Summary
This is the first study determining the three-dimensional structure of Phl p 5a which reveals a novel mechanism for high allergenic activity based on flexibly connected IgE-reactive domains which cross-link effector cell-bound IgE more efficiently than isolated rigid globular proteins. These findings may also form a basis for specific immunotherapy strategies.
Keywords: Grass pollen, allergy, allergen, protein structure, NMR spectroscopy
To the Editor
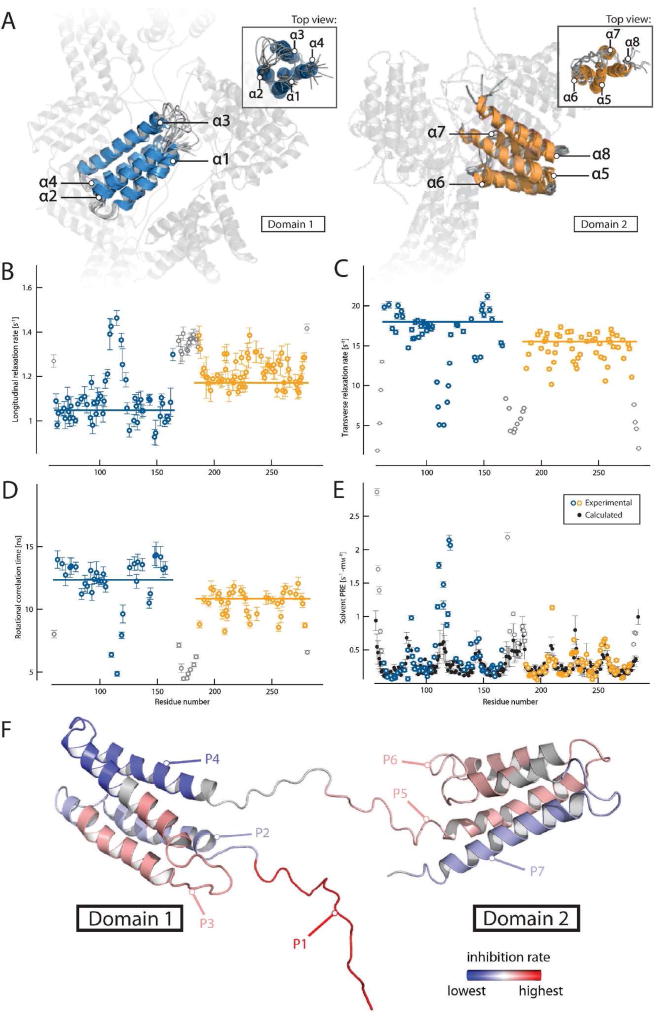
Group 5 allergens are cross-reactive allergens found in most grass species. When released from grass pollen especially after rainfalls in the form of respirable submicronic particles they cause severe asthma attacks in sensitized patients. Group 5 allergens are abundant 30 kD monomeric proteins which are localized in the amyloplasts of pollen grains and are supposed to play a role in pollen germination. One of the most striking features of group 5 allergens is that they potently trigger mast cell and basophil degranulation even at extremely low concentrations.1, 2 Attempts to obtain the full three-dimensional structure of group 5 allergens by X-ray crystallography were so far not successful and only partial structures or models were obtained.3–5 We succeeded to solve the three-dimensional structure of Phl p 5a by solution NMR spectroscopy. It indicates that Phl p 5a and the members of the family of highly cross-reactive group 5 grass pollen allergens (for sequence alignment see Fig E2) display a modular assembly (i.e., consist of domains which are similar to each other) consisting of two flexibly-connected domains of similar size which contain identical secondary structural elements, i.e., four parallel alpha helices (see Fig 1, and Fig E3 Table E1, Methods and Results section in this article’s Online Repository at www.jacionline.org). The structure of Phl p 5a (30–260) consists of two domains that are flexibly connected by a central linker region. Inspection of the NOESY-type spectra did not indicate any inter-domain contacts. Both domains display a 4-helix-bundle-like fold showing the same up-down-up-down topology (see Fig E3, A). Each helix bundle is stabilized by a hydrophobic core. The helices in domain 1 are found to be oriented almost in parallel, and the additional insert (ranging from residue 85 – 95) forms a flexible and unstructured loop. In domain 2, helix α5 is bent towards its hydrophobic core. This forces helix α6 and helix α7 apart from each other, leading to an elongated loop connecting these two helices, in contrast to the tight loops observed for the other helix-helix connections.
