Abstract
Chinese domestic pigs have experienced strong artificial selection for thousands of years. However, the molecular mechanisms underlying the selection-causing phenotypic changes in Chinese domestic pigs are still largely unknown. Here we used whole-genome resequencing data of 54 pigs from 9 Chinese diverse breeds and 16 wild boars from 7 localities across China to identify genes that show evidence of positive selection in the process of domestication. A total of 14 candidate domestication regions were detected by selective sweep analyses of genetic differentiation and variability, and a set of genes in these candidate domestication regions were found to be related to metabolic process, development, reproduction, olfactory, behavior, and nervous system. The most promising candidate gene under selection - TBX19 - probably underlies the metabolic alteration and developmental traits, and may also associate with timidity of Chinese domestic pigs. Intriguingly, we found that the haplotype at TBX19 locus shared by nearly all Chinese domestic pigs was possibly introgressed from another Sus species. We also revealed the AHR gene associated with female reproduction is under strong positive selection. These results advance our understanding of the evolutionary history of Chinese domestic pigs and shed insights into identifying functionally important genes/mutations contributing to the phenotypic diversity in pigs.
Keywords: pig, domestication, signature of selection, TBX19, interspecies introgression
Introduction
Animal domestication is an important and complicate event in human history. Understanding how domestication has shaped the patterns of genetic variation is of great importance, because it provides valuable insights into exploring the genetic mechanisms associated with disease resistance and other economical traits such as meat quality, reproduction, and production in domestic animals (Amaral etal. 2011; Rubin etal. 2012). The pigs were independently domesticated at several loci across the world - Near East, China, and several cryptic domestications in Southeast Asia (Giuffra etal. 2000; Larson etal. 2010). Since then a wide variety of distinct phenotypes have formed in diverse breeds due to strong artificial selection, including fast growth, high reproductive capacity, and altered external appearance.
Recently, several studies have reported the genetic alteration behind phenotypic changes in European and Asian pigs during/after domestication using genomic approaches. For example, Rubin etal. (2012) identified genomic selection signals at the NR6A1, PLAG1, and LOCRL loci that play important roles on elongation of body length in European domestic pigs. Fontanesi and Russo (2013) confirmed that variants at the MC1R and KIT loci determine coat color phenotypes in Mediterranean pigs. Fu etal. (2016) reported how domestication has shaped the genetic variation of Chinese black pigs, and that body size, immunity, lipid metabolism, male fertility, and developmental processes were likely the selection targets during the formation of Chinese black breeds. Wang etal. (2015) uncovered that Chinese pig breeds have high haplotype similarity around PRM1, PRM2, TNP2, GPR149, and JMJD1C genes related to reproductive traits, and MITF and EDNRB have experienced strong selection to shape the two-end black coat color phenotype in Tongcheng pigs. We identified genomic regions under selection for local adaptation in Chinese pigs (Ai etal. 2015).
Up to date, there is no systematic study (i.e., based on a large panel of Chinese breeds) aiming to detect genomic selection signals during domestication in the genomes of Chinese domestic pigs. Here, we used whole-genome resequencing data of 16 wild boars sampled across China and 54 Chinese indigenous pigs from 9 geographically distant breeds to screen signatures of selection during domestication. As a result, we highlighted a list of interesting genes such as TBX19, AHR, MSTN, P2RY1, MARCH1, OR11L1, and OR14A16 that may contribute to the adaptation to capture life and the response to artificial selection of human desired traits in Chinese domestic pigs. Our findings provide insights into the evolutionary history of Chinese pigs and uncover several promising candidate genes for economically important traits in pigs.
Materials and Methods
Samples
A total of 16 Chinese wild boars were sampled from 7 localities around China, and 54 domestic pigs were collected from 9 Chinese indigenous breeds: Bamaxiang (BMX), Erhualian (EHL), Hetao (HT), Jinhua (JH), Luchuan (LUC), Wuzhishan (WZS), Neijiang (NJ), Bamei (BM), and Baoshan (BS) (supplementary table S1, Supplementary Material online). These breeds are geographically distant and pertain to different ecotypes. Six individuals were selected from each breed. These individuals are genetically unrelated and have no common ancestors within three generations. Whole-genome resequencing data of BMX, EHL, HT, LUC, WZS pigs, and 11 wild boars have been reported in our previous study (Ai etal. 2015) and are publically available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/; last accessed September 12, 2017) under accession number SRA096093. The genome sequence data of the other five wild boars were also downloaded from the SRA (accession no. ERP001813). Genomic DNA was extracted from ear tissues of JH, NJ, BM, and BS pigs and was sequenced by a HiSeq 4000 platform (Illumina) according to the manufacturer’s standard protocols. In addition, the sequence data of 91 individuals from 16 breeds were downloaded from the NCBI SRA under accession numbers ERP001813 (Rubin etal. 2012), SRA096093 (Ai etal. 2015), SRA065461 (Li etal. 2013b), SRX473146—SRX473149, and SRX510749 (Li etal. 2014) to phase haplotypes at the TBX19 locus.
SNP Calling
Sequencing reads were aligned against the Sscrofa 10.2 reference genome (http://hgdownload.soe.ucsc.edu/goldenPath/susScr3/bigZips/; last accessed September 12, 2017) using Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009). Duplicated reads were removed using Picard (https://broadinstitute.github.io/picard/; last accessed September 12, 2017). The Genome Analyses Toolkit (GATK) (McKenna etal. 2010) was then used for base quality recalibration, local realignment around Indels (insertions/deletions), and SNP calling. By applying HaplotypeCaller in GATK, we generated a VCF file containing SNPs and short Indels. To select high-quality SNPs, we adopted SelectVariants in GATK and set the filtered standard as below:
QD > 2.0 && MQ > 40.0 && FS < 60.0 && ReadPosRankSum > −8.0 && MQRankSum > −12.5.
Phylogenetic Analyses
To better understand the genetic relationship of all individuals tested in this study, we calculated pairwise identity-by-state (IBS) distance matrix data between individuals using PLINK v.107 (Purcell etal. 2007). A neighbor-joining phylogenetic tree was constructed based on the IBS matrix using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/; last accessed September 12, 2017, Bandelt et al. 1999). Haplotypes were phased using Beagle (Browning and Browning 2007). A maximum-likelihood (ML) tree was constructed for representative haplotypes at the TBX19 locus using 1,000 times of bootstraps via MEGA (Hall 2013). Pairwise nucleotide differences per site between populations (dxy) at the TBX19 locus were calculated as previously reported (Ai etal. 2015).
