Abstract
The emergence and persistence of multidrug-resistant (MDR) diarrheagenic Escherichia coli (DEC) causing acute diarrhea is a major public health challenge in developing countries. The aim of this study was to evaluate the resistance phenotypes of DEC isolated from stool samples collected from children less than 5 years of age with acute diarrhea living in Ouagadougou/Burkina Faso. From August 2013 to October 2015, this study was carried out on 31 DEC strains of our study conducted in “Centre Médical avec Antenne Chirurgicale (CMA)” Paul VI and CMA of Schiphra. DEC were isolated and identified by standard microbiological methods and polymerase chain reaction (PCR) method was used to further characterize them. Antimicrobial susceptibility testing was done based on the disk diffusion method. DEC isolates were high resistant to tetracycline (83.9%), amoxicillin (77.4%), amoxicillin clavulanic acid (77.4%), piperacillin (64.5%), and colistin sulfate (61.3%). The most resistant phenotype represented was the extended spectrum β-lactamase (ESBL) phenotype (67.7%). Aminoglycosides were 100% active on enteroinvasive E. coli (EIEC) and enterohemorrhagic E. coli (EHEC). All the DEC isolates exhibited absolute (100%) sensitivity to ciprofloxacin. Monitoring and studying the resistance profile of DEC to antibiotics are necessary to guide probabilistic antibiotic therapy, especially in pediatric patients.
Keywords: children, diarrheagenic Escherichia coli, antibiotic, phenotype, MDR, PCR, Burkina Faso
Introduction
Infant diarrheal diseases are still a major public health problem today [1]. Among etiologic agents susceptible to incrimination in tropical areas, Escherichia coli pathovars are frequently responsible for gastroenteritis in children [2]. Generally, antibiotic therapy is not systematic in case of infantile diarrhea with bacterial etiology. However, when antibiotics are used, the choice of antibiotic is often difficult due to the emergence of resistance to first-line antibiotics (chloramphenicol, trimethoprim–sulfametoxazole, tetracycline, and penicillin A) [3]. In view of this, the treatment of gastroenteritis caused by E. coli is mainly based on the use of third generation cephalosporins (C3G), aminoglycosides, and quinolones.
However, these antibiotics also become ineffective in the management of these conditions. The current context for the emergence of bacterial resistance is of increasing concern. In fact, the fight against antibiotic resistance is the public health issue highlighted in recent years and is one of the priorities of the World Health Organization [4]. Despite progress in improving child health since the 1980s, diarrhea still accounts for 17% of deaths among children less than 5 years of age [5]. Furthermore, diarrhea continues to be one of the main reasons for consultation and hospitalization in Burkina Faso, with a still high mortality rate among children less than 5 years of age [6–8]. Given this situation, this study was undertaken to determine the resistance phenotypes of E. coli strains responsible for gastroenteritis in children less than 5 years of age in Ouagadougou, Burkina Faso.
Materials and methods
Study area and target population
Ouagadougou, the capital city of Burkina Faso, is composed of 30 districts. “Centre Médical avec Antenne Chirurgicale” (CMA) Paul VI or CMA of Schiphra received most of residents with low and middle incomes care seeking for health care services. This study was conducted between August 2013 and October 2015 in CMA Paul VI and CMA of Schiphra located in peripheral areas and in the city center of Ouagadougou (Fig. 1). The choice of these health centers is justified by the fact that they are located in different environments. The study population consisted of children less than 5 years of age with acute diarrhea and who were hospitalized or visited the health center as outpatient. Any child over the age of 5 years was excluded from the study.
Fig. 1.
Sampling sites in Ouagadougou (Source: BNDT, date: 30. 07. 2016, real: Ali Konaté)
Sample collection
Three hundred and fifteen (315) stool samples were collected in sterile containers and transported to the dedicated laboratory (“Laboratoire de Biologie Moléculaire, d’Épidémiologie et de Surveillance des Bactéries et virus Transmissibles par les Aliments (LaBESTA)/Centre de Recherche en Sciences Biologiques, Alimentaires et Nutritionnelles (CRSBAN)/Université Ouaga I, Pr Joseph KI-ZERBO”) within 24 h in a cool box at +4 °C for immediate analysis.
