Abstract
Hepatitis B virus X protein (HBx) is a multifunctional protein that interacts directly with many host proteins. For example, HBx interacts with anti-apoptotic proteins, Bcl-2 and Bcl-xL, through its BH3-like motif, which leads to elevated cytosolic calcium levels, efficient viral DNA replication and the induction of apoptosis. To facilitate sample preparation and perform detailed structural characterization of the complex between HBx and Bcl-xL, we designed and purified a recombinant HBx BH3-like motif-linker-Bcl-xL fusion protein produced in E. coli. The fusion protein was characterized by size exclusion chromatography, circular dichroism and nuclear magnetic resonance experiments. Our results show that the fusion protein is a monomer in aqueous solution, forms a stable intramolecular complex, and likely retains the native conformation of the complex between Bcl-xL and the HBx BH3-like motif. Furthermore, the HBx BH3-like motif of the intramolecular complex forms an α-helix. These observations indicate that the fusion protein should facilitate structural studies aimed at understanding the interaction between HBx and Bcl-xL at the atomic level.
Abbreviations: Bcl-2, B-cell lymphoma 2; Bcl-xL, B-cell lymphoma extra large; BH3, Bcl-2 homology 3; HBV, hepatitis B virus; HBx, hepatitis B virus X protein; HCC, hepatocellular carcinoma; HSQC, heteronuclear single quantum coherence
Keywords: BH3-like motif, Bcl-xL, Fusion protein, HBx, Purification, Structural characterization
Highlights
-
•
Soluble HBx BH3-like motif-linker-Bcl-xL fusion protein was produced in E. coli.
-
•
The fusion protein behaves as a monomer and forms a stable intramolecular complex.
-
•
The HBx BH3-like motif of the fusion protein forms an α-helix.
-
•
The fusion protein likely retains the native conformation of the complex.
1. Introduction
Hepatitis B virus X protein (HBx, 154 residues) is a multifunctional protein that is essential for viral replication and the development of liver diseases, such as hepatocellular carcinoma (HCC) [1], [2]. HBx possesses a regulatory domain at the N-terminus and a transactivation domain at the C-terminus [3]. HBx interacts directly with many host proteins through functional regions or motifs [4], [5], [6]. For example, a BH3-like motif of HBx interacts with anti-apoptotic Bcl-2 family proteins (e.g., Bcl-2 and Bcl-xL) that are important regulators of apoptotic cell death [7], [8]. This interaction between HBx and anti-apoptotic Bcl-2 family proteins results in elevated cytosolic calcium levels, efficient viral DNA replication and apoptosis.
Many structural studies of the BH3 peptide/Bcl-2 (or Bcl-xL) complex have demonstrated that the BH3 peptides form an amphipathic α-helix and hydrophobic residues of the BH3 peptides fit into a common hydrophobic groove of Bcl-2 or Bcl-xL [9], [10], [11], [12], [13]. Recently, Jiang et al. reported the structure of the HBx BH3-like motif (residues 110–135)/Bcl-2 complex and revealed that, similar to other BH3 peptides, the HBx BH3-like motif binds to the common hydrophobic groove of Bcl-2, but in a unique manner [14]. In addition, the BH3-like motif of HBx (26 residues) has a remarkably reduced binding affinity (Kd=~193 µM) to Bcl-2 [14] when compared with the binding affinities of other BH3 peptides to Bcl-xL, such as Bak (16 residues, Kd=~0.34 µM) [9], Bad (25 residues, Kd=~0.0006 µM) [10], Beclin-1 (28 residues, Kd=~1.4 µM) [12] and PUMA (25 residues, Kd=~0.003 µM) [13]. On the other hand, it is still unclear whether the BH3-like motif of HBx also binds to Bcl-xL in a similar manner, as observed in the HBx BH3-like motif/Bcl-2 complex structure. Therefore, we are focusing on structural studies of the interaction between the HBx BH3-like motif and Bcl-xL.
For structural studies of protein–protein and protein–peptide interactions, fusion proteins have often been used because these fusion proteins can form stable complexes that facilitate sample preparation and structural analysis [12], [15], [16]. In these fusion proteins, two molecules A and B are covalently connected by a flexible linker, such as consecutive glycine and serine (GS) dipeptides [12], [15]. For example, the Beclin-1 BH3 motif and Bcl-xL have been covalently linked by five repeats of the GS dipeptide, and the 1:1 complex structure was determined by NMR [12], whereas the neuropilin-1 b1 domain and VEGF-A heparin binding domain were covalently linked by three repeats of the GS dipeptide, and the structure of this complex was solved at 2.65 Å resolution by X-ray crystallography [15]. Thus, the fusion protein strategy provides a powerful tool for structural studies of protein–protein and protein–peptide interactions.