Genome-Wide Scan for Selection Signals
Two approaches were explored to identify signatures of selection differentiating Chinese domestic pigs from Chinese wild boars. Genome-wide scans were conducted using a sliding window of 40 kb with a step size of 20 kb in these two approaches. Sliding windows with less than ten SNPs were discarded. First, we calculated the genetic differentiation (Fst) value for each window using VCFtools (http://vcftools.sourceforge.net/; last accessed September 12, 2017). Then, we calculated the expected heterozygosity within each window using in-house Python scripts. Next, R “scale” function (http://stat.ethz.ch/R-manual/R-patched/library/base/html/scale.html; last accessed September 12, 2017) was used to Z-transform Fst and heterozygosity scores. BEDTools (http://nchc.dl.sourceforge.net/project/bedtools; last accessed September 12, 2017) were finally used to find overlaps between selected regions defined by these two approaches, and the continuous windows were merged into large regions.
Gene Annotation, GO Analyses and Enrichment of Known QTLs in CDRs
Fifty annotated genes were identified within candidate domestication regions (CDRs) through the Ensembl Biomart (http://www.biomart.org; last accessed September 12, 2017). Given that the gene annotation in the pig genome is far from perfect, we then adopted the UCSC Genome Browser (http://genome.ucsc.edu; last accessed September 12, 2017) to identify 5 genes that are not annotated in the pig genome but have been annotated in mice and human genomes, including MAST3, TKTL1, HMCN1, ATXN1, and SLC40A1. The GO terms and functions of these candidate genes were assessed using Panther (http://www.pantherdb.org/; last accessed September 12, 2017). To test if the known QTLs (PigQTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/SS/index; last accessed September 12, 2017; Hu etal. 2016) in the 14 defined CDRs are significantly enriched for phenotypes related to pig domestication process, we randomly sampled 14 genomic regions of size equivalent to these CDRs and repeated sampling for 100 times, avoiding the regions with low SNP density of less than ten SNPs per 40 kb. We then estimated the probability by ranking the numbers of domestication-related QTL in the CDRs against those in the randomly sampled genomic regions.
Variants Annotation
All variants located in the CDRs were annotated using Variant Effect Predictor (VEP) (McLaren etal. 2016). The command for variants annotation is shown below:
Functional significance of amino acid substitutions was predicted using SIFT (Kumar etal. 2009).
Results and Discussion
Phylogenetic Relationships of All Sequenced Individuals
In our previous study, we resequenced the genomes of 18 pigs from Tibetan, Min, and Laiwu breeds (Ai etal. 2015). Tibetan pigs are raised in a cage-free grazing way and most likely have admixture with local wild boars. Min and Laiwu pigs have clear genomic signatures of admixture with European pigs that are genetically divergent from Chinese pigs (Yang etal. 2011; Ai etal. 2013). To avoid the interference of spurious signals, we did not include the genome data of these Tibetan, Min, and Laiwu pigs. We herein made the comparison of the genomes of 54 Chinese domestic pigs from 9 diverse breeds with those of 16 Chinese wild boars.
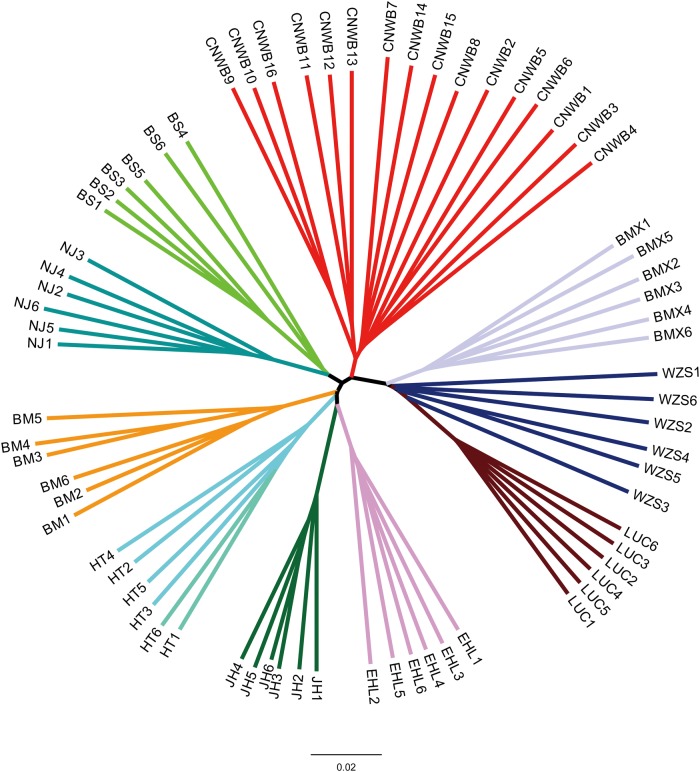
To determine the genetic relationship between 70 pigs tested in this study, we used 45,384,176 SNPs called from whole-genome sequencing data (16∼31× coverage) of these animals (supplementary table S1, Supplementary Material online) to construct a neighbor-joining phylogenetic tree. A clear divergence between breeds was observed in this tree, and different breeds and wild boars defined their own separate clades, suggesting that these breeds may have undergone different evolutionary scenario due to regional adaptation/selection or genetic drift after domestication (fig. 1), which is consistent with our previous report (Ai etal. 2015).
Fig. 1.
Neighbor joining tree of Chinese domestic pigs and wild boars based on pairwise identity-by-state (IBS) distance calculated from genome-wide SNPs. CNWB, Chinese wild boars; BMX, Bamaxiang; WZS, Wuzhishan; LUC, Luchuan; EHL, Erhualian; JH, Jinhua; HT, Hetao; BM, Bamei; NJ, Neijiang; BS, Baoshan.
