Abstract
Rhododendron delavayi Franch. is globally famous as an ornamental plant. Its distribution in southwest China covers several different habitats and environments. However, not much research had been conducted on Rhododendron spp. at the molecular level, which hinders understanding of its evolution, speciation, and synthesis of secondary metabolites, as well as its wide adaptability to different environments. Here, we report the genome assembly and gene annotation of R. delavayi var. delavayi (the second genome sequenced in the Ericaceae), which will facilitate the study of the family. The genome assembly will have further applications in genome-assisted cultivar breeding. The final size of the assembled R. delavayi var. delavayi genome (695.09 Mb) was close to the 697.94 Mb, estimated by k-mer analysis. A total of 336.83 gigabases (Gb) of raw Illumina HiSeq 2000 reads were generated from 9 libraries (with insert sizes ranging from 170 bp to 40 kb), achieving a raw sequencing depth of ×482.6. After quality filtering, 246.06 Gb of clean reads were obtained, giving ×352.55 coverage depth. Assembly using Platanus gave a total scaffold length of 695.09 Mb, with a contig N50 of 61.8 kb and a scaffold N50 of 637.83 kb. Gene prediction resulted in the annotation of 32 938 protein-coding genes. The genome completeness was evaluated by CEGMA and BUSCO and reached 95.97% and 92.8%, respectively. The gene annotation completeness was also evaluated by CEGMA and BUSCO and reached 97.01% and 87.4%, respectively. Genome annotation revealed that 51.77% of the R. delavayi genome is composed of transposable elements, and 37.48% of long terminal repeat elements (LTRs). The de novo assembled genome of R. delavayi var. delavayi (hereinafter referred to as R. delavayi) is the second genomic resource of the family Ericaceae and will provide a valuable resource for research on future comparative genomic studies in Rhododendron species. The availability of the R. delavayi genome sequence will hopefully provide a tool for scientists to tackle open questions regarding molecular mechanisms underlying environmental interactions in the genus Rhododendron, more accurately understand the evolutionary processes and systematics of the genus, facilitate the identification of genes encoding pharmaceutically important compounds, and accelerate molecular breeding to release elite varieties.
Keywords: Rhododendron delavayi, genomics, genome assembly, annotation
Background
Rhododendron L. is a genus in the family Ericaceae. It is 1 of the largest and most diverse genera in the family and is distributed predominantly throughout the Northern hemisphere, but also reaches into the Asian tropics. More than 1000 species of Rhododendron are currently recognized, of which 567 species representing 6 subgenera are known from China. Of these Chinese species, approximately 80% are endemic [1, 2]. Because of the adaptability of this genus to different environments, species such as R. arboreum and R. ferrugineum have been used to investigate the effects of different environmental factors on plant growth, development, and domestication [3–6].
Certain secondary metabolites in Rhododendron have been investigated in connection with antioxidant, anti-inflammatory, anti-carcinogen, and anti-bacterial properties; these compounds have potential in the alleviation of symptoms in conditions including diabetes, arthritis, headache, and hypertension [7–9]. Genome-level sequencing could help investigation into genes responsible for these metabolites and could facilitate the characterization of bio-active compounds and downstream production.
Most species of Rhododendron are diploid (2n = 26). The relatively low levels of ploidy and reported introgression of genetic material between species in nature might be important in the evolution and speciation of Rhododendron [10]. Hybrid varieties can be produced with relative ease by using Rhododendron as the parent because of its natural interspecific hybridization [11, 12]. Previous research on morphology, anatomy, and cytology of Rhododendron suggests that the subgenus Hymenanthes represents a basal state of this genus [13], but classification attempts that employed only a small set of gene regions were not able to resolve relationships within the subgenus [14, 15].
R. delavayi Franch. is widely distributed throughout southwest China and grows at a wide altitudinal range, between 1200 and 3200 m. The species belongs to the subgenus Hymenanthes, subsection Arborea [1, 16]. Four varieties have been described for this species. Rhododendron delavayi var. peramoenum has narrow leaves and has been reported in western Yunnan, northeast India, and Myanmar, whereas R. delavayi var. delavayi has broader leaves than the former and mainly dominates in the Chinese range of the species. Another 2 varieties, R. delavayi var. adenostylum and R. delavayi var. pilostylum, were recently shown to fall within the spectrum of morphologies observed in hybrids between R. delavayi and R. irroratum [17]. In this project, material obtained from R. delavayi var. delavayi (see as Fig. 1) was used to generate genome sequences.
Figure 1:

Rhododendron delavayi Franch. var. delavayi on Cang Shan Mountain, Dali.
Due to its very attractive flowers and good resistance to arid and cold climates, R. delavayi has become a highly profitable ornamental flower in the market, especially in China and some Southeastern Asian countries, such as Vietnam, Thailand, Burma, and India. Nevertheless, it was believed that the anthropogenic activities have significantly reduced the diversity of plants of this genus in nature [18].
The aim of this project was to obtain a genome sequence of R. delavayi. With an available genome sequence, several next-generation sequencing approaches requiring a reference will become feasible, which will enable more in-depth research into genome-environment interactions, help with marker development for phylogenetic studies, and open possibilities for genome-assisted cultivar breeding and other downstream applications.
Data Description
Sample collection
Tissue samples were obtained from a 50-year-old tree growing in Jindian National Forest Park (Kunming, Yunnan, Taxonomy ID: 321363). This tree was transplanted from Cang Shan Mountain (Dali, Yunnan) in 1995. For genome library preparation, only leaf tissue was used; for transcriptome sequencing, samples were obtained from 5 different tissues: flowers, flower buds, young leaves, mature leaves, and young stems. After collection, tissues were immediately transferred into liquid nitrogen and stored until DNA and RNA extraction.
Illumina sequencing strategy
Genomic DNA was extracted from the leaf tissue using a standard CTAB extraction [19]. Different methods were used to construct different insert size libraries. For the small insert libraries (170, 250, 500, and 800 bp), Illumina's protocols were used as follows (Illumina, San Diego, CA, USA): (i) genomic DNA was fragmented by nebulization with compressed nitrogen gas; (ii) DNA ends were polished, and an adenine was added to the ends of the fragments; (iii) DNA adaptors (Illumina) with a single “T” overhang at the 3’ end were ligated to the DNA fragments above; (iv) the ligation products were run on 2% agarose gels, and the bands corresponding to each insert size were excised. For the large insert libraries (2, 5, 10, 20, and 40 kb), Illumina's mate pair library protocols were followed: (i) genomic DNA was fragmented by nebulization with compressed nitrogen gas; (ii) DNA ends were polished using dNTPs labeled with biotin and circularized for self-ligation; (iii) circularized DNA was fragmented again by DNA Exonuclease, followed by enrichment of fragments containing biotin/streptavidin with magnetic beads; 4) fragment ends were further polished, followed by addition of an “A” base and adaptors to form the large insert libraries.