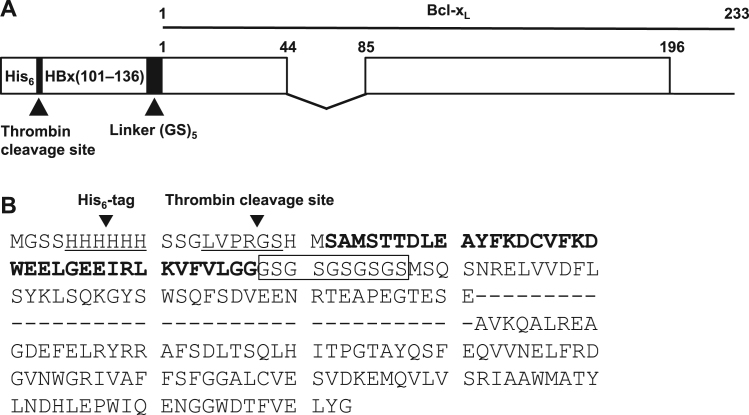
In this study, we designed, purified and characterized a fusion protein of the HBx BH3-like motif and Bcl-xL connected with five repeats of the GS dipeptide, HBx BH3-like motif-linker-Bcl-xL (Fig. 1). The BH3-like motif consists of residues 101–136 of HBx [17], and residue sequences 1–44 and 85–196 of Bcl-xL were used [12]. This fusion protein was successfully expressed in E. coli cells and purified by metal affinity and anion exchange chromatographies. Here, we demonstrate that the HBx BH3-like motif-linker-Bcl-xL fusion protein should be suitable for structural studies of this complex.
Fig. 1.
(A) Schematic representation of HBx(101−136)-(GS)5-Bcl-xL fusion protein used in this study. (B) Amino acid sequence of the fusion protein. The first sequence of underlined residues is the His6-tag and the second sequence of underlined residues is the thrombin cleavage site. The rectangle indicates the five-repeat GS dipeptide linker. The sequence of HBx(101−136) is shown in bold. Bcl-xL in this fusion protein lacks residues 45–84 of a long flexible loop (shown as dashes) and residues 197–233 at the C-terminus.
2. Materials and methods
2.1. Plasmid construction of Bcl-xL and HBx(101−136)-(GS)5-Bcl-xL
The Bcl-xL (residues 1–44 and 85–196) used in this study lacks residues 45–84 of a long flexible loop and residues 197–233 at the C-terminus [12]. First, the gene of Bcl-xL(1−196) was amplified using reverse transcribed cDNA from total HeLa RNA by the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) as a template using the F1 forward primer (5′-GGCCATATGTCTCAGAGCAACCGGGAGCTG-3′) and the R1 reverse primer (5′-GGCCTCGAGTCACCCATAGAGTTCCACAAAAGTATCCCA-3′) (underlined sequences indicate the NdeI and XhoI recognition sites, respectively). The amplified PCR product of Bcl-xL(1−196) was digested with NdeI and XhoI, and then ligated into a pET15b vector (Novagen). The resulting plasmid was named pET15b-Bcl-xL(1−196). The gene of Bcl-xL(1−196) was verified by sequencing. Next, a deletion gene of the long loop (residues 45–84) of Bcl-xL(1−196) was prepared using a KOD-Plus-Mutagenesis Kit (Toyobo) with the above plasmid, pET15b-Bcl-xL(1−196), as a template using the primers, 5′-GCAGTAAAGCAAGCGCTGAGGGAGGCA-3′ and 5′- CTCCGATTCAGTCCCTTCTGGGGCCTC-3′. The gene lacking the long loop (residues 45–84) of Bcl-xL(1−196), i.e., the gene of Bcl-xL(residues 1–44 and 85–196), was verified by sequencing. In this report, we refer to Bcl-xL(residues 1–44 and 85–196) as Bcl-xL, and the resulting plasmid was named pET15b-Bcl-xL.
The gene of HBx(101−136)-(GS)5-Bcl-xL was constructed by connecting the sequences of 101–136 of HBx and Bcl-xL with five repeats of the GS dipeptide. First, the gene of (GS)5-Bcl-xL was amplified using the pET15b-Bcl-xL plasmid as a template with the F2 forward primer (5′- GGCCATATGGGCAGCGGCAGCGGCAGCGGCAGCGGCAGCATGTCTCAGAGCAACCGGGAGCTG-3′) and the R1 reverse primer. Next, the gene of HBx(101−136)-(GS)5-Bcl-xL was amplified using the above PCR product, (GS)5-Bcl-xL, as a template with the F3 forward primer (5′-GGCCATATGTCAGCGATGTCAACGACCGACCTTGAGGCGTACTTCAAAGACTGTGTGTTTAAAGACTGGGAGGAGTTGGGGGAGGAGATTAGGTTAAAGGTCTTTGTACTAGGAGGCGGCAGCGGCAGCGGC-3′) and the R1 reverse primer. The amplified PCR product of HBx(101−136)-(GS)5-Bcl-xL was digested with NdeI and XhoI, and then ligated into the pET15b vector. The resulting plasmid was named pET15b-HBx(101−136)-(GS)5-Bcl-xL. The gene of HBx(101−136)-(GS)5-Bcl-xL was verified by sequencing.