Selective Sweep Analysis Reveals Domestication Signals
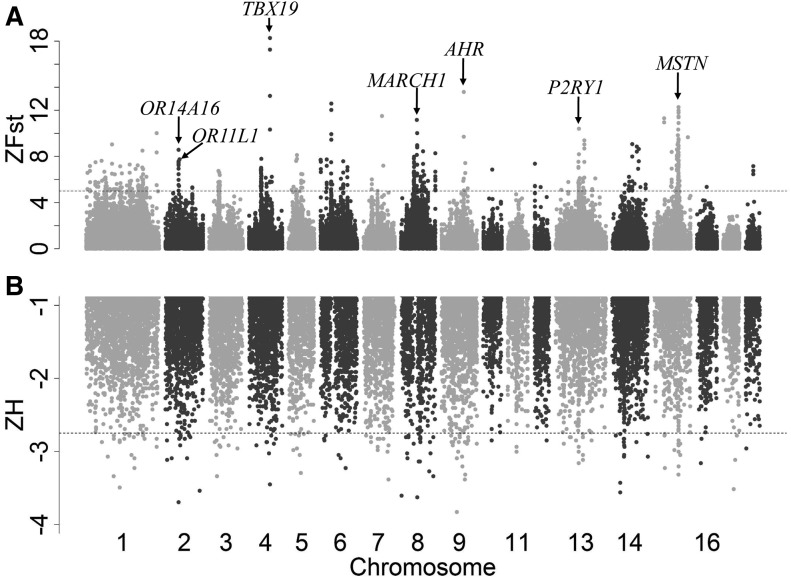
To identify domestic signals in the pig genome, we first calculated values of genetic differentiation (Fst) between domestic pigs and wild boars and values of the expected heterozygosity (H) within the two populations on autosomes. We then Z-transformed the autosomal heterozygosity (Z(H)) and Fst (Z(Fst)) values and defined the CDRs with Z(H) values of lower than −2.8 and Z(Fst) values of >5 following the similar discipline set by the previous report (Axelsson etal. 2013). By applying this threshold, we identified 14 CDRs (total size: ∼4 Mb; average size: 288 kb) containing 55 genes. Of the 55 genes, several stood out to be promising candidate genes for domestication (see below), including TBX19, AHR, MSTN, P2RY1, MARCH1, OR11L1, and OR14A16 (table 1), which are located on chromosomes 2, 2, 4, 8, 9, 13, 15, respectively (fig. 2). We noted that three genes (TBX19, SFT2D2, and ORMDL1) have been reported as candidate selected genes in Chinese native black pigs. These genes are involved in the regulation of early developmental processes and metabolic process (Fu etal. 2016). Moreover, an Asian-derived AHR haplotype is known to be introduced into European pigs during the Industrial Revolution and was preferentially selected during the development of modern European pig breeds (Bosse etal. 2014).
Table 1.
Seven Promising Candidate Genes with Strong Signatures of Selection and Their Associated Phenotypes and GO Terms
| Associated Phenotype | Gene | Z(Fst) | Z(H) | Chr | Associated GO Terms |
|---|---|---|---|---|---|
| Developmental processes | TBX19 | 17.97 | −3.52 | 4 | Anatomical structure morphogenesis (GO: 0009653); pituitary gland development (GO: 0021983); regulation of cell proliferation (GO: 0042127); cell fate commitment (GO: 0045165); regulation of cell differentiation (GO: 0045595) |
| Fertility | AHR | 13.58 | −2.98 | 9 | Xenobiotic metabolic process (GO: 0006805); apoptotic process (GO: 0006915); cell cycle (GO: 0007049); reproductive structure development (GO: 0048608); gland development (GO: 0048732); metabolic process (GO: 0008152) |
| Muscle growth | MSTN | 11.40 | −3.42 | 15 | Transforming growth factor beta receptor signaling pathway (GO: 0007179); muscle organ development (GO: 0007517); response to heat (GO: 0009408); skeletal muscle tissue regeneration (GO: 0043403); negative regulation of myoblast differentiation (GO: 0045662); negative regulation of insulin receptor signaling pathway (GO: 0046627); cell development (GO: 0048468); negative regulation of skeletal muscle satellite cell proliferation (GO: 1902723); negative regulation of satellite cell differentiation (GO: 1902725); negative regulation of myoblast proliferation (GO: 2000818); growth (GO: 0040007) |
| Nervous system | P2RY1 | 9.87 | −2.96 | 13 | Eating behavior (GO: 0042755)response to growth factor (GO: 0070848); cellular response to organic cyclic compound (GO: 0071407); protein localization to plasma membrane (GO: 0072659); relaxation of muscle (GO: 0090075); signal transduction (GO: 0007165); hemostasis (GO: 0007599) |
| Olfaction | OR14A16 | 8.12 | −3.88 | 2 | G-protein coupled receptor signaling pathway (GO: 0007186); detection of chemical stimulus involved in sensory perception of smell (GO: 0050911); signal transduction (GO: 0007165); sensory perception of smell (GO: 0007608) |
| Olfaction | OR11L1 | 7.69 | −2.99 | 2 | Detection of chemical stimulus involved in sensory perception of smell (GO: 0050911); sensory perception of smell (GO: 0007608); response to stimulus (GO: 0050896) |
| Immune system | MARCH1 | 10.24 | −3.48 | 8 | Antigen processing and presentation of peptide antigen via MHC class II (GO: 0002495); immune response (GO: 0006955); immune system process (GO: 0002376) |
Fig. 2.
Selective sweep analyses of the domestication loci in the genome of Chinese pigs. (A) Distribution of Z(Fst) values (positive end) along pig autosomes 1–18. A dashed horizontal line indicates the cut-off threshold (Z(Fst)>5). (B) Distribution of Z(H) values (negative end) along pig autosomes 1–18. A dashed horizontal line indicates the cut-off threshold (Z(H) < −2.8).
CDRs Associated with Quantitative Traits Loci in Pigs
To make a close examination on the identified CDRs, we searched for potential roles of these CDRs in the process of domestication using the pig QTL database (PigQTLdb) (Hu etal. 2016). In total, 994 (P < 0.05) QTLs overlap with the 14 CDRs, of which 159 are associated with body weight and growth (P < 0.05) and 66 with muscle growth (P = 0.08). Moreover, a proportion of the overlapping QTLs are related to fertility, food intake, eating behavior, coat color, and olfactory response, in accordance with the fact that distinct phenotypes had formed due to strong artificial selection during the pig domestication (Moon etal. 2015). Together, these findings indicate that these 14 candidate regions could have played a role in the formation of phenotypes during pig domestication.