As shown in Table 1, the read length of the large insert libraries (2, 5, 10, 20, and 40 kb) was 49 bp, and the read length of the small insert libraries (170, 500, and 800 bp) was 100 bp, with the exception of the 250 bp insert library, which had a read length of 150 bp. A total of 336.83 Gb (×482.61) raw reads were generated from all constructed libraries. Before assembly, reads with low-quality polymerase chain reaction duplication and adapter contaminations were filtered by SOAPfilter, as included in SOAPdenovo v. 2.04 (SOAPdenovo2, RRID:SCR_014986) [20], and finally 246.06 Gb (×352.55) high-quality sequences were obtained for genome assembly.
Table 1:
Sequencing libraries and data yields for whole-genome shotgun sequencing
| Library type | Lane | Read length (bp) | Insert size (bp) | Raw bases | Clean bases | ||
|---|---|---|---|---|---|---|---|
| Total bases (Gb) | Depth (×) | Total bases (Gb) | Depth (×) | ||||
| PE101 | 2 | 100 | 170 | 80.47 | 115.30 | 74.12 | 106.20 |
| PE151 | 1 | 150 | 250 | 59.69 | 85.52 | 47.20 | 67.63 |
| PE101 | 4 | 100 | 500 | 47.89 | 68.62 | 43.58 | 62.44 |
| PE101 | 3 | 100 | 800 | 42.22 | 60.49 | 36.79 | 52.71 |
| MP50 | 2 | 49 | 2000 | 30.36 | 43.50 | 19.56 | 28.03 |
| MP50 | 3 | 49 | 5000 | 23.11 | 33.11 | 9.06 | 12.98 |
| MP50 | 3 | 49 | 10 000 | 20.17 | 28.90 | 6.71 | 9.61 |
| MP50 | 2 | 49 | 20 000 | 19.01 | 27.24 | 4.35 | 6.23 |
| MP50 | 1 | 49 | 40 000 | 13.91 | 19.93 | 4.69 | 6.72 |
| Total | 21 | 336.83 | 482.61 | 246.06 | 352.55 | ||
Sequencing depth was calculated based on a genome size of 697.94 Mb. High-quality data were obtained by filtering raw data for low-quality and duplicate reads.
RNA of each tissue was extracted separately according to the TRIzol protocol (Invitrogen) and then combined in homogenized RNA concentration. Total mRNAs were purified from total RNA by Dynal Oilgo (dT) beads (Invitrogen). Random oligo-nucleotides and M-MuLV Reverse Transcriptase (RNase H) were used to synthesize the first cDNA strand, and then the second cDNA strand was synthesized using DNA Polymerase I and RNase H. The cDNA libraries with insert sizes of 200–500 base pairs (bps) were selected and purified with the AMPure XP beads system (Beckman Coulter), and subsequently sequenced on an Illumina HiSeq 2000 platform. Both cDNA library construction and Illumina sequencing were carried out by BGI-ShenZhen. Paired-end reads were generated with a read length of 90 bp. The raw reads were filtered by SOAPnuke (SOAPnuke, RRID:SCR_015025) [21], with the following criteria for being discarded: (i) reads contained adaptors; (ii) reads with unknown nucleotides larger than 5%; (iii) low-quality reads (the rate of reads at which quality value ≤ 10 is more than 20%). After filtering, 7.13 G clean reads were obtained for genome evaluation and gene annotation. All clean reads were uploaded to NCBI (SRA505613).
Genome size estimates
We characterized the genome sequence (genome size, heterozygosity, and repetitive content) using the distribution of k-mers of 17, 21, 25, and 27 lengths from the clean reads (29 Gb clean reads from 500 and 800 bp insert size libraries). This analysis was performed using KmerFreq (included in SOAPdenovo, v. 2.04). The genome size (G) of R. delavayi was estimated by the following formula: G = k-mer_number/k-mer_depth, where the k-mer_number is the total number of k-mers, and k-mer_depth refers to the most frequent peak.
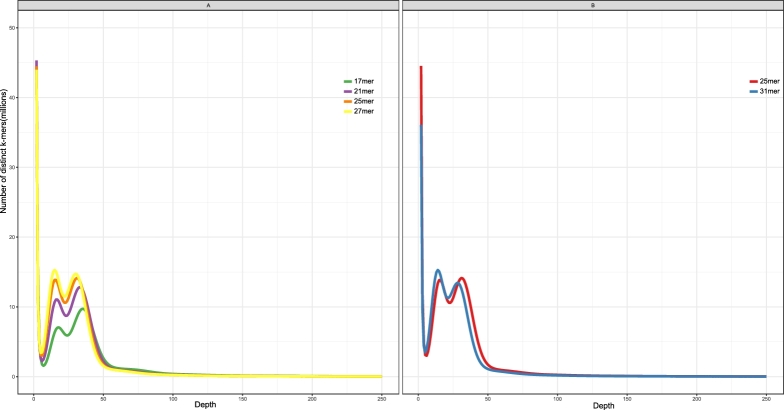
All 4 k-mer distribution curves displayed 4 distinct peaks (Fig. 2A). The first peak at k = 1 was an artifact caused by sequencing errors, each of which created a k-mer that never occurred in the genome. The remaining 3 peak distributions indicated that the genome is a slightly repetitive, heterozygous, diploid genome. The third peak was a “diploid” peak (k-mers shared between homologous chromosomes) and was twice as deep as the second “haploid” peak (k-mers unique to a haplotype due to heterozygosity). The fourth peak was a repetitive peak (k-mers duplicated due to repetition) and was twice as deep as the “diploid” peak. For k = 17, the homozygous peak (the third peak) was found at a depth of ∼×35, with a k-mer_number of 24 427 946 424 and k-mer_depth of 35. The R. delavayi genome size was estimated to be 695.94 Mb, and the data used in 17-mer analysis was about ×41.7 coverage of the genome. All the k-mer sizes yielded similar genome size estimates of ∼697–717 Mb (Table 2).
