expected functional domains, and its occurrence in the bony fish transcriptome suggest that TRT might have current or recent activity in these species. Further analysis showed that the activity of TRT elements in these species might have arisen about between 4 and 19 Ma. Interestingly, our results also implied that the widespread distribution of TRT among fishes, frog, and snakes is the result of multiple independent HT events, probably from bony fishes to snakes or frog. Finally, the mechanisms underlying horizontal transfer of TRT elements are discussed.
Keywords: Tc1/mariner transposons, DD37E, horizontal transfer
Introduction
Transposable elements (TEs) are DNA fragments that move from one host genomic site to another. TEs have been reported in almost all organisms, including plants, invertebrates, vertebrates, fungi, and bacteria, and they occupy a substantial fraction of various host genomes (Biémont 2010). TEs that integrate into a new genomic location play important roles in genome architecture as well as in genetic innovation (Feschotte and Pritham 2007).
TEs are generally classified into two classes (Classes I and II) based on their structural organization and mechanism of transposition (Feschotte and Pritham 2007). Class I or RNA elements are transposed via reverse transcription of an RNA intermediate, whereas that Class II or DNA elements transpose directly as DNA, mostly through a so-called “cut and paste” mechanism. The Tc1/mariner superfamily of TEs is the most widespread class of DNA transposons in nature (Feschotte and Pritham 2007). Tc1/mariner, which was first discovered in Drosophila mauritiana (Haymer and Marsh 1986; Jacobson et al. 1986), is ubiquitous in eukaryotes (Feschotte and Pritham 2007; Liu and Yang 2014). Tc1/mariner elements are generally 1,300–2,400 bp in size and encode a 340-amino acid transposase that is flanked by terminal inverted repeats (TIRs) and a target site duplication (TSD), TA (Lohe et al. 1996). On the basis of variations in the DDE/D signature motif, Tc1/mariner elements are further classified into various monophyletic groups (Shao and Tu 2001). Although some naturally active Tc1/mariner elements have been identified in nematodes and arthropods, most of these are inactive in vertebrates due to the occurrence of stop codons, deletions, or frameshifts. Importantly, active Tc1/mariner transposons may be potentially used as molecular tools for transgenesis and insertion mutagenesis (Pavlopoulos et al. 2007; Voigt et al. 2016). To date, only some Tc1-like elements (such as Tol1, Tol2, Passport, and Tana1) in bony fishes are known to be both intact in their native form and transpositionally active (Koga et al. 2006; Clark et al. 2009; Pujolar et al. 2013; Watanabe et al. 2014).
TEs are ubiquitous in organisms despite extensive evidence that their presence and mobility causes a variety of deleterious mutations (Craig et al. 2002). One explanation is that TEs have the ability to invade a new host by horizontal transfer (HT). A previous study demonstrated that almost all kinds of eukaryotic TEs are capable of HT (Schaack et al. 2010). Tc1/mariner elements are highly capable of invading a wide range of species because these are not dependent on host factors to mediate their mobility. Although more than 94 HT cases of Tc1/mariner elements have been reported, these phenomena mainly occur between or among invertebrates (Dotto et al. 2015). HT events occurring in vertebrates mainly involve the hAT superfamily of DNA transposons (Pace et al. 2008; Novick et al. 2010; Gilbert et al. 2010; Gilbert et al. 2012; Gilbert et al. 2013).
In the present study, we performed a detail analysis of a Tc1-like transposon in the zebrafish (Danio rario). Further analysis confirmed that TRT elements constitute a new subfamily within the Tc1 family. We also show that TRT elements are both intact in their native form and functionally active in Pundamilian yererei, Maylandia zebra, and Haplochromis burtoni. Finally, we demonstrate that TRT elements might have invaded into its hosts via multiple HT events.
Materials and Methods
Identification and Copy Number Determination of TRT
TBLASTN analysis (Altschul et al. 1990) of the zebrafish genome was performed using the amino acid sequences of Bombyx mori Bmmar1 (U47917) as query, which in turn detected a Tc1/mariner transposon that encoded a DD37E motif-harboring polypeptide. We named these elements DrTRT (an abbreviation for Danio rerio Transposon Related to Tc1) because of its several similarities to transposons of the Tc1 family, as described in Results part. To determine the distribution of DrTRT, its transposase sequence was used as query to search against 1,176 species genomes, including 437 fungus, 216 invertebrates, 226 vertebrates, 150 plants, and 147 protozoans that are available at National Center for Biotechnology Information (NCBI). This transposon was determined to exist in one species when the unique DD37E motif was detected in the catalytic domain of one transposon.
The whole-genome shotgun sequences of all studied species were downloaded from NCBI. The consensus sequences of TRT were reconstructed using a multiple alignment of full-length copies in each genome using DAMBE (Xia and Xie 2001). Then, these respective consensus sequences were employed to mask each host genome to estimate copy number. All BLAST hits with more than 100 bp in size and 80% identity were used to calculate copy number.
Sequence and Phylogenetic Analyses
Inverted repeats were manually searched by the FastPCR software (Kalendar et al. 2014). The potential open reading frame of TRT used in the present study was analyzed using GENSCAN (http://genes.mit.edu/GENSCAN.html) or getorf in the EMBOSS-6.3.1 package (Rice et al. 2000), with default parameters. A multiple alignment of these elements was created by MUSCLE (Edgar 2004), with default parameters. The secondary structure of transposase was predicted by PSIPRED (McGuffin et al. 2000), with default parameters. Putative nucleus localization signal (NLS) motifs were predicted using PSORT II Prediction as provided in the PSORT WWW server (http://psort.nibb.ac.jp/). Each pairwise identity was calculated by Bioedit (Hall 1999) after all ambiguous and gapped sites were removed.