2.2. Expression and purification of HBx(101−136)-(GS)5-Bcl-xL, Bcl-xL and HBx(101−136)
HBx(101−136)-(GS)5-Bcl-xL was expressed in E. coli BL21(DE3) pLysS cells as a hexahistidine (His6)-tagged protein at the N-terminus. LB media were used to prepare a non-labeled protein. M9 minimal media containing 15NH4Cl as the sole nitrogen source was used to prepare uniformly 15N-labeled protein. The E. coli cells were grown in LB or M9 medium containing 100 µg/mL ampicillin and 17 µg/mL chloramphenicol at 30 or 37 °C until the cells reached an optical density of 0.6–0.8. The expression of the target protein was induced with 0.2 mM isopropyl β-D-thiogalactopyranoside for 16–20 h at 25 °C. The E. coli cells were harvested and stored at −25 °C until further use.
The E. coli expressing His6-HBx(101−136)-(GS)5-Bcl-xL from 4-L culture was suspended in 120–160 mL of lysis buffer [50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1 mM dithiothreitol (DTT), 10 mM imidazole and 10% glycerol] supplemented with 0.5% Triton-X100 and 0.1 mg/mL lysozyme and incubated for 30–60 min at room temperature, followed by sonication. The supernatant containing the His6-tagged protein, which was obtained by centrifugation, was loaded onto a COSMOGEL His-accept column (3–4 mL bed volume) (Nacalai Tesque Inc.). After washing the column with 25–33 column volumes of lysis buffer, the His6-tagged protein was eluted with lysis buffer containing 0.5 M imidazole. The flow-through of the first column was further loaded onto the COSMOGEL His-accept column and the His6-tagged protein was purified as described above. The resulting protein was pooled and dialyzed against dialysis buffer [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM DTT and 10% glycerol] and then cleaved with thrombin overnight at 4 °C. The resulting sample contains four extra vector-derived residues (Gly-Ser-His-Met) at the N-terminus. To stop the cleavage reaction, serine protease inhibitor, Pefabloc SC (Roche), was added into the sample solution. The sample containing HBx(101−136)-(GS)5-Bcl-xL was concentrated to approximately 5 mL by an Amicon Ultra-15 centrifugal filter unit (NMWL 3 kDa) (Merck-Millipore), and the concentrated sample was further purified with a Resource Q anion exchange column (1 mL bed volume) (GE Healthcare) by a linear gradient from 0 to 0.6 M NaCl in a buffer [50 mM Tris-HCl (pH 8.0) and 1 mM DTT]. Finally, the buffer was exchanged to NMR buffer [50 mM potassium phosphate (pH 6.8), 50 mM NaCl and 1 mM DTT] using a PD10 column (GE Healthcare).
Both non- and 15N-labeled Bcl-xL proteins were purified using a procedure that is similar to the one used to purify HBx(101−136)-(GS)5-Bcl-xL. The Bcl-xL protein contains three extra vector-derived residues (Gly-Ser-His) at the N-terminus. In addition, HBx(101−136) was purified as described previously [17].
All the proteins and peptides purified were stored at −85 °C until further use. The protein or peptide concentration was determined by Nanodrop 2000 UV–Vis spectrometer (Thermo Scientific), using UV absorbance at 280 nm with the following extinction coefficients and molecular weights: HBx(101−136)-(GS)5-Bcl-xL, 43,430 M−1 cm−1 and 23.1 kDa; Bcl-xL, 36,440 M−1 cm−1 and 18.2 kDa; HBx(101−136), 6,990 M−1 cm−1 and 4.1 kDa.
2.3. Size exclusion chromatography
Both HBx(101−136)-(GS)5-Bcl-xL fusion protein (0.1 mg) and Bcl-xL (0.1 mg) were applied onto a Superdex-200 (10/300 GL) gel filtration column (GE Healthcare) equilibrated with gel filtration buffer [50 mM potassium phosphate (pH 6.8), 150 mM NaCl and 1 mM DTT] at a flow rate of 0.5 mL min−1 at 4 °C. The eluate was monitored at 280 nm. The molecular weights of HBx(101−136)-(GS)5-Bcl-xL and Bcl-xL were estimated using a Gel Filtration Calibration Kit LMW (GE Healthcare): conalbumin, 75 kDa: ovalbumin, 43 kDa; carbonic anhydrase, 29 kDa; ribonuclease A, 13.7 kDa; aprotinin, 6.5 kDa.
