Abstract
Brassica juncea (Indian mustard) is a commercially important oil seed crop, which is highly affected by many biotic stresses. Among them, Alternaria leaf blight and powdery mildew are the most devastating diseases leading to huge yield losses in B. juncea around the world. In this regard, genetic engineering is a promising tool that may possibly allow us to enhance the B. juncea disease resistance against these pathogens. NPR1 (non-expressor of pathogen-related gene 1) is a bonafide receptor of salicylic acid (SA) which modulates multiple immune responses in plants especially activation of induced and systemic acquired resistance (SAR). Here, we report the isolation and characterization of new NPR1 homolog (BjNPR1) from B. juncea. The phylogenetic tree constructed based on the deduced sequence of BjNPR1 with homologs from other species revealed that BjNPR1 grouped together with other known NPR1 proteins of Cruciferae family, and was nearest to B. napus. Furthermore, expression analysis showed that BjNPR1 was upregulated after SA treatment and fungal infection but not by jasmonic acid or abscisic acid. To understand the defensive role of this gene, we generated B. juncea transgenic lines overexpressing BjNPR1, and further confirmed by PCR and Southern blotting. The transgenic lines showed no phenotypic abnormalities, and constitutive expression of BjNPR1 activates defense signaling pathways by priming the expression of antifungal PR genes. Moreover, BjNPR1 transgenic lines showed enhanced resistance to Alternaria brassicae and Erysiphe cruciferarum as there was delay in symptoms and reduced disease severity than non-transgenic plants. In addition, the rate of disease spreading to uninfected or distal parts was also delayed in transgenic plants thus suggesting the activation of SAR. Altogether, the present study suggests that BjNPR1 is involved in broad spectrum of disease resistance against fungal pathogens.
Keywords: Brassica juncea, NPR1, Alternaria, Powdery mildew, systemic acquired resistance, salicylic acid, jasmonic acid
Introduction
Plants are very often exposed to a variety of biotic stresses, and thus have evolved multidimensional defense approaches to survive or retain their fitness (Roux et al., 2014). The plants display both preformed and inducible defense mechanisms to overcome pathogen challenges. However, much stronger and long lasting is inducible defense response such as systemic acquired resistance (SAR), which confers enhanced disease resistance to broad range of phytopathogens (Durrant and Dong, 2004). In plants, SAR is generally activated by local infections and immunizes the whole plant to subsequent infectious diseases (Fu and Dong, 2013; Shah and Zeier, 2013). Realistic evidences have shown that activation of SAR is reliant on the higher levels of the endogenous salicylic acid (SA), and activation of a battery of pathogen-related (PR) genes. Most of these PR proteins such as glucanases, chitinases, thaumatins, and defensins possess antifungal activities and are known to play important role in disease resistance. Exogenous application of SA or its analogs have been also revealed to activate SAR pathway in plants (Durrant and Dong, 2004; Makandar et al., 2006). Conversely, Arabidopsis thaliana plants expressing NahG transgene which codes for salicylate hydroxylase (SA-degrading enzyme) were deficient in accumulating SA, and hence failed to activate SAR (Delaney et al., 1995). In addition to SA, a group of heterogeneous proteins are crucial for the activation of SAR. Among them are the NPR1 protein, a key regulator in the SA mediated SAR signal transduction pathway. The quest to discover the SA receptor led to the discovery of a regulatory or transcription co-factor protein NPR1 (Cao et al., 1994). However, many studies have revealed that NPR1 is linked to SA signaling, however, its role as SA receptor remains largely unknown. In this context, Wu et al. (2012) has recently reported that NPR1 is the receptor for SA pathway in Arabidopsis. In addition, two NPR1 paralogs namely, NPR3 and NPR4 bind SA and control the proteasome-mediated degradation of NPR1 protein through their interaction with NPR1 (Fu et al., 2012).
After pathogen assault, plants produce a variety of phytohormones, their composition, quantity, and timing significantly varies among plant species, and depends mainly on the pathogens lifestyle and their mode of infection (De-Vos et al., 2005). SA pathway generally provides resistance to biotrophic pathogens, whereas jasmonic acid/ethylene (JA/ET) pathways are commonly associated with resistance to necrotrophic pathogens, and to herbivorous pests (Glazebrook, 2005; Bari and Jones, 2009). Generally, SA and JA signaling pathways operates antagonistically and thus, elevated resistance against biotrophs is often related with increased susceptibility to necrotrophs, and vice versa (Grant and Lamb, 2006). Many regulatory components involved in SA/JA crosstalk have been identified; among them is NPR1 which plays a crucial role in regulating SA-mediated suppression of the JA pathway (Spoel et al., 2003; Pieterse et al., 2012; Thaler et al., 2012; Van der Does et al., 2013). Furthermore, SA/JA antagonism is commonly found in many plant species under various taxonomic groups, therefore seems to be evolutionary conserved evolutionary (Thaler et al., 2012).
As first discovered in Arabidopsis, various AtNPR1 homologs have been isolated thereafter in many agriculturally important crops (Chen et al., 2013; Zhong et al., 2015). NPR1 is a multigene family in Arabidopsis with multifaceted functions. For example, AtNPR1 and AtNPR2 are notably considered as a key regulator of SAR (Cao et al., 1997, 1998; Zhang et al., 2003) while AtNPR3 and AtNPR4 are known as negative regulator of SAR (Fu et al., 2012). Moreover, another group of AtNPR1 homologs are AtBOP1, and AtBOP2, which are related with lateral organ development (Hepworth et al., 2005). However, most of the studies were carried out on Arabidopsis NPR1 (AtNPR1). Structurally, AtNPR1 and its homologs contain an ankyrin repeat, N-terminal BTB/POZ broad-complex, Tramtrack, Bric a brac/poxvirus and zinc finger domains, respectively (Cao et al., 1997; Aravind and Koonin, 1999). In non-induced state, NPR1 exists as an inactive oligomer form into cytosolic region. However, after SA accumulation, the redox status of the cell changes which leads to dissociation of the inactive oligomer NPR1 to active monomers and their translocation to the nucleus where they bind to TGA factors there by inducing PR genes (Mou et al., 2003; Tada et al., 2008). Transcriptional studies have shown that NPR1 is expressed at low levels in mock plants but is induced significantly after microbial attack or treatment with SA or its biologically active analogs. Many studies have revealed NPR1 mutant (NPR1) plants are more prone to diseases, and also show altered expression of defense marker PR genes compared to NPR1 expressing plants (Glazebrook et al., 1996; Cao et al., 1997). Furthermore, NPR1 also plays a role in cross talk of SA/JA signaling pathways and in antagonistic effect of SA on JA signaling (Spoel et al., 2003). Hence, NPR1 is considered as the positive regulator of SA mediated plant immune responses.
To explore the defense role of NPR1 against bacterial, viral and fungal pathogens, various overexpression studies have been carried out in both model and crop plant systems. NPR1 mediates the SA-induced expression of pathogenesis-related (PR) genes and SAR. Overexpression of NPR1 results in the increase of transcript levels of antifungal genes like PR1, PR2 (glucanase) and PR5 (thaumatin) which are universally known to have antifungal activity. Many studies have revealed the potential antifungal activity of these PR genes against wide range of fungal pathogens. PR gene activity is regulated at the level of redox-dependent nuclear transport of NPR1. For example, overexpression of NPR1 in Arabidopsis plants confers enhanced disease resistance to bacterial and fungal infections (Cao et al., 1998; Friedrich et al., 2001). Transgenic carrot plants overexpressing AtNPR1 exhibit high disease resistance not only to biotrophs (Erysiphe heraclei) but also to necrotrophic fungal pathogen (Botrytis cinerea, Sclerotinia sclerotiorum, and Alternaria radicina), respectively (Wally et al., 2009). Parkhi et al. (2010) also reported that cotton transgenic plants expressing AtNPR1 exhibited broad spectrum of disease resistance not only to fungal pathogens but also to nematodes. Additionally, tobacco plants overexpressing Malus hupehensis NPR1 confers resistance to Botrytis cinerea as well as induces battery of pathogen related genes. Furthermore, studies revealed have that rice and wheat plants overexpressing NPR1 gene confers broad spectrum of disease resistance against most disastrous pathogens Magnaporthe oryzae, Fusarium verticillioides, and Fusarium oxysporum, respectively (Makandar et al., 2006; Quilis et al., 2008). NPR1 overexpression in A. thaliana was reported to result an increase in the transcript levels of PR genes, hence proves that NPR1-dependent PR gene mediated disease resistance (Cao et al., 1997; Friedrich et al., 2001). Most recently, overexpression of NPR1 was also revealed to confer disease resistance against broad range of pathogens in different crops (Dutt et al., 2015; Sundaresha et al., 2016). These results revealed that NPR1 is a potential candidate gene for developing disease resistant transgenic crops against multiple pathogens.
Brassica juncea is an economically important oilseed crop in India and fulfills nearly 27% of vegetable oil requirements of the country (Giri et al., 2013), but fungal diseases have emerged as devastating factors for its poor yield and economic losses (Bal and Kumar, 2014; Bairwa et al., 2015; Chandrashekar et al., 2015). In field conditions, B. juncea are challenged by many potential fungal pathogens such as Alternaria brassicae (Alternaria leaf blight), Albugo candida (white rust), Sclerotinia sclerotiorum (Sclerotinia stem rot), Erysiphe cruciferarum (powdery mildew), and Hyaloperonospora parasitica causative agent of downy mildew. Generally, B. juncea lack sufficient innate resistance to these fungal diseases and development of resistant varieties through conventional breeding is difficult due to unavailability of disease resistant germplasm. However, fungicides are one of the tools to control fungal diseases but are environmentally detrimental, expensive and ineffective due to weather fluctuations. In this regard, B. juncea being an important source of edible oil, novel strategies of effective disease protection should be developed. Therefore, genetic transformation of defense regulatory genes (like NPR1) that controls the function of multiple defense genes are potential sources for developing broad spectrum and long lasting disease resistance against pathogens in B. juncea. In this study, we evaluated the role of NPR1 in B. juncea for improving disease resistance against biotrophic and necrotrophic fungal pathogens.