Sequences of Recombination-activating gene 1 (RAG1) were used in the comparison with transposon distance, with the purpose of testing HT hypothesis. Their accession numbers were listed in supplementary table S4, Supplementary Material Online. Multiple alignments of RAG1 and TRT were created using MUSCLE (Edgar 2004). Then, comparison distances of RAG1 and TRT were calculated using MEGA 4 (Tamura et al. 2007; pairwise deletion, maximum composite likelihood) based on two aligned files (supplementary dataset S1 and S2, Supplementary Material Online).
Transposase sequences of Tc1, mariner, and maT were downloaded from GenBank. The accession numbers are as follows: Anopheles albimanus Qeutzal (L76231), Pleuronectes platessa PplTc1 (AJ303069), Haemonchus contortus HcTc1 (AF099908), Rana pipiens RpiTc1 (BK001476), Drosophila virilis Paris (Z49253), Fusarium oxysporum Impala (AF282722), Ceratitis capitata Ccmar1 (U40493), Homo sapiens Hsmar1 (U52077), H. sapiens Hsmar2 (U49974), Drosophila mauritiana Dmmar1 (X78906), Drosophila simulans Dsmar1 (X89927), Chrysoperla plorabunda Cpmar1 (U11650), Girardia tigrina Dtmar1 (X71979), Adineta vaga Avmar1 (DQ138246), Apis mellifera Ammar1 (AY155490), Droshophila mauritiana Mos1 (AEZ51500), Forficula auricularia Famar1 (AAP51098), Shigella sonnei IS630 (X05955), Bombyx mori Bmmar1 (U47917), B. mori Bmmar6 (AF461149), Caenorhabditis briggsae CbmaT1 (AC084526); C. briggsae CbmaT4 (AC084524), C. briggsae CbmaT5 (AC084578), C. elegans CemaT1 (U41268), and Philodina roseola PrD37E (DQ138288). PrD37D (P. roseola), Tc1 (C. elegans), Tc1-1_Pm (Petromyzon marinus), Tc1-1_Lch (Latimeria chalumnae), Tc1-1_Dr (D. rerio), and Tc1-1Ory (Oryzias latipes) were downloaded from Repbase (Jurka et al. 2005). A multiple alignment of full-length transposase sequences of the above transposons and all TRT was created by MUSCLE with default parameters (Edgar 2004). Then, the appropriate amino acid (aa) substitution model was selected using the Akaike information criterion in the ProtTest 3 server (Darriba et al. 2011). The best-suited aa substitution model for these data was the WAG model. Phylogenetic trees were then built using the MRBAYES 3.1.2 software (Ronquist and Huelsenbeck 2003) until the values of the average SD of split frequencies were stably below 0.01.
Results
Identification and Characteristics of DrTRT in the Zebrafish Genome
Using the deduced protein sequence of Bmmar1, we retrieved one sequence (location: CZQB01060324 6427 7413 +) that showed significant similarity (E values: 3e–13) in a TBLASTN search against the zebrafish genome in NCBI. In contrast to the DD37D signature encoded by Bmmar1, the spacing of the DDE motif within the catalytic domain of DrTRT was DD37E (fig. 1). Although DrTRT (named Mariner-14_DR in Repbase) has been deposited in Repbase (http://www.girinst.org/repbase/), knowledge and evolutionary history of DrTRT remain largely unknown. Therefore, DrTRT was utilized in subsequent analyses of Tc1/mariner elements in the present study. The nucleotide sequence of DrTRT was used as query for BLASTN analysis of the zebrafish genome. All obtained significant hits were extracted with 1,000-bp flanking sequences using our Perl script, and these were aligned to determine their boundaries. The consensus sequence of DrTRT was reconstructed, which was determined to be 1,563 bp in length (fig. 1). The transposase was flanked by TIRs that were 38 bp in length (fig. 1). DrTRT also contained a 23-bp 5'-subterminal inverted repeat (SIR) (TGTGCATAATTATTAGGCAACTT) that pairs with a reverse complementary 3'-SIR (AAGTTGCCTAATAATTATGCACA) (fig. 1). A previous study suggested that the subterminal region of DrTRT plays critical structural or functional roles during transposition (Tu, 2000). DrTRT was detected as about 44 copies (table 1). Further analyses showed that 50% (22/44) of DrTRT were verified full-length elements that showed >95% sequence identity to the consensus sequence (supplementary table S1, Supplementary Material Online).
Fig. 1.—
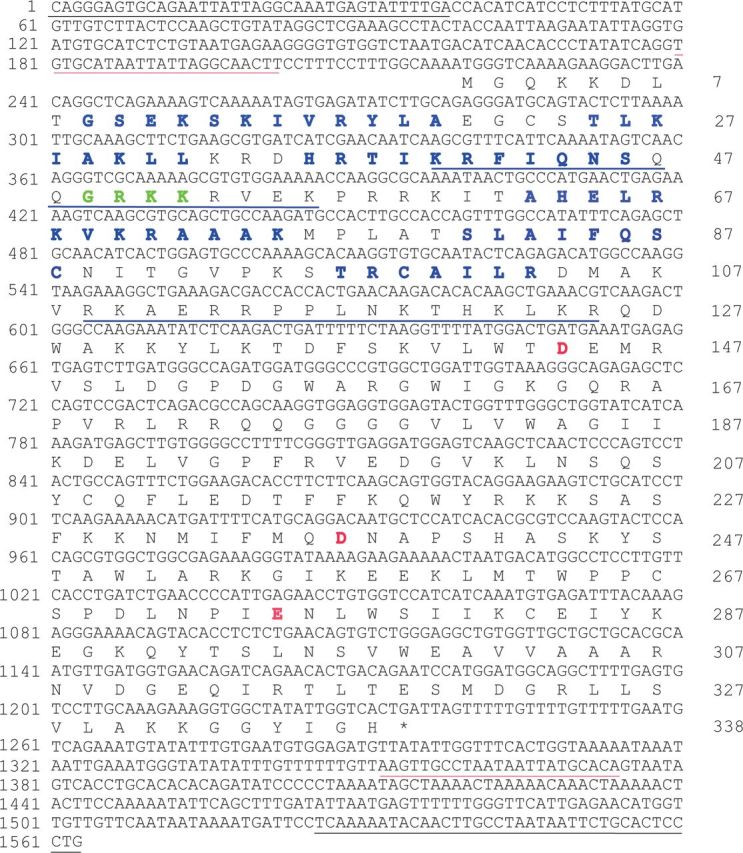
Nucleotide sequence and amino acid translation of DrTRT. Its TIRs and SIR are underlined in black and red, respectively. The stop codon is indicated by an asterisk. The six α-helices at the N-terminal of the transposase are indicated by blue letters. A GRKK putative AT-hook like motif is shown by a green letter. Two bipartite NLS at the N-terminal of the transposase are underlined in blue. The catalytic triad DD37E motif within the catalytic domain is indicated by red letters.