2.4. CD spectroscopy
CD spectra were measured on a JASCO-820 spectropolarimeter at 25, 35 and 45 °C with a 1-mm path length cuvette. The temperatures were selected based on the high thermal stability of the Bcl-xL protein: the Tm values of Bcl-xL (residues 1–207) are from 70–77 °C at pH 4–8 [18]. CD data were recorded from 260 to 198 nm using approximately 9 µM HBx(101−136)-(GS)5-Bcl-xL or Bcl-xL in a buffer composed of 5 mM potassium phosphate (pH 6.8), 5 mM NaCl and 0.1 mM DTT. All data were obtained by the subtraction of a blank corresponding to the buffer. Mean residue molar ellipticity [θ’] was calculated according to the protein concentration, the number of residues in the protein and the cuvette path length. The helical contents of HBx(101−136)-(GS)5-Bcl-xL and Bcl-xL were estimated using the average of two experiments by the Chen and Yang method [19], the Helix-coil model method [10], [20], [21] and the program K2D3 [22]. In the Chen and Yang method, the following equation was used:
| (1) |
where [θ’]222 is the mean residue molar ellipticity [θ’] at 222 nm.
In the Helix-coil model method, the following equation was used:
| (2) |
where [θ’]helix and [θ’]coil represent the mean residue molar ellipticity of a complete helix [–42,500(1−(3/n)) where n=number of residues in the protein] and a complete random coil (+640), respectively.
The helical contents of both molecules were also estimated by the program K2D3 [22], using θ’ values in a range between 200 and 240 nm.
2.5. NMR spectroscopy
NMR experiments were acquired using a Bruker AVANCE III 500 spectrometer. The NMR sample contained 0.3 mM 15N-labeled HBx(101−136)-(GS)5-Bcl-xL, 0.3 mM 15N-lableled Bcl-xL, or 0.3 mM 15N-lableled Bcl-xL with 0.45 mM non-labeled HBx(101−136), in NMR buffer supplemented with 0.5 mM DSS and 10% D2O. The 2D 1H–15N HSQC spectra [23], [24] were recorded with the following acquisition times 128 ms (1H) and 56 ms (15N) at 25, 35 and 45 °C. Proton chemical shifts were referenced directly to DSS at 0 ppm, and 15N chemical shifts were referenced indirectly to the absolute frequency ratio 15N/1H =0.101329118 [25]. These data were processed using TopSpin 3.2 and analyzed by Sparky ver. 3.115 [26].
3. Results and discussion
3.1. Construction and purification of HBx(101−136)-(GS)5-Bcl-xL
The construct of HBx(101−136)-(GS)5-Bcl-xL used in this study contains the following features (Fig. 1). There is a His6-tag before HBx(101−136), and a five repeat GS dipeptide linker is located between HBx(101−136) and Bcl-xL(residues 1–44 and 85–196). Additionally, to remove the His6-tag from His6-HBx(101−136)-(GS)5-Bcl-xL, a thrombin cleavage site is placed between the His6-tag and HBx(101−136).
Fig. 2 shows the SDS-PAGE result of each purification step of HBx(101−136)-(GS)5-Bcl-xL. The amount of His6-tagged protein obtained was 3–4 mg per liter of LB or M9 culture after purification by the COSMOGEL His-accept column (lane 3). In the SDS-PAGE analysis, the fusion protein was detected as a molecular weight of ~25 kDa, which is consistent with its theoretical molecular weight (25 kDa). After dialysis, the His6-tag was cleaved from His6-HBx(101−136)-(GS)5-Bcl-xL by thrombin overnight digestion at 4 °C. Removal of the His6-tag was confirmed (lane 5), and then the resulting sample was purified using the Resource Q anion exchange column with a linear gradient from 0 to 0.6 M NaCl. The HBx(101−136)-(GS)5-Bcl-xL fusion protein eluted at ~0.3 M NaCl and the yield of the fusion protein was ~1 mg per liter of LB or M9 culture. The pooled protein was passed through a PD10 column to exchange the buffer to NMR buffer. Finally, non- and 15N-labeled samples were obtained with a final yield of ~0.7 mg per liter of culture. The HBx(101−136)-(GS)5-Bcl-xL fusion protein was purified to >95% homogeneity, as judged by SDS-PAGE (lanes 7–9).
Fig. 2.