Gene Ontology Analyses of Candidate Genes
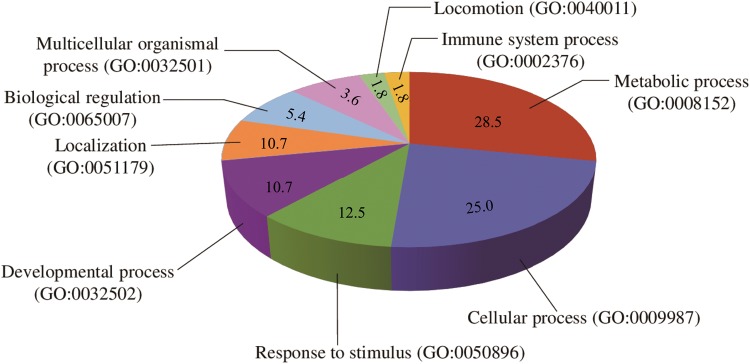
We adopted Panther (Mi etal. 2013) to understand gene ontology and function of the 55 genes within the CDRs in more detail. These genes were involved in multiple function terms, such as developmental system (GO: 0021983; TBX19, ASS1, ATXN1, GPR161, SLC40A1, ORMDL1), fertility (GO: 0048608; AHR, FNDC3A), nervous system (GO: 0009612; P2RY1, KCNMA1, PRDM12, SLC5A5, STMN3), muscle growth (GO: 0048632; MSTN, MBNL1), immune response (GO: 0006955; MARCH1, IL12RB1, IFI30, PIK3R2), olfactory response (GO: 0050911; OR11L1, OR14A16, OR6F1), and stimulus response (GO: 0050896; HMCN1, HMCN2, NUP35) (supplementary table S2, Supplementary Material online). These genes were also participated in multiple biological processes, such as “metabolic process,” “cellular process,” “response to stimulus,” “developmental process,” and “immune system process” (fig. 3 and supplementary table S3, Supplementary Material online), indicating that these processes could be the targets of selection during the domestication of Chinese pigs. Intriguingly, seven genes including TBX19, AHR, MSTN, P2RY1, MARCH1, OR11L1, and OR14A16 (table 1) exhibited the highest Z(Fst) value and overlapped with the peaks of selective signals. These genes were explored for further analyses.
Fig. 3.
The enriched biological processes for the 55 candidate genes.
Seven Strong Candidate Genes Involved in the Domestication of Chinese Pigs
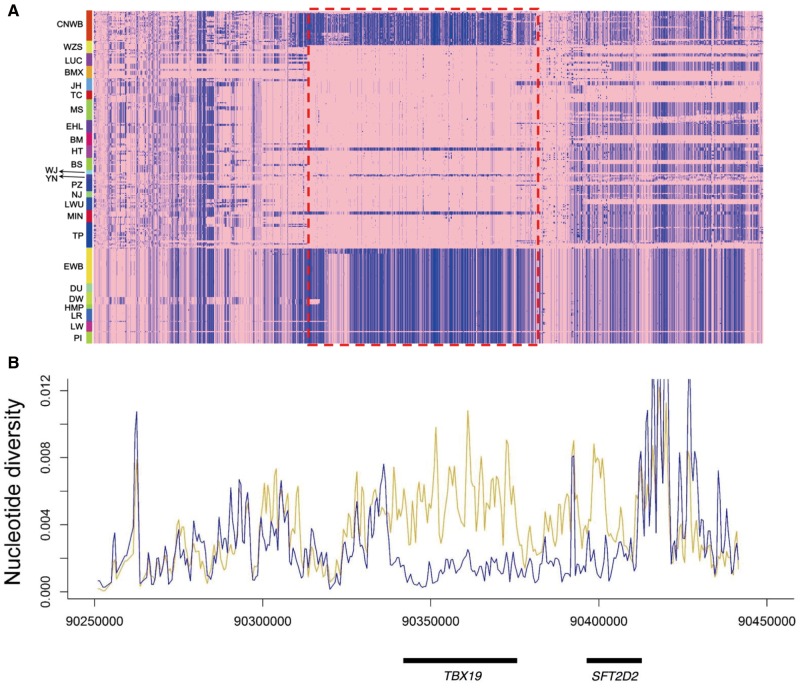
The TBX19 (a member of the T-box gene family and a transcription factor) gene showed the most striking selection signal in this study, which was also reported in Chinese black pigs (Fu etal. 2016). TBX19 is a key regulator to activate the differentiation of the pituitary cell lineage expressing pro-opiomelanocortin (POMC) genes, which is in charge of producing adrenocorticotropic hormone (ACTH) and plays an important role in the hypothalamic–pituitary–adrenal (HPA) axis (Liu etal. 2001). Mutations in TBX19 have been implicated in patients with isolated deficiency of pituitary POMC-derived ACTH (Liu etal. 2001; Lefebvre etal. 2016). In the HPA axis, ACTH simulates glucocorticoid synthesis in the adrenal cortex to regulate metabolic functions. ACTH-deficient patients are characterized by low plasma ACTH and low or absent cortisol production and are associated with hypoglycaemia, low blood pressure, severe obesity, weight loss, and lack of appetite (Metherell etal. 2004; Lefebvre etal. 2016). This again suggests that development and metabolic process was likely under strong positive selection in Chinese domestic pigs, which was probably required by the adaptation to rice or grain rich food in domestic condition. We note that TBX19 also regulates behavior through modulating the dosage of adrenocorticoids and androgens produced by adrenal cortex in human and other animals. Wasserman etal. (2007) reported that genetic variants in the TBX19 gene were associated with the angry/hostility personality trait (and potentially suicidal behavior) in human. Compared with aggressive wild boars, Chinese domestic pigs behave very tamely and timidly. Given that TBX19 plays a role in regulating behavior traits, we argue that the selective sweep around TBX19 were likely related to the timidity trait in Chinese domestic pigs. Timidity was a common feature of Chinese indigenous pigs. To validate this domestic sweep, we then constructed haplotypes at the TBX19 locus using a worldwide panel of 168 pigs from 17 Chinese breeds and 7 European breeds, 18 Asian wild boars, and 17 European wild boars. The result clearly showed that nearly all individuals from Chinese breeds shared a 70-kb haplotype, which was distinct from those of European pigs and all wild boars (fig. 4A), indicating that the locus had undergone strong selection during and after domestication of Chinese wild boars. Statistics of nucleotide diversity (fig. 4B) further proved that the TBX19 region were nearly devoid of genetic variety in Chinese domestic pigs in comparison with Chinese wild boars enriched for SNPs, indicative of a strong selective sweep. Altogether, the top signature of selection centered at the TBX19 gene likely underlies the altered metabolism and timidity traits in Chinese domestic pigs.