Figure 2:
k-mer analysis of the R. delavayi genome. (A) Histograms of k-mer frequencies in the clean read data for k = 17 (green), k = 21 (purple), k = 25 (orange), and k = 27 (yellow) by KmerFreq. (B) Histograms of k-mer frequencies in clean data for k = 25 (red) and k = 31 (blue) by jellyfish. The x-axis shows the number of times a k-mer occurred; e.g., the peaks near x = 31 indicate the number of k-mers that occurred 31 times in the data.
Table 2:
Statistics of genome size estimation by KmerFreq with k = 17, 21, 25, and 27
| Genome | K-mer length (bp) | K-mer numbers | K-mer depths | Estimated genome size | Read numbers | Genome coverage |
|---|---|---|---|---|---|---|
| R. delavayi | 17 | 24 427 946 424 | 35 | 697 941 326 | 290 808 886 | ×41.8 |
| 21 | 23 264 710 880 | 33 | 704 991 238 | 290 808 886 | ×41.25 | |
| 25 | 22 101 475 336 | 31 | 712 950 817 | 290 808 886 | ×40.79 | |
| 27 | 21 519 857 564 | 30 | 717 328 585 | 290 808 886 | ×40.54 |
The genome size was estimated according to the formula Genome size = k-mer_numbers/k-mer_depths.
We also used jellyfish v. 2.0 (jellyfish, RRID:SCR_005491) [22] to make k-mer histograms for k-mers 25 and 31 (Fig. 2B), and genome size estimates were 693 and 703 Mb, respectively (Table 3). The k-mer distribution obtained by jellyfish showed a similar trend to KmerFreq. Using the result from jellyfish as input for GenomeScope [23], heterozygosity estimates for the R. delavayi genome were in the range of ∼0.9–1.1%.
Table 3:
Properties of the R. delavayi k-mer distributions for k = 25 and k =31 using jellyfish
| k-mer length | k = 25 | k = 31 |
|---|---|---|
| Total k-mers | 22 120 556 922 | 20 373 342 031 |
| Error k-mers | 615 612 427 | 688 273 368 |
| Haploid coverage depth | 16 | 14 |
| Diploid coverage depth | 31 | 28 |
| Diploid genome size | 693 707 887 | 703 038 167 |
The genome size was estimated according to the formula Genome size = (Total k-mers—Error k-mers)/Diploid coverage depth.
Genome and transcriptome assembly
The Rhododendron delavayi genome was assembled using Platanus v. 1.2.4 (Platanus, RRID:SCR_015531) [20], employing the 3 following steps: contig assembly, scaffolding, and gap closing. For the contig assembly step, the command line parameters “platanus assemble -t 20 -m 300 -u 0.2 -d 0.5 -k 41 -s 10” were specified to construct de Bruijn graphs for small insert size libraries (170, 250, 500, and 800 bp), to modify the graphs, and to display the output sequences. With these options, Platanus increased the k-mer size by the step size kstep (default 10) and iteratively reconstructed the graphs. Assembled contigs and bubbles in the graphs were obtained from this step. In the scaffolding step, the bubbles and reads from the libraries with small insert sizes (170, 250, 500, and 800 bp) and large insert sizes (2, 5, 10, 20, and 40 kb) were mapped onto the assembled contigs for scaffold construction. The command used for this was “platanus scaffold -t 20 -u 0.2 -c contigs.fasta -b bubble.fasta -IP <reads from small insert size libraries>” -OP <reads from large insert size libraries.> In the gap-filling step, the command used was “platanus gap_close –t 20 –IP <reads from small insert size libraries,>” and gaps within scaffolds were filled by reads from small insert size libraries where 1 end could be mapped to 1 contig and the other end extended into a gap. Two more gap-filling steps were performed based on the assembly results, first utilizing KGF (v. 1.06) [24], followed by GapCloser v. 1.12-r6 (GapCloser, RRID:SCR_015026) [24].
To remove a probable redundant sequence in the genome, we used jellyfish v. 2.0 to calculate the 17-mer frequency table from all short insert libraries, then passed the result to trimDup, which comes as part of Rabbit (the software is also archived in the Gigascience repository, GigaDB) [25, 26, 27]. The following command was used “trimDup 17-mer_table 17 1.5*main_peak genome.fa 0.3.” Hence, k-mers were excluded if their frequency was higher than 1.5 times the main peak. Each k-mer was defined as either a “repeat” or a “unique” k-mer, depending on whether its occurrence frequency was greater or less than twice the average frequency. Rabbit uses a Poisson-based k-mer model to establish a 17-mer frequency table from each scaffold of the genome sequences and then determines unique k-mers belonging to each scaffold and common k-mers shared by the scaffolds. The 17-mer frequency table generated in jellyfish is then used to filter the scaffolds so that the ratio of common to unique k-mers reaches 0.3. After the removal of 57.52 Mb of redundant scaffolds, a total scaffold length of 695 Mb was generated (Table 4). The contig N50 was 61.81 Kb, and the scaffold N50 was 637.82 Kb, while scaffolds with lengths of less than 100 bp were excluded. Meanwhile, we also ran another de novo assembler, SOAPdenovo2 (SOAPdenovo2, RRID:SCR_014986), with various modifications of parameters, but the results (Table 5) from SOAPdenovo2 were not better than those generated above.
Table 4:
The genome assembly and completeness of R. delavayi
| Contig | Scaffold | |||
|---|---|---|---|---|
| Size (bp) | Number | Size (bp) | Number | |
| N50 | 61 801 | 2871 | 637 826 | 313 |
| Minimum length | 13 | 79 | ||
| Maximum length | 581 429 | 3 407 404 | ||
| Total size | 657 780 215 | 695 092 854 | ||
| Number (≥100 bp) | 209 926 | 193 086 | ||
| Number (≥2 kb) | 20 175 | 4972 | ||
| Number (≥100 kb) | 1315 | 1230 | ||
| Number (≥1 Mb) | 140 | |||
| CEGMA completeness | 95.87% [238] | |||
| CEGMA partial | 98.39% [244] | |||
| BUSCO completeness | 92.8% [1337] | |||
| BUSCO fragment | 1.8%% [26] | |||
Numbers of genes that match CEGMA or BUSO are shown in square brackets.
