Abstract
Over the past few years, exosomes and their RNA cargo have been extensively studied because of the fascinating biological roles they play in cell-to-cell communication, including the signal exchange among cancer, stromal, and immune cells, leading to modifications of tumor microenvironment. RNAs, especially miRNAs, stored within exosomes, seem to be among the main determinants of such signaling: their sorting into exosomes appears to be cell-specific and related to cellular physiopathology. Accordingly, the identification of exosomal miRNAs in body fluids from pathological patients has become one of the most promising activity in the field of biomarker discovery. Several analyses on the qualitative and quantitative distribution of RNAs between cells and their secreted exosomes have given rise to questions on whether and how accurately exosomal RNAs would represent the transcriptomic snapshot of the physiological and pathological status of secreting cells. Although the exact molecular mechanisms of sorting remain quite elusive, many papers have reported an evident asymmetric quantitative distribution of RNAs between source cells and their exosomes. This phenomenon could depend both on passive and active sorting mechanisms related to: (a) RNA turnover; (b) maintaining the cytoplasmic miRNA:target equilibrium; (c) removal of RNAs not critical or even detrimental for normal or diseased cells. These observations represent very critical issues in the exploitation of exosomal miRNAs as cancer biomarkers. In this review, we will discuss how much the exosomal and corresponding donor cell transcriptomes match each other, to better understand the actual reliability of exosomal RNA molecules as pathological biomarkers reflecting a diseased status of the cells.
Keywords: exosomes, RNAs, RNA sorting, biomarkers, cancer, asymmetric molecular distribution
Introduction
The complex functional coordination among different cell types, tissues and organs in Metazoans is made possible thanks to cell-to-cell communication (Gerdes and Pepperkok, 2013). Cells are able to communicate by soluble factors (e.g., hormones, cytokines), adhesion molecules mediating cell-to-cell interactions, and specialized cellular structures (e.g., cytonemes, nanotubules), which connect neighboring cells and enable the transfer of surface components and cytoplasmic molecules (Majka et al., 2001; Rustom et al., 2004; Sherer and Mothes, 2008). Cell communication is critical also in specific pathological processes, including tumorigenesis. Indeed, cancer cells need to cross-talk to each other, normal cells, and immune system to survive, proliferate, and metastasize. Communication among tumor and stromal cells leads to modifications of tumor microenvironment favoring tumor growth, survival, immune-escape, and invasion (Brucher and Jamall, 2014). Moreover, long-distance communication with stroma located at distant non-cancerous sites facilitates the formation of metastatic niches and promotes metastatic processes (Ungefroren et al., 2011). Although the molecular mechanisms promoting cell-to-cell communication have not been fully understood yet, recent studies have shown that cells may also communicate by secreting and exchanging small membranous particles named extracellular vesicles (EV). Initially, EVs were considered to be cellular debris released by cells, following cell damage or dynamic plasma membrane turnover (Siekevitz, 1972). Inconsistent with this initial hypothesis, in 1977 it has been shown that EVs can also be the result of a specific and active cellular process and that they may carry functional membrane enzymes (De Broe et al., 1977). Cells can secrete different types of EVs that are classified according to their subcellular origin (Colombo et al., 2014). EVs may come from the cell plasma membrane as shedding vesicles by direct budding of plasmatic membrane. Such EVs display a wide range of diameter sizes (100–1,000 nm) and are generally known as microvesicles; depending on their cellular origin, EVs also have been named ectosomes (from polymorphonuclear leukocytes), microparticles (from platelets), or argosomes (secreted during the morphogenesis of multicellular organisms from basolateral epithelial membranes; George et al., 1982; Hess et al., 1999; Greco et al., 2001). Another source of EVs is represented by apoptotic bodies, which originate from apoptotic cells and range from 1 to 5 μm in diameter (Hristov et al., 2004). Otherwise, EVs may arise from multivesicular bodies from the endosomal compartment (EC): after its fusion with the plasma membrane, EC releases intraluminal vesicles into the extracellular space as exosomes (Thery et al., 2002). Exosomes are vesicles spanning from 30 to 100 nm in diameter, which are enriched in endosomal molecules as tetraspanin proteins CD9, CD63, CD81, and CD82 (Lotvall et al., 2014). Therefore, EVs circulating in body fluids are an extremely heterogeneous population, which differ in cellular origin, size, and surface molecules, and are secreted by nearly all cell types in both physiological and pathological conditions. Over the past 10 years, researchers have mainly focused on unveiling the functional role and pathological involvement of EVs (above all, exosomes), especially after the discovery by Valadi et al. that exosomes carry fully functional mRNAs and miRNAs able to modify the physiological state of recipient cells (Valadi et al., 2007). The RNA contents of exosomes is heterogeneous and may depend on different biomolecular factors, including the specific cellular status. Some miRNAs are highly expressed in exosomes, while others are barely present in them; moreover, some miRNAs are enriched only in exosomes secreted by distinct cell types (Pigati et al., 2010; Nolte-'T Hoen et al., 2012; Villarroya-Beltri et al., 2013). All these clues suggest that RNA molecules are not randomly loaded into exosomes, but that rather a machinery actively regulating the sorting of specific sets of RNAs into exosomes does exist. Similar to other molecular apparati, this molecular machinery may be functionally perturbed by drugs, diseases and infections (Purrello et al., 1998; Di Pietro et al., 2009).
Exosomes carry distinctive sets of mRNAs, rRNAs, miRNAs, other small non-coding RNAs (ncRNAs; e.g., piRNAs, snRNAs, snoRNAs, scaRNAs, Y-RNAs), and long non-coding RNAs (Li M. et al., 2014; Van Balkom et al., 2015). The exosomal cargo is critical to determine the outcome of cell communication (Mittelbrunn et al., 2011; Hergenreider et al., 2012; Halkein et al., 2013). As reported above, cancer-derived exosomes are able to modulate the immune response against the tumor, and promote angiogenesis, invasion, and metastasis (Milane et al., 2015; Silva and Melo, 2015). Much evidence has suggested that miRNA signatures of tumor-derived exosomes may be used as potential circulating biomarkers for the diagnosis of several types of cancers (Munson and Shukla, 2015; Soung et al., 2017). Moreover, circulating exosomal miRNAs have also been reported as candidate biomarkers for pregnancy disorders, liver damage and inflammation, cardiovascular diseases, and neurodegeneration (Cosme et al., 2013; Masyuk et al., 2013; Tsochandaridis et al., 2015; Soria et al., 2017). Intriguingly, some disease-related exosomal miRNAs mirror pathway dysfunctions of their source cells (Haug et al., 2015; Reclusa et al., 2016; Zhong et al., 2016). The discovery of exosomal miRNAs circulating in body fluids and their potential exploitation as non-invasive disease biomarkers have given rise to the following question (Ma R. et al., 2012; Wang J. et al., 2016): how exactly exosomal RNAs represent an accurate transcriptomic snapshot of the physiological and pathological status of their source cells? To date, many studies have dealt with such question directly or indirectly, but results from them are conflicting and not conclusive. In this review, we will summarize the most critical issues on the sorting of RNA molecules into exosomes and we will discuss how much the exosomal and corresponding donor cell transcriptomes match each other, in order to better understand the actual reliability of exosomal RNA molecules as pathological biomarkers reflecting the diseased status of cells.