Table 1.
Characteristics of TRT and Tc1 transposons identified in this study
| Species | TEs | Length (bp) | TIRs (bp) | Tpase (aa) | Number of copies | Representatives |
|---|---|---|---|---|---|---|
| Vertebrates | ||||||
| Elopocephala (bony fishes) | ||||||
| Amphilophus citrinellus | AcTRT | 1561 | 27 | 263 | 48 | emb|CCOE01000287.1|52741355275697+ |
| Anguilla japonica | AnjTRT | 1444 | 27 | 217 | 6 | gb|AVPY01162937.1| 2525 3968 – |
| Anoplopoma fimbria | AnofTRT | 1045 | — | — | 27 | gb|AWGY01162360.1| 2696 3736 - |
| Astyanax mexicanus | AsmTRT | 1549 | 28 | 202 | 27 | gb|APWO01056927.1| 208 1648 + |
| Boleophthalmus pectinirostris | BpTRT | 1592 | 44 | 265 | 19 | gb|JACK01022919.1| 5353 6944 + |
| Cynoglossus semilaevis | CsTRT | 1552 | 27 | 297 | 150 | gb|AGRG01027172.1| 7453 9002 + |
| Cyprinodon variegatus | CypvTRT | 1570 | 27 | 338 | 254 | gb|JPKM01031331.1| 7025 8581 - |
| Danio rerio | DrTRT | 1563 | 38 | 338 | 44 | chr2 13613290 13614851 - |
| Dicentrarchus labrax | DilTRT | 1282 | — | 317 | 12 | emb|CABK01017223.1| 2118 3399 - |
| Esox lucius | ElTRT | 1561 | 27 | 323 | 258 | gb|AZJR01046952.1| 21014 22554 + |
| Haplochromis burtoni | HbTRT | 1563 | 27 | 338 | 252 | gb|AFNZ01059715.1| 6228 7790 + |
| Larimichthys crocea | LcTRT | 1574 | 30 | 335 | 13 | gb|JPYK01009954.1| 5998 7571 - |
| Maylandia zebra | MzTRT | 1563 | 27 | 338 | 251 | gb|AGTA02022835.1| 1150 2701 + |
| Neolamprologus brichardi | NbTRT | 885 | — | 294 | 22 | gb|AFNY01026505.1| 2222 3106 + |
| Nothobranchius furzeri | NofTRT | 1011 | — | 336 | 6 | gb|ABLO01004310.1| 411 1421 + |
| Oryzias latipes | OlTRT | 1513 | 28 | 231 | 10 | dbj|BAAF04056582.1| 7120 8632 + |
| Pampus argenteus | PaTRT | 982 | — | 224 | 46 | gb|JHEK01284027.1| 235 1216 + |
| Periophthalmus magnuspinnatus | PemTRT | 1559 | 22 | 338 | 34 | gb|JACL01047866.1| 9655 11168 + |
| Pseudopleuronectes yokohamae | PsyTRT | 1398 | — | 250 | 31 | dbj|BBOV01014196.1| 3143 4536 + |
| Pundamilian yererei | PunTRT | 1563 | 27 | 338 | 257 | gb|AFNX01048038.1| 1455 3017 + |
| Salmo salar | SsTRT | 1561 | 27 | 338 | 435 | gb|AGKD04000285.1| 913827 915387 + |
| Scartelaos histophorus | SchTRT | 1562 | 27 | 264 | 183 | gb|JACN01050586.1| 12429 13966 - |
| Stegastes partitus | StpTRT | 1562 | 27 | 338 | 56 | gb|JMKM01022513.1| 24666 26230 + |
| Takifugu flavidus | TfTRT | 1552 | 27 | 338 | 40 | gb|AOOT01008521.1| 14902 16436 + |
| Takifugu rubripes | TrTRT | 1551 | 27 | 338 | 36 | scaffold_12 809695810893 + |
| Xiphophorus maculatus | XmTRT | 1556 | 27 | 250 | 250 | gb|AGAJ01016295.1| 39713 41243 + |
| Amphibians (clawed frog) | ||||||
| Xenopus (Silurana) tropicalis | XtTRT | 1573 | 202 | 326 | >45 | gb|AAMC02022243.1| 71710 73283 + |
| Lepidosauria (snakes) | ||||||
| Crotalus mitchellii pyrrhus | CmpTRT | 1562 | 27 | 338 | 216 | gb|JPMF01075523.1| 3636 5142 + |
| Ophiophagus hannah | OhTRT | 1561 | 26 | 234 | 220 | gb|AZIM01005391.1| 25631 27415 + |
| Python bivittatus | PbTRT | 1729 | 27 | 320 | 251 | gb|AEQU02140404.1| 2144 3872 + |
| Protozoa | ||||||
| Perkinsus marinus | PmTRT | 1229 | — | 289 | 48 | gb|AAXJ01001025.1| 1452 2678 + |
| Fungi (Mucorales) | ||||||
| Cokeromyces recurvatus | CreTc1 | 1672 | 218 | 286 | 60 | gb|JNEH01002547.1| 21772 23445 - |
| Cunninghamella bertholletiae | CbeTc1 | 1488 | 126 | 343 | 1 | gb|JNEG01000333.1| 1 1488 + |
| Lichtheimia corymbifera | LcoTc1 | 1368 | 59 | 303 | 2 | gb|JNEE01000902.1| 112 1479 + |
| Mucor circinelloides | McTc1 | 1579 | — | 343 | 53 | gb|JNDM01001565.1| 655 2233 – |
| Mucor racemosus | MrTc1 | 1647 | 38 | 343 | 230 | gb|JNEI01003901.1| 14238 15884 - |
| Mucor ramosissimus | MraTc1 | 1647 | 216 | 242 | 123 | gb|JNEF01002670.1| 804 2448 - |
| Mucor velutinosus | MvTc1 | 1646 | 33 | 343 | 125 | gb|JNDK01001598.1| 1060 2704 + |