SDS-PAGE analysis of the protein fractions collected at each step of purification of HBx(101−136)-(GS)5-Bcl-xL. Lane 1, molecular weight marker; lane 2, supernatant; lane 3, His6-HBx(101−136)-(GS)5-Bcl-xL purified by a COSMOGEL His-accept column; lane 4, His6-HBx(101−136)-(GS)5-Bcl-xL after dialysis; lane 5, HBx(101−136)-(GS)5-Bcl-xL after cleavage of the His6-tag by thrombin; lane 6, HBx(101−136)-(GS)5-Bcl-xL purified by a Resource Q anion exchange column; lanes 7–9, purified HBx(101−136)-(GS)5-Bcl-xL (1-, 3- and 5-µg loadings, respectively).
3.2. Size exclusion chromatography analysis
To determine whether the purified HBx(101−136)-(GS)5-Bcl-xL fusion protein is monomeric or forms oligomers in solution, we performed size exclusion chromatography of the fusion protein and Bcl-xL (Fig. 3). HBx(101−136)-(GS)5-Bcl-xL showed a single peak (Fig. 3A, middle), and its molecular weight was determined to be ~30.5 kDa (Fig. 3B), which is 1.3 times larger than the theoretical molecular weight (23.1 kDa) of this fusion protein. On the other hand, Bcl-xL was estimated to have a molecular weight of ~22.2 kDa, which is 1.2 times larger than its theoretical molecular weight (18.2 kDa). These results suggest that the purified HBx(101−136)-(GS)5-Bcl-xL and Bcl-xL exist as monomers under these conditions. This is consistent with the results that the Beclin-1-(GS)5-Bcl-xL fusion protein and Bcl-xL were determined to be monomeric in solution based on size exclusion chromatography and analytical ultracentrifugation, respectively [12], [27].
Fig. 3.
Size exclusion chromatography analysis of HBx(101−136)-(GS)5-Bcl-xL and Bcl-xL using a Superdex-200 gel filtration column. (A) The chromatographic profiles of the five standard proteins (top), HBx(101−136)-(GS)5-Bcl-xL (middle) and Bcl-xL (bottom) are shown. (B) The molecular weights of HBx(101−136)-(GS)5-Bcl-xL (H) and Bcl-xL (B) were estimated to be ~30.5 kDa and ~22.2 kDa, respectively, from the elution profiles of the standard proteins used: conalbumin (C), 75 kDa; ovalbumin (O), 43 kDa; carbonic anhydrase (CA), 29 kDa; ribonuclease A (R), 13.7 kDa; aprotinin (A), 6.5 kDa.
3.3. CD analysis
At each temperature the CD spectra of the HBx(101−136)-(GS)5-Bcl-xL fusion protein and Bcl-xL exhibited well-defined double minima at 208 and 222 nm (Fig. 4), which is characteristic of an α-helical conformation. These results are in good agreement with the structures of Bcl-xL[27] and the BH3-peptide/Bcl-xL complex [9], [10], [11], [12], [13], both of which mainly consist of α-helices. The overall structures of the fusion protein and Bcl-xL were not affected largely by high temperature, i.e., 45 °C. The helical contents of the fusion protein and Bcl-xL estimated by three different methods (see Materials and methods) are summarized in Table 1. For example, using the program K2D3, the helical content of HBx(101−136)-(GS)5-Bcl-xL was estimated to be 46.7% (which corresponds to 96 residues in α-helices) at 25 °C, 45.3% (93 residues in α-helices) at 35 °C and 43.3% (89 residues in α-helices) at 45 °C, while that of Bcl-xL was estimated to be 51.0% (81 residues in α-helices) at 25 °C, 49.6% (79 residues in α-helices) at 35 °C and 47.8% (76 residues in α-helices) at 45 °C. The helical content of the fusion protein is lower than that of Bcl-xL at each temperature and by each method. This is probably because the HBx(101−136)-(GS)5 part of the fusion protein adopts a non-helical conformation more than a helical conformation in this fusion protein. On the other hand, the number of residues in α-helices in the fusion protein increases by 11–15 (Table 1). It should be noted that this increase in number of residues in α-helices is similar to the number of residues of the HBx BH3-like motif (17 residues from D114 to K130), which form an α-helix in the HBx BH3-like motif/Bcl-2 complex structure [14]. In addition, many structures of BH3 peptides/Bcl-xL have revealed that the BH3 peptides form an α-helical conformation in the Bcl-xL bound state [9], [10], [11], [12], [13] and that α-helix formation of the Bcl-xL protein is not induced by binding the BH3 peptides [9], [10], [11], [12], [13]. Therefore, these results strongly suggest that the HBx(101−136) part of the fusion protein binds to the Bcl-xL part and forms an α-helix.
Fig. 4.
CD spectra of 9 µM HBx(101−136)-(GS)5-Bcl-xL (A) and 9 µM Bcl-xL (B) in 5 mM potassium phosphate (pH 6.8), 5 mM NaCl and 0.1 mM DTT at 25 (black), 35 (blue) and 45 °C (red).
