To the Editor
APDS (activated PI3Kδ syndrome (1)) or PASLI (PI3Kδ-activating mutations causing senescent T cells, lymphadenopathy, and immunodeficiency (2)) disease is a relatively prevalent primary immunodeficiency disorder (PID) characterized by recurrent sinopulmonary infections with associated lung damage, susceptibility to Epstein-Barr virus (EBV) and cytomegalovirus, and lymphoproliferative disease. It is caused by heterozygous, gain-of-function mutation in the PIK3CD (1, 2) or PIK3R1 (3, 4) genes encoding the p110δ catalytic or p85α regulatory subunit of the phosphoinositide 3-kinase complex PI3Kδ. Augmented PI3Kδ signaling causes terminal differentiation and senescence of T cells, increased transitional B cells, and immunoglobulin derangements (5, 6).
The leukocyte-restricted p110δ subunit consists of an adaptor-binding domain (ABD) that binds p85, a Ras-binding domain (RBD), a C2 domain, a helical domain, and a lipid kinase domain. The regulatory p85 subunit makes inhibitory contacts with the C2, helical, and kinase domains of p110δ, and it is these three domains that are affected by previously described APDS1 mutations. Specifically, heterozygous PIK3CD mutations causing amino acid changes N334K and C416R in the C2 domain, E525K and E525A in the helical domain, and E1021K in the kinase domain have been reported (5, 6). Additionally, the p110 ABD makes a putative intramolecular inhibitory contact with the kinase domain (7). Indeed, mutations in the related PIK3CA gene affecting the p110α ABD or the ABD-RBD linker abolish this inhibitory contact and cause hyperactivation (8).
We identified two families with three individuals having clinical features of APDS but no previously reported APDS mutations (5). The proband, patient A.II.1, was suspected of having humoral defects and lymphoproliferative disease at six months of age and suffered from severe susceptibility to pneumonia (at least 19 episodes) and airway disease throughout childhood. He experienced recurrent otitis media and eczema and was treated for Clostridium difficile colitis and vaccination-induced varicella infection. He also had lymphadenopathy and splenomegaly, as well as poor responses to polysaccharide, tetanus, and mumps vaccinations. In addition, he suffered from mild thrombocytopenia, was splenectomized, and at 11 years of age succumbed to EBV lymphoproliferative disease (Tables 1, E1). His mother, Patient A.I.1, is a 41-year-old female who presented with severe pneumonia at six years of age, has a history of lymphadenopathy and EBV lymphadenitis, and has had recurrent sinopulmonary infections with bronchiectasis and left lung resection. She has a reduced CD4:CD8 T cell ratio, low naïve CD4+ T cells, and a preponderance of senescent effector CD8 T cells (Tables 1, E1 and Figure E1). Patient B.1 in a second, unrelated family is a 13-year old male who presented within the first year of life with an abscess, severe diaper rash, recurrent otitis media, and eczema. At 18 months of age, he had pneumonia, and at four years of age, he began having bloody stools associated with lesions suspicious for lymphoma. Upon bowel resection, pathological examination revealed marginal zone hyperplasia. Later episodes of lymphadenopathy prompted additional biopsies that confirmed EBV lymphadenitis. His growth has been poor since the age of four years, and his measured bone age is more than two standard deviations below his chronologic age. Clinical immune studies on patient B.1 revealed hypergammaglobulinemia, lymphocytopenias, and elevated transitional B cells (Tables 1, E1). NK cell numbers were low or normal in these patients (Table E1), and the CD4 T cell lymphopenia and hyper-IgM are both consistent with findings in other cohorts of APDS patients (5).
Table 1. Patient characteristics.