| Rhizopus delemar | RdTc1 | 1671 | 211 | 343 | 167 | gb|AACW02000276.1| 2545 4216 + |
| Rhizopus oryzae | RoTc1 | 1672 | 211 | 343 | 177 | gb|JNDV01011981.1| 454 2125 - |
| Rhizopus microsporus | RmTc1 | 861 | — | 195 | 24 | gb|JNEJ01004884.1| 638 1498 – |
The coding domain of the transposase was 1,014 bp (338 amino acids) in length. Several conserved motifs that were characteristics of Tc1-like transposons (Plasterk et al. 1999) were also observed in the DrTRT sequence (fig. 1). First, there were two helix-turn-helix (HTH) motifs at the N-terminal of the transposase, and each motif consisted of three α-helices. Second, a GRKK putative AT hook-like motif was present between the two HTH motifs. Third, two bipartite NLS were identified at the N-terminal of the transposase. Finally, a catalytic triad DDE motif within their catalytic domain, with 37 amino acids between the second aspartic acid (D) and glutamic acid (E).
Distribution of TRT in Other Sequenced Species
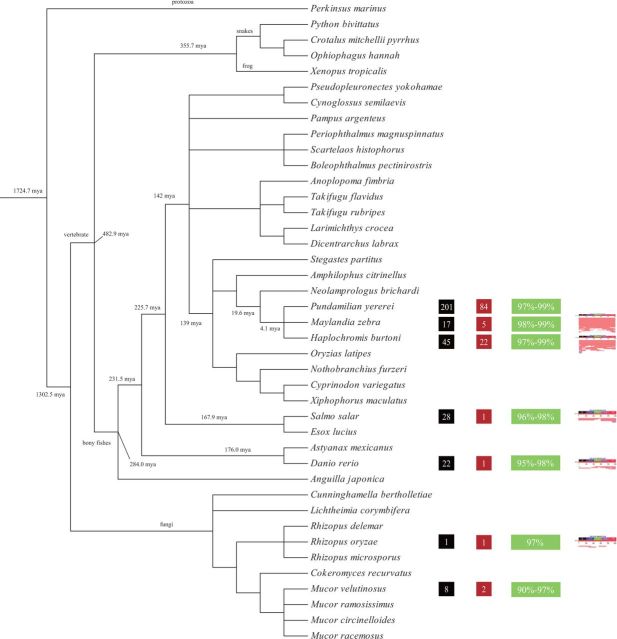
To determine the species distribution of TRT, a TBLASTN search against 1,176 species (437 fungus, 216 invertebrates, 226 vertebrates, 150 plants, and 147 protozoans) whose complete or nearly complete genome sequences are available at NCBI was performed using the protein encoded by DrTRT as query. Significant hits encoding the conserved DD37E motifs were detected in bony fishes, clawed frog, snakes, protozoans, and fungi (table 1 and supplementary table S2, Supplementary Material Online). We should notice that TRT copies may reside within unassembled portions of a genome because many genomes available in public databases are not truly complete in the sense that they are highly fragmented and transposons are more likely to be interrupted by gaps in the sequence. Their copy number per genome extensively varied among species, changing from 1 copy in the fungi Cunninghamella bertholletiae to 435 copies in the Atlantic salmon Salmo salar (table 1), thereby suggesting that these transposons underwent species-specific proliferation in their host genomes. Interestingly, the transposases of several full-length copies of TRT that were identified in bony fishes and fungi did not harbor internal stop codons or frameshift mutations and presented all expected functional domains, as well as an intact TIR (fig. 2 and supplementary table S1, Supplementary Material Online), suggesting that TRT might have current or recent activity in these host genomes. For example, TRT identified in the bony fish P. yererei had 257 copies, and almost all copies (201/257) were present in full length (table 1 and supplementary table S1, Supplementary Material Online and fig. 2). Meanwhile, all complete copies revealed a strikingly high level of sequence identity (97–99% pairwise nucleotide identity) to the entire length of TRT elements, of which 84/201 encoded intact transposases (fig. 2). To further investigate the activity of TRT, the nucleotide sequences of the transposases were used as a BLASTN query against the species Transcriptome Shotgun Assembly (TSA) database. Interestingly, when the putative complete transposase sequences were aligned against the full transcriptome database of the bony fishes M. zebra and H. burtoni, the references were completely covered (100% in length) (fig. 2). Our results also showed that the active TRT elements might be restricted to members of Haplochromini (P. yererei, M. zebra, and H. burtoni).The bony fishes P. yererei, M. zebra, and H. burtoni shared the last common ancestor about 4 Ma (Hedges et al. 2006) and they were diverged from the lyretail cichlid Neolamprologus brichardi (no active TRT was found in this species) about 19.6 Ma, thereby suggesting that the activity of TRT elements in these species might have begun at about between 4 and 19 Ma (fig. 2).