Isolation and identification of DEC
Stool samples were plated on eosin methylene blue agar (Liofilchem, Italy) and the plates incubated at +37 °C for 18–24 h. After incubation, the suspected E. coli colonies (dark purple colonies with a metallic sheen) were selected and streaked onto Mueller Hinton agar plate (Liofilchem, Italy). Confirmation was carried out by a biochemical microbiology method based on negative urease (Bio-Rad, France), negative citrate (Liofilchem, Italy), positive indole (Bio-Rad, France), positive lactose (Lioflchem, Italy), and positive orthonitrophenyl-β-D-galactopyranoside (ONPG) (bioMerieux, France). E. coli strains isolated were confirmed by API 20E (bioMérieux, France). The 16-plex polymerase chain reaction (PCR) was used to characterize the five main pathogroups of E. coli as described by Antikainen et al. [9].
Antimicrobial susceptibility testing
Thirty-one (31) diarrheagenic Eschericia coli (DEC) were subjected to the antimicrobial susceptibility testing. It was carried out by disk diffusion method on Müller-Hinton agar (Liofilchem, Italy) according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [10]. After depositing the antibiotics, the plates were incubated at +37 °C for 18–24 h. The diameters of the antibiotic sensitivity halos were recorded according to the recommendations of EUCAST. Intermediary (I) susceptibility of pathovars was classified as resistant (R). According to EUCAST recommendations on antibiotics used in case of infection with enterobacteria and in view of the multidrug resistance observed in recent years, 19 antibiotics divided into 7 different families were tested. These include amoxicillin (25 µg), amoxicillin–clavulanic acid (20/10 µg), ceftriaxone (30 µg), cefotaxime (30 µg), cefepime (30 – g), cefixime (10 µg), piperacillin (75 µg), piperacillin–tazobactam (100 +10 µg), imipenem (10 µg), tetracycline (30 µg), chloramphenicol (30 µg), trimethoprim–sulfametoxazole (1.25 ± 23.75 µg), aztreonam (30 µg), colistin sulfate (50 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), gentamycin (15 µg), netilmicin (10 µg), and tobramycin (10 µg) (Bio-Rad, France).
Antibiotyping method
The antibiotyping method involves the simultaneous presence of one or more antibiotic resistance markers. A strain may not wear a resistance marker or wear one or more [11]. When studying the susceptibility of a strain to several antibiotics, its resistance phenotype to antibiotics was determined. If the strain expresses only natural resistances, it is said to belong to the “wild” or sensitive phenotype. If its acquired resistances have changed its sensitivity, it expresses a “phenotype of resistance” that can be identified and whose mechanism must be determined. This phenotype is often referred to as initials of antibiotics that have become inactive. A strain is described as multidrug resistant when it is resistant to three antibiotics of different families [12–14].
Phenotypic detection of ESBL
Strains that were β-lactams resistant were subjected to investigation of extended spectrum β-lactamase (ESBL) activity according to the recommendations of EUCAST [10]. A disk of amoxicillin–clavulanic acid and two disks of third generation cephalosporins (C3G) (ceftriaxone and cefotaxime) were placed on the bacterial plate separated by a distance of 2 to 3 cm from one another. The presence of ESBL is indicated by a syngergetic effect between the disks, giving rise to an extended halo with the appearance of a “champagne cork” of keyhole.
Statistical analysis
The Fisher’s exact test with two-tailed p of Open Epi version 7.1.2.0 was used to determine the statistical significance of the results. A p value of <0.05 was considered statistically significant.
Ethical considerations
Permission to conduct the study was obtained from the hospital authorities of Burkina Faso, and informed verbal consent was obtained from the parents/guardians of every child before sample collection. The study protocol was approved by the National Ethical Committee(s) of Burkina Faso (N° 2009-39).
Results
Epidemiology of antibiotics resistance
A total of 31 E. coli pathovars (enteropathogenic E. coli [EPEC] [n = 8; 25.8%], enterohemorrhagic E. coli [EHEC] [n = 3; 9.67%], enteroinvasive E. coli [EIEC] [n = 4; 12.9%], enteroaggregative E. coli [EAEC] [n = 15; 48.4%], enterotoxigenic E. coli [ETEC] [n = 1; 3.2%]) were characterized. Antimicrobial susceptibility testing revealed that E. coli pathovars were the most resistant to the antibiotics tested. However, according to the results, DEC strains exhibited high-level multidrug resistance. E. coli strains were resistant to tetracycline (83.9%), amoxicillin–clavulanic acid (77.4%), amoxicillin (77.4%), piperacillin (64.5%), colistin sulfate (61.3%), and aztreonam (45.2%) (Fig. 2). Resistances were noted for C3G (ceftriaxone [41.9%]) and aminoglycosides (netilmicin [12.9%] and tobramycin [22.6%]). Resistance to quinolones (nalidixic acid [19.4%]) and carbapenems (imipenem [16.1%]) was also noted. However, aminoglycosides (gentamicin, netilmicin, and tobramycin) were 100% active on EHEC and EIEC. All the DEC strains showed absolute (100%) sensitivity to ciprofloxacin (Fig. 2). The prevalence of antibiotics resistance was particularly high in children less than 2 years of age (Table 1).