Table 5:
Statistics of the assembly with different parameters
| Assembler | Assembly size (bp) | Contig N50 (bp) | Scaffold N50 (bp) | K-mer (bp) | Gapcloser | Rabbit |
|---|---|---|---|---|---|---|
| SOAPdenovo2 | 854 390 781 | 900 | 3380 | 63 | No | No |
| SOAPdenovo2 | 543 175 156 | 1118 | 5946 | 37 | No | No |
| SOAPdenovo2 | 1 231 272 241 | 19 792 | 67 539 | 87 | Yes | No |
| SOAPdenovo2 | 796 221 798 | 25 301 | 104 917 | 87 | Yes | Yes |
| Platanus | 750 231 563 | 13 232 | 583 084 | 41 | No | No |
| Platanus | 809 870 271 | 7886 | 383 826 | 47 | No | No |
| Platanus | 752 607 346 | 54 782 | 584 190 | 41 | Yes | No |
| Platanus | 695 092 854 | 61 801 | 637 826 | 41 | Yes | Yes |
Transcript assembly was carried out in Trinity release-20130225 (Trinity, RRID:SCR_013048) [28] with the following parameters: minimum contig length 200 bp, min glue 3, group pair distance 280, path reinforcement distance 85, and min kmer covage 3. The TGI Clustering Tool (TGICL) v. 2.1 [29] was used to remove redundancies and merge the Unigenes with overlaps of at least 40 bp. Finally, a total of 83 515 Unigenes were obtained, with a mean length of 1014 bp and an N50 of 1727 bp.
Genome evaluation
We evaluated the completeness of the genome assembly using Core Eukaryotic Genes Mapping Approach (CEGMA) v. 2.5 (CEGMA, RRID:SCR_015055) [30] and Benchmarking Universal Single-Copy Orthologs v. 2.0 (BUSCO, RRID:SCR_015008) [31], which assess genome completeness using the conserved genes from the NCBI Eukaryotic Clusters of Orthologous Groups (KOGs) and BUSCO databases, respectively. CEGMA results indicated that 95.97% of core eukaryotic genes were contained in our assembly (238 out of 248 core eukaryotic genes). BUSCO analysis resulted in 92.8% of plants set (embryophyta_odb9, downloaded from BUSCO) identified as complete (1337 out of 1440 BUSCOs). More detailed information is given in Table 4. The Unigenes were aligned to the R. delavayi genome using BLAT v. 0.36 (BLAT, RRID:SCR_011919) [32] with default parameters. The alignment indicated that the assembled genome of R. delavayi covered 96.98% of the Unigenes, 89.57% of the Unigenes with at least 90% coverage in 1 scaffold, and 98.90% of the Unigenes with at least 50% coverage in 1 scaffold, suggesting a high level of coverage (Table 6).
Table 6:
The unigene coverage of transcriptome data by R. delavayi assembly
| Data | Number | Total | Base coverage | >90% sequence in | >50% sequence in |
|---|---|---|---|---|---|
| set | unigenes | length (bp) | by assembly (%) | 1 scaffold (%) | 1 scaffold (%) |
| >200 bp | 83 515 | 84 701 674 | 96.98 | 89.57 | 98.90 |
| >500 bp | 46 582 | 73 471 401 | 96.90 | 85.64 | 99.03 |
| >1000 bp | 29 816 | 61 377 043 | 96.80 | 82.85 | 99.08 |
Repeat annotation
To identify tandem repeats, TRF v. 4.07 [33] was used with the following parameters: Match = 2, Mismatch = 7, Delta = 7, PM = 80, PI = 10, Minscore = 50, MaxPerid = 2000. In total 29 073 954 bp of tandem repeat sequences were detected, representing 4.18% of the R. delavayi genome. Transposable elements were identified by using homology and de novo methods. Homology: RepeatMasker v. 4.0.5 (RepeatMasker, RRID:SCR_012954) [34] was employed to identify transposable elements with RepBase library (version 20.04) [35], while RepeatProteinMask (v. 4.05) [36] was used to identify transposable elements against the TE protein database in RepBase. De novo: (i) RepeatModeler v. 1.07 (RepeatModeler, RRID:SCR_015027) [37] and LTR_FINDER v. 1.05 (LTR_Finder, RRID:SCR_015247) [38] were used to identify transposable elements; (ii) the results from RepeatModeler and LTR_FINDER were merged into a de novo repeat library; (iii) RepeatMasker was employed to categorize the genome sequence against the de novo repeat library. Finally, transposable elements identified by homology or de novo library within the same category were merged by overlap. Transposable elements accounted for 51.77% of the R. delavayi genome, while long terminal repeat elements (LTRs) represented the largest fraction (37.48%) of transposable elements (Table 7). The most abundant subtypes were Copia and Gypsy, representing 6.84% and 25.49% of the assembly genome, respectively.
Table 7:
Transposable elements in the R. delavayi genome
| Repbase TE length | Protein TE length | De novo TE length | Combined TEs | ||
|---|---|---|---|---|---|
| Length | Percentage | ||||
| DNA | 7 882 501 | 7 328 645 | 69 812 249 | 77 776 557 | 11.19 |
| LINE | 4 811 976 | 12 454 813 | 31 065 638 | 36 834 088 | 5.30 |
| SINE | 125 792 | 0.00 | 869 547 | 991 785 | 0.14 |
| LTR | 34 884 681 | 52 469 776 | 257 040 066 | 260 532 496 | 37.48 |
| Other | 552 | 0.00 | 0.00 | 552 | 0.00 |
| Unknown | 0.00 | 0.00 | 4 565 754 | 4 565 754 | 0.67 |
| Total | 47 001 844 | 72 016 848 | 350 372 642 | 359 874 503 | 51.77 |
Repbase TEs means RepeatMask against Repbase; Protein TEs means RepeatProteinMask result against Repbase protein; De novo TEs means RepeatMask against the de novo library; Combined TEs means the combined result of the 3 steps.