Fig. 2.—
Phylogeny of various species and analysis of potential transposition activity of TRT and Tc1 elements. Numbers in a black box represent numbers of full-length copies in one species. Numbers in a purple box represent numbers of full-length copies encoding intact transposases in one species. Numbers in a green box indicate the sequence identity between full-length copies and consensus sequences in one species. The figures obtained from NCBI show the BLASTN results when searching for transpose nucleotide sequences in the species TSA database.
Despite the observation that transposase sequences have consensus sequences with similar lengths, those of TIRs significantly differed between metazoans and fungi (table 1). Generally, TRT elements in metazoans except for the clawed frog had a short TIR sequence (26–44 bp) at their termini. TRT elements in fungi was flanked by much longer TIRs (generally >100 bp) compared to those of metazoan TRT elements. TIRs played important roles (e.g., providing a cleavage signal sequence and binding site for transposases) in the transposition of Tc1/mariner transposons (Brillet et al. 2007). Tc1/mariner transposons could be divided into different groups based on variations in their TIRs, including length and motif content (Brillet et al. 2007). Therefore, it is important to analyze the functional domain of these length-variable TIRs during transposition by biochemical techniques, as well as compare their structural and functional parts with other transposons of the Tc1/mariner superfamily. Phylogenetic analysis of transposases based on their full length indicated that the TRT elements of metazoans and fungi could be classified into two independent groups (fig. 3). Interestingly, phylogeny also suggested that TRT elements from fungi and reported Tc1 elements showed much closer relationship when compared with TRT elements from metazoans, implying that they might be derived from a more recent ancestor. Indeed, the C-terminal domains of TRT elements from fungi and reported Tc1 elements were very conserved (supplementary fig. S1, Supplementary Material Online). The above results suggest that the TRT elements from fungi are members of Tc1. Therefore, it is not appropriate to call these elements TRT. In order to solve this, all elements from fungi were renamed Tc1.
Fig. 3.—
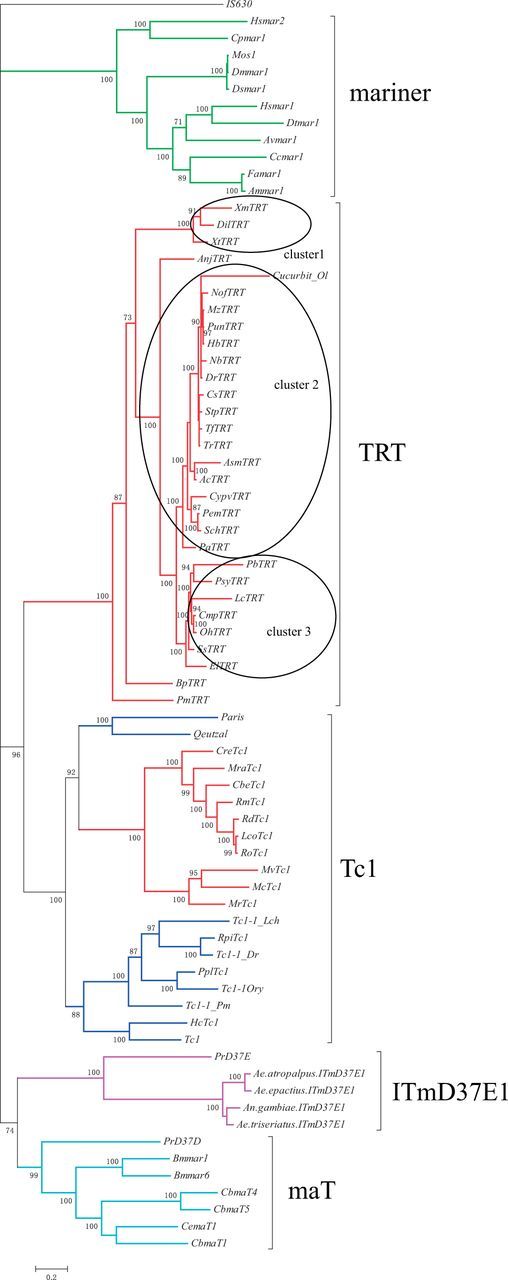
Phylogenetic relationships of all TRT and Tc1 elements identified in this study with other members of the Tc1/mariner superfamily based on their transposases. Bootstrap values of <70% are not shown. Three clusters of TRT elements might be involved in HTs were circled up in circles. TRT and Tc1 elements identified in this study were shown using red color.
Although more than 437 fungal genomes have been sequenced, Tc1 elements were only detected in fungi belonging to the Mucoromycotina (fig. 4). Tc1 elements were also extensively distributed in Mucoromycotina, although only 24 the Mucoromycotina species have been completely sequenced. Although the Dikarya embrace two large phyla (the Ascomycota and the Basidiomycota) that account for the vast majority of fungi and most (379/437) of the sequenced fungi, no Tc1 elements were detected in this taxonomic cluster. We should notice that sampling bias of the available databases may influence our level of detection for Tc1 elements in fungi. Sequencing of additional fungal genomes may facilitate in the identification of Tc1 elements. Such expansion will also facilitate in the elucidation of the evolutionary dynamics or patterns of Tc1 elements in a wide range of fungal species. Several different strains of fungal species have been completely sequenced, and thus we investigated the presence/absence of polymorphisms in the Tc1 elements of these strains. The fungi Lichtheimia corymbifera and Cunninghamella bertholletiae harbored polymorphisms in their TRT elements (supplementary fig. S2, Supplementary Material Online), thereby suggesting that Tc1 elements recently invaded their host and currently have not undergone fixation. Alternatively, Tc1 polymorphisms might result from cut-and-paste excision of the element.