Fig. 4.
The selective sweep region around the TBX19 gene. (A) Heatmap of haplotype sharing in pairwise comparisons between worldwide panels of pig populations. The major allele in Chinese domestic pigs is indicated in pink and the minor allele in blue. The 70-kb haplotype shared by Chinese domestic pigs is indicated by a red dashed rectangle. (B) Distribution of nucleotide diversity in a window size of 1000 bp on chromosome 4 within Chinese wild boars (dark yellow) and Chinese domestic pigs (blue). CNWB, Chinese wild boars; TC, Tongcheng; MS, Meishan; WJ, Wujin; YN, Yunnan; PZ, Penzhou; LWU, Laiwu; MIN, Min; TP, Tibetan pigs; PI, Pietrain; LW, Large White; LR, Landrace; HMP, Hampshire; DU, Duroc; DW, Duroc White; EWB, Europe wild boars. The abbreviations of other populations are same as in figure 1.
The second strongest signal was observed around the AHR gene (aryl hydrocarbon receptor), which has a critical function in reproduction by regulating the expression of ovarian P450 aromatase (Cyp19) gene (Cochez etal. 2016) and also serves as a key factor in ovary development by affecting the regulation of hypothalamus–pituitary–gonadal (HPG) axis (Baba etal. 2005). Several reports support that this gene is related to female reproduction (Hernández-Ochoa etal. 2009; Jablonska etal. 2011; Onteru etal. 2012). Given that most of Chinese breeds are known for desirable reproduction performance (supplementary table S1, Supplementary Material online), the selection on the AHR gene has most likely contributed to improved fertility traits in Chinese domestic pigs as compared with wild boars. A recent study also showed that Chinese AHR haplotypes had been introgressed to Europe breeds to improve their fertility (Bosse etal. 2014).
The MSTN gene (growth/differentiation factor 8) is well-known for controlling muscle hypertrophy in a range of mammalian species, most notably in cattle, dogs, mice, sheep, and humans (Grobet etal. 1997; Meng etal. 2008; Gong etal. 2009; Boman etal. 2010; Stefaniuk etal. 2014; Tang etal. 2016). The strong selection signal at the MSTN locus indicates that muscle growth was one of the target traits during Chinese pig domestication, and MSTN functional mutations may potentially underlie the physiognomic difference between Chinese wild boars and Chinese domestic pigs.
The P2RY1 gene (P2 Purinoceptor Subtype Y1) acts as the dominant receptor for neural-mediated smooth muscle relaxation in the gastrointestinal tract and is involved in eating behavior (GO: 0042755) and response to growth factor (GO: 0070848). Purinergic responses are abolished in P2RY1-deficient mice, suggesting the importance of P2RY1 in purinergic neurotransmission in regulating colonic motility (Sanders etal. 2012). We hence deduce that P2RY1 may be implicated in altering eating habits in Chinese domestic pigs by regulating neuronal system, which is in agreement with the previous report that neuronal development had often been targeted during domestication (Carneiro etal. 2014).
Immunity-related genes are known to be actively evolved in domestication process. Here we observed a clear selection signal around the MARCH1 gene (E3 ubiquitin-protein ligase). This gene has been repeatedly associated with immunity in mice and human (Li etal. 2013a). Thibodeau etal. (2008) showed that the immunosuppressive effect of IL-10 on antigen presentation is mainly mediated through the induced expression of MARCH1. IL-10 is an anti-inflammatory cytokine interfering with MHC class II (MHC-II) in presenting pathogen-derived peptides to CD4+ T helper cells. The interaction between MHC-II and MARCH1 showed by coimmunoprecipitation assays also indicated that the expression of MARCH1 is strongly induced by IL-10 in monocytes. Thus, we assume that the sweeping signal at the MARCH1 locus in Chinese domestic pigs might be involved in immune traits.
Two olfactory receptor genes (OR11L1 and OR14A16) are likely under selection during domestication in Chinese indigenous pigs. Wild boars need a sharp sense of smell to find food and water, look for females in breeding seasons and define their home range. In contrast, domestic pigs are raised in a well-managed in-door condition with a stable supply of feed, leading to a general less sensitive sense of smell than wild boars. However, it has been reported that adding garlic to feed as sweetener can improve feed intake and pork yield (Horton etal. 1991). We thus assume that selection on specific olfactory receptors could enable domestic pigs to have a certain food preference for producing human-desired meat under captivity. This may explain the signatures of selection at these two olfactory receptor genes.
Altogether, the process of domestication, accompanied by selection on genes related to development and growth (TBX19 and MSTN), nervous system or behavior (TBX19 and P2RY1), fertility (AHR), immune response (MARCH1), and olfactory response (OR11L1, OR14A16) during captive breeding, could have dramatically effects on the gene pools of Chinese domestic pigs. However, further investigations are required to better understand the molecular mechanisms of how these candidate genes contribute to these selected traits.