Gene prediction
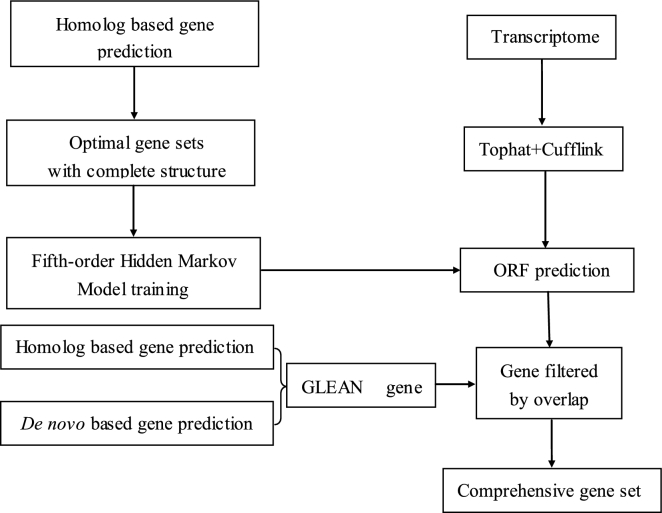
We combined homology-based, de novo, and transcript alignment methods to predict protein-coding genes in the R. delavayi genome. Four major steps were employed, and a detailed pipeline is given in Fig. 3.
Figure 3:
The gene prediction pipeline.
For gene prediction based on homology, we obtained gene sets from Arabidopsis thaliana [37], Actinidia chinensis [39], Capsicum annuum [40], Mimulus guttatus [41], Solanum tuberosum [42], and Solanum lycopersicum [43]. For genes with alternative splicing variants, the longest transcript was selected to represent the gene. We aligned these homologous protein sequences to the R. delavayi genome using TBLASTN (v. 2.2.26) [44], employing an E-value threshold of 1e-5. The resulting BLAST hits were linked to candidate gene loci using solar (v. 0.9.6) [45] with options “-a prot2genome2 –z.” Then, we extracted the candidate gene locus sequences including 1 kb of flanking DNA upstream and downstream and used Genewise v. 2.2.0 (GeneWise, RRID:SCR_015054) [46] to define the intron-exon boundary. Genes with lengths of less than 150 bp or with erroneous structure (premature stop codon or frame shifts) were excluded from further analysis.
For the de novo prediction step, the repeat masked genome was used as input for 2 programs, AUGUSTUS v. 3.03 (Augustus: Gene Prediction, RRID:SCR_008417) [47] and GENSCAN v. 1.0 (GENSCAN, RRID:SCR_012902) [48]. To obtain a training set for AUGUSTUS, we randomly selected 5919 full-length genes that had been predicted based on homology, while for GENSCAN, Arabidopsis parameters were used. For the final non-redundant gene set, genes predicted based on both homology and de novo methods were combined with GLEAN (v. 1.0) [49], setting options “-gff -minlen 150 -minintron 11 -maxintron 15 000.” Genes with erroneous structure or short length were again excluded based on the same thresholds used for homology prediction.
For the transcript alignment prediction step, the short reads from the transcriptome data set generated in the previous step were mapped to the R. delavayi genome using Tophat v. 2.1.1 (TopHat, RRID:SCR_013035) [50] to identify the splice junctions. Cufflinks v. 2.2.1 (Cufflinks, RRID:SCR_014597) [51] was then used to assemble transcripts from the Tophat outputs. The coding potential of these transcripts was identified by using the same gene sets with a fifth-order Hidden Markov Model, which was achieved by the same gene sets used in the training of AUGUSTUS.
In the gene set combination step, outputs from GLEAN were combined with transcript assemblies as follows: first, translated sequences of both sets were cross-matched with an all-to-all BLASTP using an E-value cutoff of 1e-10. The matching transcript assemblies were then added to the GLEAN results as either untranslated regions (UTRs) or alternative splice forms, based on whether coverage and identity of the alignment results was larger than 0.9. The transcript assemblies that had no BLAST hit with the GLEAN results were added to the final set as novel genes.
As a result of these steps, a total of 32 938 non-redundant genes were predicted in the R. delavayi genome (Table 8). These genes were scattered over 2149 scaffolds, averaging 15.33 genes per scaffold.
Table 8:
Summary of R. delavayi genome annotation
| Gene | Gene numbers | Average gene | Average CDS | Average exons | Average exon | Average intron | |
|---|---|---|---|---|---|---|---|
| set | of prediction | length (bp) | length (bp) | per gene | length (bp) | length (bp) | |
| De novo | AUGUSTUS | 42 672 | 2623.41 | 974.42 | 4.76 | 204.56 | 438.16 |
| GENSCAN | 35 859 | 11 242.68 | 1186.91 | 6.35 | 186.87 | 1879.03 | |
| Homolog | A. chinensis | 45 449 | 3501.48 | 846.20 | 3.21 | 263.43 | 1200.29 |
| A. thaliana | 31 950 | 3724.50 | 994.90 | 4.07 | 244.30 | 888.41 | |
| C. annuum | 47 672 | 2558.30 | 805.26 | 3.01 | 267.50 | 872.00 | |
| M. guttatus | 34 616 | 3454.51 | 963.76 | 3.95 | 244.21 | 845.35 | |
| S. lycopersicum | 38 800 | 3324.95 | 917.11 | 3.74 | 245.47 | 880.01 | |
| S. tuberosum | 39 085 | 2958.21 | 850.18 | 3.22 | 263.79 | 948.30 | |
| GLEAN | 29 585 | 4126.65 | 1150.32 | 4.84 | 237.78 | 775.53 | |
| RNA-seq | 38 273 | 2989.97 | 828.78 | 3.45 | 240.07 | 881.29 | |
| Final set | 32 938 | 4434.22 | 1153.21 | 4.62 | 249.70 | 785.08 |
We also used Maker-P [52] to predict gene model with current homolog, de novo, and transcriptome results by using the parameter “est_gff, protein_gff, pred_gff” according to the Maker-P manual. The CEGMA assessment showed that our current pipeline identified 97.09% (234 of 241) of core eukaryotic genes, while the Maker-P pipeline identified only 86.72% (209 of 241) core eukaryotic genes. The BUSCO evaluation demonstrated that 87.4% and 6.4% of 1440 expected plant genes were identified as completeness and fragment, respectively (Table 9). Both assessment methods suggested that for the R. delavayi genome our current pipeline performed better than the Maker-P pipeline.