Fig. 4.—
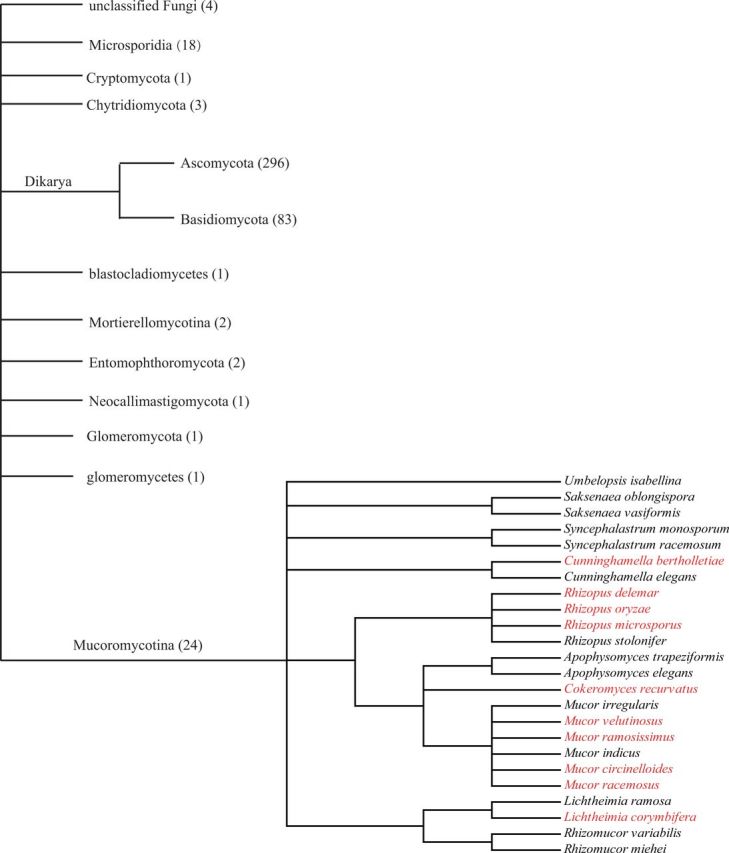
Phylogeny of fungal species with complete or nearly complete genome sequences are available at NCBI. Species harboring Tc1 elements are shown in red.
Comparison of TRT Elements with Other Known Tc1/Mariner Transposons
To determine the relationship between TRT elements and other members of the Tc1/mariner superfamily, the transposase sequences of each TRT element were aligned to 11 mariner, 12 Tc1, and 7 maT elements. The reported DD37E transposons (ITmD37E) were also included in the analysis. The consensus amino acid sequence of the element (IS630) from Shigella sonnei was used as out-group. The phylogenetic tree showed that TRT elements were more closely related to Tc1 than to mariner and maT (fig. 3). The bootstrap values obtained using two different methods were all >99%. Our results also showed that TRT elements and the reported DD37E transposons formed independent clades, thereby suggesting that these belong to two different subfamilies of the Tc1/mariner superfamily.
Next, we compared the functional motifs in the catalytic domain of Tc1, mariner, maT, ITmD37E, and TRT elements. The results showed that three conserved motifs in the catalytic domain could separate mariner (TXDE, HDNA, and SPDLAP(S/T/I)DY) from Tc1 ((W/F)(S/T)DE, QDND and SPDLNPIE). The three conserved functional motifs of TRT elements included (W/F)TDE, Q/HDNA, and SPD/HLNPIE (supplementary fig. S3, Supplementary Material Online), which was similar to those of Tc1. However, the ITmD37E conserved functional motifs (MDDE, PDLA, and V/CPQA/FRPIE) clearly differed from those of other members of the Tc1/mariner superfamily, thereby suggesting that ITmD37E could be organized into a distinct family of the Tc1/mariner superfamily.
Evidence for Multiple HTs of TRT
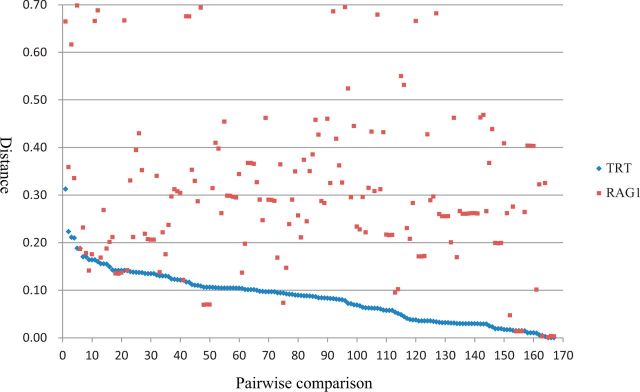
The above phylogeny showed that TRT elements identified in this study could be classified into three major clusters (fig. 3): Cluster 1 includes 3 species (1 frog and 2 bony fishes); Cluster 2 includes 17 bony fishes; Cluster 3 includes 8 species (3 snakes and 5 bony fishes). Phylogenetic analysis also suggested that the host and TRT phylogenies were incongruent. This result might imply that TRT elements have been exposed to multiple episodes of HTs. To further demonstrate this conclusion, pairwise distances between all consensus sequences were calculated. Distances of almost all pairwise comparisons were extremely low (average = 0.084; SD = 0.053; range = 0.000–0.313) (fig 5 and supplementary table S3, Supplementary Material Online). Meanwhile, most of species involved in TRT pairwise distances shared last common ancestor more than 110 Ma (supplementary table S3, Supplementary Material Online). Considering these deep divergence time and the strikingly low-pairwise TRT distances seem incompatible with the possibility that these transposons were obtained from parents to offspring. Indeed, for almost all pairwise comparisons (153/167), the distances computed for TRT are much lower than those calculated for RAG1 (average = 0.302; SD = 0.160; range = 0.001–0.698) (supplementary table S3, Supplementary Material Online), which was usually used to infer the HT events of transposons in vertebrates (Gilbert et al. 2010; Gilbert et al. 2012). Two additional lines of evidence ruled out the scenario that TRT elements were vertically inherited from the last common ancestor of these species. First, TRT elements identified in closely related bony fishes showed a higher level of nucleotide sequence divergence than those of bony fishes and snakes (data not shown). Second, the taxonomic distribution of TRT elements was highly discontinuous. Although more than 226 vertebrates have been sequenced, TRT elements were only detected in bony fishes, frog, and snakes. Together, our results showed that the presence of TRT in most of species reported in this study is as a result of HT events.