FIG 1.
Structure of Phl p 5a and domain mobility. (A) Overlay of the 10 lowest energy models of both structured regions (left: domain 1 in blue, right: domain 2 in orange) showing the unstructured domain-linking loop and counter-domain in transparent form. Labeling of the helices is performed in a sequential manner starting at the N-terminal domain. Backbone dynamics of Phl p 5a are shown as 15N longitudinal relaxation rates (B), transverse relaxation rates (C), overall correlation times (D) and solvent PRE (paramagnetic relaxation enhancement) rates (E), experimental values of structured domains in color, flexible termini and linker in grey, calculated values in black, outliers may arise from high solvent exposure and increased hydrogen exchange with water). Values are plotted as a function of residue number and trimmed mean values of structured regions are indicated by horizontal bars. (F) Position of peptides within the Phl p 5a structure. The regions of the peptides are labeled in different colors using a linear gradient ranging from red, displaying the highest average inhibition rate of IgE binding, to blue, with the lowest inhibition. Gray regions were not part of the study. The flexible N-terminus containing P1 was modeled into the structured part of the molecule.
The modular assembly of two flexibly-connected domains is a novel structural feature among allergens which usually assume a compact structure containing several, often spatially-oriented binding sites for patients’ IgE antibodies. Earlier epitope mapping data indicated that group 5 allergens contain several IgE binding sites in their N- as well as C-terminal portion.2, 6 Furthermore, it has been shown that a recombinant fragment of Phl p 5a which resembles the N-terminal domain reacted strongly with allergic patients IgE and was extremely potent in inducing cross-linking of basophil bound IgE antibodies.2 In order to investigate if both Phl p 5a domains demonstrate IgE reactivity, we expressed two Phl p 5 fragments, NTD and CTD, representing the isolated N-terminal and C-terminal allergen domain, respectively (see Fig E3, E4, in this article’s Online Repository at www.jacionline.org). Purity, correct mass and fold comparable to Phl p 5a were demonstrated for NTD and CTD by SDS-PAGE, mass spectrometry and circular dichroism (see Fig E1, Methods and Results section in this article’s Online Repository at www.jacionline.org). Direct IgE binding assays as well as antibody probes raised against peptides from each domain in IgE inhibition experiments showed that each of the isolated domains is important for the binding of allergic patients’ IgE antibodies (see Fig E4, E5, Table E2, Methods and Results section in this article’s Online Repository at www.jacionline.org). Equivalent IgE binding capacity of the Phl p 5a and the equimolar mix of the isolated domains NTD and CTD was demonstrated for sera from patients tested in basophil activation experiments (Fig 2A, Table E4, Fig E8, Methods and Results section in this article’s Online Repository at www.jacionline.org). Sequence and structural comparison of the two Phl p 5a domains showed that they contain similar sequences as well as structural elements pointing to the possibility that they not only contain independent IgE epitopes but possibly also harbor repetitive IgE epitopes (see Fig E3, in this article’s Online Repository at www.jacionline.org) which was confirmed by antibody binding and IgE inhibition assays (see Fig E4, E5, Table E2, Methods and Results section in this article’s Online Repository at www.jacionline.org). In fact, the presence of repetitive antibody binding sites has so far only been observed recently for high molecular weight glutenins, a family of wheat food allergens containing identical repetitive sequence elements. Importantly, we show that the Phl p 5a allergen in which the two domains are connected by the flexible linker is more potent in cross-linking IgE on basophils and in inducing basophil degranulation as compared to an equimolar mix of the isolated domains (see Fig 2A, Fig E8 and Methods and Results section in this article’s Online Repository at www.jacionline.org). The results obtained in the basophil experiments may need confirmation by in vivo testing in patients but this goes beyond the scope of this study.