Table 9:
BUSCO assessment of gene prediction with different pipelines
| Current pipeline | Maker-P | |||
|---|---|---|---|---|
| BUSCO benchmark | Number | Percentage | Number | Percentage |
| Total BUSCO groups searched | 1440 | 1440 | ||
| Complete single copy BUSCOs | 1188 | 82.5 | 1056 | 73.3 |
| Complete duplicated BUSCOs | 70 | 4.9 | 67 | 4.7 |
| Fragmented BUSCOs | 92 | 6.4 | 152 | 10.6 |
| Missing BUSCOs | 90 | 6.2 | 165 | 11.4 |
Functional annotation
Gene function annotation was assigned based on sequence and domain conservation. (i) Assignment based on sequence conservation: protein sequences of R. delavayi were aligned to KEGG (v. 76) [53] and SwissProt and TrEMBL (Uniprot release 201406) [54] by BLASTP (v. 2.2.26) using an E-value threshold of 1e-5. Best-hit BLAST results were then used to define the gene functions. (ii) Assignment based on domain conservation: InterProScan-5.11–51.0 (InterProScan, RRID:SCR_005829) [55] was employed to identify motifs and domains by matching against public databases Pfam [56], PRINTS [57], ProDom [58], SMART [59], and PANTHER [60]. Gene ontology identities [61] for each gene were then obtained from the corresponding InterPro entry [62]. Overall, 85.91% of genes were functionally annotated by at least 1 of the 5 databases above, with 22 946 InterPro entries, 16 471 GO entries, 21 210 KEGG entries, 22 693 SwissProt entries, and 27 975 TrEMBL entries (Table 10).
Table 10:
Statistics for functional annotations in corresponding InterPRro entry
| Numbers of | Percent of | |
|---|---|---|
| matching genes | annotated genes | |
| InterPro | 22 946 | 69.66 |
| GO | 16 471 | 50.00 |
| KEGG | 21 210 | 64.39 |
| Swissprot | 22 693 | 68.90 |
| TrEMBL | 27 975 | 84.93 |
| Annotated | 28 296 | 85.91 |
| Unannotated | 4642 | 14.09 |
Gene family construction
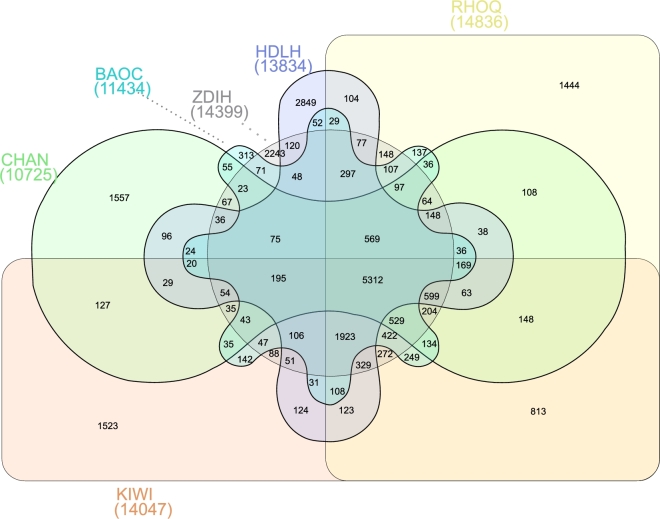
As references, protein sequences of 10 angiosperms (Actinidia chinensis, Primula veris, Catharanthus roseus, Dendrobium officinale, Phalaenopsis equestris, Tarenaya hassleriana, Solanum tuberosum, Solanum lycopersicum, Arabidopsis thaliana, and Oryza sativa) were downloaded (see the Supplementary Data). For genes with alternative splicing variants, the longest transcript was selected to represent the gene. Similarities between sequence pairs were calculated using BLASTP with an E-value threshold of 1e-5. Additionally, OrthoMCL (OrthoMCL DB: Ortholog Groups of Protein Sequences, RRID:SCR_007839) [63] was used with default parameters to identify gene family membership based on overall gene similarity combined with Markov Chain Clustering (MCL). Of all annotated genes, 77.60% were assigned to a family. A total of 14 836 families were represented, of which 1097 were specific of Rhododendron delavayi (Table 11). Figure 4 showed the number of orthologous gene families shared between 6 flower plant genomes, which have 5312 orthologous gene families in common with ancestral functions.
Table 11:
The statistic results of gene family clusters
| Number of | Genes in | Unclustered | Number of | Unique | Average number of | |
|---|---|---|---|---|---|---|
| Species | genes | families | genes | families | families | genes per family |
| R. delavayi | 32 938 | 25 560 | 7378 | 14 836 | 1097 | 1.72 |
| A. chinensis | 39 040 | 26 061 | 12 979 | 14 047 | 1100 | 1.86 |
| P. veris | 18 269 | 15 080 | 3189 | 11 434 | 180 | 1.32 |
| C. roseus | 28 172 | 15 122 | 13 050 | 10 725 | 1231 | 1.41 |
| D. officinale | 35 474 | 25 525 | 9949 | 14 416 | 1091 | 1.77 |
| P. equestris | 29 413 | 21 086 | 8327 | 13 834 | 705 | 1.52 |
| T. hassleriana | 39 881 | 38 100 | 1781 | 14 399 | 623 | 2.65 |
| S. tuberosum | 34 879 | 28 093 | 6786 | 16 118 | 667 | 1.74 |
| S. lycopersicum | 33 585 | 25 623 | 7962 | 17 139 | 532 | 1.50 |
| A. thaliana | 26 637 | 23 007 | 3630 | 14 482 | 539 | 1.59 |
| O. sativa | 38 942 | 26 644 | 12 298 | 13 632 | 2020 | 1.95 |
Figure 4:
Groups of orthologues shared among the angiosperms Rhododendron delavayi (RHOQ), Actinidia chinensis (KIWI), Primula veris (BAOC), Catharanthus roseus (CHAN), Phalaenopsis equestris (HDLH), and Tarenaya hassleriana (ZDIH). Venn diagram generated by http://www.interactivenn.net/.
Phylogenetic analysis
For a phylogenetic analysis, 326 single copy orthologs were selected from the gene family step, and translated protein sequences were aligned in MUSCLE v. 3.8.31 (MUSCLE, RRID:SCR_011812) [64]. Next, the protein alignments were converted to corresponding coding sequences (CDS) using an in-house Perl script. Afterwards, the coding sequences of each single copy family were concatenated to form 1 supergene for each species. The nucleotides at positions 2 (phase 1 site) and 3 (4-fold degenerate site) of each codon were extracted separately and were used to construct 2 separate phylogenetic trees in PhyML3.0 (PhyML, RRID:SCR_014629) [65] specifying a HKY85 substitution model with a gamma distribution across sites. The tree using the phase 1 site was consistent with the tree using the 4 degenerate sites.