Fig. 5.—
Graph illustrating the pairwise distances of TRT and RAG1 between species included in this study. The distances are obtained from all possible pairwise comparisons (n = 167; labeled on the x axis) between the 3 (Cluster 1), 17 (Cluster 2), and 8 (Cluster 3) species in which TRT was identified.
However, for 14 pairs of species, we also noticed that the distance computed between TRT was greater than the distances calculated for RAG1 (supplementary table S3, Supplementary Material Online). Two hypotheses may explain this phenomenon: First, TRT was transferred into the common ancestor of comparison species by HTs and these species got TRT by vertical transfers; Second, TRT was introduced in each species by HTs but the happening time of these HT events and the species diverged was very close. This phenomenon was also observed in other vertebrates (Gilbert et al. 2012).
In conclusion, our findings indicate that vertical and HTs may not be mutually exclusive and may have concurred in the evolution of TRT elements.
Discussion
Description of a Tc1-like Transposon
The present study identified a Tc1-like transposon in zebrafish. DrTRT presented all the hallmark features of Tc1-like elements, including the existence of a transposase of about 340 amino acids in length, a DDE motif, two HTH motifs in the DNA-binding domains, TIRs, and TSD TA at each end. The spacing of the DDE motif within the catalytic domain of DrTRT was unique, with 37 amino acids (DD37E) separating the second aspartic acid and the glutamic residues. Although ITmD37E has been previously reported (Shao and Tu 2001), phylogenetic analysis and functional domain comparison demonstrated that TRT elements and ITmD37E belong to two distinct subfamilies of the Tc1/mariner superfamily. Phylogenetic analysis also showed that TRT elements belong to the Tc1 family (fig. 3). However, the observed high genetic distance with the rest of the Tc1 elements (DD34/35E) and the presence of a unique DDE spacing (DD37E) suggest that TRT elements constitute a novel subfamily within the Tc1 family.
The present study observed different species distributions for TRT and ITmD37E elements. TRT elements were detected in the genomes of bony fishes, clawed frog, snakes and protozoans following a search in GenBank. No TRT elements were observed in insects. In contrast, previous studies and our results (data not shown) indicated that ITmD37E extensively occurred in various species, including Arthropoda, Rotifera, flatworms, Hydrozoa, and ciliates (Shao and Tu 2001; Arkhipova and Meselson 2005; Biedler et al. 2007). On the other hand, ITmD37E was not detected in vertebrates.
TRT Elements Are Complete and Potentially Active in Haplochromini
To date, only some Tc1-like elements (such as Tol1, Tol2, Passport, and Tana1) that were identified in bony fishes have been demonstrated to be transpositionally active (Koga et al. 2006; Clark et al. 2009; Pujolar et al. 2013; Watanabe et al. 2014). Previous studies showed that most Tc1-like elements in fishes are inactive because of stop codons, deletions, or frameshifts within their sequences (Radice et al. 1994; Reed 1999). In contrast, TRT elements were determined to be intact in the genomes of P. yererei, M. zebra, and H. burtoni. These transposases showed no internal stop codons or frameshift mutations, and their TIRs were completely identical, thereby suggesting that TRT elements have current or recent activity in these species. Generally, inactive transposons are expected to rapidly accumulate random mutations not only in the transposase but also in the TIRs, thereby disrupting their complementarity (Pujolar et al. 2013). Recent activity of TRT elements is also supported by the observations of a very high sequence identity that was observed between full-length copies and consensus sequences, high copy number, and the presence of several elements in the TSA databases (fig. 2). Although intact transposases of TRT or Tc1 elements were also identified in other species, including Salmo salar, D. rerio, and fungi, no complete transposases was detected in their transcriptomes (fig. 2). These findings suggest that TRT elements might not be active in these species. Further analysis suggested that the activity of TRT elements in P. yererei, M. zebra, and H. burtoni might have begun about between 4 and 19 Ma.
Homology-based analysis indicates that TRT elements may have occur as approximately 250 copies in P. yererei, M. zebra, and H. burtoni (table 1). Similar copy numbers were also observed for the other two active Tc1-like elements, Passport (300 copies) and Tana1 (764–1,568 copies) (Leaver 2001; Pujolar et al. 2013). Furthermore, the detection of Passport and Tana1 in divergent species also suggests that HT played important roles in their dissemination (Leaver 2001; Pujolar et al. 2013).
Mechanisms and Direction of HT Events Involving TRT Elements
Although we have demonstrated that TRT elements underwent HT events to bony fishes, snakes and frogs, their underlying mechanisms remain unknown. However, it should be noted that DNA viruses are well recognized as potential intermediates for HT of transposons among various animal species (Schaack et al. 2010). Interestingly, it is reported that a snake retroposon integrated into the genome of the taterapox virus, a poxvirus that infects West African rodents, by HT (Piskurek and Okada 2007). Therefore, we speculate that poxviruses may be an efficient vector for the HT of TRT elements that were identified in snakes. Parasitism might also facilitate the HT of transposons. Recently, some cases of HT events involving transposons between vertebrate and their parasites were reported (Kuraku et al. 2012; Walsh et al. 2013; Zhang et al. 2014; Suh et al. 2016). In the present study, a high sequence identity of TRT elements was also observed in several bony fishes, indicating that parasitism might have served as an effective delivery system for the HT of TRT elements.
A second important issue that needs to be addressed is the direction of HT. In the present study, the continuous distribution of TRT elements across bony fish genomes and TRT elements from snakes and frog are nested within the bony fishes (fig. 3), thereby suggesting HT events from the bony fishes to the snakes or frog.