Table 1.
Estimation of the secondary structure of the HBx(101–136)-(GS)5-Bcl-xL fusion protein and Bcl-xL.
| Helical Content (%) |
α-Helical residuesa |
||||||
|---|---|---|---|---|---|---|---|
| 25 °C | 35 °C | 45 °C | 25 °C | 35 °C | 45 °C | ||
| Chen & Yang | Fusion protein | 51.3 | 49.2 | 47.1 | 106 | 101 | 97 |
| Bcl-xL | 57.9 | 56.2 | 54.1 | 92 | 89 | 86 | |
| HBx(101–136)b | − | − | − | 14 | 12 | 11 | |
| helix-coil model | Fusion protein | 43.6 | 42.1 | 40.6 | 90 | 87 | 84 |
| Bcl-xL | 48.5 | 47.3 | 45.8 | 77 | 75 | 73 | |
| HBx(101–136)b | − | − | − | 13 | 12 | 11 | |
| K2D3 | Fusion protein | 46.7 | 45.3 | 43.3 | 96 | 93 | 89 |
| Bcl-xL | 51.0 | 49.6 | 47.8 | 81 | 79 | 76 | |
| HBx(101–136)b | − | − | − | 15 | 14 | 13 | |
The number of residues in α-helices of each protein was calculated by multiplying the helical content by the residue number of the HBx(101–136)-(GS)5-Bcl-xL fusion protein (206 residues) or Bcl-xL (159 residues).
The number of residues in α-helices of HBx(101–136) in the Bcl-xL bound state was estimated by subtraction of the number of α-helical residues of Bcl-xL from that of the HBx(101–136)-(GS)5-Bcl-xL fusion protein.
3.4. NMR analysis
To examine whether the HBx(101−136)-(GS)5-Bcl-xL fusion protein is suitable for structural studies of the interaction between HBx(101−136) and Bcl-xL, we acquired NMR data of 15N-labeled HBx(101−136)-(GS)5-Bcl-xL alone and 15N-labeled Bcl-xL in the absence and presence of non-labeled HBx(101−136) at a molar ratio of 1:1.5.
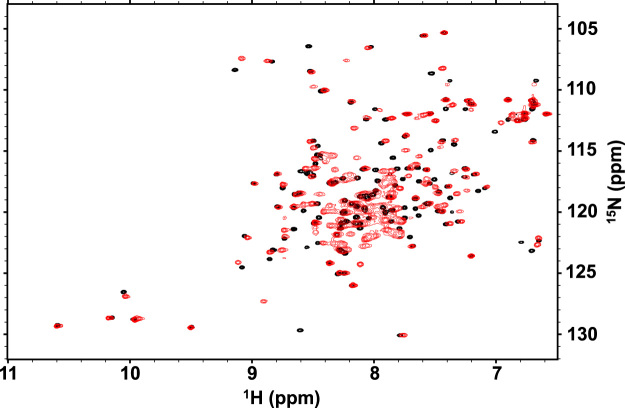
Fig. 5 shows the 2D 1H–15N HSQC spectra of the fusion protein at different temperatures. Most of the NMR signals of the fusion protein were broadened at 25°C, but became sharper with increasing temperature, i.e., at 35 and 45 °C, indicating that structural analysis at higher temperature is feasible. In addition, the well-dispersed NMR signals indicated that the fusion protein is folded even at 45 °C. This is also demonstrated by the similar percentages of helical contents at three different temperatures revealed by the CD analysis (Table 1). The NMR signals at ~110 ppm for 15N and ~8.6 ppm for 1H were observed in the 2D 1H–15N HSQC spectra of the fusion protein at 25 and 35 °C (dotted black circles in Fig. 5A and B), while corresponding signals were not observed at the same temperature in the 2D 1H–15N HSQC spectrum of the 15N-labeled Bcl-xL in the presence of non-labeled HBx(101−136) (data not shown). This suggests that these peaks are derived from the (GS)5-linker portion, which is supported by the results of the Beclin-1-linker-Bcl-xL fusion protein [12]. In contrast, these peaks hypothesized to be from the (GS)5-linker portion of the fusion protein were not observed at 45 °C (Fig. 5C), presumably owing to rapid solvent exchange at this temperature.
Fig. 5.
2D 1H–15N HSQC spectra of 0.3 mM 15N-labeled HBx(101−136)-(GS)5-Bcl-xL at 25 (A), 35 (B) and 45 °C (C). The NMR signals predicted to arise from the backbone amide groups of the (GS)5-linker are shown in dotted black circles in panels A and B.