Mechanisms of molecular sorting into exosomes
Different mechanisms for sorting molecules into exosomes have been described, although the precise molecular signaling controlling them are unsatisfactorily known (Villarroya-Beltri et al., 2014). Endosomal Sorting Complexes Required for Transport (ESCRT) controls the sorting of ubiquitinated proteins into Intraluminal Vesicles (ILVs) through a molecular cascade involving several ESCRT sub-complexes (Henne et al., 2011). Specifically, ESCRT-0 binds ubiquitinated proteins and is associated to the endosomal compartment thanks to its interaction with phosphatidylinositol 3-phosphate (PI3P). ESCRT-0 recruits ESCRT-I, which in turn recruits ESCRT-II proteins, which lastly activate the ESCRT-III machinery. Snf7 protein (an ESCRT-III component) forms oligomeric assemblies inducing vesicle budding and recruits the adaptor protein ALG-2-Interacting Protein X (Alix) to stabilize the ESCRT-III complex (Henne et al., 2011). ESCRT-independent mechanisms of sorting into exosomes have been also reported (Stuffers et al., 2009). Proteolipid-positive exosomes are enriched in cholesterol and ceramide and their secretion is closely related to the production of ceramide by neutral sphingomyelinase 2 (nSMase2; Trajkovic et al., 2008). Indeed, nSMase2 controls the secretion of Aβ-peptide-exosomes in neurons, whereas the ceramide induces a curvature of the endosomal membranes and the coalescence of microdomains, leading to the budding of intraluminal vesicles (Yuyama et al., 2012). Another process independent from the ESCRT machinery could be regulated by tetraspanins, integral membrane proteins that are highly abundant on the exosome surface. Tetraspanins are able to form intra-membrane tetraspanin-enriched domains by interacting with other membrane proteins and lipids (Escola et al., 1998; Yanez-Mo et al., 2009): for instance, CD81 structurally organizes the membranes in microdomains, while CD63 regulates the loading of LMP1 protein into exosomes and PMEL into intraluminal vesicles during melanogenesis (Van Niel et al., 2011; Verweij et al., 2011; Perez-Hernandez et al., 2013). The specific mechanisms of RNA sorting into exosomes are still poorly characterized and represent a matter of debate. The sorting of RNA molecules within mammalian cells appears to be independent of ESCRT and dependent on ceramide (Kosaka et al., 2010). It has been proposed that RNA loading into exosomes occurs before the budding process, when RNA molecules bind to raft-like regions of multivesicular body membranes creating intraluminal vesicles through inward budding (Janas and Janas, 2011; Janas et al., 2012). RNA binding to membranes is determined by hydrophobic modifications, lipid structures, and sphingosine at physiological concentration in rafted membranes (Janas et al., 2015). It has also been reported that specific nucleotide sequences show enhanced affinity to phospholipid bilayers (Khvorova et al., 1999; Vlassov et al., 2001; Janas and Yarus, 2003; Janas et al., 2004). Bolukbasi et al. suggested that the loading of mRNAs into exosomes could be mediated by a specific zipcode-like sequence, present within the 3′UTR of mRNAs that are enriched in exosomes, and by the presence of binding sites for miRNAs that are highly expressed in source cells (Bolukbasi et al., 2012). Computational analysis of over-represented motifs in the sequence of miRNAs that are enriched in exosomes, along with mutagenesis experiments, led to the identification of specific nucleotide motifs (named EXOmotifs) that may regulate the loading of miRNAs into exosomes. EXOmotifs are recognized and bound by the heterogeneous ribonucleoprotein A2B1 (hnRNPA2B1). Specifically, the sumoylated form of hnRNPA2B1 performs miRNA sorting into exosomes (Villarroya-Beltri et al., 2013). Previously, hnRNPA2B1had been reported to be involved in the intracellular transport of specific mRNAs in neurons and HIV genomic RNAs to packaging sites (Munro et al., 1999; Levesque et al., 2006). Intriguingly, Signal Recognition Particle RNAs (SRP-RNAs), also present within exosomes, are bound by Signal Recognition Particle (SRP) proteins through a GGAG tetraloop, which shares the same sequence of one of the two EXOmotifs previously identified (Wild et al., 2001). Recently, a MALDI-TOF/TOF mass spectrometry analysis of proteins specifically binding to exosome-enriched miRNAs in an in vitro hepatocyte model revealed a new molecular player in the sorting of miRNAs into exosomes: synaptotagmin-binding cytoplasmic RNA-interacting protein (SYNCRIP; Santangelo et al., 2016). Knockdown of SYNCRIP impaired the exosomal loading of specific exosome-enriched miRNAs, whereas RNA immunoprecipitation showed that SYNCRIP binds directly to some EXO-miRNAs thanks to a short common sequence (the hEXO motif) that is shared by 60% of exosome-enriched miRNAs (Santangelo et al., 2016). Similarly to hnRNPA2B1, SYNCRIP is also sumoylated, but the hEXO motif which it binds to is different from the EXOmotifs bound by hnRNPA2B1. This suggests that the exosome sorting machinery might involve different ribonucleoproteins, each controlling the exosomal loading of a specific set of miRNAs. Bang et al. found that about one-quarter of miRNAs found in fibroblast-derived exosomes is represented by miRNAs derived from the passenger strand (also named star miRNAs), which usually undergo intracellular degradation (Bang et al., 2014). This suggests the existence of a preferential transport mechanism of star miRNAs into exosomes. Intriguingly, Squadrito et al. showed that miRNA distribution in macrophages and their exosomes depends on the cellular levels of their target transcripts (Squadrito et al., 2014). They found a negative correlation between miRNA:target interactions in cells and miRNA enrichment in exosomes. Physiological or artificial overexpression of either specific miRNAs or their target mRNAs promoted a bidirectional miRNA redistribution from the cell cytoplasm/P bodies to the multivesicular bodies and, accordingly, a controlled miRNA sorting into exosomes (Squadrito et al., 2014). In other words, when the mRNA target of a specific miRNA is abundant in the cytoplasm, that miRNA needs to stay in the cytoplasm in order to bind to it, and, as a consequence, its amount decreases within exosomes. On the contrary, a miRNA, more abundant in the cytoplasm than its target, will have an increased level also in the exosomes. These findings would suggest that secretion of miRNAs into exosomes is a mechanism through which cells, at least in part, can arrange miRNAs in excess respect to their targets in order to adjust the physiological miRNA:mRNA homeostasis (Squadrito et al., 2014). Altogether, these observations would suggest that RNA sorting into exosomes is the complex result of: (a) active mechanisms of molecular loading, based on specific sequence features of RNA molecules; (b) partially passive processes, which depend on the availability of RNA molecules free and unrestrained from functional binding mechanisms inside the cells. However, these considerations could be challenged by a study showing that miRNAs, even the most abundant, are present at far less than one copy per exosome, suggesting that the most of exosomes circulating in biological fluids are almost empty (Chevillet et al., 2014). These results do not underplay the role of exosomes in cell-to-cell communication, but rather provide some interesting scenarios on exosome sorting and uptake mechanisms. Indeed, the authors propose two models: (1) a low-occupancy/low-miRNA concentration model, in which a sub-population of exosomes carries a low amount of miRNAs; or (2) a low-occupancy/high-miRNA concentration model, in which rare exosomes inside the same nanovesicle population carry many copies of a given miRNA. This latter appears to be the most consistent with exosome-mediated communication, if exosome uptake is a selective and infrequent event. However, low-occupancy/low-miRNA concentration model could be considered valid if cellular uptake of exosomes is rapid, and therefore miRNAs can accumulate within the cell in functionally sufficient quantities. Both these models lead to the hypothesis that an exosome population from the same cell source exhibits a dramatic quantitative heterogeneity of its miRNA cargo (Chevillet et al., 2014). The molecular determinants of such heterogeneity of exosome loading remains still uncharacterized.
Asymmetric distribution of RNA molecules between source cells and their exosomes
Several papers on in vitro cellular models have analyzed the qualitative and quantitative distribution of RNAs between cells and their secreted exosomes, both at steady states and after biological stimuli. In one of the earliest reports, Pigati et al. found that nearly 30% of exosomal miRNAs released by normal and malignant mammary epithelial cells does not reflect the cellular profile, suggesting that some miRNAs are selectively retained or released (Pigati et al., 2010). Nevertheless, they observed that about 66% of the released miRNAs had an amount that closely reflected the corresponding abundance inside the cells. These findings would suggest that the majority of miRNAs is passively secreted through exosomes depending on miRNA mass. Intriguingly, the authors also reported that miRNAs with well-characterized roles in mammary biology tend to be selectively retained by cells and nearly absent in the extracellular population. This category of miRNAs included: miR-196a-1, which is overexpressed in breast cancer cells; miR-210, a hypoxia sensor with prognostic value in breast cancer; other miRNAs associated with metastasis (e.g., miR-148a, miR-335, miR-373, and miR-520c). In flat opposition to this group of retained miRNAs, the authors found miR-451 to be one of the most disproportionately exported miRNAs: more than 90% of it was secreted into the extracellular space (Pigati et al., 2010). MiR-451 functions as a tumor suppressor in breast cancer: its active removal from cells might represent a convenient mechanism to promote cancer progression (Liu et al., 2015). Furthermore, the excessive release of miR-451 might also explain the interstitial accumulation of miR-451 reported in breast tumors (Sempere et al., 2007). This asymmetric distribution of miR-451 has also been reported in another study on normal and malignant mammary epithelial cells and their exosomes, in which the authors observed the selective encapsulation of miRNAs inside the exosomes and characterized different miRNA profiles between the exosomes secreted by cancer cells and those produced by normal cells (Hannafon et al., 2016). Among the exosome-enriched miRNAs, those that were more abundant in exosomes from the breast cancer cell lines MCF7 and MDA-MB-231 than in exosomes from the normal cell line MCF10A were miR-21, miR-122, miR-451, and miR-1246 (Hannafon et al., 2016). Interestingly, miR-21 and miR-1246 were also highly expressed in plasma exosomes from patients affected by breast cancer. Although both miRNAs are highly abundant in breast cancer exosomes, they are also in large measure retained inside the cells because of their oncogenic role in breast cancer. On the contrary, miR-122 and miR-451 are downregulated in breast cancer cells and have tumor suppressive properties (Wang et al., 2012; Liu et al., 2015). The observations from these two studies on mammary epithelial cells further strengthen the idea that miRNAs secreted via exosomes may represent a mixture of: (a) highly expressed cellular miRNAs, which passively pass through the endosomes for an osmotic-like effect; (b) selectively secreted miRNAs, whose function inside the cytoplasm could be unnecessary or unfavorable for the cells in certain time frames and in specific physiological or pathological conditions (Pigati et al., 2010; Hannafon et al., 2016). This hypothesis has also been proved by studies on other cell types. Unequal distribution of RNAs between endothelial cells