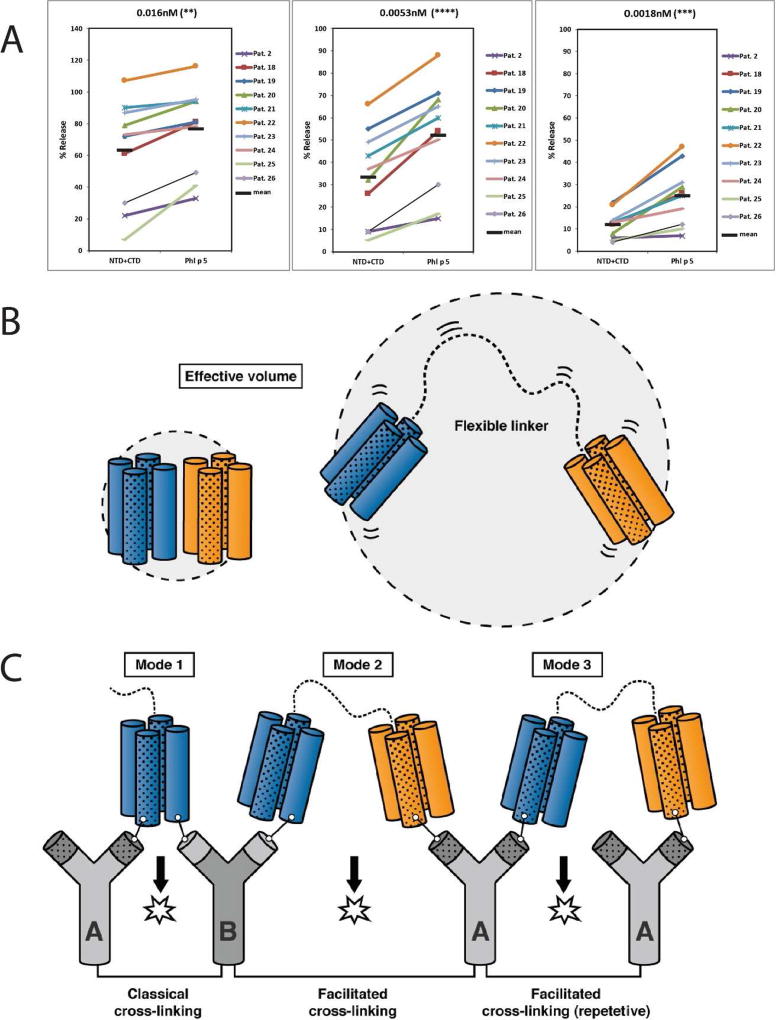
FIG 2.
(A) Basophil degranulation experiments. RBL cells expressing the human FcεRI were sensitized with sera from ten grass pollen allergic patients (#2, 18–26) and then incubated with different concentrations of Phl p 5 or with an equimolar mix of NTD+CTD (0.016nM, 0.0053nM, 0.0018nM, from left to right). The release of β-hexosaminidase is shown as percentage of the total β–hexosaminidase contents of the cells. The horizontal black bars denote the mean values. Asterisks indicate p-values of 0.0022 (**), < 0.0001 (****) and 0.0007 (***), respectively. (B) Facilitated cross-linking of IgE by Phl p 5a through presence on different domains and repetitive epitopes. In Phl p 5a the two domains (domain 1 blue, domain 2 orange) are connected by an unstructured linker region (right) and are able to sample a much larger volume than a rigid single domain molecule (left). The presence of repetitive IgE epitopes is indicated by the fillings (blank and spotted). (C) The unstructured linking region between the domains of Phl p 5a facilitates binding to a second IgE antibody on the surface of mast cells and effector cells and thus may induce more rapid initial cross-linking of FcεRI and degranulation (IgE antibody in gray). Additional IgE molecules with specificity for either a different epitope on the same domain or for a repetitive epitope on the other domain can be engaged leading to enhanced cross-linking and degranulation.