Nonsynonymous Variants in the Sweep Genes
To decipher possible mutations underlying selected traits during domestication, we further annotated all variants located in the CDRs. In total, 618 variants in coding sequences were found, of which 271 were nonsynonymous, including 6 stop-gain, 21 frame-shift, and 239 missense variants. All stop-gain mutations occurred at low frequency (< 0.10) in few pig breeds. This suggests that gene loss had not contributed to rapid evolution in domestic pigs, similar to the finding in European pigs (Rubin etal. 2012). The nonsynonymous variants were further filtered by demanding a nearly fixation of derived alleles (allele frequency > 0.80) in domestic pigs but at low frequency (< 0.25) in wild boars. Intriguingly, ten such nonsynonymous mutations were observed in the TBX19 gene (table 2). Four out of the ten nonsynonymous mutations were within the evolutionary conserved T-box domain (fig. 5) (48th to 216th amino acid) (Liu etal. 2001), and three of those mutations causing amino acid substitutions (N105T, M158V, and T189A) were predicted as function-altering variants by SIFT (Vaser etal. 2016). In human, deleterious mutations within the T-box domain cause isolated ACTH deficiency, a rare and life-threatening disorder characterized by weight loss, lack of appetite, muscle weakness, nausea and vomiting, and hypotension (Pulichino etal. 2003a, 2003b; Lefebvre etal. 2016). The symptoms of ACTH deficiency seem to be inversely related to the preferably selected phenotypes in domestic pigs. Hence, it raises a possibility that these three candidate selected functional variants (N105T, M158V, and T189A) are gain-of-function mutations, contributing to improved metabolism and tame behaviors in Chinese domestic pigs.
Table 2.
Nonsynonymous Substitutions Having a Significant Imbalance of Allele Distribution in Chinese Domestic Pigs and Wild Boars
| Chr | Position (bp) | ID | Ancestral Allele | Derived Allele | Variant | Gene | AFa in Domestic Pigs | AFa in Wild Boars | SIFT Predictionb |
|---|---|---|---|---|---|---|---|---|---|
| 4 | 90338649 | rs80957510 | A | G | Missense | TBX19 | 0.96 | 0.07 | Tolerated_low_confidence (0.06) |
| 4 | 90338654 | rs80905569 | C | G | Missense | TBX19 | 0.96 | 0.07 | Deleterious_low_confidence (0) |
| 4 | 90338694 | rs80825297 | C | T | Missense | TBX19 | 0.97 | 0.07 | Tolerated (0.1) |
| 4 | 90338781 | rs344983154 | A | G | Missense | TBX19 | 0.96 | 0.06 | Tolerated (0.58) |
| 4 | 90338853 | rs80866535 | C | T | Missense | TBX19 | 0.97 | 0.15 | Tolerated (0.34) |
| 4 | 90354828 | rs341417992 | T | C | Missense and splice_region | TBX19 | 0.96 | 0.06 | Tolerated (0.13) |
| 4 | 90358483 | rs80951600 | T | C | Missense | TBX19 | 0.96 | 0.03 | Deleterious (0.01) |
| 4 | 90358576 | rs343615016 | T | C | Missense | TBX19 | 0.96 | 0.06 | Deleterious (0.02) |
| 4 | 90360139 | rs346121845 | T | G | Missense | TBX19 | 0.96 | 0.06 | Deleterious (0) |
| 4 | 90371704 | rs320504853 | C | T | Missense | TBX19 | 0.96 | 0.06 | Tolerated (1) |
| 9 | 95536780 | rs345994568 | C | A | Missense | AHR | 0.95 | 0.16 | Tolerated (1) |
| 9 | 95537674 | rs339939442 | T | G | Missense | AHR | 0.95 | 0.16 | Tolerated_low_confidence (0.31) |
| 9 | 95537932 | rs324951230 | C | T | Missense | AHR | 0.95 | 0.16 | Tolerated_low_confidence (1) |
| 9 | 95540357 | rs339238482 | C | A | Missense | AHR | 0.95 | 0.17 | Tolerated_low_confidence (1) |
| 8 | 55660954 | rs339707100 | C | A | Missense | MARCH1 | 0.90 | 0.23 | Tolerated (0.46) |
The alternative allele frequency in wild boars and domestic pigs.
The deleterious variants are better interpreted as function-altering here.
Fig. 5.
Multispecies alignment of the T-box region of TBX19 protein. Dots indicate identity to the master sequence and dashes indicate missing data. The amino acid changes N105T, M158V, T189A, and I202V correspond to rs346121845, rs343615016, rs80951600, and rs341417992, respectively.
There are four missense mutations in AHR and one missense mutation in MARCH1 (table 2) that also showed a significant imbalance of allele distribution in Chinese domestic pigs and wild boars. However, these missense variants are all tolerated alterations predicted by SIFT and are located in nonconserved regions (supplementary figs. S1–S3, Supplementary Material online). This suggests that regulatory rather than protein-alternating variants may underlie these selection signals. However, we cannot rule out the possibility that large structure variants may underlie these loci.
It has been suggested that most phenotypic differences in mammals largely result from regulatory variations. As an evidence, our and previous studies found a considerable proportion of regions under selection that lack obvious coding variants or are in gene desert. To understand to what extent regulatory elements such as enhancers and promoters influence the process of domestication in pigs, we will need to generate comparable ENCODE (Gerstein etal. 2012; Thurman etal. 2012) -like data sets to systematically identify regulatory elements. The international consortium of Functional Analyses of Animal Genome (http://www.animalgenome.org/community/FAANG/; last accessed September 12, 2017) has been established for generating such data sets for farm animals. We anticipate that the release of the ENCODE-like data in the animal field will speed up pinpointing the genetic basis of domestication, adaptation, and complex traits.