F: female; M: male; N.D.: Not determined. Numerical data indicate ranges of patient values listed above age-matched reference ranges in parentheses. *CD62L also included for this stain
| p110δ Adaptor-Binding Domain/Linker | |||
|---|---|---|---|
| A.I.1 | A.II.1 | B.1 | |
| Amino acid substitution | G124D | G124D | E81K |
| Age, sex | 41, F | 11, M (deceased) | 13, M |
| EBV | EBV lymphadenitis | EBV lymphoproliferative disease | EBV lymphadenitis |
| Sinopulmonary bacterial infections | ✔ | ✔ | ✔ |
| Lymph node findings | N.D. | N.D. | Marginal zone hyperplasia |
| Lymphadenopathy | ✔ | ✔ | ✔ |
| CD4:CD8 ratio | 0.53–0.7 ↓ (1.11–5.17) | 0.2–0.3 ↓ (0.7–2.7) | 1.41–1.77 (0.7–2.4) |
| CD4+ T cells | 236–349/μL ↓
(359–1565/μL) 27.9–35.3% ↓ (31.9–62.2%) |
319–564/μL
(300–2000/μL) 12–20% ↓ (27–53%) |
296–403/μL ↓
(538–1569/μL) 27.7–33.8% (23–50%) |
| CD8+ T cells | 367–655/μL
(178–853/μL) 48.9–52.4% ↑ (11.2–34.8%) |
1726/μL
(300–1800/μL) 65% ↑ (19–34%) |
172–250/μL ↓
(371–436/μL) 16–21% (15–35%) |
| Naïve CD4+ T cells (CD45RA+) | *16–24/μL
↓
(102–1041/μL) *1.9–2.3% ↓ (7.6–37.7%) |
N.D. | 82–100/μL ↓
(134–969/μL) 7–8.1% (3–33%) |
| Naïve CD8+ T cells (CD62L+CD45RA+) | 89–110/μL
(85–568/μL) 8.8–11.9% (5.7–19.7%) |
N.D. | N.D. |
| Effector memory CD8+ T cells (CD62L-CD45RA−) | 92–274/μL ↑
(24–175/μL) 12.3–21.9% ↑ (1.1–9.2%) |
N.D. | N.D. |
| Senescent CD8+ T cells (CD57+) | 192–320/μL
(0–397/μL) 21.6–25.6% ↑ (0–16.2%) |
N.D. | N.D. |
| CD19+ B cells | 17–42/μL ↓
(59–329/μL) 1.9–5.6% ↓ (3–19%) |
27–141/μL ↓
(200–1600/μL) 1–5% ↓ (10–31%) |
387–601/μL
(204–703/μL) 35–46.9% ↑ (11–25%) |
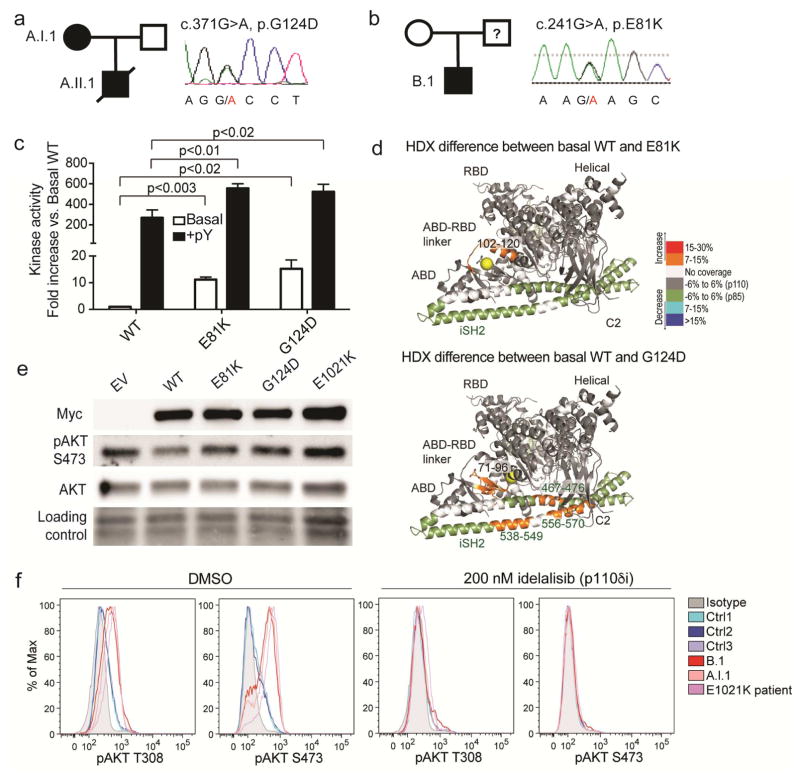
Whole-exome sequencing revealed a heterozygous mutation resulting in a G124D amino acid substitution in p110δ in both patients A.I.1 and A.II.1 but not the healthy father (Figure 1a). In patient B.1 (but not his healthy mother), a heterozygous mutation resulting in an E81K amino acid substitution was identified (Figure 1b). The father of patient B.1 was not available for analysis. Both G124D and E81K are more N-terminal than previously reported APDS1 mutations (Figure E2a). The conserved G124 residue of p110δ lies between two helices in the ABD-RBD linker, and the presence of a glycine or proline in this position in related p110 proteins maintains proper ABD orientation (Figure E2b–c). E81 of p110δ lies in the ABD and forms a salt bridge with K111 in the ABD-RBD linker, which is also predicted to help orient the ABD (Figure E2b–c) (8). Moreover, there is evidence in cancers and overgrowth syndromes that mutations at the equivalent E81 and P124 residues in p110α are activating (Table E2).
Figure 1. G124D and E81K dominantly activate p110δ.
(a–b) Pedigrees and PIK3CD Sanger chromatograms. (c) Lipid kinase activity with (+pY) or without (Basal) phosphopeptide. (d) HDX-MS differences greater than 0.7 Da and 7% compared to WT PI3Kδ. (e) Immunoblot of indicated proteins in healthy T cells overexpressing Myc-tagged forms of p110δ. (f) Phospho-AKT (T308 or S473) in indicated T cells without (left) or with (right) idelalisib.