Materials and Methods
Plant Materials
The plant material used in the present study is B. juncea var. Varuna, and plants were grown in pots containing a mixture of soil and organic manure (2:1) in a growth chamber under a 16 h day/8 h dark photoperiod at a temperature of 22–24°C, with irradiance of 100–125 μmol/m2s and a relative humidity of 80%. For cDNA library construction, B. juncea plants were sprayed with 2 mM SA (pH 7.0) and control plants were similarly treated with sterile distilled water. Leaf samples for RNA isolation were harvested from control and SA treated plants at different time points.
Isolation and In Silico Analysis of NPR1 Gene from B. juncea
Brassica juncea cDNA library was constructed using total RNA (2 μg) from SA treated leaves as described (Ali et al., 2017). The full length cDNA of BjNPR1 was isolated from B. juncea cDNA library through colony hybridization using radiolabeled Arabidopsis NPR1 (AtNPR1) probe. Primers used to amplify AtNPR1 probe are presented in Table 1. A BLAST homology search against the NCBI database was carried out to confirm whether the obtained sequence encoded BjNPR1. The multiple sequence alignment was performed using ClustalW1. To determine the evolutionary relationship of BjNPR1 protein with other NPR1 homologs from monocots and dicots, phylogenetic tree was constructed using MEGA 7.1 with 1000 bootstraps. Conserved domain structure of this protein was analyzed by Pfam database2. The 3D (three-dimensional) structure of BjNPR1 was obtained using Phyre2 server3. Molecular weight and isoelectric point of BjNPR1 protein were obtained using Compute PI/MW tool of ExPASy. In silico subcellular localization of this protein was predicted using Cell-PLoc 2.0 program4.
Table 1.
List of primers used in this study.
| Gene | Primer |
|---|---|
| 35SPro | F-5′ CGGATTCCATTGCCCAGCTA 3′ |
| NPR1pro | R-5′ GAGAGTGCTGCTTTGGTTGC 3′ |
| RT-NPR1 | F-5′ GGAAGGAGCCGAGTTTGATAG 3′ R-5′ GTTATACTCACCCGCCTTAGTG 3′ |
| NPTII | F-5′ AGGCGATAGAAGGCGATGCGC 3′ R-5′ CAATCGGCTGCTCTGATGCCG 3′ |
| RT-PR1 | F-5′ GAACACGTGCAATGGAGAATG 3′ R-5′ CCATTGTTACACCTCGCTTTG 3′ |
| RT-PR2 | F-5′ CGTCTCTCTACAATTCGCTCTG 3′ R-5′ CGATATTGGCGTCGAATAGGT 3′ |
| RT-PR3 | F-5′ AAGACCAGGTTCTTGCCTTC 3′ R-5′ TCCGGTACACTCCCTACTATTC 3′ |
| RT-PR5 | F-5′ GCAGAACAATTGCCCTTACAC 3′ R-5′ GCGCCTGGATTCAGTTGATA 3′ |
| RT-PR12 | F-5′ CAATGGTGAAAGCGCAGAAG 3′ R-5′ AGGTTGATGCACTGGTTCTT 3′ |
| RTPR13 | F-5′ GAGAAGCAATGGCAGGTTCTA 3′ R-5′ CGCACTCCGTGTTGTAGTT 3′ |
| Alpha tubulin | F-5′ GCCTCGTCTCTCAGGTTATTTC 3′ R-5′ TGAAGTGGATTCTTGGGTATGG 3′ |
Expression Pattern of BjNPR1 under Hormonal Treatments and Fungal Infection
To investigate the induction of BjNPR1 gene, 40 days old B. juncea plants were sprayed with 2 mM SA, 100 μM JA and 50 μM abscisic acid (ABA) individually. Control plants for each treatment were treated with sterile distilled water containing equal amount of solvent used for hormone preparation. Leaf samples for RNA isolation were harvested from control and hormone treated plants after 0, 2, 4, 8, 12, 24, 48, and 72 h. For fungal infection, A. brassicae strain (I.D. No. 81651) was grown on radish dextrose agar (RDA) medium (Thakur and Kolte, 1985) at 22°C for 15 days and spores were collected to prepare inoculum. The 40 days old B. juncea plants were infected with A. brassicae as described by Ali et al. (2017). The inoculated plants were maintained at 100% relative humidity at 25°C. For powdery mildew infection, E. cruciferarum (H.C.I.O-ID: No.52067) previously isolated and identified in our lab was used as inoculum. Forty days old B. juncea plants were infected by physically dislodging conidia of E. cruciferarum from infected leaves on healthy leaves of B. juncea plants. The inoculated plants were kept in growth chamber at 100% relative humidity and 25°C. Control B. juncea plants were mock inoculated with sterile distilled water and incubated separately to prevent cross contamination. Leaf samples from control and fungal infected plants were harvested at different time points flash frozen in liquid nitrogen and stored at -80°C.
RNA Isolation and Reverse Transcription Quantitative PCR
To analyze the expression of BjNPR1 after hormonal treatments and fungal infection, reverse transcription quantitative PCR (RT-qPCR) was performed using BjNPR1 gene specific primers. Total RNA was isolated from 100 mg of leaf sample collected from treated and control plants using Ambion RNA isolation kit as described by manufacturer’s protocol (Life Technologies). Complementary DNA (cDNA) was synthesized from 2 μg of purified total RNA by reverse transcriptase in 20 μl reaction volume containing oligo(dT) 18 primers, 10 mM deoxynucleotide (dNTPS) and water following the manufacturer’s instructions (Invitrogen, Canada). RT-qPCR reaction mixture contains 2 μl of cDNA, 5 μl of SYBR green real-time PCR master mix (Takara, Japan) and 0.5 μl (10 pmol) of each primer (BjNPR1). The RT-qPCR thermocycling program was following: 95°C for 5 min, followed by 40 cycles at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 30 s. All primers used in this study were designed by Oligoanalyzer software (Table 1). The alpha tubulin gene (GenBank accession no-NM_100360.) was used as reference gene for normalization of expression values. The relative expression levels of BjNPR1 were quantified by 2-ΔΔCt method (Livak and Schmittgen, 2001). All reactions were conducted with three biological replicates. Fold changes with p-values less than 0.05 were considered significant.
Binary Vector Construction and Agrobacterium Transformation
The full length cDNA of BjNPR1 was cloned in sense direction into pBI121 at Sma1 and Sac1 site downstream of constitutive promoter 35S CaMV (cauliflower mosaic virus). The correct orientation of BjNPR1 fragment in the recombinant plasmid was further identified by PCR and sequencing. The resulting recombinant binary vector pBI121-BjNPR1 was mobilized into Agrobacterium tumefaciens EHA105 by freeze-thawing method (Holsters et al., 1978).
Brassica juncea Transformation
Brassica juncea cv. Varuna seeds were germinated on half strength Murashige and Skoog (MS) medium in Magenta boxes (Magenta vessel Corp., United States) at 24 ± 2°C under cool white florescent light (90–150 μmol photons/m2s) in a 16/8 h (light/dark) photoperiod (Murashige and Skoog, 1962). BjNPR1 transgenic plants were generated through Agrobacterium mediated co-cultivation method according to protocol with some modifications (Sharma et al., 2009). Seeds obtained from T0 transformed B. juncea plants were primary screened on kanamycin selection medium and then transferred into pots for further analysis.
Molecular Screening of BjNPR1 Transformants
Genomic DNA was isolated from putative BjNPR1 transformed and non-transformed plants following CTAB method. For molecular screening of BjNPR1 transgenic plants PCR and Southern blot analysis was used. PCR detection of BjNPR1 transgene was carried out using 35S promoter (forward) and BjNPR1 (reverse) primers. Southern blot analysis was performed to detect the transgene insertion and the copy number using DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Science, Mannheim, Germany). For integration, a 600 bp fragment of 35S promoter (forward primer) and NPR1 (reverse primer) were used as a probe in BjNPR1 transgenics. To detect copy number of BjNPR1 in transgenic plants, NPTII probe (500 bp) was used. Details of gene specific primers of BjNPR1, 35S promoter, and NPTII used for PCR analysis and probe synthesis are given in Table 1.
Expression Analysis of NPR1 and PR Genes in BjNPR1 Transgenics
RT-qPCR was performed to monitor the transcript levels of the BjNPR1, PR1, PR2 (β 1-3 glucanse), PR3 (chitinase), PR5 (thaumatin), PR12 (defensin), and PR13 (thionin) in the leaf tissue of the transgenic and non-transgenic plants. RNA isolation, cDNA synthesis and RT-qPCR experiments were executed as described in the above section. All primers used in this study are listed in Table 1.
Phenotypic Characterization of BjNPR1 Transgenic Plants
Different agronomic traits namely, size and shape of leaf, siliques, flowers, number of pods, number of seeds as well plant height were investigated for any phenotypic abnormalities between BjNPR1 transgenic and wild-type B. juncea plants.
Necrotrophic and Biotrophic Resistance Assay
BjNPR1 transgenic lines were evaluated for disease resistance to both necrotrophic and biotrophic fungal pathogens. Alternaria infection in BjNPR1 transgenic lines and non-transformed plants were carried out as described in our previous work (Ali et al., 2017). For disease scoring, three components of partial resistance, including lesion appearance, number of lesions per leaf, lesion diameter (mm) and percentage of disease leaf area (%DLA) were measured and compared between BjNPR1 transgenic lines and control plants after inoculation. Disease severity was calculated after 15 days inoculation (DAI), with a 10-point disease rating scale. For, E. cruciferarum infection, 40 days old healthy BjNPR1 transgenic and wild-type B. juncea plants were infected as described in above section. The inoculated plants were maintained at 22°C with 100% RH in an inoculation chamber. Disease phenotype was examined at 7 days after powdery mildew inoculation. For disease scoring, different parameters were used such as colony appearance, number of colonies or spots, disease index, percentage disease leaf area (%DLA) between BjNPR1 transgenic lines and control plants. Disease index including six grades: 0, 1, 3, 5, 7, and 9, which correspond, respectively, to disease incidence levels of 0, ≤5%, 6–10%, 11–20%, 21–40%, and ≥41%). The experiments were carried out in three biological replicates.