and their exosomes has also been reported (Van Balkom et al., 2015). More specifically, the authors observed a partially overlapping distribution of small RNAs in exosomes and corresponding donor cells. As a matter of fact, the most abundant miRNAs within the cells matched the most numerous ones in exosomes. However, about half of the identified miRNAs and 5p-, 3p-, and stem-loop fragments were differentially distributed. Just as an example: (1) miR-30d, miR-30e, miR-92b, and miR-125a were among the most abundant miRNAs in cells, but they were barely present in exosomes; (2) miR-25, miR-27a, miR-186, and miR-4485 were the most abundant miRNAs in exosomes, but were scarcely present inside the cells (Van Balkom et al., 2015). Interestingly, the authors focused their attention also on other RNA molecules than miRNAs: they observed that mRNA, lncRNA, vRNA, mtRNA, and yRNA fragments were more enriched in exosomes than in cells. These data on RNA molecule fragments in exosomes would suggest a link between RNA turnover and exosome biogenesis: endothelial endosomes could selectively sequester cytoplasmic RNA-degrading machineries and release degraded RNAs via exosomes (Van Balkom et al., 2015). In a previous work from our group, we have analyzed the miRNA transcriptome of two different colorectal carcinoma (CRC) cell lines and their secreted exosomes before and after treatment with Cetuximab, a monoclonal antibody that binds and blocks the Epidermal Growth Factor Receptor (EGFR; Ragusa et al., 2014, 2015b). About 90% of cellular miRNAs were also present inside the exosomes for both cell lines. However, in contrast with such a qualitative miRNA overlapping, we found a strong quantitative asymmetry of miRNAs between secreted exosomes and their source cells. Some miRNAs were mainly released into exosomes (e.g., miR-127, miR-136*, miR-144*, miR-432, miR-433, miR-487b, and miR-495, in Caco-2 exosomes; miR-136*, miR-223*, miR-380-5p, miR-432, and miR-672, in HCT-116 exosomes). Exosomes from both cell lines were enriched in miRNAs with a potential tumor suppressive role in CRC or with immunosuppressive properties (e.g., miR-142-5p, miR-150, miR-223, and miR-433; Ragusa et al., 2014). This finding was consistent with our functional data showing a decreased proliferation of Caco-2 cells after transfection with HCT-116-derived exosomes and vice versa (Ragusa et al., 2014). This anticancer effect of tumor-derived exosomes on tumor cells had been previously reported for pancreatic cancer cells (Ristorcelli et al., 2008). Asymmetric distribution of miRNA contents in exosomes and their source cells was highly accentuated by Cetuximab treatment in Caco-2 (Cetuximab sensitive), but less pronounced in HCT-116 (Cetuximab unresponsive). The sets of differentially expressed miRNAs in exosomes and their source cells were different for these two CRC cell lines. However and interestingly, for both of them the exosomal miRNAs that were most dysregulated after treatment were involved in many cancer-related functions, as oncomirs or tumor suppressors, and in inflammatory processes (Ragusa et al., 2014). We concluded that cellular miRNAs, both at steady state and after pharmacological treatment, are selectively released into the tumor microenvironment through exosomes in order to: (a) throw functionally disadvantageous miRNAs out of the cells, or (b) favorably influence the immune system response (Ragusa et al., 2014). We obtained similar data on the asymmetric distribution of retained cellular miRNAs and secreted exosomal ones in an in vitro model of non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH; Di Mauro et al., 2016). Interestingly, in HepG2 cells at steady state we found that: (1) 104 miRNAs were specifically expressed within the cells; (2) 20 miRNAs were exclusively secreted in the culture medium; (3) 284 miRNAs were both present inside the cells and secreted in the medium. In the same cell line analyzed after lipotoxic and non-lipotoxic stimuli we found quantitative, but not qualitative, alterations for many secreted miRNAs, and just few and relatively minor changes for those intracellular (Di Mauro et al., 2016). The majority of extracellular miRNAs were involved in inflammatory-related pathways. This finding led us to propose the following hypothesis: in response to lipogenic stimuli, some miRNAs could be selectively secreted to either act in an endocrine manner and determine a cross-talk among NAFLD-associated inflamed tissues or act in a paracrine/autocrine manner and regulate inflammation in hepatocytes or innate immune liver cells (Di Mauro et al., 2016). Our results suggest that the innate disequilibrium between retained and secreted miRNAs could be perturbed by biochemical stimuli (Ragusa et al., 2014; Di Mauro et al., 2016). A similar rearrangement of the cellular environment by exosomes has been observed in a study on HIV infection (Aqil et al., 2014). In this work, it has been demonstrated that transfection of U937 cells with the HIV Nef protein (a multifunctional virulence factor) leads to the selective secretion through exosomes of 47 miRNAs and retention of 2 miRNAs in Nef-expressing cells. Intriguingly, exosomes secreted in response to the intracellular upregulation of Nef were enriched in miRNAs that target proinflammatory cytokines and can potentially attack HIV-1 (Aqil et al., 2014). In this way, Nef expression would reduce the intracellular levels of miRNAs responsible for innate immune responses and targeting viral transcripts by throwing them out of the cell via the exosomes. This mechanism could potentially be used by the virus to modify the host cell environment in favor of its replication and dissemination. In general, miRNA sorting into exosomes has been demonstrated to be largely influenced by virus infection. Just to cite a different and peculiar example, lymphoma cell lines infected by Kaposi sarcoma-associated herpes virus (KSHV) have been found to produce exosomes that contained about 48% of miRNAs of viral origin (Hoshina et al., 2016). This phenomenon is due to the expression of viral small RNAs in the host cells: some of these viral miRNAs may possess EXOmotifs, like CCCT or CCCG, that influence the exosome loading of the host cells. Even more interesting is the finding that exosomes from virus-infected cells more frequently contained non-exact mature miRNAs (i.e., mutated miRNAs) than the corresponding infected source cells (Hoshina et al., 2016). This observation would suggest the presence of a mechanism that preferentially sorts non-exact mature miRNAs to the exosomes. Moreover, this would inspire the idea that exosomes might have the function of removing the mutated mature miRNAs from cells and, accordingly, concentrating wild-type and functional miRNAs into the cytoplasm. Otherwise, the delivery of non-exact miRNAs to target cells via exosomes could have other unexpected and unknown roles and functions (Hoshina et al., 2016). All these data on the asymmetrical distribution of RNAs between exosomes and source cells lead us to some critical considerations on the biological meaning of RNAs released by exosomes and to potential general sorting criteria. The first evident issue is that cells tend to retain and accumulate into the cytoplasm RNAs that exert critical functions for the cell itself in both physiological and pathological conditions. We note that a normal epithelial cell expresses and retains RNAs involved in its correct specific physiology and homeostasis. Nevertheless, when the same cell undergoes cancer transformation, it will tend to retain RNAs that promote cell growth and inhibit cell death. Cells preferentially hold back RNAs that are advantageous for cell functioning in specific conditions (Kanlikilicer et al., 2016). This consideration leads us to a second question: which RNAs do cells secrete via exosomes? Different from RNAs retained within the cytoplasm, those encapsulated in exosomes are preferentially RNAs that could be judged to be not critical for the appropriate cell performance or detrimental in normal or diseased conditions. Moreover, also mutated miRNAs and other species of degraded RNAs are largely abundant in exosomes (Van Balkom et al., 2015; Hoshina et al., 2016). The expulsion of these useless, dysfunctional, or potentially deleterious RNAs out of the cells would represent a convenient cellular process to accelerate the turnover of non-effective RNAs. Intriguingly, when secreted exosomes are captured by recipient cells, this apparent waste RNA cargo can in some cases be functionally effective: the main effect reported for some pathological conditions is the modification of the cell environment to favor cancer dissemination or virus replication (Meckes et al., 2010; Pegtel et al., 2010; Neviani and Fabbri, 2015). However, stating that cells retain into the cytoplasm functionally useful RNAs and throw out useless or disadvantageous ones via exosomes would be a largely misleading and inappropriate over-simplification. Indeed, miRNAs harboring EXOmotifs or hEXO motifs are selectively transported by specific RNA-binding proteins into exosomes, regardless of their amount or mutated form (Villarroya-Beltri et al., 2013; Santangelo et al., 2016). Moreover, the most abundant cytoplasmic miRNAs are passively incorporated into the exosomes, especially when they exceed the amount of their mRNA targets and, accordingly, are not engaged in Ago2 binding (Squadrito et al., 2014). In this way, these miRNAs become unconstrained from any functional commitment in the cytoplasm and can be passively stored inside the exosomes. This observation could appropriately explain the presence of oncomir-enriched exosomes in the blood of cancer patients, as discussed in the next paragraph.
Exosomal miRNAs appear to be promising biomarkers in cancer patients, but caveats are needed
Our knowledge on the exact relationship between the profile of exosomal miRNAs circulating in body fluids and the pathological condition of cancer patients is admittedly incomplete: this notwithstanding, the potential of exosomal miRNAs and other ncRNAs as cancer biomarkers is very promising. First of all, miRNAs encapsulated in exosomes are very stable because they are protected against RNase degradation by the lipid bilayer (Valadi et al., 2007). Moreover, exosomal miRNA expression may be easily analyzed by using whatever liquid of our body, even though the most of studies have focused on the exosomes purified from systemic blood circulation (Keller et al., 2011; Revenfeld et al., 2014; Boukouris and Mathivanan, 2015). As stated in the previous paragraph, RNA molecules stored in exosomes represent the molecular outcome of both active and passive sorting mechanisms, which seem to be cell-specific. Exosomes circulating in the blood are a heterogeneous population of nanovesicles derived from a plethora of different cell types, but principally coming from blood cells (Chen et al., 2008; Hunter et al., 2008; Pritchard et al., 2012b; Zhou et al., 2017). Accordingly, exosomal miRNAs purified from serum or plasma are a mixture of molecules of different cellular origins and with different roles in extracellular communication. Given these premises, it is hardly to be expected that in the whole population of secreted exosomes, dysfunctional exosomes derived from diseased cells may possess an RNA cargo able to markedly modify the relative amounts of circulating miRNAs. Nevertheless, multiple evidence shows that: (1) exosomal miRNAs from serum or plasma of cancer patients are quantitatively altered; (2) interestingly, some of these dysregulations mirror those detected in source tumor cells (Table 1). A fitting example is the one about miR-21. MiR-21 is among the most commonly upregulated miRNAs in cancer: its genetic locus at 17q23 is amplified in many solid tumors and its expression is stimulated by a variety of cancer-associated phenomena, such as inflammation and hypoxia (Griffin et al., 1988; Wu