Possible Interspecies Introgression at the TBX19 Locus
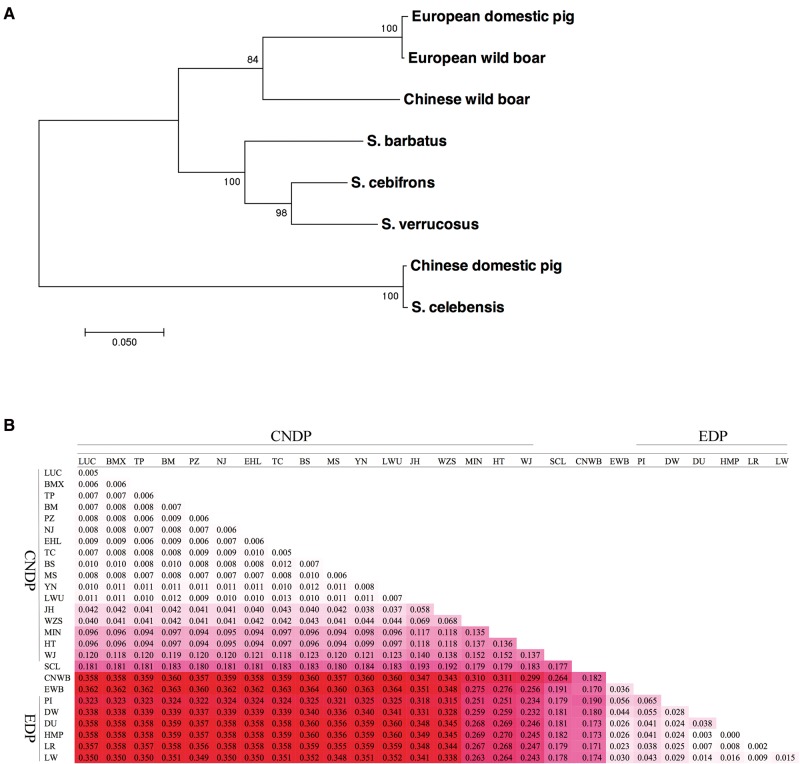
The unusual haplotype pattern and the excess of highly differentiated protein-altering mutations at the TBX19 locus in Chinese domestic pigs led us to assume that this distinct haplotype could be originated from another species of the genus Sus. To test this assumption, we first constructed a ML tree for representative haplotypes of 70 kb at this locus in pigs and other four Sus species. Surprisingly, the Chinese domestic haplotype was closely related to that of S. celebensis and showed a clear divergence as compared with those of Euroasian wild boars and three Island Southeast Asia Sus species (S. barbatus, S. cebifrons, and S. verrucosus) (fig. 6A). This phylogenetic relationship was further confirmed by an IBS distance based phylogenetic tree and a haplotype network using the SNP data of 161 individuals (supplementary figs. S4 and S5, Supplementary Material online). We also calculated pairwise nucleotide distances (dxy) per site between populations at the TBX19 locus, confirming that Chinese domestic pigs are genetically close to S. celebensis in comparison with Chinese wild boars, European wild boars, and domestic pigs (fig. 6B). A reasonable explanation for this observation is that the TXB19 haplotype of Chinese domestic pigs was the product of gene-flow from S. celebensis or S. celebensis-like species. It has been reported that S. celebensis was kept captive in Southeast Asia, where pigs from European and Asia mainland were introduced in the past few hundred years (Groves 1984; Larson etal. 2007; Frantz etal. 2013). Therefore, it also raised a possibility that the ancestor of Chinese domestic pigs (Sus scrofa) received this TBX19 haplotype from an extinct Sus species, and then was introgressed to S. celebensis by recent admixture events. However, the nucleotide distance between S. celebensis and any one of Chinese domestic pig breeds is obviously larger than those among Chinese domestic pigs (fig. 6B). Thus, we favor the hypothesis that the TXB19 haplotype of Chinese domestic pigs was originated from introgression of DNA from S. celebensis-like species. In addition, given that the 70-kb haplotype was shared by almost all Chinese domestic breeds (fig. 4A) and the TBX19 locus showed strong evidence of reduced nucleotide diversity in Chinese domestic pigs (fig. 4B), we suspect that the introgression events occurred at the very early domestication stage or even before domestication and had undergone strong selection after introgression. It should be mentioned that several Chinese domestic pigs had European TBX19 haplotypes (figs. 4A and 6B). These pigs are from Hetao, Wujin, and Min breeds that show a clear signal of admixture with European pigs at the genome level (Ai etal. 2013; Ai etal. 2015). Hence, we speculate that these haplotypes were most likely introgressed from European pigs by recent admixture events. Taken together, the TBX19 haplotype shared by Chinese domestic pigs was likely originated from an interspecies introgression event and was maintained by strong selections after introgression. We have previously reported such interspecies introgression on swine chromosome X that contributed to local adaptation to high-latitude environments (Ai etal. 2015). However, the introgression on chromosome X affected pigs from Northern China and domestic pigs and wild boars from Europe, which is clearly different from the introgression pattern found in this study. The present study provides another clear-cut example for the interspecies introgression that is being evidenced to play a role in the evolution of mammals and humans.
Fig. 6.
The haplotypes at the TBX19 locus in Chinese domestic pigs was possibly introgressed from an extinct Sus species. (A) The maximum-likelihood (ML) tree for representative haplotypes at the TBX19 locus in pigs and four other Sus species. This tree was built for eight haplotypes of 70 kb each one representing one population using 1,000 times of bootstraps. Scale bars represent the number of nucleotide substitutions per SNP site. (B) Average number of nucleotide differences per site between populations. dxy values are highlighted with different density colors, where darker colors correspond to larger values. SCL, S. celebensis; CNDP, Chinese domestic pigs; EDP, European domestic pigs. Other abbreviations of European and Chinese domestic pig breeds and wild boars are same as in the legend of figures 1 and 4.
Conclusions
In this study, we identified selective sweeps associated with domestication of pigs in China and highlighted several important genes and biological processes that may contribute to the pig domestication. Fifty-five genes in the CDRs are enriched in metabolic process, development, fertility, behavior, and immune system process, and adaptation to different environment and the desire of human. For the first time, we show that the TBX19 gene, the strongest selection locus, is most likely associated with altered metabolism and timidity of Chinese domestic pigs and the “domestic” haplotype was possibly introgressed from another Sus species. We also highlight the AHR gene under strong selection for female fertility. These results advance our understanding of the evolutionary history of the pig, and provide a valuable resource for genetic analyses, particularly for identifying functionally important genes contributing to the phenotypic diversity of Chinese pigs.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author’s Contributions
W.L., J.R., and L.H. designed this study; Y.Z. and W.L. performed data analysis; B.Y., Z.Z., and H.A. contributed to data processing; Y.Z., W.L., and J.R. wrote the manuscript, L.H. revised the article. All authors have read and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank sample collectors for their contributions. This work was supported by the National Natural Science Foundation of China [grant numbers 31402058, 31525023].