Trypan Blue Staining and Microscopy
Trypan blue staining was used for observing dead cells and fungal biomass in control and BjNPR1 transgenic plants. Briefly, control and transgenic infected leaves were stained with trypan blue staining solution [containing 40 mg of trypan blue, 10 mL lactic acid (85% w:w), 10 mL phenol (pH 7.5–8.0), 10 mL glycerol (≥99%) and 10 mL of distilled water] for 30 min at room temperature. The samples were washed with sterile water to remove the excess stain and then immersed in 70% ethanol solution overnight to remove the chlorophyll. The solution was then removed and tissue samples were immersed in 60% glycerol and photographed. The fungal biomass and cell death was visualized under light microscope (10 xs, Nikon).
Statistical Analysis
For all experiments, three biological replicates were used and each repeated three times. A student’s t-test was carried out to determine significant differences in BjNPR1 gene expression in control and treated samples as well transgenic and non-transgenic plants. The differences between two groups of data for comparisons in all the experiments were evaluated as statistically significant (∗p < 0.05) or extremely significant (∗∗p < 0.01).
Results
Isolation and In Silico Analysis of BjNPR1
The full-length cDNA of BjNPR1 gene was isolated from SA treated B. juncea library, and submitted to the Genbank with accession number DQ359129. In silico analysis of BjNPR1 cDNA revealed that it is comprised of 1781 bp with an open reading frame of 1857 bp, encoding a protein of 593 amino acids with a molecular mass of 65.77 kDa, and a theoretical PI of 5.25. Phylogenetic analysis showed that BjNPR1 is very close homolog to NPR1 of B. napus, B. oleracea, B. rapa, and A. thaliana, respectively, but was largely diverged from NPR1 of Poaceae family (Figure 1). Alignment of deduced amino acid sequence of BjNPR1 (accession no. ABC94642) revealed 92% identity with BnNPR1 (accession no. XP013725724), 78% with BolNPR1 (accession no. XP013605797), 78% with BrNPR1 (accession no. XP009109186), and 66% with AtNPR1 (accession no. ABR46023) (Figure 2A). To investigate the typical domain structure of BjNPR1, its protein sequence were analyzed using Pfam software which revealed the predicted BTB, ANK conserved domain as shown in Figure 2B.
FIGURE 1.
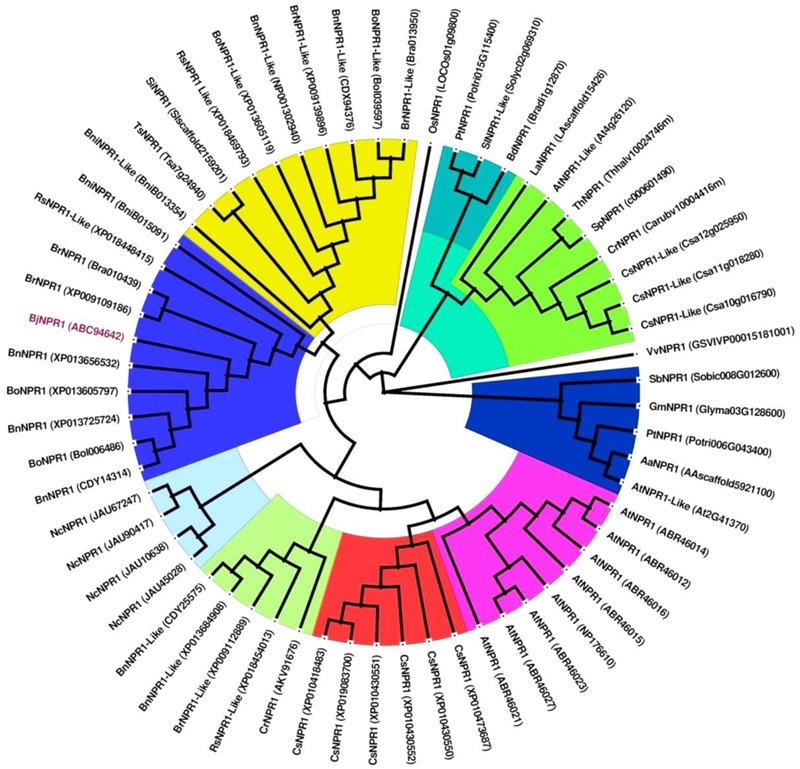
Phylogenetic analysis of BjNPR1 with other NPR1 proteins from different plant species. The deduced amino acid sequences of BjNPR1 was retrieved from NCBI GenBank and were further aligned with ClustalW using MEGA7.1 bioinformatic tool. The tree was generated using Maximum-Likelihood (ML) method with 1000 bootstrap replicates. GenBank IDs of each NPR1 protein sequence are given in the brackets behind the species names.
FIGURE 2.
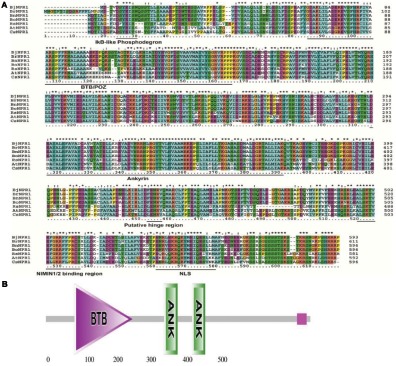
A multiple sequence alignment and in silico analysis of BjNPR1 protein sequence. (A) Alignments of the deduced amino acid sequences of BjNPR1 (accession no. ABC94642) revealed 92% identity with BnNPR1 (accession no. XP013725724), 78% with BolNPR1 (accession no. XP013605797), 78% with BrNPR1 (accession no. XP009109186), and 66% with AtNPR1 (accession no. ABR46023). (B) The conserved domains (BTB-ANK) of the BjNPR1 protein after analyzed by Pfam.
Expression Analysis of BjNPR1 in B. juncea after Hormonal Treatments and Fungal Infection
The expression pattern of BjNPR1 was investigated under various hormonal stresses (SA, JA, and ABA) as well as inoculation with A. brassicae and E. cruciferarum through real-time PCR. Upon SA treatment, transcript levels of BjNPR1 started to increase at 2 h (2.9-fold) and reached a peak at 12 h with a sharp decline at later time points (Figure 3A). On the other hand, no significant induction of BjNPR1 was seen in JA-treated plants and remains the same as control (Figure 3B). Treatment of B. juncea plants with ABA decreases the transcript levels of BjNPR1 at 2 h (0.65-fold) and remained low until 72 h time interval (Figure 3C). It has been well documented that NPR1 plays critical role in disease resistance in plants. To further study the defensive role of BjNPR1, we inoculated B. juncea plants with both necrotrophic (A. brassicae) and biotrophic (E. cruciferarum) fungal pathogens. After Alternaria inoculation, the expression of BjNPR1 was slightly increased at 4 h, reaching maximum at 12 h (2.7-fold) of post inoculation (Figure 3D). On the other hand, inoculation of B. juncea plants with E. cruciferarum led higher up-regulation of BjNPR1, and the highest expression levels were observed after 72 h (6.11-fold) to 96 h (6.33-fold) compared to control (Figure 3E). Hence, these results suggest that BjNPR1 is induced by fungal pathogens and might play important role in B. juncea disease resistance.
FIGURE 3.
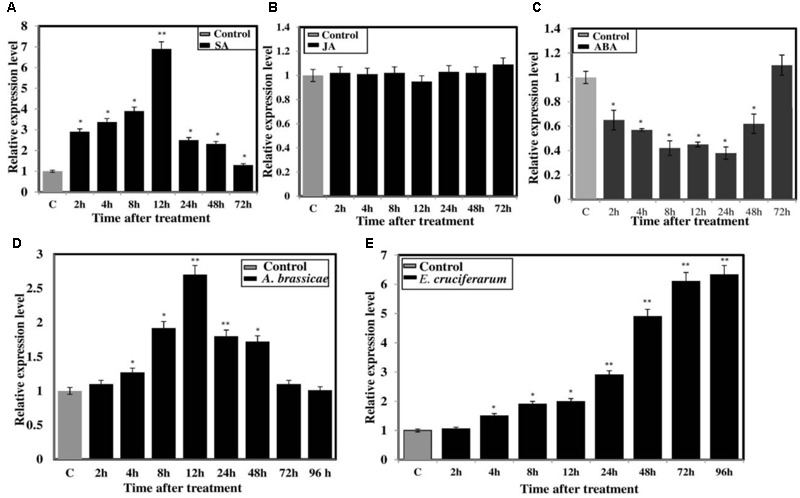
Expression analysis of BjNPR1 gene after hormonal treatment and fungal infection. Forty days old B. juncea plants were treated with different defense stimulators (SA, JA, and ABA) and also infected with fungal pathogens (A. brassicae and E. cruciferarum). Leaf samples were harvested at different time points for RNA isolation. Control plants for each treatment were treated with sterile distilled water containing equal amount of solvent used for hormone preparation. (A) Expression analysis of BjNPR1 after SA (B) JA (C) ABA (D) Alternaria infection and (E) E. cruciferarum, respectively. SE for each bar is shown. The asterisks indicate statistically significant difference relative to control and was calculated by student’s t-test (∗P < 0.05; ∗∗P < 0.01).