et al., 2001; Loffler et al., 2007; Fujita et al., 2008; Ribas et al., 2009). Notably, several studies have reported increased levels of miR-21 in both serum and plasma exosomes from patients affected by tumors showing a cellular miR-21 upregulation (Skog et al., 2008; Taylor and Gercel-Taylor, 2008; Que et al., 2013; Ogata-Kawata et al., 2014; Hannafon et al., 2016; Lai et al., 2017; Tsukamoto et al., 2017). Taylor et al. showed that the levels of 8 miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205, and miR-214) were increased in exosomes from serum of ovarian cancer patients, as well as in tumor tissues from the same patients (Iorio et al., 2007; Nam et al., 2008; Taylor and Gercel-Taylor, 2008; Niu et al., 2015; Azizmohammadi et al., 2016; Xiaohong et al., 2016; Li J. et al., 2017; Wei et al., 2017). Liu et al. observed elevated levels of exosomal miR-23b-3p, miR-10b-5p, and miR-21-5p in plasma of non-small-cell lung cancer (NSCLC) patients and their association with poor overall patient survival (Liu et al., 2017). Interestingly, these three miRNAs have been reported to be also overexpressed in NSCLC cells and furthermore their intracellular expression has been found to be associated with poor prognosis (Begum et al., 2015; Huang et al., 2015; Tian et al., 2016; Xue et al., 2016; Li C. et al., 2017). In a previous work of our group, we showed the constant and consistent upregulation of miR-146a in: (1) vitreous humor, (2) vitreal exosomes, (3) serum, (4) serum exosomes, and (5) cancerous tissues of uveal melanoma patients (Ragusa et al., 2015a). We suggested that miR-146a might be released by melanoma cells inside the ocular chamber and then conveyed to the systemic circulation through tumor blood vessels. Specifically, we observed this fixed expression trend among tissues and body fluids only for miR-146a: this would suggest the existence of specific mechanisms of retention, secretion, and filtering of exosomal miRNAs through the cellular and extracellular compartments (Ragusa et al., 2015a). In contrast with these examples of exosomal miRNA alterations that match the corresponding cellular ones in cancer patients, there are other reports in which the miRNA expression trend in blood exosomes was inconsistent with or even the opposite to the one observed in the corresponding cancer cells (Table 1). Profiling of serum exosomal miRNAs in primary CRC patients showed the upregulation of let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a (Ogata-Kawata et al., 2014). It is interesting to note that the overexpression of exosomal miR-23a, miR-223, and miR-1246 conformed with their upregulation reported in CRC tissues and linked to their oncogenic role (Yong et al., 2013, 2014; Della Vittoria Scarpati et al., 2014; Li Z. W. et al., 2014; Zhang et al., 2014; Neerincx et al., 2015; Wang S. et al., 2016; Wang F.F. et al., 2017). On the contrary, augmented serum levels of let-7a and miR-150 were inconsistent with their documented downregulation and tumor suppressive property in CRC (Ma Y. et al., 2012; Li et al., 2016). Recently, another study on serum exosomes from CRC patients reported the upregulation of miR-486-5p and miR-3180-5p, and the downregulation of miR-548c-5p, miR-638, miR-5787, miR-6869-5p, miR-8075 (Yan et al., 2017). Among these differentially expressed miRNAs, miR-638 was the most relevant concerning the clinics: its downregulation has been previously reported to be related to invasion and mesenchymal-like transition in CRC cells (Ma et al., 2014). This is not the case for miR-486-5p: its exosomal upregulation was diametrically opposed to its marked downregulation in CRC tissues, previously reported by Liu et al. (2016). For all the other dysregulated exosomal miRNAs identified in this study the corresponding alterations in CRC tissues have not been reported yet. Wang et al. found miR-125a-3p and miR-320c to be significantly upregulated in plasma exosomes from patients with early stage CRC (Wang J. et al., 2017): there is no experimental evidence of miR-125a-3p alteration in CRC, while miR-320c has been reported to be frequently downregulated in CRC tissues together with the other members of miR-320 family (Tadano et al., 2016; Vishnubalaji et al., 2016). Interestingly, the upregulation of miR-320 was also detected in serum exosomes from Glioblastoma Multiforme (GBM) patients (Manterola et al., 2014), in contrast with miR-320 decreased levels reported in GBM tissues (Guo et al., 2014). Taken together, these data on exosomes circulating in the blood of cancer patients would be an in vivo indirect proof that exosomes secreted by cancer cells might carry a cargo of RNA molecules that does not exactly reflect the cytoplasmic RNA alterations of their source cells. This could be considered inessential from a diagnostic point of view. For instance, the upregulation of exosomal miR-21 and miR-320 in serum of CRC patients could represent for clinicians a very useful evidence to diagnose CRC in a non-invasive manner, regardless of miR-21 overexpression and miR-320 downregulation described in CRC source cells (Slaby et al., 2007; Vishnubalaji et al., 2016). However, many works on the differential expression of exosomal miRNAs in cancer have attempted to computationally reconstruct miRNA-based networks in order to infer pathways that might be involved in a specific tumor (Eldh et al., 2014; Alhasan et al., 2016). This approach could be considered misleading: a part of altered circulating miRNAs in body fluids of cancer patients has a relative expression that is largely different from the one in the corresponding diseased tissues. The tumor suppressor miRNA let-7a inhibits tumor cell growth and metastasis in CRC and its levels are significantly decreased in CRC tissues and cell lines (Li et al., 2016); nevertheless, its exosomal levels in serum of CRC patients are increased (Ogata-Kawata et al., 2014). This apparent incongruity might be the result of a strategy adopted by the cancer to remove from cells miRNAs that are disadvantageous for viability and dissemination of CRC. This scenario is made even trickier by the presence in circulation of exosomes from other cellular sources that could contribute to the cancer-related miRNA dysregulation reported in literature (Figure 1). The presence of exosome populations of different origin could dilute the actual expression alterations deriving from cancer cells. However, it is worth mentioning that exosome concentrations are increased in cancer patients compared to normal controls (Taylor and Gercel-Taylor, 2008; Logozzi et al., 2009; Caivano et al., 2015; Alegre et al., 2016), suggesting the hypothesis that cancer cells release huge quantity of exosomes due to their intrinsic cancer-related mutations (Yu et al., 2006). Moreover, exosomes coming from immune cells could have an important role in changing the expression of miRNAs within the whole exosomal population. Cancer development elicits an immune response, triggered by tumor antigens distinguishing the cancerous cells from the other non-cancerous ones (Grivennikov et al., 2010). On one hand, tumor-infiltrating lymphocytes act by slowing or arresting the development of the tumor. On the other hand, cancer cells are able to favorably influence the tumor microenvironment by inducing signaling cascades, leading to immune-suppression and thus facilitating evasion of immune surveillance and cancer dissemination (Gardner and Ruffell, 2016; Jang et al., 2017). This fight between tumor and immune system is also fought by means of exosome secretion by both cancer cells and regulatory immune-cells (Bobrie and Thery, 2013; Greening et al., 2015). Indeed, tumor-derived exosomes directly suppress the anti-tumor responses of cytotoxic T lymphocytes and NK-cells, and also induce the generation, expansion, and suppressive function of T regulatory cells (Treg). Okoye et al. demonstrated that Treg cell-derived exosomes are able to transfer both in vitro and in vivo a specific set of miRNAs to T cells (e.g., miR-155, let-7b, and let-7d), thus suppressing T helper cell proliferation and interferon-γ production (Okoye et al., 2014). On the contrary, several reports demonstrated that activated dendritic cells secrete exosomes and induce a T-cell-mediated anti-tumor immune response (Zitvogel et al., 1998; Viaud et al., 2010; Tian and Li, 2017). These observations lead us to consider that part of dysregulated exosomal miRNAs in blood of cancer patients could derive from different activated regulatory immune cells and act by suppressing or activating the immune system. In agreement with such hypothesis of the over-representation of cancer- and immune-derived exosomes in the systemic circulation of cancer patients, it should not be surprising that a large number of exosomal miRNAs proposed as cancer biomarkers also has a critical functional role in the differentiation and activation of immune cells (e.g., miR-10b, miR-17, miR-20a, miR-146a, miR-150, miR-181a, miR-223; Table 1; Paladini et al., 2016). This could explain why in some cases exosomal miRNAs differentially expressed in the blood of cancer patients have no links to altered cytoplasmic miRNAs of the native tumor. There is a final technical warning that may be worth mentioning to fully understand the effectiveness of exosome as cancer biomarkers. One of potential pitfalls of studies on exosome functions and their molecular cargo is the lack of standard methods to obtain highly pure exosome populations. Several methods are reported in scientific literature, including ultracentrifugation, density gradient centrifugation, chromatography, filtration, polymer-based precipitation, and immunoaffinity (Taylor and Shah, 2015). It has been demonstrated that these methods could lead to co-isolate contaminating non-exosomal material or to the loss of exosomes due to damaged membrane integrity, thus, resulting in significant artifacts of recovery, quality and molecular content of exosomes (Whiteside, 2017). Just as an example: ultracentrifugation is the “gold standard” exosome isolation method, but the type, quantity and quality of the vesicles isolated is extremely sensitive to parameters, including g force, rotor type, angle of rotor sedimentation, radius of centrifugal force, solution viscosity, and vesicle density (Taylor and Shah, 2015). Needless to say, accounting for, controlling and standardizing all of these parameters is quite impossible. Van Deun et al. performed a comparative study of 4 exosome isolation protocols (i.e., ultracentrifugation, OptiPrep density gradient centrifugation, and two commercial polymer-based precipitation methods) to evaluate their reliability for downstream molecular analyses (Van Deun et al., 2014). The four methods provided different qualitative and quantitative results, but OptiPrep density gradient ultracentrifugation outperformed the other 3 methods and revealed a unique and reproducible mRNA profile. In a similar work, Tauro et al. compared ultracentrifugation, density gradient centrifugation and immunoaffinity to purify exosomes from biological fluids and conditioned media from in vitro cell cultures (Tauro et al., 2012). Their proteomic analyses revealed that immunoaffinity capture was the most effective method to isolate exosomes. Indeed, the immunoaffinity capture is considered the better alternative for exosome isolation, because there is no damage to the nanovesicle structure and the loss of exosomes is negligible. All these considerations strongly suggest that the choice of specific isolation method severely affects the purity and quality of exosomes and, accordingly, the reliability of molecular profiles of their RNA content. Moreover, also the use of disparate methods to profile RNA expression from exosomes could add further bias to final data and cause a scanty reproducibility among different studies (Git et al., 2010; Chugh and Dittmer, 2012; Pritchard et al., 2012a; Moldovan et al., 2014).
Table 1.