Potential Applications
The discovery of a widespread Tc1-like transposon in organisms may have potentially important applications. Molecular tools are being developed for the genetic manipulation of organisms. Active Tc1/mariner transposons could be potentially used as molecular tools for transgenesis and insertion mutagenesis (Pavlopoulos et al. 2007; Voigt et al. 2016). The integrity of the native form (with an intact transposase), the presence of all functional domains, and their occurrence in the M. zebra and H. burtoni transcriptomes are suggestive of current or recent activity of TRT elements. Meanwhile, TRT elements are widespread in organisms. Therefore, we speculate that TRT elements identified in this study might be used as a molecular tool.
Supplementary Material
Acknowledgments
The National Natural Science Foundation of China (31560308 to Z.H.H., 31260632 to Z.X.G., and 31401106 to H.M.J.), the Natural Science Foundation of Jiangxi Province (20161BAB214151 to Z.H.H.) and the Hi-Tech Research and Development (863) Program of China (2013AA102507) supported this study. We also thank the language revision of this manuscript by the LetPub company.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Arkhipova IR, Meselson M. 2005. Diverse DNA transposons in rotifers of the class Bdelloidea. Proc Natl Acad Sci U S A. 102:11781–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler JK, Shao H, Tu Z. 2007. Evolution and horizontal transfer of a DD37E DNA transposon in mosquitoes. Genetics 177: 2553–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C. 2010. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics 186:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillet B, Bigot Y, Augé-Gouillou C. 2007. Assembly of the Tc1 and mariner transposition initiationcomplexes depends on the origins of their transposase DNAbinding domains. Genetica 2007 130:105–120. [DOI] [PubMed] [Google Scholar]
- Clark KJ, Carlson DF, Leaver MJ, Foster LK, Fahrenkrug SC. 2009. Passport, a native Tc1 transposon from flatfish, is functionally active in vertebrate cells. Nucleic Acids Res. 37:1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL, Craigie R, Gellert M, Lambowitz AM. 2002. Mobile DNA II. Washington, DC: American Society for Microbiology Press. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto BR, et al. 2015. HTT-DB: horizontally transferred transposable elements database. Bioinformatics 31:2915–2917. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41:331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Hernandez SS, Flores-Benabib J, Smith EN, Feschotte C. 2012. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol Biol Evol. 29:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. 2010. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 464:1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Waters P, Feschotte C, Schaack S. 2013. Horizontal transfer of OC1 transposons in the Tasmanian devil. BMC Genomics 14:134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program forWindows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. [Google Scholar]
- Haymer DS, Marsh JL. 1986. Germ line and somatic instability of a white mutation in Drosophila mauritiana due to a transposable genetic element. Dev Genetics 6:281–291. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledgebase of divergence times among organisms. Bioinformatics 22:2971–2972. [DOI] [PubMed] [Google Scholar]
- Jacobson JW, Medhora MM, Hartl DL. 1986. Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci U S A. 83:8684–8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Lee D, Schulman AH. 2014. FastPCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol Biol. 1116:271–302. [DOI] [PubMed] [Google Scholar]
- Koga A, Iida A, Hori H, Shimada A, Shima A. 2006. Vertebrate DNA transposon as a natural mutator: the medaka fish Tol2 element contributes to genetic variation without recognizable traces. Mol Biol Evol. 23:1414–1419. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Qiu H, Meyer A. 2012. Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol Evol. 4:929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver MJ. 2001. A family of Tc1-like transposons from the genomes of fish and frogs: evidence from horizontal transmission. Gene 271: 203–214. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yang G. 2014. Tc1-like transposable elements in plant genomes. Mob DNA 5:17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A, Sullivan D, Hartl D. 1996. Genetic evidence for subunit interactions in the transposase of the transposable element mariner. Genetics 144:1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. [DOI] [PubMed] [Google Scholar]
- Novick P, Smith J, Ray D, Boissinot S. 2010. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene 449:85–94. [DOI] [PubMed] [Google Scholar]
- Pace JK, 2nd, Gilbert C, Clark MS, Feschotte C. 2008. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci U S A. 105:17023–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A, Oehler S, Kapetanaki MG, Savakis C. 2007. The DNA transposon Minos as a tool for transgenesis and functional genomic analysis in vertebrates and invertebrates. Genome Biol. 8:S2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek O, Okada N. 2007. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci U S A. 104:12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH, Izsvák Z, Ivics Z. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15:326–332. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, et al. 2013. Tana1, a new putatively active Tc1-like transposable element in the genome of sturgeons. Mol Phylogenet Evol. 66:223–232. [DOI] [PubMed] [Google Scholar]
- Radice AD, Bugaj B, Fitch DH, Emmons SW. 1994. Widespread occurrence of the Tc1 transposon family: Tc1-like transposons from teleost fish. Mol Gen Genet. 244:606–612. [DOI] [PubMed] [Google Scholar]
- Reed KM. 1999. Tc1-like transposable elements in the genome of lake trout (Salvelinus namaycush). Mar Biotechnol. 1:60–67. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Tu Z. 2001. Expanding the diversity of the IS630-Tc1-mariner superfamily: discovery of a unique DD37E transposon and reclassification of the DD37D and DD39D transposons. Genetics 159:1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, et al. 2016. Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat Commun. 7:11396.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionarygenetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- Tu Z. 2000. Molecular and evolutionary analysis of two divergent subfamilies of a novel miniature inverted repeat transposable element in the yellow fever mosquito, Aedes aegypti. Mol Biol Evol. 17:1313–1325. [DOI] [PubMed] [Google Scholar]
- Voigt F, et al. 2016. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat Commun. 7:11126. doi: 10.1038/ncomms11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. 2013. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A. 110:1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, et al. 2014. Spontaneous germline excision of Tol1, a DNA-based transposable element naturally occurring in the medaka fish genome. Genome 57:193–199. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 92:371–373. [DOI] [PubMed] [Google Scholar]
- Zhang HH, Feschotte C, Han MJ, Zhang Z. 2014. Recurrent horizontal transfers of Chapaev transposons in diverse invertebrate and vertebrate animals. Genome Biol Evol. 6:1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.