Development and Molecular Analysis of Transgenic B. juncea Lines Overexpressing BjNPR1
In order to further confirm the defensive role of the BjNPR1, transgenic B. juncea lines with constitutive expression of BjNPR1 were generated through tissue culture method to enhance the immunity (Supplementary Figures S1A–J). For this purpose, full length cds of BjNPR1 was cloned into pBI121 binary construct under the control of CaMV 35S constitutive promoter (Figure 4A). Further, BjNPR1 transgenic plants were generated using Agrobacterium hypocotyls co-cultivation method (Supplementary Figure S1). In the present study, overall transformation efficacy was found to be 2.7% using co-cultivation method (Supplementary Table S1). A total of 10 transgenic lines were obtained through kanamycin screening, and T-DNA integrations were confirmed by PCR amplification (Figure 4B). Overexpression of BjNPR1 was examined in transgenic lines by q-RT PCR. The mRNA levels of the BjNPR1 gene varied greatly in different lines such as lines 2, 5, 7, and 8 showed high accumulation of BjNPR1, while remaining lines (1, 3, 4, 6, 9, and 10) revealed relatively low expression levels of BjNPR1 (Figure 4C). To further investigate the integration and copy number of BjPR1 transgene in selected highly expression lines (2, 5, 7, and 8), Southern blotting was performed using 35S-NPR1 and NPTII probes, and all the four lines showed 35S-BjNPR1 integration, respectively (Figure 4D). In addition, single copy insertion was observed in T-DNA lines 2 and 5, where as two copies were found in lines 7 and 8 (Figure 4E). Finally, single copy number and high expression lines of BjNPR1 transgenic (lines 2 and 5) were selected for disease screening.
FIGURE 4.
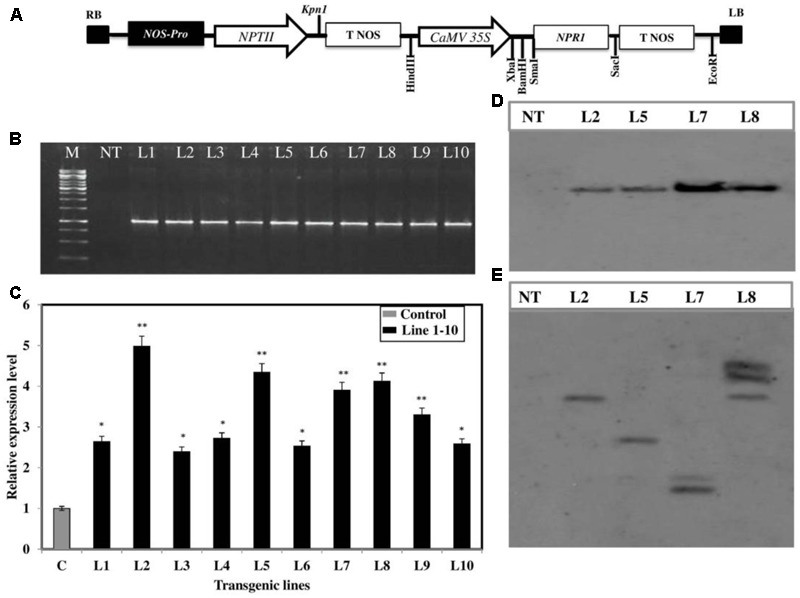
Development of 35S-BjNPR1 transgenic lines and molecular analysis. (A) Schematic representation of BjNPR1-pBi121 construct and T-DNA map of pBi121 binary vector. (B) T-DNA integration in 10 transgenic lines was confirmed by PCR amplification, using 35S promoter (forward) and BjNPR1 (reverse) gene specific primers. (C) Real-time PCR quantification of BjNPR1 in wild type and 35S-NPR1 overexpressed B. juncea plants. The asterisks indicate statistically significant differences between control (non-transgenic) and BjNPR1 transgenic plants (∗P < 0.05; ∗∗P < 0.01). (D) Southern blotting analysis showing BjNPR1 transgene integration, DNA was digested with HindIII restriction enzyme and hybridized with DIG labeled 35S-NPR1 probe. (E) Southern blotting analysis showing copy number in selected highly expressed transgenic line (L2, L5, L7, and L8), respectively, DNA was digested with Sac1 and hybridized with DIG labeled NPTII gene probe.
BjNPR1 Transgenic Plants Modulates the Expression of SA and JA Signature (PR) Genes
Pathogen-related genes are not only known as molecular signatures of the SA and MeJA signaling pathways but also widely used as diagnostic markers in pathogen resistance assays. Generally, overexpression of NPR1 genes has been shown to enhance the immune response (so called priming) through the activation of SAR marker or PR genes. Here, we examined whether B. juncea plants overexpressing BjNPR1 gene will lead to induction of PR genes. For this, we studied the expression of SA and JA signaling diagnostic genes (PR1, PR2, PR3, PR5, PR12, and PR13) in selected highly expressed BjNPR1 transgenic lines and non-transgenic plants under non-stressed conditions. The expression levels of SA marker genes PR1, PR2, and PR5 in BjNPR1 transgenic lines (L2 and L5) was found to be comparatively higher than in control (non-transgenic) plants (Figure 5). On the other hand, low expression levels of JA signature genes (PR3, PR12, and PR13) were observed in BjNPR1 transgenic lines when compared to SA signature genes but were higher than in control (non-transgenic) plants. These results showed that the constitutive expression of BjNPR1 was associated with the faster and stronger activation of PR genes which could enhance disease resistance in B. juncea to multiple pathogens.
FIGURE 5.
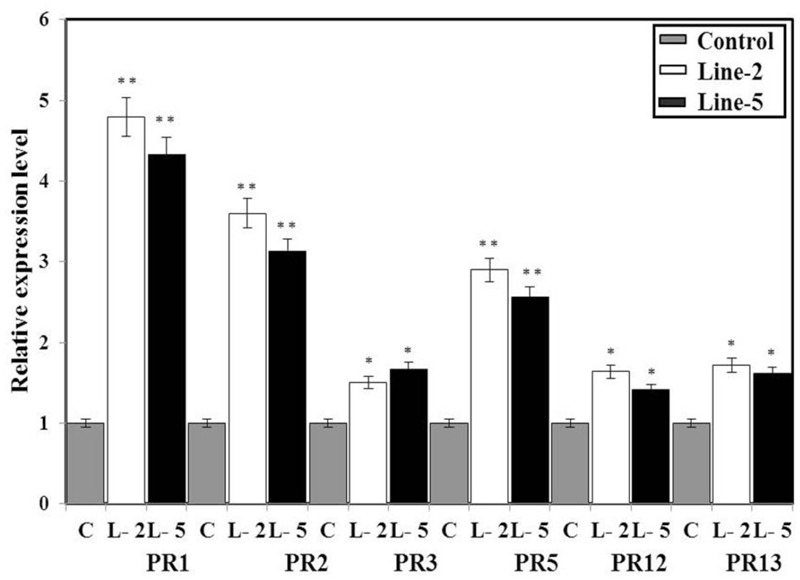
Quantification of mRNA levels of SA- and JA-dependent PR genes in wild type and BjNPR1 transgenic plants. Expression analysis of PR1, PR2, PR3, PR5, PR12, and PR13 in control and BjNPR1 transgenic plants. The relative expression levels of SA and JA dependent BjPR genes in transgenic and wild-type plants were compared with that of a control alpha tubulin gene. The data are the mean ± SE of three biological replicates. SE for each bar is shown. The asterisks indicate statistically significant differences between the BjNPR1 transgenic and control (non-transgenic) plants (∗P < 0.05; ∗∗P < 0.01).
Phenotypic Analysis of BjNPR1 Transgenic Plants
In this study, comparative analyses on phenotypic abnormalities in BjNPR1 transgenic plants were systematically evaluated. Our results revealed that all the studied agronomic traits namely, shape and size of leaves, siliques, flower morphology, seed shape, number of pods, number of seeds and plants height in BjNPR1 transgenic plants were similar with that of wild-type plants (Figure 6 and Table 2). Hence, these results provides the evidence that BjNPR1 transgenic lines did not show any phenotypic abnormalities as was observed in other crop plants after overexpressing AtNPR1. Altogether, this data indicated that BjNPR1 transgenic plants showed normal growth and development.
FIGURE 6.

Agronomic traits of wild type and BjNPR1 transgenic plants. Different agronomic traits such as size and shape of leaf, silique, flower, number of pods, number of seeds as well plant height were studied in BjNPR1 transgenic and control plants. BjNPR1 transgenic lines did not show any phenotypic abnormalities. Scale bar 1 cm (A,B), 0.1 cm (C,D), 0.5 cm (E,F), 0.1 cm (G,I), and 0.05 cm (H,J).
Table 2.
Phenotypic analysis of BjNPR1 transgenic lines and wild-type plants did not show significant variations in terms of plant height, no of seeds and pods.
| S. No. | Plants | Plant height (cm) | No. of seeds/pod | No. of pods/plant |
|---|---|---|---|---|
| 1 | C | 134.3 ± 1.5 | 13 ± 0.3 | 77 ± 0.3 |
| 2 | L2 | 135.7 ± 1.5 | 12 ± 0.3 | 76 ± 1.8 |
| 3 | L5 | 135 ± 1.2 | 12 ± 0.6 | 78 ± 0.7 |
Overexpression of BjNPR1 in B. juncea Transgenic Plants Confers Partial Disease Resistance to Necrotrophic Fungal Pathogen
To explore the role of BjNPR1 in disease resistance, we evaluated resistance level in BjNPR1 transgenic plants against necrotrophic (A. brassicae) fungal pathogen, which is the most serious pathogen of B. juncea. We selected two transgenic lines (L2 and L5) for this study based on expression levels of BjNPR1 transgene. For Alternaria infection, BjNPR1 transgenic and control plants were infected and disease scoring was assessed at different time intervals. After inoculation, small necrotic lesions began after 3 days in non-transgenic plants and size of the necrotic lesions increased significantly after disease progression. In contrast, BjNPR1 transgenic plants also showed lesions but the lesion size or diameter was comparatively lower than non-transgenics (Figure 7A). Furthermore, our results revealed that disease severity was very high in non-transgenic plants, and covering approximately 30% of the total leaf area than the BjNPR1 transgenic lines at 15th dpi (Figure 7B). The disease resistance was assessed by measuring the average lesion diameter in the Alternaria infected leaves for both BjNPR1 transgenic and non-transformed plants, and lesion diameter was 50% reduced in the former compared to non-transgenics (Figure 7B). We also observed that increased number of lesions spread on distal or non-infected leaves in non-transformed plants than BjNPR1 transgenic plants after Alternaria infection (Figure 7C). Based on disease index (0–10 scale), highest disease incidence (3–4) was seen in control plants, while disease incidence 1–2 was seen in BjNPR1 transgenic lines following infection (Figure 7D). Additionally, we also monitor the cell death and fungal biomass in transgenic and wild-type plants after Alternaria infection using trypan blue staining. Based on the microscopic examinations, the infection increased the number of dead cells with larger and expanding cell death areas observed beyond the inoculation site in non-transgenic plants compared to transgenic lines (Figure 8A). Moreover, the fungal biomass and spore load after 6th, 12th, and 15th of Alternaria infection were significantly low in transgenic lines than that of non-transgenic B. juncea plants (Figure 8B). Therefore, our results revealed that BjNPR1 transgenic plants display partial resistance to Alternaria leaf blight as there was delay in lesion appearance, size, and spread of infection in comparison to non-transgenic plants.