Expression of miRNA cancer biomarkers in exosomes and their relative tumor tissues.
| miRNAs | Cancer | Exosome origin | Expression within exosomes | PMID EX | Expression within cells | PMID CELL |
|---|---|---|---|---|---|---|
| let-7a | Colon cancer | Serum | Up | 24705249 | Down | 27498032 |
| let-7a | Pancreatic ductal adenocarcinoma | Plasma | Down | 28232049 | Down | 19323605 |
| miR-10b | Pancreatic ductal adenocarcinoma | Plasma | Up | 28232049 | Up | 22018284 |
| miR-10b-5p | Non-small-cell lung cancer | Plasma | Up | 28055956 | Up | 25988292 |
| miR-17-5p | Pancreatic adenocarcinoma | Serum | Up | 24007214 | Up | 27400681 |
| miR-20a | Pancreatic ductal adenocarcinoma | Plasma | Up | 28232049 | Down | 25485236 |
| miR-21 | Breast cancer | Plasma | Up | 27608715 | Up | 16103053, 18812439, 26549725 |
| miR-21 | Colon cancer | Serum; plasma | Up | 24705249; 28376502 | Up | 18196926, 22844381, 28376502 |
| miR-21 | Glioblastoma | Serum | Up | 19011622 | Up | 21895872 |
| miR-21 | Glioma | Cerebrospinal fluids | Up | 26284486 | Up | 16024602, 24326156 |
| miR-21 | Ovarian cancer | Serum | Up | 18589210 | Up | 17875710, 18451233 |
| miR-21 | Pancreatic adenocarcinoma | Serum; plasma | Up | 24007214; 28232049 | Up | 20093556, 22850622, 28239461 |
| miR-21 | Uveal melanoma | Vitreus | Up | 25951497 | Up | 25951497 |
| miR-21-5p | Non-small-cell lung cancer | Plasma | Up | 28055956 | Up | 26453197, 27811366, 28445945 |
| miR-21-5p | Prostate cancer | Urine | Up | 26417675 | Up | 16461460, 27040772 |
| miR-23a | Colon cancer | Serum | Up | 24705249 | Up | 23758639, 24992592 |
| miR-23b-3p | Non-small-cell lung cancer | Plasma | Up | 28055956 | Up | 26314549 |
| miR-30c | Pancreatic ductal adenocarcinoma | Plasma | Up | 28232049 | / | / |
| miR-34a | Uveal melanoma | Vitreus | Up | 25951497 | Up | 25951497 |
| miR-106b | Pancreatic ductal adenocarcinoma | Plasma | Up | 28232049 | / | / |
| miR-122 | Pancreatic ductal adenocarcinoma | Plasma | Down | 28232049 | Down | 22850622, 28239461 |
| miR-125a-3p | Colon cancer | Plasma | Up | 28646161 | / | / |
| miR-140-3p | Chronic myeloid leukemia | Serum | Up | 28197964 | / | / |
| miR-141 | Ovarian cancer | Serum | Up | 18589210 | Up | 17875710, 18451233 |
| miR-141-5p | Prostate cancer | Urine | Up | 26417675 | / | / |
| miR-146a | Uveal melanoma | Serum | Up | 25951497 | Up | 25951497 |
| miR-146a | Uveal melanoma | Vitreus | Up | 25951497 | Up | 25951497 |
| miR-150 | Colon cancer | Serum | Up | 24705249 | Down | 22052060 |
| miR-181a | Pancreatic ductal adenocarcinoma | Plasma | Up | 28232049 | Up | 17473300 |
| miR-200a | Ovarian cancer | Serum | Up | 18589210 | Up | 17875710, 18451233 |
| miR-200b | Ovarian cancer | Serum | Up | 18589210 | Up | 17875710, 18451233 |
| miR-200c | Ovarian cancer | Serum | Up | 18589210 | Up | 17875710, 18451233 |
| miR-203 | Ovarian cancer | Serum | Up | 18589210 | Up | 27347348, 27655286 |
| miR-205 | Ovarian cancer | Serum | Up | 18589210 | Up | 26275944, 28145479, 28476165 |
| miR-214 | Ovarian cancer | Serum | Up | 18589210 | Up; down | 17875710; 18451233 |
| miR-223 | Colon cancer | Serum | Up | 24705249 | Up | 24819398, 25270282, 27916606 |
| miR-320 | Glioblastoma multiforme | Serum | Up | 24435880 | Down | 25117070 |
| miR-320c | Colon cancer | Plasma | Up | 28646161 | Down | 27119506, 27559432 |
| miR-486-5p | Colorectal cancer | Serum | Up | 28636562 | Down | 27284245 |
| miR-548c-5p | Colorectal cancer | Serum | Down | 28636562 | / | / |
| miR-574-3p | Glioblastoma multiforme | Serum | Up | 24435880 | / | / |
| miR-574-3p | Prostate cancer | Urine | Up | 26417675 | Down | 23554959 |
| miR-638 | Colorectal cancer | Serum | Down | 28636562 | Down | 24885288 |
| miR-1229 | Colon cancer | Serum | Up | 24705249 | / | / |
| miR-1246 | Breast cancer | Plasma | Up | 27608715 | / | / |
| miR-1246 | Colon cancer | Serum | Up | 24705249 | Up | 25143946, 26436952, 26573378 |
| miR-3180-5p | Colorectal cancer | Serum | Up | 28636562 | / | / |
| miR-6869-5p | Colorectal cancer | Serum | Down | 28636562 | / | / |
| miR-8075 | Colorectal cancer | Serum | Down | 28636562 | / | / |
| miR-5787 | Colorectal cancer | Serum | Down | 28636562 | / | / |
This table reports the trend of expression of exosomal miRNA biomarkers in cancer and in tumor parental cells. PMID EX, Pubmed IDs of papers about exosomal miRNAs; PMID CELL, Pubmed IDs of papers about cancer cellular miRNAs. Slash: no data available.
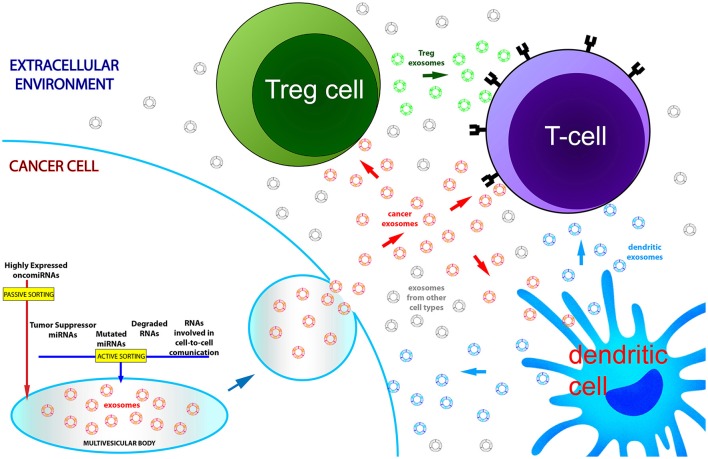
Figure 1.
Exosomal RNA dysregulation partially reflects transcriptomic alterations of parental cancer cells. Active and passive sorting mechanisms are responsible for the native RNA quantitative asymmetry existing between cells and exosomes. In blood of cancer patients the nanovesicle population is the complex outcome of exosome production by multiple cell types: cancer cells and immune cells, through exosomal secretion, regulate their molecular homeostasis and communicate with each other during cancer development and progression. For these reasons, exosomal RNAs recovered from the systemic circulation only partially mirror the transcriptome of tumor cells.
Conclusions
The key concepts reviewed in this paper may be summarized in the three following points. (1) RNA molecules secreted via exosomes are encapsulated thanks to both active, scarcely characterized, cell-specific mechanisms of sorting and also passive processes depending on the amount of cytoplasmic RNAs. The biological meaning of the active processes remains largely elusive: however, it appears to be related to the maintenance of the correct molecular equilibrium between RNA molecules that are biologically important, ineffective, or potentially deleterious for the cell in a certain physiological or pathological condition. Moreover, secreted RNAs might have a role in the homeostasis of the extracellular microenvironment. (2) This co-occurrence of both active and passive transport processes leads to a partial match between exosome and source cell transcriptomes, as observed in the in vitro models reviewed in this paper. (3) Differentially expressed exosomal miRNAs in the circulation of cancer patients only partly reflect miRNA dysregulations found in corresponding cancer tissues and, as a matter of fact, divergent expression trends between exosomes and native tumors have been documented for many miRNAs. These differences could be the assorted result of the asymmetric distribution of miRNAs between exosomes and cells, but also the confounding effect of the presence in the blood of exosomes from different cellular origins. Specifically, the regulatory cells of the immune system can induce the activation or suppression of T cells in the tumor microenvironment by producing regulatory exosomes and, thus, they significantly influence the total relative expression of circulating miRNAs.
Finally, exosomal miRNAs could be considered as good biomarkers in cancer, even if no standard reproducible method to isolate them has been proposed. Nevertheless, they do not represent an accurate reflection of miRNA intracellular expression in the diseased cells, but they are the complex outcome of a mixed exosome production from multiple cell types, which through exosomal secretion regulate their molecular homeostasis and communicate with each other during cancer development and progression.
Author contributions
All authors contributed to the writing of the paper, performed a critical revision of the review and approved the final manuscript.