Then, we compared the 2D 1H–15N HSQC spectra of 15N-labeled HBx(101−136)-(GS)5-Bcl-xL alone (red) and 15N-labeled Bcl-xL alone (black) at 45 °C (Fig. 6). The spectrum of the fusion protein differs from that of Bcl-xL, suggesting that the HBx(101−136) portion binds to the Bcl-xL portion in the fusion protein. To confirm this and whether the fusion protein retains the native conformation of this complex, we next compared the 2D 1H–15N HSQC spectra of 15N-labeled HBx(101−136)-(GS)5-Bcl-xL alone (red) and 15N-labeled Bcl-xL in the presence of non-labeled HBx(101−136) at a molar ratio of 1:1.5 (blue) at 45 °C (Fig. 7). As expected, the overall spectral pattern of the HBx(101−136)-(GS)5-Bcl-xL fusion protein is similar to that of Bcl-xL with non-labeled HBx(101−136), indicating that the HBx(101−136)-(GS)5-Bcl-xL fusion protein forms an intramolecular complex and likely retains the native conformation of the complex. In addition, we could observe approximately 16–18 NMR signals (dotted black circles) and a region of broadened signals (dotted green circle) in the 2D 1H–15N HSQC spectrum of the fusion protein, whereas similar signals were not observed in the spectrum of 15N-labeled Bcl-xL in the presence of non-labeled HBx(101−136) (Fig. 7). Therefore, these NMR signals observed in the 2D 1H–15N HSQC spectrum of the fusion protein are likely derived from the 15N-labeled HBx(101−136) portion of the fusion protein. Our NMR results demonstrated that the HBx(101−136)-(GS)5-Bcl-xL fusion protein should be useful for structural studies to understand the interaction between HBx and Bcl-xL in atomic detail.
Fig. 6.
Overlaid 2D 1H–15N HSQC spectra of 0.3 mM 15N-labeled Bcl-xL (black) and 0.3 mM 15N-labeled HBx(101−136)-(GS)5-Bcl-xL (red) at 45 °C.
Fig. 7.
Overlaid 2D 1H–15N HSQC spectra of 0.3 mM 15N-labeled Bcl-xL with 0.45 mM non-labeled HBx(101−136) (blue) and 0.3 mM 15N-labeled HBx(101−136)-(GS)5-Bcl-xL (red) at 45 °C. NMR signals only observed in the spectrum of the HBx(101−136)-(GS)5-Bcl-xL fusion protein are indicated by black and green dotted circles.
HBx interacts with anti-apoptotic Bcl-2 family proteins through its BH3-like motif [7], [8]. The modulation of apoptotic pathways by HBx is suggested to contribute to the development of (HBV-associated) HCC [5]. Therefore, the complex structure between the HBx BH3-like motif and Bcl-xL might provide clues for preventing or suppressing the development of HCC. This complex structure also gives us valuable information about the protein-protein interactions involved in the network of apoptosis. The strategy using the fusion protein will be of great advantage, in particular, for understanding the recognition mechanisms of BH3 peptides/Bcl-2 family proteins having relatively weak affinities, which also play important roles in apoptosis.
4. Conclusions
The HBx(101−136)-(GS)5-Bcl-xL fusion protein was expressed successfully in E. coli cells and purified to >95% homogeneity, as judged by SDS-PAGE analysis. The purified fusion protein behaved as a monomer in aqueous solution and formed a stable intramolecular complex. In addition, our results indicate that this fusion protein retains the native conformation of the complex of Bcl-xL with the HBx BH3-like motif and that the HBx(101−136) portion of the fusion protein forms an α-helix. Taken together, HBx(101−136)-(GS)5-Bcl-xL is a useful fusion protein for structural studies of the interaction between HBx and Bcl-xL.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgement
This work was partially supported by JSPS KAKENHI Grant Number JP15K09462 and a Grant-in-Aid from the Japan Agency for Medical Research and Development (16fk0108119j0101).
Contributor Information
Hideki Kusunoki, Email: kusunoki@niid.go.jp.
Toshiyuki Tanaka, Email: ttanaka@tara.tsukuba.ac.jp.
Toshiyuki Kohno, Email: tkohno@med.kitasato-u.ac.jp.
Hirokazu Kimura, Email: kimhiro@niid.go.jp.
Kazuo Hosoda, Email: khosoda1968@oak.gunma-u.ac.jp.
Kaori Wakamatsu, Email: kwakamats@gunma-u.ac.jp.
Isao Hamaguchi, Email: 130hama@niid.go.jp.