FIGURE 7.
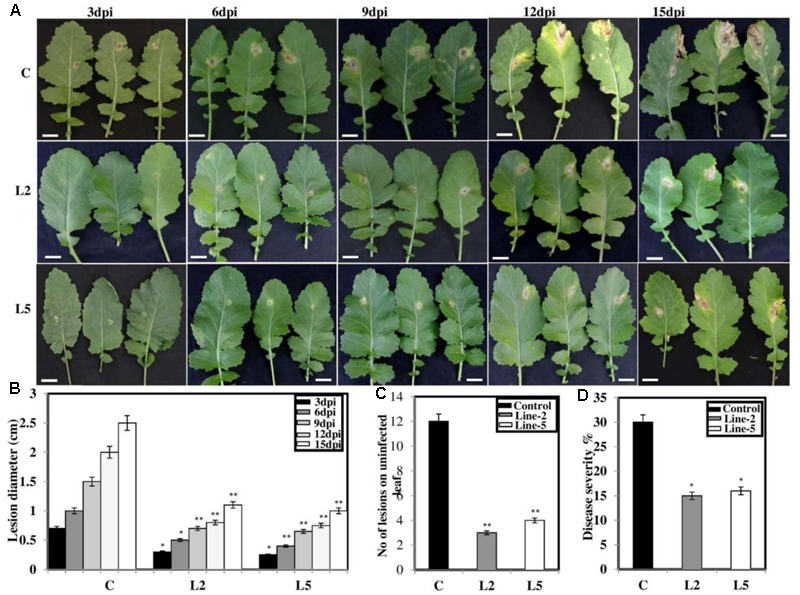
Overexpression of BjNPR1 improves disease resistance in B. juncea. (A) Disease resistance screening of BjNPR1 transgenic lines (L2 and L5) after Alternaria infection. L2 and L5 showed delay and reduction in disease severity than wild-type plants. (B) Lesions diameter of control (wild type), L2 and L5 BjNPR1 lines after Alternaria infection at different dpi. Bar = 40 mm. (C) Evaluation of disease resistance to Alternaria in BjNPR1 transgenic lines using six-point disease severity index. (D) Alternaria disease severity was monitored in BjNPR1 transgenic lines and control plants at 15th dpi based on total leaf area infected. Three biological replicates were used for infection. SD are the means of three biological replicates and asterisks shows statistically significant difference (∗P < 0.05; ∗∗P < 0.01) between BjNPR1 transgenic lines and wild-type plants.
FIGURE 8.
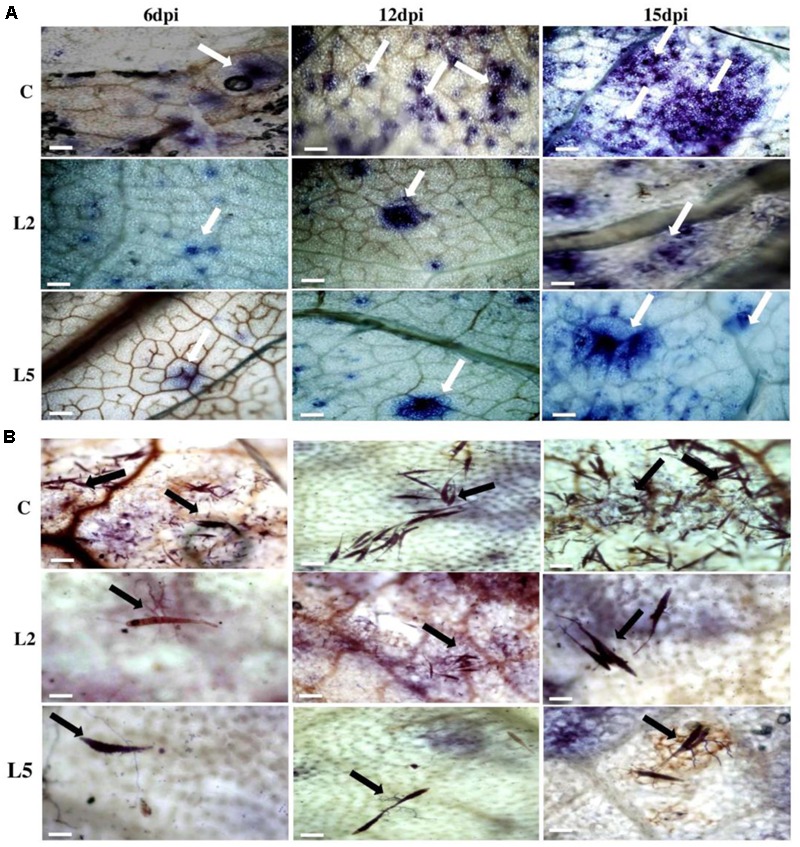
Analysis of cell death and fungal biomass in BjNPR1 transgenic and wild-type plants using trypan blue staining. (A) Microscopic examination of Alternaria mediated cell death in BjNPR1 lines and wild-type plants are shown with white color arrows. (B) Trypan blue staining of BjNPR1 transgenic and control plants after Alternaria infection. Alternaria spore load or biomass in BjNPR1 transgenic lines and wild-type plants at various dpi are shown with black color arrows. BjNPR1 transgenic lines showed reduced cell death and spore count as compared to wild-type plants. Bar = 40 μm.
BjNPR1 Plants Showed Improved Resistance against Powdery Mildew
In the present study, we also examined the resistance level of the BjNPR1 transgenic lines against powdery mildew disease, another important fungal disease of B. juncea caused by biotrophic pathogen (E. cruciferarum) which is entirely different in mode of infection style and signaling pathways from A. brassicae. To assess the resistance level of BjNPR1 transgenic lines against powdery mildew, plants were infected and disease scoring was done for 1–3 weeks. In non-transgenic plants, higher number of E. cruciferarum colonies was observed than transgenic lines on 7th, 12th, and 17th day after infection (Figure 9A). Based on colony count, there was approximately 50% reduction in newly formed colonies between transgenic lines and non-transgenic plants (Figure 9B). At 17th day of infection, transgenic plants showed powdery mildew infection with a disease scale of 3–4 (30–40%), while as non-transformed leaves (wild type) revealed 7–8 (70–80%) of disease incidence, respectively (Figure 9C). In addition, E. cruciferarum mediated cell death was examined in BjNPR1 transgenic and wild-type plants at different time points using trypan blue staining and light microscopy. Based on microscopic observations, more cell death was observed in control than that of transgenic plants (Figure 10A). To further investigate the role of BjNPR1 in improving powdery mildew disease resistance, the growth or fungal biomass of E. cruciferarum in BjNPR1 transgenic lines with wild-type plants was compared using light microscopy. As shown in Figure 10B, overexpression lines (L2 and L5) showed significant reduction of fungal biomass of E. cruciferarum than wild-type plants at 7th, 12th, and 17th dpi. However, overexpression of BjNPR1 could not inhibit the growth of E. cruciferarum completely, thereby providing only partial resistance to powdery mildew disease. Also, more number of leaves and pods were infected in non-transgenic plants as compared to transgenic lines. These results indicate that BjNPR1 transgenic plants exhibited partial resistance to powdery mildew infection, which was sufficient to delay the spread of infection in non-infected leaves or other parts.
FIGURE 9.
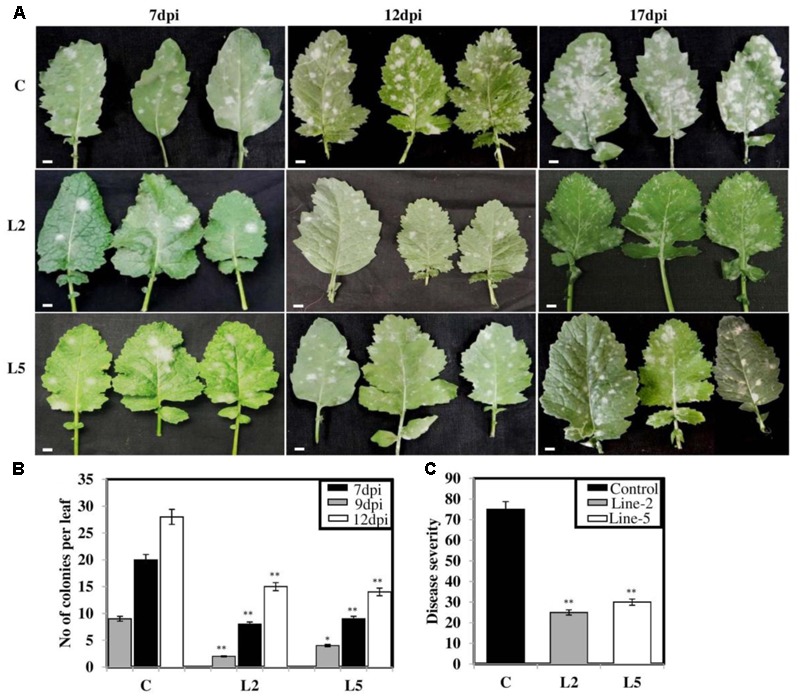
Screening of BjNPR1 transgenic lines for powdery mildew disease resistance. Forty days old wild-type plants and BjNPR1 transgenic plants were infected with E. cruciferarum and disease scoring was done at different time intervals. (A,B) BjNPR1 transgenic lines (L2 and L5) showed reduced number of E. cruciferarum colonies than wild-type plants at 7th, 12th, and 18th dpi. (C) E. cruciferarum disease severity in BjNPR1 transgenic lines and wild-type plants. Bar = 35 μm. The asterisks indicate statistically significant differences between the BjNPR1 transgenic and control (non-transgenic) plants after powdery mildew infection (∗P < 0.05; ∗∗P < 0.01).