Conflict of interest statement
The authors declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alegre E., Zubiri L., Perez-Gracia J. L., Gonzalez-Cao M., Soria L., Martin-Algarra S., et al. (2016). Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta 454, 28–32. 10.1016/j.cca.2015.12.031 [DOI] [PubMed] [Google Scholar]
- Alhasan A. H., Scott A. W., Wu J. J., Feng G., Meeks J. J., Thaxton C. S., et al. (2016). Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 10655–10660. 10.1073/pnas.1611596113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqil M., Naqvi A. R., Mallik S., Bandyopadhyay S., Maulik U., Jameel S. (2014). The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J. Extracell. Vesicles 3. 10.3402/jev.v3.23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizmohammadi S., Azizmohammadi S., Safari A., Kosari N., Kaghazian M., Yahaghi E., et al. (2016). The role and expression of miR-100 and miR-203 profile as prognostic markers in epithelial ovarian cancer. Am. J. Trans. Res. 8, 2403–2410. [PMC free article] [PubMed] [Google Scholar]
- Bang C., Batkai S., Dangwal S., Gupta S. K., Foinquinos A., Holzmann A., et al. (2014). Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 124, 2136–2146. 10.1172/JCI70577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum S., Hayashi M., Ogawa T., Jabboure F. J., Brait M., Izumchenko E., et al. (2015). An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: clinical significance of miR-23b-3p deregulation. Sci. Rep. 5:13236. 10.1038/srep13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A., Thery C. (2013). Exosomes and communication between tumours and the immune system: are all exosomes equal? Biochem. Soc. Trans. 41, 263–267. 10.1042/BST20120245 [DOI] [PubMed] [Google Scholar]
- Bolukbasi M. F., Mizrak A., Ozdener G. B., Madlener S., Strobel T., Erkan E. P., et al. (2012). miR-1289 and “Zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids 1, e10. 10.1038/mtna.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouris S., Mathivanan S. (2015). Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin. Appl. 9, 358–367. 10.1002/prca.201400114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucher B. L., Jamall I. S. (2014). Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell. Physiol. Biochem. 34, 213–243. 10.1159/000362978 [DOI] [PubMed] [Google Scholar]
- Caivano A., Laurenzana I., De Luca L., La Rocca F., Simeon V., Trino S., et al. (2015). High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumour Biol. 36, 9739–9752. 10.1007/s13277-015-3741-3 [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- Chevillet J. R., Kang Q., Ruf I. K., Briggs H. A., Vojtech L. N., Hughes S. M., et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U.S.A. 111, 14888–14893. 10.1073/pnas.1408301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P., Dittmer D. P. (2012). Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA 3, 601–616. 10.1002/wrna.1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Thery C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Cosme J., Liu P. P., Gramolini A. O. (2013). The cardiovascular exosome: current perspectives and potential. Proteomics 13, 1654–1659. 10.1002/pmic.201200441 [DOI] [PubMed] [Google Scholar]
- De Broe M. E., Wieme R. J., Logghe G. N., Roels F. (1977). Spontaneous shedding of plasma membrane fragments by human cells in vivo and in vitro. Clin. Chim. Acta 81, 237–245. 10.1016/0009-8981(77)90054-7 [DOI] [PubMed] [Google Scholar]
- Della Vittoria Scarpati G., Calura E., Di Marino M., Romualdi C., Beltrame L., Malapelle U., et al. (2014). Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. Biomed. Res. Int. 2014:40921. 10.1155/2014/840921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mauro S., Ragusa M., Urbano F., Filippello A., Di Pino A., Scamporrino A., et al. (2016). Intracellular and extracellular miRNome deregulation in cellular models of NAFLD or NASH: clinical implications. Nutr. Metab. Cardiovasc. Dis. 26, 1129–1139. 10.1016/j.numecd.2016.08.004 [DOI] [PubMed] [Google Scholar]
- Di Pietro C., Ragusa M., Barbagallo D., Duro L. R., Guglielmino M. R., Majorana A., et al. (2009). The apoptotic machinery as a biological complex system: analysis of its omics and evolution, identification of candidate genes for fourteen major types of cancer, and experimental validation in CML and neuroblastoma. BMC Med. Genom. 2:20. 10.1186/1755-8794-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldh M., Olofsson Bagge R., Lasser C., Svanvik J., Sjostrand M., Mattsson J., et al. (2014). MicroRNA in exosomes isolated directly from the liver circulation in patients with metastatic uveal melanoma. BMC Cancer 14:962. 10.1186/1471-2407-14-962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. (1998). Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127. 10.1074/jbc.273.32.20121 [DOI] [PubMed] [Google Scholar]
- Fujita S., Ito T., Mizutani T., Minoguchi S., Yamamichi N., Sakurai K., et al. (2008). miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 378, 492–504. 10.1016/j.jmb.2008.03.015 [DOI] [PubMed] [Google Scholar]
- Gardner A., Ruffell B. (2016). Dendritic cells and cancer immunity. Trends Immunol. 37, 855–865. 10.1016/j.it.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J. N., Thoi L. L., McManus L. M., Reimann T. A. (1982). Isolation of human platelet membrane microparticles from plasma and serum. Blood 60, 834–840. [PubMed] [Google Scholar]
- Gerdes H. H., Pepperkok R. (2013). Cell-to-cell communication: current views and future perspectives. Cell Tissue Res. 352, 1–3. 10.1007/s00441-013-1590-1 [DOI] [PubMed] [Google Scholar]
- Git A., Dvinge H., Salmon-Divon M., Osborne M., Kutter C., Hadfield J., et al. (2010). Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 16, 991–1006. 10.1261/rna.1947110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V., Hannus M., Eaton S. (2001). Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell 106, 633–645. 10.1016/S0092-8674(01)00484-6 [DOI] [PubMed] [Google Scholar]
- Greening D. W., Gopal S. K., Xu R., Simpson R. J., Chen W. (2015). Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 40, 72–81. 10.1016/j.semcdb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Griffin C. A., Hawkins A. L., Packer R. J., Rorke L. B., Emanuel B. S. (1988). Chromosome abnormalities in pediatric brain tumors. Cancer Res. 48, 175–180. [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R., Karin M. (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Feng Y., Liu Q., Yang X., Jiang T., Chen Y., et al. (2014). MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1R. Tumour Biol. 35, 11269–11275. 10.1007/s13277-014-2283-4 [DOI] [PubMed] [Google Scholar]
- Halkein J., Tabruyn S. P., Ricke-Hoch M., Haghikia A., Nguyen N. Q., Scherr M., et al. (2013). MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J. Clin. Invest. 123, 2143–2154. 10.1172/JCI64365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannafon B. N., Trigoso Y. D., Calloway C. L., Zhao Y. D., Lum D. H., Welm A. L., et al. (2016). Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 18, 90. 10.1186/s13058-016-0753-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug B. H., Hald O. H., Utnes P., Roth S. A., Lokke C., Flaegstad T., et al. (2015). Exosome-like extracellular vesicles from MYCN-amplified Neuroblastoma cells contain oncogenic miRNAs. Anticancer Res. 35, 2521–2530. Available online at: http://ar.iiarjournals.org/content/35/5/2521.long [PubMed] [Google Scholar]
- Henne W. M., Buchkovich N. J., Emr S. D. (2011). The ESCRT pathway. Dev. Cell 21, 77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Hergenreider E., Heydt S., Treguer K., Boettger T., Horrevoets A. J., Zeiher A. M., et al. (2012). Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 14, 249–256. 10.1038/ncb2441 [DOI] [PubMed] [Google Scholar]
- Hess C., Sadallah S., Hefti A., Landmann R., Schifferli J. A. (1999). Ectosomes released by human neutrophils are specialized functional units. J. Immunol. 163, 4564–4573. [PubMed] [Google Scholar]
- Hoshina S., Sekizuka T., Kataoka M., Hasegawa H., Hamada H., Kuroda M., et al. (2016). Profile of Exosomal and Intracellular microRNA in Gamma-Herpesvirus-Infected Lymphoma Cell Lines. PLoS ONE11:e0162574. 10.1371/journal.pone.0162574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristov M., Erl W., Linder S., Weber P. C. (2004). Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood 104, 2761–2766. 10.1182/blood-2003-10-3614 [DOI] [PubMed] [Google Scholar]
- Huang J., Sun C., Wang S., He Q., Li D. (2015). microRNA miR-10b inhibition reduces cell proliferation and promotes apoptosis in non-small cell lung cancer (NSCLC) cells. Mol. Biosyst. 11, 2051–2059. 10.1039/C4MB00752B [DOI] [PubMed] [Google Scholar]
- Hunter M. P., Ismail N., Zhang X., Aguda B. D., Lee E. J., Yu L., et al. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3:e3694. 10.1371/journal.pone.0003694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Visone R., Di Leva G., Donati V., Petrocca F., Casalini P., et al. (2007). MicroRNA signatures in human ovarian cancer. Cancer Res. 67, 8699–8707. 10.1158/0008-5472.CAN-07-1936 [DOI] [PubMed] [Google Scholar]
- Janas T., Janas T. (2011). The selection of aptamers specific for membrane molecular targets. Cell. Mol. Biol. Lett. 16, 25–39. 10.2478/s11658-010-0023-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas T., Yarus M. (2003). Visualization of membrane RNAs. RNA 9, 1353–1361. 10.1261/rna.5129803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas T., Janas M. M., Sapon K., Janas T. (2015). Mechanisms of RNA loading into exosomes. FEBS Lett. 589, 1391–1398. 10.1016/j.febslet.2015.04.036 [DOI] [PubMed] [Google Scholar]
- Janas T., Janas T., Yarus M. (2004). A membrane transporter for tryptophan composed of RNA. RNA 10, 1541–1549. 10.1261/rna.7112704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas T., Janas T., Yarus M. (2012). Human tRNA(Sec) associates with HeLa membranes, cell lipid liposomes, and synthetic lipid bilayers. RNA 18, 2260–2268. 10.1261/rna.035352.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. E., Hajdu C. H., Liot C., Miller G., Dustin M. L., Bar-Sagi D. (2017). Crosstalk between regulatory T cells and tumor-associated dendritic cells negates anti-tumor immunity in pancreatic cancer. Cell Rep. 20, 558–571. 10.1016/j.celrep.2017.06.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanlikilicer P., Rashed M. H., Bayraktar R., Mitra R., Ivan C., Aslan B., et al. (2016). Ubiquitous release of Exosomal tumor suppressor miR-6126 from Ovarian Cancer cells. Cancer Res. 76, 7194–7207. 10.1158/0008-5472.CAN-16-0714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S., Ridinger J., Rupp A. K., Janssen J. W., Altevogt P. (2011). Body fluid derived exosomes as a novel template for clinical diagnostics. J. Trans. Med. 9:86. 10.1186/1479-5876-9-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Kwak Y. G., Tamkun M., Majerfeld I., Yarus M. (1999). RNAs that bind and change the permeability of phospholipid membranes. Proc. Natl. Acad. Sci. U.S.A. 96, 10649–10654. 10.1073/pnas.96.19.10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452. 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X., Wang M., McElyea S. D., Sherman S., House M., Korc M. (2017). A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 393, 86–93. 10.1016/j.canlet.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque K., Halvorsen M., Abrahamyan L., Chatel-Chaix L., Poupon V., Gordon H., et al. (2006). Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 7, 1177–1193. 10.1111/j.1600-0854.2006.00461.x [DOI] [PubMed] [Google Scholar]