Literature Cited
- Ai H, et al. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 47(3):217–225. [DOI] [PubMed] [Google Scholar]
- Ai H, Huang L, Ren J.. 2013. Genetic diversity, linkage disequilibrium and selection signatures in chinese and Western pigs revealed by genome-wide SNP markers. PLoS One 8(2):e56001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AJ, et al. 2011. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One 6(4):e14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, et al. 2013. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495(7441): 360–364. [DOI] [PubMed] [Google Scholar]
- Baba T, et al. 2005. Intrinsic function of the aryl hydrocarbon (dioxin) receptor as a key factor in female reproduction. Mol Cell Biol. 25(22):10040–10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Boman IA, Klemetsdal G, Nafstad O, Blichfeldt T, Våge DI.. 2010. Impact of two myostatin (MSTN) mutations on weight gain and lamb carcass classification in Norwegian White Sheep (Ovis aries). Genet Sel Evol. 42:4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M, et al. 2014. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat Commun. 5:4392.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81(5):1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, et al. 2014. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345(6200):1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochez PM, et al. 2016. AHR modulates the IL-22-producing cell proliferation/recruitment in imiquimod-induced psoriasis mouse model. Eur J Immunol. 46(6):1449–1459. [DOI] [PubMed] [Google Scholar]
- Fontanesi L, Russo V.. 2013. Molecular genetics of coat colour in pigs. Acta Agric Slovenica 4:16. [Google Scholar]
- Frantz LA, et al. 2013. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol. 14(9):R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, et al. 2016. Genomic analysis reveals selection in Chinese native black pig. Sci Rep 6: 36354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489(7414):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra E, et al. 2000. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154(4):1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong YF, et al. 2009. SNP detection and haplotype analysis in partial sequence of MSTN gene in sheep. Genetika 45(12):1646–1649. [PubMed] [Google Scholar]
- Grobet L, et al. 1997. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 17(1):71–74. [DOI] [PubMed] [Google Scholar]
- Groves CP. 1984. Of mice and men and pigs in the Indo-Australian Archipelago. Canberra Anthropol. 7(1–2):1. [Google Scholar]
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30(5):1229–1235. [DOI] [PubMed] [Google Scholar]
- Hernández-Ochoa I, Karman BN, Flaws JA.. 2009. The role of the aryl hydrocarbon receptor in the female reproductive system. Biochem Pharmacol. 77(4):547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton G, Blethen D, Prasad B.. 1991. The effect of garlic (Allium sativum) on feed palatability of horses and feed consumption, selected performance and blood parameters in sheep and swine. Can J Anim Sci. 71(2):607–610. [Google Scholar]
- Hu Z-L, Park CA, Reecy JM.. 2016. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 44(D1):D827–D833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska O, et al. 2011. The expression of the aryl hydrocarbon receptor in reproductive and neuroendocrine tissues during the estrous cycle in the pig. Anim Reprod Sci. 126(3–4):221–228. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC.. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 4(7):1073–1081. [DOI] [PubMed] [Google Scholar]
- Larson G, et al. 2007. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc Natl Acad Sci U S A. 104(12):4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, et al. 2010. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci U S A. 107(17):7686–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre H, Thomas M, Duparc C, Bertherat J, Louiset E.. 2016. Role of ACTH in the interactive/paracrine regulation of adrenal steroid secretion in physiological and pathophysiological conditions. Front Endocrinol (Lausanne) 7:98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Lu JY, Liu Q, Wang HW, Guo H.. 2013a. Altered MARCH1 ubiquination-regulated dendritic cell immune functions during the early stage of zymosan-induced multiple organ dysfunction syndrome (MODS) in mice. Immunol Lett. 150:105–115. [DOI] [PubMed] [Google Scholar]
- Li M, et al. 2013b. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 45:1431–1438. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. 2014. Genome-wide scans to detect positive selection in Large White and Tongcheng pigs. Anim Genet. 45(3):329–339. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. 2001. TBX19, a tissue-selective regulator of POMC gene expression. Proc Natl Acad Sci U S A. 98(15):8674–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, et al. 2016. The Ensembl variant effect predictor. Genome Biol. 17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XR, et al. 2008. Variation of MSTN gene UTR in eleven sheep breeds. Yi Chuan 30:1585–1590. [DOI] [PubMed] [Google Scholar]
- Metherell L, et al. 2004. TPIT mutations are associated with early-onset, but not late-onset isolated ACTH deficiency. Eur J Endocrinol. 151(4):463–465. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD.. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41(Database issue):D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, et al. 2015. A genome-wide scan for signatures of directional selection in domesticated pigs. BMC Genomics 16:130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onteru SK, et al. 2012. A whole-genome association study for pig reproductive traits. Anim Genet. 43(1):18–26. [DOI] [PubMed] [Google Scholar]
- Pulichino AM, et al. 2003a. Human and mouse TPIT gene mutations cause early onset pituitary ACTH deficiency. Genes Dev. 17:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulichino AM, et al. 2003b. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 17(6):738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CJ, et al. 2012. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci U S A. 109(48):19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders KM, Koh SD, Ro S, Ward SM.. 2012. Regulation of gastrointestinal motility-insights from smooth muscle biology. Nat Rev Gastroenterol Hepatol. 9(11):633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefaniuk M, Kaczor U, Kulisa M.. 2014. MSTN gene polymorphism in livestock animals. Postepy Hig Med Dosw (Online) 68:633–639. [DOI] [PubMed] [Google Scholar]
- Tang L, et al. 2016. Combination of weight-bearing training and anti-MSTN polyclonal antibody improve bone quality in rats. Int J Sport Nutr Exerc Metab. 26(6):516–524. [DOI] [PubMed] [Google Scholar]
- Thibodeau J, et al. 2008. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur J Immunol. 38(5):1225–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489(7414):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC.. 2016. SIFT missense predictions for genomes. Nat Protoc. 11(1):1–9. [DOI] [PubMed] [Google Scholar]
- Wang C, et al. 2015. Genome-wide analysis reveals artificial selection on coat colour and reproductive traits in Chinese domestic pigs. Mol Ecol Resour. 15(2):414–424. [DOI] [PubMed] [Google Scholar]
- Wasserman D, Geijer T, Sokolowski M, Rozanov V, Wasserman J.. 2007. Genetic variation in the hypothalamic–pituitary–adrenocortical axis regulatory factor, T-box 19, and the angry/hostility personality trait. Genes Brain Behav. 6(4):321–328. [DOI] [PubMed] [Google Scholar]
- Yang S, et al. 2011. The local origin of the Tibetan pig and additional insights into the origin of Asian pigs. PLoS One 6(12):e28215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.