FIGURE 10.
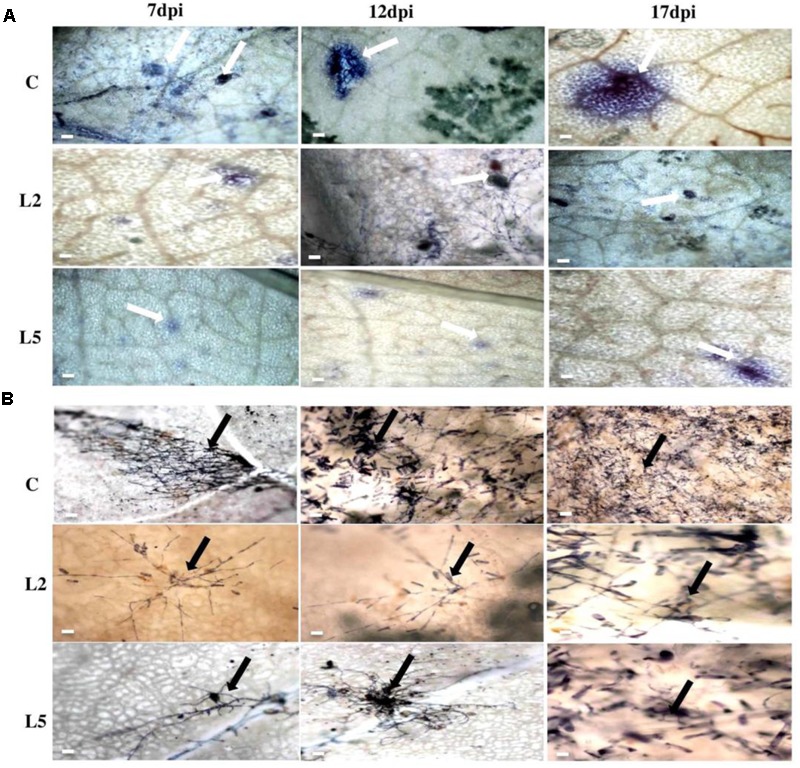
Microscopic examination of cell death and fungal biomass in BjNPR1 transgenic and wild-type plants using trypan blue staining. (A) Microscopic examination of E. cruciferarum mediated cell death in BjNPR1 lines and wild-type plants are shown with bold white arrows. (B) E. cruciferarum spore load or biomass in BjNPR1 transgenic lines and wild-type plants at various dpi after trypan blue staining are highlighted with bold black arrows. Bar = 30 μm.
Discussion
Identification and understanding the role of defense regulatory genes is necessary to develop disease resistant transgenic crops in agricultural system. Manipulation of regulatory genes has many beneficial roles such as activation of multiple defense genes or pyramids which provides effective and long-lasting protection compared with a single gene approach. Hence, in the present study BjNPR1 a regulatory gene was isolated and characterized, and phylogenetic analysis of the predicted BjNPR1 protein with other known NPR1-like sequences revealed that they are grouped into distinct clades. However, BjNPR1 fall within the same clade as other Brassica genus NPR1 proteins (Figure 1). Based on structural analysis, BjNPR1 protein contains domains such as an ankyrin repeat domain and a BTB/POZ domain, which are highly conserved in all NPR1 proteins (Cao et al., 1997; Kinkema et al., 2000; Mou et al., 2003). These domains are essential components of NPR1 and provide functions relating to NPR1-dependant co-activation of TGA transcription factors and protein–protein binding (Cao et al., 1997; Rochon et al., 2006). A time course expression analysis of BjNPR1 after defense hormonal treatments and fungal infections were carried out, and finally BjNPR1 was overexpressed in B. juncea to exhibit disease resistance to Alternaria blight and powdery mildew.
Phytohormones, including SA, MeJA, ET, and ABA play an essential role in the regulation of plant immune responses to microbial pathogens. However, each signal molecule or pathogen has its specific mechanism (Kunkel and Brooks, 2002). It is well documented that SA signaling pathway is involved in the induction of SAR while as JA/ET are involved in the activation of induced systemic resistance. NPR1 (a regulatory protein) is not only a bonafide receptor of SA but also a positive regulator of SAR, and plays a vital role in SA/JA signaling crosstalk (Spoel et al., 2003; Li et al., 2004; Wu et al., 2012). Mutant npr1-1 plants are not only compromised in SAR but also in basal resistance against many types of pathogens that are sensitive to SA dependent defenses (Dong, 2004). Previous studies have revealed that NPR1 plays a central role in the induced defense signaling network that is controlled by SA, JA, and ET (Dong, 2004; Pieterse and Van Loon, 2004). There is plethora of studies on defense signaling cascades, but most of them have been carried out in model plants. Therefore, uncovering the role of SA/JA master regulator (BjPNR1) in B. juncea will provide novel insights at molecular level. Previously, the expression kinetics of NPR1 or its homologs was found to be increased significantly after exogenous application of SA that leads to the activation of SAR (Mou et al., 2003; Yuan et al., 2007). Exogenous application of SA not only increases NPR1 transcript accumulation but also changes its protein architecture in the nucleus, mainly through posttranslational modifications (Mou et al., 2003; Tada et al., 2008). NPR1 and TGA1 are crucial redox-controlled regulators of SAR in plants. Generally, NPR1 is found as an oligomer within the cytoplasm of uninduced cells and changes in SA concentration lead to an altered redox environment within the cell, leading the nuclear localization of NPR1 in its monomeric form (Mou et al., 2003). NPR1 monomers interact with the reduced form of TGA1, which targets the activation sequence-1 (as-1) element of the promoter region of defense proteins (Després et al., 2003). In addition, SA-mediated redox modulation also plays an important role in the SA-mediated attenuation of the JA signaling pathway (Koornneef and Pieterse, 2008). In this study, we found that SA increased transcript levels of BjNPR1 in B. juncea (Figure 3A), consistent with results observed in different crop plants (Zhang et al., 2008). On the other hand, exogenous treatment with JA did not alter the expression of BjNPR1 (Figure 3B), similar results were also observed in avocado plants (Backer et al., 2015). However, contradictory results were seen in rice and banana seems to be host specific interactions (Yuan et al., 2007; Endah et al., 2008). Generally, ABA not only plays a central role in abiotic stress signal transduction, but also has been known to have positive or negative impact on plant immune system (Fan et al., 2005; de Torres–Zabala et al., 2007). Previous reports have shown that ABA promotes NPR1 degradation in Arabidopsis. In this study, exogenous application of ABA decreases the expression of BjNPR1 when compared to mock treated plants (Figure 3C). Many reports have shown that ABA appears upstream of NPR1 and suppresses the expression of both NPR1-dependent and independent signaling signatures. Our results further provide the evidence that ABA negatively regulates NPR1, positive regulator of SAR pathway in B. juncea. Altogether, our results revealed that NPR1 is distinctly regulated by defense stimulators, and also confirms that BjNPR1 is likely to be dependent on SA signaling which was consistent with the sequence analysis data that BjNPR1 contained a nuclear localization signal (NLS1) that was critical for SA-mediated expression of PR genes.
There is growing body of evidences that SA signaling triggers resistance to biotrophic pathogens, whereas a JA/ET pathway induces resistance to necrotrophic pathogens (Glazebrook, 2005). In the present study, BjNPR1 was moderately induced by necrotrophic pathogen (A. brassicae) (Figure 3D) and the expression seems to be JA independent as it was not induced during JA treatment. Mazumder et al. (2013) also reported that A. brassicicola, a necrotrophic pathogen increases SA accumulation in B. juncea during early stages of disease development which might suppress the JA pathway for successful infection. In our previous study, we have also observed the induction of SA marker gene PR1 after A. brassicae infection (data not shown). These results suggest that there is a hormonal crosstalk in B. juncea during Alternaria infection which could trigger the expression of NPR1 or SA dependent genes. As expected, transcript levels of BjNPR1 were significantly increased during E. cruciferarum infection as compared to uninfected plants (Figure 3E), similar to that observed by Dai et al. (2016). Furthermore, the expression levels of BjNPR1 in powdery mildew infected plants were relatively higher than that of Alternaria infected plants because pathogens causing powdery mildew disease are known as strict biotrophic pathogens which rely on SA pathway (Oliver and Ipcho, 2004).