- Li B., Chen P., Chang Y., Qi J., Fu H., Guo H. (2016). Let-7a inhibits tumor cell growth and metastasis by directly targeting RTKN in human colon cancer. Biochem. Biophys. Res. Commun. 478, 739–745. 10.1016/j.bbrc.2016.08.018 [DOI] [PubMed] [Google Scholar]
- Li C., Yin Y., Liu X., Xi X., Xue W., Qu Y. (2017). Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget 8, 24564–24578. 10.18632/oncotarget.15596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hu K., Gong G., Zhu D., Wang Y., Liu H., et al. (2017). Upregulation of MiR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci. Rep. 7:41330. 10.1038/srep41330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zeringer E., Barta T., Schageman J., Cheng A., Vlassov A. V. (2014). Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369. 10.1098/rstb.2013.0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. W., Yang Y. M., Du L. T., Dong Z., Wang L. L., Zhang X., et al. (2014). Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med. Oncol. 31, 256. 10.1007/s12032-014-0256-5 [DOI] [PubMed] [Google Scholar]
- Liu C., Li M., Hu Y., Shi N., Yu H., Liu H., et al. (2016). miR-486-5p attenuates tumor growth and lymphangiogenesis by targeting neuropilin-2 in colorectal carcinoma. Oncol. Targets Ther. 9, 2865–2871. 10.2147/OTT.S103460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yu Z., Yuan S., Xie W., Li C., Hu Z., et al. (2017). Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 8, 13048–13058. 10.18632/oncotarget.14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Miao T., Feng T., Jiang Z., Li M., Zhou L., et al. (2015). miR-451a inhibited cell proliferation and enhanced Tamoxifen sensitive in breast cancer via macrophage migration inhibitory factor. Biomed. Res. Int. 2015:207684. 10.1155/2015/207684 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Loffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermuller J., Kretzschmar A. K., et al. (2007). Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 110, 1330–1333. 10.1182/blood-2007-03-081133 [DOI] [PubMed] [Google Scholar]
- Logozzi M., De Milito A., Lugini L., Borghi M., Calabro L., Spada M., et al. (2009). High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 4:e5219. 10.1371/journal.pone.0005219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall J., Hill A. F., Hochberg F., Buzas E. I., Di Vizio D., Gardiner C., et al. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell Vesicles 3:26913. 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Pan X., Fan P., He Y., Gu J., Wang W., et al. (2014). Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Mol. Cancer 13:118. 10.1186/1476-4598-13-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R., Jiang T., Kang X. (2012). Circulating microRNAs in cancer: origin, function and application. J. Exp. Clin. Cancer Res. 31:38. 10.1186/1756-9966-31-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang P., Wang F., Zhang H., Yang J., Peng J., et al. (2012). miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 61, 1447–1453. 10.1136/gutjnl-2011-301122 [DOI] [PubMed] [Google Scholar]
- Majka M., Janowska-Wieczorek A., Ratajczak J., Ehrenman K., Pietrzkowski Z., Kowalska M. A., et al. (2001). Numerous growth factors, cytokines, and chemokines are secreted by human CD34(+) cells, myeloblasts, erythroblasts, and megakaryoblasts and regulate normal hematopoiesis in an autocrine/paracrine manner. Blood 97, 3075–3085. 10.1182/blood.V97.10.3075 [DOI] [PubMed] [Google Scholar]
- Manterola L., Guruceaga E., Gállego Pérez-Larraya J., González-Huarriz M., Jauregui P., Tejada S., et al. (2014). A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 16, 520–527. 10.1093/neuonc/not218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk A. I., Masyuk T. V., Larusso N. F. (2013). Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J. Hepatol. 59, 621–625. 10.1016/j.jhep.2013.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes D. G., Jr., Shair K. H., Marquitz A. R., Kung C. P., Edwards R. H., Raab-Traub N. (2010). Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. U.S.A. 107, 20370–20375. 10.1073/pnas.1014194107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milane L., Singh A., Mattheolabakis G., Suresh M., Amiji M. M. (2015). Exosome mediated communication within the tumor microenvironment. J. Control. Release 219, 278–294. 10.1016/j.jconrel.2015.06.029 [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M., Gutierrez-Vazquez C., Villarroya-Beltri C., Gonzalez S., Sanchez-Cabo F., Gonzalez M. A., et al. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. 10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan L., Batte K. E., Trgovcich J., Wisler J., Marsh C. B., Piper M. (2014). Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 18, 371–390. 10.1111/jcmm.12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro T. P., Magee R. J., Kidd G. J., Carson J. H., Barbarese E., Smith L. M., et al. (1999). Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274, 34389–34395. 10.1074/jbc.274.48.34389 [DOI] [PubMed] [Google Scholar]
- Munson P., Shukla A. (2015). Exosomes: potential in cancer diagnosis and therapy. Medicines 2, 310–327. 10.3390/medicines2040310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E. J., Yoon H., Kim S. W., Kim H., Kim Y. T., Kim J. H., et al. (2008). MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 14, 2690–2695. 10.1158/1078-0432.CCR-07-1731 [DOI] [PubMed] [Google Scholar]
- Neerincx M., Sie D. L., Van De Wiel M. A., Van Grieken N. C., Burggraaf J. D., Dekker H., et al. (2015). MiR expression profiles of paired primary colorectal cancer and metastases by next-generation sequencing. Oncogenesis 4, 170. 10.1038/oncsis.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neviani P., Fabbri M. (2015). Exosomic microRNAs in the tumor microenvironment. Front. Med. 2:47. 10.3389/fmed.2015.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu K., Shen W., Zhang Y., Zhao Y., Lu Y. (2015). MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene 574, 330–336. 10.1016/j.gene.2015.08.017 [DOI] [PubMed] [Google Scholar]
- Nolte-'T Hoen E. N., Buermans H. P., Waasdorp M., Stoorvogel W., Wauben M. H., Hoen P. A. (2012). Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40, 9272–9285. 10.1093/nar/gks658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata-Kawata H., Izumiya M., Kurioka D., Honma Y., Yamada Y., Furuta K., et al. (2014). Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9:e92921. 10.1371/journal.pone.0092921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye I. S., Coomes S. M., Pelly V. S., Czieso S., Papayannopoulos V., Tolmachova T., et al. (2014). MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41, 89–103. 10.1016/j.immuni.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladini L., Fabris L., Bottai G., Raschioni C., Calin G. A., Santarpia L. (2016). Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 35, 103. 10.1186/s13046-016-0375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., Van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., et al. (2010). Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333. 10.1073/pnas.0914843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Hernandez D., Gutierrez-Vazquez C., Jorge I., Lopez-Martin S., Ursa A., Sanchez-Madrid F., et al. (2013). The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 288, 11649–11661. 10.1074/jbc.M112.445304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigati L., Yaddanapudi S. C., Iyengar R., Kim D. J., Hearn S. A., Danforth D., et al. (2010). Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 5:e13515. 10.1371/journal.pone.0013515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard C. C., Cheng H. H., Tewari M. (2012a). MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 13, 358–369. 10.1038/nrg3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard C. C., Kroh E., Wood B., Arroyo J. D., Dougherty K. J., Miyaji M. M., et al. (2012b). Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. 5, 492–497. 10.1158/1940-6207.CAPR-11-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purrello M., Di Pietro C., Viola A., Rapisarda A., Stevens S., Guermah M., et al. (1998). Genomics and transcription analysis of human TFIID. Oncogene 16, 1633–1638. [DOI] [PubMed] [Google Scholar]
- Que R., Ding G., Chen J., Cao L. (2013). Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 11:219. 10.1186/1477-7819-11-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M., Barbagallo C., Statello L., Caltabiano R., Russo A., Puzzo L., et al. (2015a). miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications. Cancer Biol. Ther. 16, 1387–1396. 10.1080/15384047.2015.1046021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M., Barbagallo D., Purrello M. (2015b). Exosomes: nanoshuttles to the future of biomedicine. Cell Cycle 14, 289–290. 10.1080/15384101.2015.1006535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M., Statello L., Maugeri M., Barbagallo C., Passanisi R., Alhamdani M. S., et al. (2014). Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: biomolecular, genetic and translational implications. Oncoscience 1, 132–157. 10.18632/oncoscience.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reclusa P., Sirera R., Araujo A., Giallombardo M., Valentino A., Sorber L., et al. (2016). Exosomes genetic cargo in lung cancer: a truly Pandora's box. Transl. Lung Cancer Res. 5, 483–491. 10.21037/tlcr.2016.10.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenfeld A. L., Baek R., Nielsen M. H., Stensballe A., Varming K., Jorgensen M. (2014). Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin. Ther. 36, 830–846. 10.1016/j.clinthera.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Ribas J., Ni X., Haffner M., Wentzel E. A., Salmasi A. H., Chowdhury W. H., et al. (2009). miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 69, 7165–7169. 10.1158/0008-5472.CAN-09-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristorcelli E., Beraud E., Verrando P., Villard C., Lafitte D., Sbarra V., et al. (2008). Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J. 22, 3358–3369. 10.1096/fj.07-102855 [DOI] [PubMed] [Google Scholar]