References
- 1.Ganem D., Varmus H.E. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 2.Feitelson M.A., Duan L.X. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am. J. Pathol. 1997;150:1141–1157. [PMC free article] [PubMed] [Google Scholar]
- 3.Murakami S., Cheong J.H., Kaneko S. Human hepatitis virus X gene encodes a regulatory domain that represses transactivation of X protein. J. Biol. Chem. 1994;269:15118–15123. [PubMed] [Google Scholar]
- 4.Tang H., Oishi N., Kaneko S., Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawat S., Clippinger A.J., Bouchard M.J. Modulation of apoptotic signaling by the hepatitis B. virus X protein, Viruses. 2012;4:2945–2972. doi: 10.3390/v4112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie N., Chen X., Zhang T., Liu B., Huang C. Using proteomics to identify the HBx interactome in hepatitis B virus: how can this inform the clinic? Expert Rev. Proteom. 2014;11:59–74. doi: 10.1586/14789450.2014.861745. [DOI] [PubMed] [Google Scholar]
- 7.Geng X., Harry B.L., Zhou Q., Skeen-Gaar R.R., Ge X., Lee E.S., Mitani S., Xue D. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2012;109:18465–18470. doi: 10.1073/pnas.1204652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng X., Huang C., Qin Y., McCombs J.E., Yuan Q., Harry B.L., Palmer A.E., Xia N.S., Xue D. Hepatitis B virus X protein targets Bcl-2 proteins to increase intracellular calcium, required for virus replication and cell death induction. Proc. Natl. Acad. Sci. USA. 2012;109:18471–18476. doi: 10.1073/pnas.1204668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sattler M., Liang H., Nettesheim D., Meadows R.P., Harlan J.E., Eberstadt M., Yoon H.S., Shuker S.B., Chang B.S., Minn A.J., Thompson C.B., Fesik S.W. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 10.Petros A.M., Nettesheim D.G., Wang Y., Olejniczak E.T., Meadows R.P., Mack J., Swift K., Matayoshi E.D., Zhang H., Thompson C.B., Fesik S.W. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petros A.M., Olejniczak E.T., Fesik S.W. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta. 1644;2004:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Feng W., Huang S., Wu H., Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J. Mol. Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 13.Follis A.V., Chipuk J.E., Fisher J.C., Yun M.K., Grace C.R., Nourse A., Baran K., Ou L., Min L., White S.W., Green D.R., Kriwacki R.W. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat. Chem. Biol. 2013;9:163–168. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang T., Liu M., Wu J., Shi Y. Structural and biochemical analysis of Bcl-2 interaction with the hepatitis B virus protein HBx. Proc. Natl. Acad. Sci. USA. 2016;113:2074–2079. doi: 10.1073/pnas.1525616113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker M.W., Xu P., Li X., Vander Kooi C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012;287:11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata T., Shirakawa K., Kobayashi N., Shiheido H., Tabata N., Sakuma-Yonemura Y., Horisawa K., Katahira M., Doi N., Yanagawa H. Structural basis for inhibition of the MDM2: p53 interaction by an optimized MDM2-binding peptide selected with mRNA display. PLoS One. 2014;9:e109163. doi: 10.1371/journal.pone.0109163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusunoki H., Tanaka T., Kohno T., Wakamatsu K., Hamaguchi I. Structural characterization of the BH3-like motif of hepatitis B virus X protein. Biochem. Biophys. Res. Commun. 2014;450:741–745. doi: 10.1016/j.bbrc.2014.06.042. [DOI] [PubMed] [Google Scholar]
- 18.Vargas-Uribe M., Rodnin M.V., Ladokhin A.S. Comparison of membrane insertion pathways of the apoptotic regulator Bcl-xL and the diphtheria toxin translocation domain. Biochemistry. 2013;52:7901–7909. doi: 10.1021/bi400926k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y.H., Yang J.T. A new approach to the calculation of secondary structures of globular proteins by optical rotatory dispersion and circular dichroism. Biochem. Biophys. Res. Commun. 1971;44:1285–1291. doi: 10.1016/s0006-291x(71)80225-5. [DOI] [PubMed] [Google Scholar]
- 20.Rohl C.A., Chakrabartty A., Baldwin R.L. Helix propagation and N-cap propensities of the amino acids measured in alanine-based peptides in 40 volume percent trifluoroethanol. Protein Sci. 1996;5:2623–2637. doi: 10.1002/pro.5560051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers J.K., Pace C.N., Scholtz J.M. Helix propensities are identical in proteins and peptides. Biochemistry. 1997;36:10923–10929. doi: 10.1021/bi9707180. [DOI] [PubMed] [Google Scholar]
- 22.Louis-Jeune C., Andrade-Navarro M.A., Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 23.Bodenhausen G., Ruben D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 24.Piotto M., Saudek V., Sklenář V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 25.Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 26.T. Goddard, D. Kneller, SPARKY 3, University of California, San Francisco
- 27.Muchmore S.W., Sattler M., Liang H., Meadows R.P., Harlan J.E., Yoon H.S., Nettesheim D., Chang B.S., Thompson C.B., Wong S.L., Ng S.C., Fesik, S.W., X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]