To illustrate the dynamic properties of group 5 proteins, Fig 2B provides a schematic diagram comparing a rigid molecule (left) with two flexibly-connected domains (right). Due to the flexibility of the two IgE epitope-containing domains, the Phl p 5a allergen should be more potent than other allergens in initiating the cross-linking of effector cell bound IgE antibodies. Upon binding of one domain, the other domain is still mobile and able to sample a much larger volume than for instance an 8-helix-bundle, or a rigid dimer of a four-helix-bundle, making it more efficient in binding to a second IgE antibody. The elongated conformation of the two folded domains, including the flexible linker, is able to expand up to 120 Å (see Fig E6, Methods and Results section in this article’s Online Repository at www.jacionline.org) which allows for efficient interaction to other IgE molecules on the cell surface. Another important structural feature for respiratory allergens that could be identified for Phl p 5a is the observation of recurring sequential and structural elements. Group 5 proteins can thus be considered as highly repetitive molecules that possess similar sequences not only on both domains but also within the same four-helix-bundle.
According to our results, different possibilities for effector cell degranulation by Phl p 5a can therefore be envisaged (see Fig 2C). First, Phl p 5a may crosslink IgE antibodies by IgE epitopes present on one or both of the domains which would correspond to the classical mode of cross-linking known so far (mode 1). However, there are at least two additional novel possibilities of cross-linking. Cross-linking may be achieved by different (mode 2) or repetitive IgE epitopes (mode 3) on each of the domains. This process is facilitated by the flexible domains and their high rotational and translational freedom to recruit additional IgE antibodies into the emerging complexes. In agreement with this assumption, we know that the Phl p 5a allergen indeed accounts for a high percentage of grass pollen-specific IgE antibodies.1 Furthermore, we know from experimental models with artificial allergen constructs that the intensity of effector cell degranulation increases with the number of engaged IgE molecules.7
More importantly, the structural analysis of Phl p 5a not only provides an explanation why Phl p 5a is such a potent allergen, it also explains why it has been difficult to produce recombinant Phl p 5a variants with reduced allergenic activity to be safely administered as vaccines into patients. It is known that allergen-specific immunotherapy especially with grass pollen allergens can induce severe side effects due to the allergenic activity of allergens in the vaccines. Therefore, it is a long-sought goal to engineer recombinant and synthetic vaccines with reduced allergenic activity. Attempts to construct hypoallergenic Phl p 5a variants have so far proven difficult, because it becomes obvious from the structural analysis of the protein that larger fragments comprising each of the domains or point mutations will not be sufficient for reducing the allergenic activity. In fact, each of the isolated domains showed strong IgE reactivity. According to the structural data, one possibility for constructing a safe and effective vaccine for Phl p 5a is to incorporate several non-allergenic small peptide elements from both domains in a carrier-bound vaccine.8, 9
In summary, our study reveals a novel mechanism responsible for the high allergenic activity of an allergen and provides the structural basis for the engineering of safe candidate vaccines to treat patients sensitized to group 5 allergens.
Supplementary Material
Acknowledgments
C.G. and E. S. thank the Austrian Academy of Sciences for a DOC and DOCfFORTE fellowship, respectively. Financial support to K.Z. by the Austrian Science Foundation (FWF) (DK Molecular Enzymology W901) is gratefully acknowledged. This work was supported by the Intramural Research Program of National Heart, Lung, and Blood Institute of the NIH to N.T. and by grants F4605 and F4607 of the Austrian Science Fund (FWF) to R.V. and S.F., respectively. We thank Motoshi Suzuki and Kang Chen for technical assistance, Vanessa Morris for carefully reading the manuscript and Verena Hameter for help with protein purification.
COMPETING FINANCIAL INTERESTS
R.V. has received research grants from BIOMAY AG, Vienna, Austria and Thermofisher, Uppsala, Sweden and serves as a consultant for these companies.
Abbreviations used
- IgE
Immunoglobulin E
- FcεRI
high affinity IgE receptor
- kD
kilodalton
- SAXS
small-angle X-ray scattering
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- RDC
residual dipolar couplings
- NOESY
nuclear Overhauser effect spectroscopy
- Å
Angstrom
- RMSD
root-mean-square deviation
- ns
nanosecond
- PRE
paramagnetic relaxation enhancement
- Rg
radius of gyration
- Dmax
maximum particle size
- CTD
C-terminal domain
- NTD
N-terminal domain
- P1–7
peptide 1–7
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes. Protein Data Bank: Atomic coordinates of Phl p 5a (30–260) have been deposited under accession code 2M64. BioMagResBank: Chemical shift data has been deposited with entry number 19107.