Previous studies have revealed that Arabidopsis NPR1 when transformed into different crop plants showed enhanced disease resistance which make NPR1 a promising and potential candidate gene for developing disease resistant transgenic plants. In this study, B. juncea transgenic plants were generated by overexpressing BjNPR1 using 35S promoter through Agrobacterium mediated plant transformation. It was earlier reported that overexpression of AtNPR1 or its homolog OsNH1 in rice transgenic lines although improved resistance to pathogens but showed many detrimental effects such as chlorotic lesions, hypersensitive to light and produced higher amount of reactive oxygen species which leads cell death (Fitzgerald et al., 2004; Chern et al., 2005). However, in our study BjNPR1 transgenic plants exhibited normal phenotypes and did not showed any abnormalities, and similar findings were also observed in wheat, tobacco, and apple NPR1 transgenic lines. We next addressed whether BjNPR1 transgenic lines could activate SA or JA defense pathways by increasing the accumulation of defense marker genes (PR genes). As many studies have revealed that transgenic plants overexpressing AtNPR1 activates PR gene expression in tomato, grape, tobacco, and rice (Lin et al., 2004; Chern et al., 2005; Zhang et al., 2010; Le Henanff et al., 2011). However, contradictory results were also seen in carrot plants where NPR1 overexpression lines did not increase the transcript levels of PR genes under normal conditions. In present study, overexpression of BjNPR1 significantly increases the transcript levels of SA dependent PR genes like BjPR1, BjPR2, and BjPR5. However, low induction of JA signature (PR) genes was observed in BjNPR1 transgenic plants (Figure 5). NPR1 has been demonstrated to be an important transducer of the SA signal in the SA-mediated activation of PR gene expression and broad-spectrum resistance (Cao et al., 1994). Most of these PR proteins possess antifungal activity, and contribute effective and broad spectrum of disease resistance in BjNPR1 transgenic lines. The expression of BjPR genes in BjNPR1 transgenics further reveals that NPR1 activates SAR in B. juncea, an immune response effective against multiple pathogens. Interestingly, microarray analysis in Arabidopsis revealed that among SA-induced defense genes, more than 90% were NPR1-dependent genes (Wang et al., 2006; Blanco et al., 2009). On the other hand, Arabidopsis npr1 mutants are not responsive to SA, are compromised in their ability to express PR genes like PR1, PR2, and PR5 (Liu et al., 2005).
Many reports have revealed that NPR1 confers resistance to both necrotrophs and biotrophs (Cao et al., 1997; Friedrich et al., 2001; Makandar et al., 2006; Malnoy et al., 2007; Zhang et al., 2010) which led to the proposal of introduce and overexpression of BjNPR1 as a promising candidate gene for engineering broad-spectrum disease resistance in B. juncea. To evaluate the role of BjNPR1 in disease resistance, two lines (L2 and L5) were chosen for disease screening against Alternaria and powdery mildew infection. Recently, overexpression of NPR1 in peanut was reported to lead enhanced disease resistance against fungal pathogens (Sundaresha et al., 2016). Consistent with these reports, the results of our study revealed that overexpression of BjNPR1 in B. juncea leads partial disease resistance against both necrotrophic (A. brassicae) and biotrophic (E. cruciferarum) fungal pathogen, as transgenic plants showed delayed symptoms, reduced mean lesion diameter, number of colonies and disease spreading to distal/non-infected parts of the plant (Figures 7, 8). These results provide the evidence that constitutive expression of NPR1 in B. juncea showed high alertness in distal leaves (in the form of SAR) for subsequent infections at least in the early stages of infection. Our results also revealed that overexpression of BjNPR1 delayed the onset of Alternaria and powdery mildew disease therefore displays partial not complete resistance in B. juncea. Consistent with our reports, overexpression of AtNPR1 in carrot plants led enhanced disease resistance to biotrophic and necrotrophic fungal pathogens (Wally et al., 2009; Zhang et al., 2010). Many studies have shown that NPR1 or NPR1-like proteins confer resistance against fungal and bacterial pathogens, and this resistance is related to PR gene expression in transgenic plants. In present study, overexpression of BjNPR1 significantly increases the transcript levels of SA dependent PR genes like BjPR1, BjPR2, and BjPR5 which are universally known to posses potential antifungal activity. Previous studies have also shown that constitutive high-level expression of PR1, PR2, and PR5 in transgenic plants conferred tolerance to infection (Alexander et al., 1993; Liu et al., 2012; Gupta et al., 2013).
In summary, overexpression of BjNPR1 into B. juncea imparts disease resistance to two economically important fungal pathogens, thereby supporting results from previous studies on NPR1 transgenic carrot (Wally et al., 2009); cotton (Parkhi et al., 2010), and peanut (Sundaresha et al., 2016). Differences in the degree and development of disease symptoms caused by A. brassicae and E. cruciferarum between BjNPR1 and untransformed plants were clearly observed. The disease resistance of BjNPR1 transgenic B. juncea exposed to fungal pathogens may be due to the protection conferred by the accumulation of PR genes and SA mediated activation of SAR. However, in Arabidopsis, AtNPR1 also is associated with the activation of systemic defenses that are independent of SA (Pieterse et al., 1998). Future studies will be carried out to ensure the overall efficiency of disease resistance of overexpression of BjNPR1 transgenic plants under field conditions. Our future study would also focus on exploring the role of BjNPR1 against combined biotic and abiotic stresses in B. juncea which is the theme of future research. The results described in this study point to the need to further dissect the signaling pathways or knockout mutant studies will provide new insights into the precise functions of the BjNPR1 gene in regulating responses to biotic as well as in abiotic stress in B. juncea. Indeed, constitutive expression of AtNPR1 in many crops was associated with dwarfing and the spontaneous development of lesions which was not observed in BjNPR1 transgenic plants which could have limit their commercial qualities. However, considering the detrimental phenotypes associated with AtNPR1 expression in rice, the exploitation of NPR1 for improving disease resistance in economic important crops should take into consideration the physiology of the transgenic plants. In conclusion, BjNPR1 may serve as a potential candidate gene for developing disease resistant transgenic crops by genetic engineering.
Author Contributions
AG conceived and designed the research. SA has performed all the experiments and wrote the manuscript. ZM, PY, and PP contributed in data analysis. AT and JB has contributed in bioinformatic analysis. HM, RM, and SR contributed in tissue culture. AG contributed in manuscript proofreading. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the Project Director, National Research Centre on Plant Biotechnology for providing all the facilities and also ICAR-NPTC for financial support to complete this work.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01693/full#supplementary-material
Shoot regeneration from the calli of hypocotyls in Brassica juncea. (A) B. juncea plantlets grown in magenta boxes. (B) Hypocotyls on pre-culture media. (C) Explants on co-cultivation media. (D) Explants on selective media. (E) Explants grown on shoot induction media. (F,G) Explants grown in shoot regeneration media. (H,I) Explants grown in rooting media. (J) Transgenic to plant grown in soil-rite for hardening. Scale bars = 1 cm.
References
- Alexander D., Goodman R. M., Gut-Rella M. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. U.S.A. 90 7327–7331. 10.1073/pnas.90.15.7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Chandrashekar N., Rawat S., Nayanakantha N. M. C., Mir Z. A., Manoharan A., et al. (2017). Isolation and molecular characterization of pathogenesis related PR2 gene and its promoter from Brassica juncea. Biol. Plant. 61 763–773. 10.1007/s10535-017-0726-7 [DOI] [Google Scholar]
- Aravind L., Koonin E. V. (1999). Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285 1353–1361. 10.1006/jmbi.1998.2394 [DOI] [PubMed] [Google Scholar]
- Backer R., Mahomed W., Reeksting B. J., Engelbrecht J., Ibarra–Laclette E., van den Berg N. (2015). Phylogenetic and expression analysis of the NPR1-like gene family from Persea americana (Mill). Front. Plant Sci. 6:300 10.3389/fpls.2015.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa S. K., Godara S. L., Meena S. (2015). In vitro efficacy of fungicides on Sclerotinia sclerotiorum and their potential for control of stem rot in Indian Mustard (Brassica juncea L.). Int. J. Bioresour. Stress Manage. 6 497–502. 10.5958/0976-4038.2015.00073.1 [DOI] [Google Scholar]
- Bal R. S., Kumar A. (2014). Studies on the epidemiology of white rust and Alternaria leaf blight and their effect on the yield of Indian mustard. Afr. J. Agric. Res. 9 302–306. 10.5897/AJAR2013.7352 [DOI] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Blanco F., Salinas P., Cecchini N. M., Jordana X., Van Hummelen P., Alvarez M. E., et al. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70 79–102. 10.1007/s11103-009-9458-1 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J. D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. 10.1016/S0092-8674(00)81858 [DOI] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X. N. (1998). Generation of broad–spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. U.S.A. 95 6531–6536. 10.1073/pnas.95.11.6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar N., Ali S., Rawat S., Grover A. (2015). Gene expression profiling of Arabidopsis thaliana chitinase genes in response to Alternaria brassicae challenge. Indian Phytopathol. 68 106–111 [Google Scholar]
- Chen X., Barnaby J. Y., Sreedharan A., Huang X., Orbović V., Grosser J. W., et al. (2013). Over–expression of the citrus gene CtNH1 confers resistance to bacterial canker disease. Physiol. Mol. Plant Pathol. 84 115–122. 10.1016/j.pmpp.2013.07.002 [DOI] [Google Scholar]
- Chern M., Fitzgerald H. A., Canlas P. E., Navarre D. A., Ronald P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 18 511–520. 10.1094/MPMI-18-0511 [DOI] [PubMed] [Google Scholar]
- Dai L., Wang D., Xie X., Zhang C., Wang X., Xu Y., et al. (2016). The novel gene VpPR4–1 from Vitis pseudoreticulata increases powdery mildew resistance in transgenic Vitis vinifera L. Front. Plant Sci. 7:695 10.3389/fpls.2016.00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres–Zabala M., Truman W., Bennett M. H., Lafforgue G., Mansfield J. W., Rodriguez Egea P., et al. (2007). Pseudomonas syringae pv.tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 26 1434–1443. 10.1038/sj.emboj.7601575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T. P., Friedrich L., Ryals J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92 6602–6606. 10.1073/pnas.92.14.6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191. 10.1105/tpc.012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Vos M., Van-Oosten V. R., van-Poecke R. M. P., Van-Pelt J. A., Pozo M. J., Mueller M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18 923–937. 10.1094/MPMI-18-0923 [DOI] [PubMed] [Google Scholar]