Divergence time
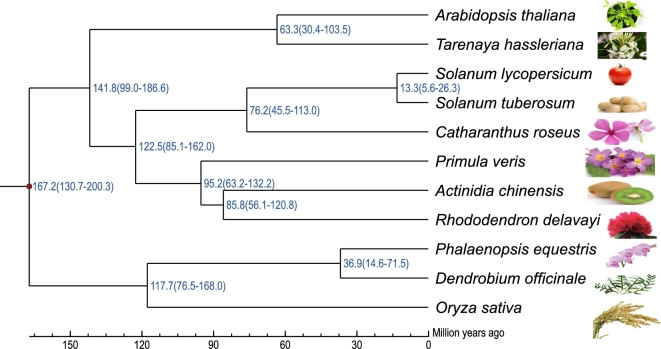
A Bayesian relaxed molecular clock approach was used to estimate species divergence time using MCMCTREE in PAML (PAML, RRID:SCR_014932) [66] based on the 4 degenerate sites data set used in phylogenetic analysis. When using previously published calibration times (split of Oryza sativa and Arabidopsis thaliana fixed as 130∼200 Mya) [67], the divergence time between R. delavayi and Actinidia chinensis was estimated to be in the range of 56.1–120.8 million years ago (Fig. 5).
Figure 5:
Estimation of divergence time. The blue numbers on the nodes are the divergence times from present; the red node indicates the calibrated split.
Conclusion
Now the order Ericales has 3 draft genome sequences of 3 economically important species (kiwi fruit [Actinidia chinensis], American cranberry [Vaccinium macrocarpon], and R. delavayi), 2 of which (V. macrocarpon and R. delavayi) also belong to the family Ericaceae. The availability of the R. delavayi genome sequence should facilitate de novo genome assembly of other species in this genus and, moreover, allow scientists to investigate interactions between environmental factors and related species at a molecular level. Furthermore, phylogenetic research can now draw on a genome as a resource to identify regions providing suitable resolution in this taxonomically difficult group, and it may become easier to identify the genes involved in metabolite pathways that have potential pharmaceutical importance.
Availability of supporting data
Supporting data and the Rabbit software are available in the GigaDB database [27]. The raw data were deposited in the SRA527514 with project accession PRJNA361437 for the Rhododendron delavayi genome.
Actinidia chinensis (ftp://bioinfo.bti.cornell.edu/pub/kiwifruit/)
Catharanthus roseus (http://bioinformatics.psb.ugent.be/orcae/overview/Catro)
Primula veris (http://datadryad.org/resource/doi:10.5061/dryad.2s200)
Dendrobium officinale (ftp://202.203.187.112/genome/dendrobe/)
Phalaenopsis equestris (ftp://ftp.genomics.org.cn/from_BGISZ/20130120/)
Solanum tuberosum: phytozome12.0 (https://phytozome.jgi.doe.gov/pz/portal.html)
Solanum lycopersicum: phytozome12.0 (https://phytozome.jgi.doe.gov/pz/portal.html)
Arabidopsis thaliana: phytozome12.0 (https://phytozome.jgi.doe.gov/pz/portal.html)
Oryza sativa: phytozome12.0 (https://phytozome.jgi.doe.gov/pz/portal.html)
Abbreviations
Gb: gigabase; GO: gene ontology; PE: paired-end; TE: transposable element.
Competing interests
The authors declare that they have no competing interests.
Funding
This project was supported by the Program of Science and Technology Talents Training in Yunnan province (2016HA005), the Program of Innovative Talents Promotion by the Chinese Ministry of Science and Technology (2014HE002), the Applied Basic Research Project of Yunnan Province (2016FB058), and the National Natural Science Foundation of China (31460217, 31560225).
Author contributions
L.Z., J.W., Y.C., L.M., and Q.G. conceived the project. S.L., F.L., W.X., J.S., L.P., and H.Y. designed sample collection and extracted the genomic DNA. P.X. led the genome analysis, conducted the genome assembling, and predicted gene structure and repeat sequences. All of the authors listed above participated in discussions of the project and data. P.X., L.Z. (Lu Zhang), Q.G., and J.W. co-drafted the manuscript, and L.Z. (Ling Zou), Y.M., and C.Z. helped with manuscript revision. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgements
We thank to Tobias Marczewski for his gracious help polishing the language.
References
- 1. Chamberlain D, Hyam R, Argent G et al. The Genus Rhododendron: Its Classification and Synonymy. Edinburgh, UK: Royal Botanic Garden Edinburgh; 1996. [Google Scholar]
- 2. Fang M, Fang R, He M et al. Flora of China – Apiaceae through Ericaceae, vol. 14 China: Science Press and Missouri Botanic Garden Press; 2005:260–455. [Google Scholar]
- 3. Gaira KS, Rawal RS, Rawat B et al. Impact of climate change on the flowering of Rhododendron arboreum in central Himalaya, India. Curr Sci 2014;106:12. [Google Scholar]
- 4. Ranjitkar S, Luedeling E, Shrestha KK et al. Flowering phenology of tree Rhododendron along an elevation gradient in two sites in the Eastern Himalayas. Int J Biometeorol 2013;57(2):225–40. [DOI] [PubMed] [Google Scholar]
- 5. Bi Y, Xu J, Yang J et al. Ring-widths of the above tree-line shrub Rhododendron reveal the change of minimum winter temperature over the past 211 years in Southwestern China. Climate Dynam 2016;1–15. [Google Scholar]
- 6. Komac B, Esteban P, Trapero L et al. Modelization of the current and future habitat suitability of Rhododendron ferrugineum using potential snow accumulation. PLoS One 2016;11(1):e0147324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao Y, Chu Q, Ye J. Chromatographic and electrophoretic methods for pharmaceutically active compounds in Rhododendron dauricum. J Chromatog B 2004;812(1):231–40. [DOI] [PubMed] [Google Scholar]
- 8. Zhou W, Oh J, Li W et al. Chemical constituents of the Korean endangered species Rhododendron brachycarpum. Biochem Syst Ecol 2014;56:231–6. [Google Scholar]
- 9. Qiang Y, Zhou B, Gao K. Chemical constituents of plants from the genus Rhododendron. Chemistry Biodiversity 2011;8(5):792–815. [DOI] [PubMed] [Google Scholar]
- 10. Zha HG., Milne RI, Sun H.. Morphological and molecular evidence of natural hybridization between two distantly related Rhododendron species from the sino-himalaya. Botan J Linnean Soc 2008;156(1):119–29. [Google Scholar]
- 11. Yu SX. Research on the problem on the problems of classification of the genus Rhododendron. J Wuhan Botan Res 1986;24(3):161–4. [Google Scholar]
- 12. Zha HG, Milne RI, Sun H. Asymmetric hybridization in Rhododendron agastum: a hybrid taxon comprising mainly F1s in Yunnan, China. Ann Botany 2010;105(1):89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ming TL, Fang RC. The phylogeny and evolution of genus Rhododendron. Acta Botanica Yunnanica 1990;12(4):353–65. [Google Scholar]
- 14. Milne RI, Davies C, Prickett R et al. Phylogeny of Rhododendron subgenus Hymenanthes based on chloroplast DNA markers: between-lineage hybridisation during adaptive radiation? Plant Syst Evol 2010;285(3–4):233–44. [Google Scholar]
- 15. Eckert AJ, Carstens BC. Does gene flow destroy phylogenetic signal? The performance of three methods for estimating species phylogenies in the presence of gene flow. Mol Phylogenet Evol 2008;49(3):832–42. [DOI] [PubMed] [Google Scholar]
- 16. Zha HG, Milne RI, Sun H. Morphological and molecular evidence of natural hybridization between two distantly related Rhododendron species from the Sino-Himalaya. Botan J Linnean Soc 2008;156(1):119–29. [Google Scholar]