In vitro kinase assays revealed an approximately 10-fold and 20-fold increase in basal activity of E81K and G124D, respectively, while phospho-tyrosine-induced activity of both mutants was increased by 2-fold compared to WT (Figure 1c). To probe changes in protein conformation, hydrogen deuterium exchange mass spectrometry (HDX-MS) was used to measure the exchange rate of amide hydrogens with solvent for WT, E81K, and G124D PI3Kδ complexes (Figure 1d). Compared to WT p110δ, E81K and G124D displayed increased exchange at the interface of the ABD and kinase domain. The G124D mutation also disrupted the inhibitory contact between p110δ C2 domain and p85α (Figure 1d, E3, E4).
To confirm dominant PI3K activation, we overexpressed G124D and E81K p110δ protein in healthy T cells and found increased phospho-AKT (Figure 1e, E5a–b). Furthermore, we directly observed markedly increased levels of phospho-AKT in T cells from patients A.I.1, B.1, and a patient with the E1021K mutation compared to healthy subjects (Figure 1f left). At least two clinical trials of p110δ-specific inhibitors for APDS1 have been announced (NCT02435173 and NCT02593539). We tested idelalisib (9), a p110δ inhibitor that is FDA-approved for chronic lymphocytic leukemia, in cultured T cells from patients A.I.1 and B.1 and found robust inhibition of hyperactive signaling (Figures 1f right, E5c–e). Consistent with milder structural changes in E81K, the extent of AKT and especially S6 hyperphosphorylation was lesser in patient B.1 compared to patient A.I.1 (Figure E5c–e). Supporting an intramolecular activation mechanism, we found no difference in association of ectopically expressed WT, E81K, or G124D p110δ with endogenous p85α in healthy T cells (Figure E5f). Thus, the novel E81K and G124D variants of p110δ are hyperactive and can be targeted by p110δ inhibitors.
More broadly, our findings highlight the utility of biochemical information about protein changes in paralogs (e.g., p110α and p110δ) regardless of whether or not the disease phenotypes (e.g., cancer and PID) overlap. The list of APDS1 mutation sites is likely to expand and, based on frequency and impact in p110α, we predict additional ABD changes, including at R88 and R38, may be discovered (Table E2). Importantly, our findings emphasize that the entirety of the PIK3CD coding sequence should be sequenced in suspected APDS patients.
Supplementary Material
Acknowledgments
We thank the patients and their families as well as referring clinicians and their teams. We also thank Nisha Momin, RN, BSN from Texas Children’s Hospital and Monica Konstantino, RN, BSN from the Yale Pediatric Genomics Discovery Program, for their clinical support and assistance, and Tom Fleisher, MD, Sergio Rosenzweig, MD, and Julie Niemela, MS for support through the Department of Laboratory Medicine at the Clinical Center, NIH. CLL is supported by Yale University and NHBLI R00HL125668 award. JEB is supported by a new investigator grant from CIHR and a discovery research grant from the Natural Sciences and Engineering Research Council of Canada (NSERC-2014-05218). This work was supported in part by the Intramural Research Program of the NIAID, NIH.
Abbreviations
- APDS
Activated PI3Kd Syndrome
- PASLI
PI3Kd activation with senescence, lymphadenopathy, and immunodeficiency
- PID
primary immunodeficiency disorder
- PI3K
phosphoinositide 3-kinase
- ABD
adaptor-binding domain
- RBD
Ras-binding domain
- AKT
protein kinase B
Footnotes
Disclosures
CLL collaborates with Novartis on related studies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Angulo I, Vadas O, Garcon F, Banham-Hall E, Plagnol V, Leahy TR, et al. Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342(6160):866–71. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucas CL, Kuehn HS, Zhao F, Niemela JE, Deenick EK, Palendira U, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deau MC, Heurtier L, Frange P, Suarez F, Bole-Feysot C, Nitschke P, et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J Clin Invest. 2014;124(9):3923–8. doi: 10.1172/JCI75746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucas CL, Zhang Y, Venida A, Wang Y, Hughes J, McElwee J, et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J Exp Med. 2014;211(13):2537–47. doi: 10.1084/jem.20141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulter TI, Chandra A, Bacon CM, Babar J, Curtis J, Screaton N, et al. Clinical spectrum and features of activated phosphoinositide 3-kinase delta syndrome: A large patient cohort study. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas CL, Chandra A, Nejentsev S, Condliffe AM, Okkenhaug K. PI3Kdelta and primary immunodeficiencies. Nat Rev Immunol. 2016 doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke JE, Vadas O, Berndt A, Finegan T, Perisic O, Williams RL. Dynamics of the phosphoinositide 3-kinase p110delta interaction with p85alpha and membranes reveals aspects of regulation distinct from p110alpha. Structure. 2011;19(8):1127–37. doi: 10.1016/j.str.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA) Proc Natl Acad Sci U S A. 2012;109(38):15259–64. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman SE, Gordon AL, Wagner AJ, Heerema NA, Zhao W, Flynn JM, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078–88. doi: 10.1182/blood-2010-02-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.