- Dong X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7 547–552. 10.1016/j.pbi.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Durrant W. E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. 10.1146/annurev.phyto.42.040803.140421 [DOI] [PubMed] [Google Scholar]
- Dutt M., Barthe G., Irey M., Grosser J. (2015). Transgenic citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against huanglongbing (HLB; citrus greening). PLOS ONE 10:e0137134 10.1371/journal.pone.0147657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endah R., Beyene G., Kiggundu A., van–den Berg N., Schluter U., Kunert K., et al. (2008). Elicitor and Fusarium–induced expression of NPR1–like genes in banana. Plant Physiol. Biochem. 46 1007–1014. 10.1016/j.plaphy.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Fan J., Hill L., Crooks C., Doerner P., Lamb C. (2005). Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiol. 150 1750–1761. 10.1104/pp.109.137943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald H. A., Chern M. S., Navarre R., Ronald P. C. (2004). Overexpression of (At) NPR1 in rice leads to a BTH– and environment induced lesion–mimic/cell death phenotype. Mol. Plant Microbe Interact. 17 140–151. 10.1094/MPMI.2004.17.2.140 [DOI] [PubMed] [Google Scholar]
- Friedrich L., Lawton K., Dietrich R., Willits M., Cade R., Ryals J. (2001). NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol. Plant Microbe Interact. 14 1114–1124. 10.1094/MPMI.2001.14.9.1114 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Dong X. (2013). Systemic acquired resistance: turning local infection into Global defense. Annu. Rev. Plant Biol. 64 839–863. 10.1146/annurev-arplant-042811-105606 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486 228–232. 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri P., Taj G., Kumar A. (2013). Comparison of artificial inoculation methods for studying pathogenesis of Alternaria brassicae (Berk.) Sacc on Brassica juncea (L.) Czern. (Indian mustard). Afr. J. Biotechnol. 12 2422–2426. [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E. E., Ausubel F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M., Lamb C. (2006). Systemic immunity. Curr. Opin. Plant Biol. 9 414–420. 10.1016/j.pbi.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Gupta P., Ravi I., Sharma V. (2013). Induction of β-1,3-glucanase and chitinase activity in the defense response of Eruca sativa plants against the fungal pathogen Alternaria brassicicola. J. Plant Interact. 8 155–161. 10.1080/17429145.2012.679705 [DOI] [Google Scholar]
- Hepworth S. R., Zhang Y., McKim S., Li X., Haughn G. W. (2005). BLADE-ON-PETIOLE–dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M., Waele D., Depicker A., Messens E., van Montagu M., Schell J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163 181–187. 10.1007/BF00267408 [DOI] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350. 10.1105/tpc.12.12.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C. M. (2008). Cross talk in defense signaling. Plant Physiol. 146 839–844. 10.1104/pp.107.112029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B. N., Brooks D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5 325–331. 10.1016/S1369-5266(02)00275-3 [DOI] [PubMed] [Google Scholar]
- Le Henanff G., Farine S., Kieffer-Mazet F., Miclot A. S., Heitz T., Mestre P., et al. (2011). Vitis vinifera VvNPR1.1 is the functional ortholog of AtNPR1 and its overexpression in grapevine triggers constitutive activation of PR genes and enhanced resistance to powdery mildew. Planta 234 405–417. 10.1007/s00425-011-1412-1 [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331. 10.1105/tpc.016980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. C., Lu C. F., Wu J. W., Cheng M. L., Lin Y. M., Yang N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13 567–581. 10.1007/s11248-004-2375-9 [DOI] [PubMed] [Google Scholar]
- Liu D. Q., He X., Li W. X., Chen C. Y., Ge F. (2012). Molecular cloning of a thaumatin-like protein gene from Pyrus pyrifolia and overexpression of this gene in tobacco increased resistance to pathogenic fungi. Plant Cell Tissue Organ Cult. 111 29–39. 10.1007/s11240-012-0167-0 [DOI] [Google Scholar]
- Liu G., Holub E. B., Alonso J. M., Ecker J. R., Fobert P. R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41 304–318. 10.1111/j.1365-313X.2004.02296x [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real–time quantitative PCR and the 2-ΔΔCT method. Methods 25 4402–4408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Makandar R., Essig J. S., Schapaugh M. A., Trick H. N., Shah J. (2006). Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 19 123–129. 10.1094/MPMI-19-0123 [DOI] [PubMed] [Google Scholar]
- Malnoy M., Jin Q., Borejsza-Wysocka E. E., He S. Y., Aldwinckle H. S. (2007). Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol. Plant Microbe Interact. 20 1568–1580. 10.1094/MPMI-20-12-1568 [DOI] [PubMed] [Google Scholar]
- Mazumder M., Das S., Saha U., Chatterjee M., Bannerjee K., Basu D. (2013). Salicylic acid mediated establishment of the compatibility between Alternaria brassicicola and Brassica juncea is mitigated by abscisic acid in Sinapis alba. Plant Physiol. Biochem. 70 43–51. 10.1016/j.plaphy.2013.04.025 [DOI] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR function through redox changes. Cell 113 935–944. 10.1016/S0092-8674(03)00429-X [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Plant 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Oliver R. P., Ipcho S. V. S. (2004). Arabidopsis pathology breathes new life into the necrotrophs-vs.-biotrophs classification of fungal pathogens. Mol. Plant Pathol. 5 347–352. 10.1111/j.1364-3703.2004.00228.x [DOI] [PubMed] [Google Scholar]
- Parkhi V., Kumar V., Campbell L. M., Bell A. A., Shah J., Rathore K. S. (2010). Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 19 959–975. 10.1007/s11248-010-9374-9 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., Van Loon L. C. (2004). NPR1: the spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7 456–464. 10.1016/j.pbi.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., van Wees S. C., van Pelt J. A., Knoester M., Laan R., Gerrits H., et al. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10 1571–1580. 10.1105/tpc.10.9.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilis J., Penas G., Messeguer J., Brugidou C., Segundo B. S. (2008). The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant Microbe Interact. 21 1215–1231. 10.1094/MPMI-21-9-1215 [DOI] [PubMed] [Google Scholar]
- Rochon A., Boyle P., Wignes T., Fobert P. R., Després C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C–terminal cysteines. Plant Cell 18 3670–3685. 10.1105/tpc.106.046953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F., Voisin D., Badet T., Balagué C., Barlet X., Huard-Chauveau C., et al. (2014). Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol. Plant. Pathol. 15 427–432. 10.1111/mpp.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Zeier J. (2013). Long–distance communication and signal amplification in Systemic acquired resistance. Front. Plant Sci. 4:30 10.3389/fpls.2013.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M. K., Solanke A. U., Jani D., Singh Y., Sharma A. K. (2009). A simple and efficient Agrobacterium–mediated procedure for transformation of tomato. J. Biosci. 34 423–433. 10.1007/s12038-009-0049-8 [DOI] [PubMed] [Google Scholar]
- Spoel S. H., Koornneef A., Claessens S. M., Korzelius J. P., Van-Pelt J. A., Mueller M. J., et al. (2003). NPR1 modulates cross–talk between salicylate– and jasmonate–dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770. 10.1105/tpc.009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresha S., Rohini S., Appanna V. K., Arthikala M. K., Shanmugam N. B., Shashibhushan N. B., et al. (2016). Co-overexpression of Brassica juncea NPR1 (BjNPR1) and Trigonella foenum-graecum defensin (Tfgd) in transgenic peanut provides comprehensive but varied protection against Aspergillus flavus and Cercospora arachidicola. Plant Cell Rep. 35 1189–1203. 10.1007/s00299-016-1945-7 [DOI] [PubMed] [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321 952–956. 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur R., Kolte S. J. (1985). Radish root extract agar, a suitable medium for the growth and sporulation of Alternaria brassicae. News Lett. 10 117–118. [Google Scholar]
- Thaler J. S., Humphrey P. T., Whiteman N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17 260–270. 10.1016/j.tplants.2012.02.010 [DOI] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A., Koornneef A., Van Verk MC, Rodenburg N., Pauwels L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25 744–761. 10.1105/tpc.112.108548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wally O., Jayaraj J., Punja Z. K. (2009). Comparative resistance to foliar fungal pathogens in transgenic carrot plants expressing genes encoding for chitinase, beta–1, 3– glucanase and peroxidase. Eur. J. Plant Pathol. 123 331–342. 10.1007/s10658-008-9370-6 [DOI] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLOS Pathog. 2:e123 10.1371/journal.ppat.0020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1 639–647. 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Zhong S., Li Q., Zhu Z., Lou Y., Wang L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5 313–324. 10.1111/j.1467-7652.2007.00243.x [DOI] [PubMed] [Google Scholar]
- Zhang X., Francis M. I., Dawson W. O., Graham J. H., Orbović V. (2010). Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 128 91–100. 10.1007/s10658-010-9633-x [DOI] [Google Scholar]
- Zhang Y., Tessaro M. J., Lassner M., Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. 10.1105/tpc.014894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang M., Li Z., Li Q., He Z. (2008). Arabidopsis GH3.5 regulates salicylic acid–dependent and both NPR1– dependent and independent defense responses. Plant Signal. Behav. 3 537–542. 10.4161/psb.3.8.5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Xi L., Lian Q., Luo X., Wu Z., Seng S., et al. (2015). The NPR1 homolog GhNPR1 plays an important role in the defense response of Gladiolus hybridus. Plant Cell Rep. 34 1063–1074. 10.1007/s00299-015-1765-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shoot regeneration from the calli of hypocotyls in Brassica juncea. (A) B. juncea plantlets grown in magenta boxes. (B) Hypocotyls on pre-culture media. (C) Explants on co-cultivation media. (D) Explants on selective media. (E) Explants grown on shoot induction media. (F,G) Explants grown in shoot regeneration media. (H,I) Explants grown in rooting media. (J) Transgenic to plant grown in soil-rite for hardening. Scale bars = 1 cm.