- 17. Marczewski T, Ma YP, Zhang XM et al. Why is population information crucial for taxonomy? A case study involving a hybrid swarm and related varieties. AoB Plants 2016;8:plw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang MY, Fang RZ, He MY et al. Rhododendron. In: Wu ZY, Raven PH, eds. Flora of China, vol. 14 Beijing and St Louis: Science Press and Missouri Botanical Garden; 2005:260–455. [Google Scholar]
- 19. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 1980;8:4321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo R, Liu B, Xie Y et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2012;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Developmented by BGI-flexlab, A MapReduce Acceleration supported Software for integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. https://github.com/BGI-flexlab/SOAPnuke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011;27(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vurture G, Sedlazeck M, Nattestad M et al. GenomeScope: Fast reference-free genome prodiling from short reads. Bioinformatics 2017;14(15):2202–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kajitani R, Toshimoto K, Noguchi H et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 2014;24(8):1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. You M, Yue Z, He W et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet 2013;45(2):220–5. [DOI] [PubMed] [Google Scholar]
- 26. Rabbit Genome Assembler: https://github.com/gigascience/rabbit-genome-assembler. [Google Scholar]
- 27. Zhang L, Cai Y, Xu P et al. Supporting data for “The draft genome assembly of Rhododendron delavayi Franch. var. delavayi.” GigaScience Database. 2017. http://dx.doi.org/10.5524/100331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grabherr MG, Haas BJ, Yassour M et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011;29(7):644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pertea G, Huang XQ, Liang F et al. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 2003;19(5):651–2. [DOI] [PubMed] [Google Scholar]
- 30. Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate coregenes in eukaryotic genomes. Bioinformatics 2007;23:1061–7. [DOI] [PubMed] [Google Scholar]
- 31. Simão FA, Waterhouse RM, Ioannidis P et al. BUSCO: assessing genome assembly and annotation completeness with single copy orthologs. Bioinformatics 2015;31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 32. Kent WJ. BLAT the BLAST like alignment tool. Genome Res 2002;12:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benson G, Tandem repeats finder: a program to analyze DNA sequence. Nucleic Acid Res 1999;27:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen NS. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinform 2004;Chapter 4:Unit 4.10. [DOI] [PubMed] [Google Scholar]
- 35. Jurka J, Kapitonov VV, Pavlicek A et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 2005;110:462–7. [DOI] [PubMed] [Google Scholar]
- 36. Abrusán G, Grundmann N, DeMester L et al. TEclass–a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 2009;25:1329–30. [DOI] [PubMed] [Google Scholar]
- 37. Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 2007;35:W265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaul S, Koo HL, Jenkins J et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000;408:796–815. [DOI] [PubMed] [Google Scholar]
- 39. Huang S, Ding J, Deng D et al. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun 2013;4:2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qin C, Yu C, Shen Y et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci U S A 2014;111(14):5135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelly JK, Koseva B, Mojica JP. The genomic signal of partial sweeps in Mimulus guttatu. Genome Biol Evol 2013;5(8):1457–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. The Potato Genome Sequencing Consortium Genome sequence and analysis of the tuber crop potato. Nature 2011;475:189–95. [DOI] [PubMed] [Google Scholar]
- 43. The Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012;485:635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altschul SF, Madden TL, Schaffer AA et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li R, Fan W, Tian G et al. The sequence and de novo assembly of the giant panda genome. Nature 2010;463:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res 2004;14:988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stanke M, Keller O, Gunduz I et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acid Res 2006;34:W435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol 1997;268:78–94. [DOI] [PubMed] [Google Scholar]
- 49. Elsik CG, Mackey AJ, Reese JT et al. Creating a honey bee consensus gene set. Genome Biol 2007;8:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trapnell C, Williams BA, Pertea G et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Campbell MS, Holt C, Moore B et al. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinform 2014;48:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ogata H, Goto S, Sato K et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 1999;27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acid Res 2000;28:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zdobnov EM, Apweiler R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001;17:847–8. [DOI] [PubMed] [Google Scholar]
- 56. Bateman A, Birney E, Durbin R et al. The Pfam protein families database. Nucleic Acids Res 2000;28:263–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Attwood TK, Cronig MD, Flower DR et al. PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res 2000;28:225–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Corpet F, Gouzy J, Kahn D. Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res 1999;27:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schult J, Copley RR, Doerks T et al. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 2000;28:231–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mi HY, Lazareva-Ulitsky B, Loo R et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 2005;33:284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ashburner M, Ball CA, Blake JA et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Burge S, Kelly E, Lonsdale D et al. Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation. Database 2012;2012:257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li L, Stoeckert CJ Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13(9):2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guindon S, Dufayard JF, Lefort V et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010;59(3):307–21. [DOI] [PubMed] [Google Scholar]
- 66. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 2007;24:1586–91. [DOI] [PubMed] [Google Scholar]
- 67. Tuskan GA, Difazio S, Jansson S et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006;313:1596–604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.