- Rustom A., Saffrich R., Markovic I., Walther P., Gerdes H. H. (2004). Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010. 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., et al. (2016). The RNA-binding protein SYNCRIP Is a component of the hepatocyte Exosomal machinery controlling MicroRNA sorting. Cell Rep. 17, 799–808. 10.1016/j.celrep.2016.09.031 [DOI] [PubMed] [Google Scholar]
- Sempere L. F., Christensen M., Silahtaroglu A., Bak M., Heath C. V., Schwartz G., et al. (2007). Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 67, 11612–11620. 10.1158/0008-5472.CAN-07-5019 [DOI] [PubMed] [Google Scholar]
- Sherer N. M., Mothes W. (2008). Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends Cell Biol. 18, 414–420. 10.1016/j.tcb.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz P. (1972). Biological membranes: the dynamics of their organization. Annu. Rev. Physiol. 34, 117–140. 10.1146/annurev.ph.34.030172.001001 [DOI] [PubMed] [Google Scholar]
- Silva M., Melo S. A. (2015). Non-coding RNAs in Exosomes: new players in cancer biology. Curr. Genomics 16, 295–303. 10.2174/1389202916666150707154719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Wurdinger T., Van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O., Svoboda M., Fabian P., Smerdova T., Knoflickova D., Bednarikova M., et al. (2007). Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72, 397–402. 10.1159/000113489 [DOI] [PubMed] [Google Scholar]
- Soria F. N., Pampliega O., Bourdenx M., Meissner W. G., Bezard E., Dehay B. (2017). Exosomes, an unmasked culprit in neurodegenerative diseases. Front. Neurosci. 11:26. 10.3389/fnins.2017.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung Y. H., Ford S., Zhang V., Chung J. (2017). Exosomes in cancer diagnostics. Cancers 9:8. 10.3390/cancers9010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito M. L., Baer C., Burdet F., Maderna C., Gilfillan G. D., Lyle R., et al. (2014). Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 8, 1432–1446. 10.1016/j.celrep.2014.07.035 [DOI] [PubMed] [Google Scholar]
- Stuffers S., Sem Wegner C., Stenmark H., Brech A. (2009). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10, 925–937. 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- Tadano T., Kakuta Y., Hamada S., Shimodaira Y., Kuroha M., Kawakami Y., et al. (2016). MicroRNA-320 family is downregulated in colorectal adenoma and affects tumor proliferation by targeting CDK6. World J. Gastrointest. Oncol. 8, 532–542. 10.4251/wjgo.v8.i7.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro B. J., Greening D. W., Mathias R. A., Ji H., Mathivanan S., Scott A. M., et al. (2012). Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304. 10.1016/j.ymeth.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Taylor D. D., Gercel-Taylor C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. 10.1016/j.ygyno.2008.04.033 [DOI] [PubMed] [Google Scholar]
- Taylor D. D., Shah S. (2015). Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 87, 3–10. 10.1016/j.ymeth.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Thery C., Zitvogel L., Amigorena S. (2002). Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579. 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- Tian H., Li W. (2017). Dendritic cell-derived exosomes for cancer immunotherapy: hope and challenges. Ann. Transl. Med. 5, 221. 10.21037/atm.2017.02.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Shan W., Zhang Y., Lv X., Li X., Wei C. (2016). Up-regulation of miR-21 expression predicate advanced clinicopathological features and poor prognosis in patients with non-small cell lung cancer. Pathol. Oncol. Res. 22, 161–167. 10.1007/s12253-015-9979-7 [DOI] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., et al. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Tsochandaridis M., Nasca L., Toga C., Levy-Mozziconacci A. (2015). Circulating microRNAs as clinical biomarkers in the predictions of pregnancy complications. Biomed. Res. Int. 2015:294954. 10.1155/2015/294954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto M., Iinuma H., Yagi T., Matsuda K., Hashiguchi Y. (2017). Circulating Exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 92, 360–370. 10.1159/000463387 [DOI] [PubMed] [Google Scholar]
- Ungefroren H., Sebens S., Seidl D., Lehnert H., Hass R. (2011). Interaction of tumor cells with the microenvironment. Cell Commun. Signal. 9:18. 10.1186/1478-811X-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Van Balkom B. W., Eisele A. S., Pegtel D. M., Bervoets S., Verhaar M. C. (2015). Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell Vesicles 4:26760. 10.3402/jev.v4.26760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun J., Mestdagh P., Sormunen R., Cocquyt V., Vermaelen K., Vandesompele J., et al. (2014). The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell Vesicles 3. 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., et al. (2011). The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 21, 708–721. 10.1016/j.devcel.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij F. J., Van Eijndhoven M. A., Hopmans E. S., Vendrig T., Wurdinger T., Cahir-McFarland E., et al. (2011). LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-kappaB activation. EMBO J. 30, 2115–2129. 10.1038/emboj.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S., Thery C., Ploix S., Tursz T., Lapierre V., Lantz O., et al. (2010). Dendritic cell-derived exosomes for cancer immunotherapy: what's next? Cancer Res. 70, 1281–1285. 10.1158/0008-5472.CAN-09-3276 [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Baixauli F., Gutierrez-Vazquez C., Sanchez-Madrid F., Mittelbrunn M. (2014). Sorting it out: regulation of exosome loading. Semin. Cancer Biol. 28, 3–13. 10.1016/j.semcancer.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980. 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnubalaji R., Hamam R., Yue S., Al-Obeed O., Kassem M., Liu F. F., et al. (2016). MicroRNA-320 suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1. Oncotarget 7, 35789–35802. 10.18632/oncotarget.8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov A., Khvorova A., Yarus M. (2001). Binding and disruption of phospholipid bilayers by supramolecular RNA complexes. Proc. Natl. Acad. Sci. U.S.A. 98, 7706–7711. 10.1073/pnas.141041098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang H., Yang Z. (2012). MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS ONE 7:e47053. 10.1371/journal.pone.0047053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. F., Zhang X. J., Yan Y. R., Zhu X. H., Yu J., Ding Y., et al. (2017). FBX8 is a metastasis suppressor downstream of miR-223 and targeting mTOR for degradation in colorectal carcinoma. Cancer Lett. 388, 85–95. 10.1016/j.canlet.2016.11.031 [DOI] [PubMed] [Google Scholar]
- Wang J., Chen J., Sen S. (2016). MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 231, 25–30. 10.1002/jcp.25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yan F., Zhao Q., Zhan F., Wang R., Wang L., et al. (2017). Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 7, 4150. 10.1038/s41598-017-04386-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zeng Y., Zhou J. M., Nie S. L., Peng Q., Gong J., et al. (2016). MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction. Mol. Med. Rep. 13, 273–280. 10.3892/mmr.2015.4557 [DOI] [PubMed] [Google Scholar]
- Wei J., Zhang L., Li J., Zhu S., Tai M., Mason C. W., et al. (2017). MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J. Ovarian Res. 10, 33. 10.1186/s13048-017-0328-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside T. L. (2017). Extracellular vesicles isolation and their biomarker potential: are we ready for testing? Ann. Transl. Med. 5:54. 10.21037/atm.2017.01.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild K., Sinning I., Cusack S. (2001). Crystal structure of an early protein-RNA assembly complex of the signal recognition particle. Science 294, 598–601. 10.1126/science.1063839 [DOI] [PubMed] [Google Scholar]
- Wu G., Sinclair C., Hinson S., Ingle J. N., Roche P. C., Couch F. J. (2001). Structural analysis of the 17q22-23 amplicon identifies several independent targets of amplification in breast cancer cell lines and tumors. Cancer Res. 61, 4951–4955. Available online at: http://cancerres.aacrjournals.org/content/61/13/4951.long [PubMed] [Google Scholar]
- Xiaohong Z., Lichun F., Na X., Kejian Z., Xiaolan X., Shaosheng W. (2016). MiR-203 promotes the growth and migration of ovarian cancer cells by enhancing glycolytic pathway. Tumour Biol. 37, 14989–14997. 10.1007/s13277-016-5415-1 [DOI] [PubMed] [Google Scholar]
- Xue X., Liu Y., Wang Y., Meng M., Wang K., Zang X., et al. (2016). MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 7, 84508–84519. 10.18632/oncotarget.13022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Han B., Gao S., Wang X., Wang Z., Wang F., et al. (2017). Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 8, 60149–60158. 10.18632/oncotarget.18557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Mo M., Barreiro O., Gordon-Alonso M., Sala-Valdes M., Sanchez-Madrid F. (2009). Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446. 10.1016/j.tcb.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Yong F. L., Law C. W., Wang C. W. (2013). Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 13:280. 10.1186/1471-2407-13-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong F. L., Wang C. W., Roslani A. C., Law C. W. (2014). The involvement of miR-23a/APAF1 regulation axis in colorectal cancer. Int. J. Mol. Sci. 15, 11713–11729. 10.3390/ijms150711713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Harris S. L., Levine A. J. (2006). The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 66, 4795–4801. 10.1158/0008-5472.CAN-05-4579 [DOI] [PubMed] [Google Scholar]
- Yuyama K., Sun H., Mitsutake S., Igarashi Y. (2012). Sphingolipid-modulated exosome secretion promotes clearance of amyloid-beta by microglia. J. Biol. Chem. 287, 10977–10989. 10.1074/jbc.M111.324616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Luo X., Li H., Yue X., Deng L., Cui Y., et al. (2014). MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol. Rep. 32, 115–120. 10.3892/or.2014.3173 [DOI] [PubMed] [Google Scholar]
- Zhong S., Chen X., Wang D., Zhang X., Shen H., Yang S., et al. (2016). MicroRNA expression profiles of drug-resistance breast cancer cells and their exosomes. Oncotarget 7, 19601–19609. 10.18632/oncotarget.7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiao Z., Ji J., Li S., Huang X., Lu X., et al. (2017). Characterization of mouse serum exosomal small RNA content: The origins and their roles in modulating inflammatory response. Oncotarget 8, 42712–42727. 10.18632/oncotarget.17448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Regnault A., Lozier A., Wolfers J., Flament C., Tenza D., et al. (1998). Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 4, 594–600. 10.1038/nm0598-594 [DOI] [PubMed] [Google Scholar]