AUTHORS CONTRIBUTIONS
C. G., E. S.,C.M., M.F-T., Y.D. and S.K. produced proteins, M.F-T. and E. S. performed cloning, C.M., N.N., S.F. and M.F-T. performed immunological experiments, C.G., E. S., and S.K. performed NMR assignment and C. G. and N.T. performed structure calculation. T.M. performed SAXS measurements and modeling. C. G., N. T., S.F., R.V., and K. Z. designed experiments and wrote the manuscript. All authors analyzed data, discussed the results and provided input for the manuscript. The RS-ATL8 cell line was kindly provided by K. Nakamura, Division of Novel Foods and Immunochemistry, National Institute of Health Science, Setagaya-ku, Tokyo, Japan.
References
- 1.Vrtala S, Sperr WR, Reimitzer I, van Ree R, Laffer S, Müller WD, et al. cDNA cloning of a major allergen from timothy grass (Phleum pratense) pollen; characterization of the recombinant Phl pV allergen. J Immunol. 1993;151:4773–4781. [PubMed] [Google Scholar]
- 2.Flicker S, Vrtala S, Steinberger P, Vangelista L, Bufe A, Petersen A, et al. A human monoclonal IgE antibody defines a highly allergenic fragment of the major timothy grass pollen allergen, Phl p 5: molecular, immunological, and structural characterization of the epitope-containing domain. J Immunol. 2000;165:3849–3859. doi: 10.4049/jimmunol.165.7.3849. [DOI] [PubMed] [Google Scholar]
- 3.Bufe A, Betzel C, Schramm G, Petersen A, Becker WM, Schlaak M, et al. Crystallization and preliminary diffraction data of a major pollen allergen. J Biol Chem. 1996;271:27193–27196. doi: 10.1074/jbc.271.44.27193. [DOI] [PubMed] [Google Scholar]
- 4.Maglio O, Saldanaha JW, Vrtala S, Spitzauer S, Valenta R, Pastore A. A major IgE epitope-containing grass pollen allergen domain from Phl p 5a folds as a four-helix bundle. Protein Eng. 2002;15:635–642. doi: 10.1093/protein/15.8.635. [DOI] [PubMed] [Google Scholar]
- 5.Rajashankar K, Bufe A, Weber W, Eschenburg S, Lindner B, Betzel C. Structure of the functional domain of the major grass-pollen allergen Phlp 5b. Acta Crystallogr D Biol Crystallogr. 2002;58:1175–1181. doi: 10.1107/s0907444902007254. [DOI] [PubMed] [Google Scholar]
- 6.Focke-Tejkl M, Campana R, Reininger R, Lupinek C, Blatt K, Valent P, et al. Dissection of the IgE and T-cell recognition of the major group 5 grass pollen allergen Phl p 5. J Allergy Clin Immunol. 2014;133:836–845. doi: 10.1016/j.jaci.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gieras A, Linhart B, Roux KH, Dutta M, Khodoun M, Zafred D, et al. IgE epitope proximity determines immune complex shape and effector cell activation capacity. J Allergy Clin Immunol. 2016;137:1557–65. doi: 10.1016/j.jaci.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Focke-Tejkl M, Weber M, Niespodziana K, Neubauer A, Huber H, Henning R, et al. Development and characterization of a recombinant, hypoallergenic, peptide-based vaccine for grass pollen allergy. J Allergy Clin Immunol. 2015;135:1207–17. doi: 10.1016/j.jaci.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zieglmayer P, Focke-Tejkl M, Schmutz R, Lemell P, Zieglmayer R, Weber M, et al. Mechanisms, safety and efficacy of a B cell epitope-based vaccine for immunotherapy of grass pollen allergy. EBioMedicine. 2016;11:43–57. doi: 10.1016/j.ebiom.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.