Fig. 2.
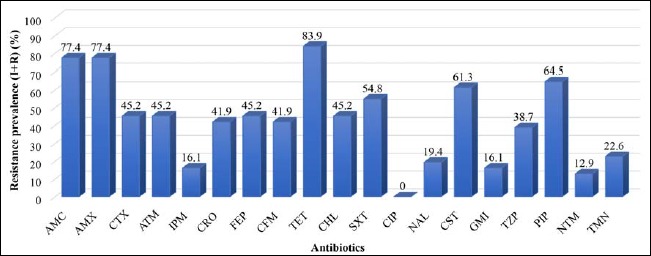
Antibiotic resistance profile of the DEC strains. Legend: AMC = amoxicillin–clavulanic acid, AMX = amoxicillin, CTX = cefotaxime, ATM = aztreoname, IPM = imipenem, CRO = ceftriaxone, FEP = cefepime, CFM = cefixime, TET = tetracycline, CHL = chloramphenicol, SXT = trimethoprim–sulfametoxazole CIP = ciprofloxacin, NAL = nalidixic acid, CST = colistin sulfate, GMI = gentamicin, TZP = piperacillin–tazobactam, PIP = piperacillin, NTM = netilmicin, TMN = tobramycin, I = intermediate, and R = resistant
Table 1.
Distribution of resistance rates by age group
| Antibiotics families | Antibiotics | Resistance N (%) | ||||
|---|---|---|---|---|---|---|
| Age groups (years) | ||||||
| (1–2) | (2–3) | (3–4) | (4–5) | |||
| β-lactams | Penicillins | AMC | 15 (48.4) | 6 (19.4) | 1 (3.2) | 2 (6.5) |
| AMX | 15 (48.4) | 6 (19.4) | 1 (3.2) | 2 (6.5) | ||
| PIP | 13 (41.9) | 4 (12.9) | 1 (3.2) | 2 (6.5) | ||
| TZP | 10 (32.3) | 2 (6.5) | 0 (0) | 0 (0) | ||
| C3G | CRO | 9(29) | 4 (12.9) | 0 (0) | 0 (0) | |
| CFM | 9(29) | 4 (12.9) | 0 (0) | 0 (0) | ||
| CTX | 10 (32.3) | 4 (12.9) | 0 (0) | 0 (0) | ||
| C4G | FEP | 10 (32.3) | 4 (12.9) | 0 (0) | 0 (0) | |
| Monobactam | ATM | 10 (32.3) | 4 (12.9) | 0 (0) | 0 (0) | |
| Carbapenems | IPM | 4 (12.9) | 1 (3.2) | 0 (0) | 0 (0) | |
| Quinolones | NAL | 5 (16.1) | 1 (3.2) | 0 (0) | 0 (0) | |
| Fluoroquinolones | CIP | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Cyclins | TET | 17 (54.8) | 6 (19.4) | 1 (3.2) | 2 (6.5) | |
| Phenicols | CHL | 9(29) | 3 (9.7) | 0 (0) | 2 (6.5) | |
| Sulfamides | SXT | 11 (35.5) | 3 (9.7) | 0 (0) | 3 (9.7) | |
| Polymyxins | CST | 13 (41.9) | 4 (12.9) | 1 (3.2) | 1 (3.2) | |
| Aminoglycosides | GMI | 4 (12.9) | 1 (3.2) | 0 (0) | 0 (0) | |
| NTM | 3 (9.7) | 1 (3.2) | 0 (0) | 0 (0) | ||
| TMN | 6 (19.4) | 1 (3.2) | 0 (0) | 0 (0) | ||
Legend: AMC = amoxicillin–clavulanic acid, AMX = amoxicillin, CTX = cefotaxime, ATM = aztreoname, IPM = imipenem, CRO = ceftriaxone, FEP = cefepime, CFM = cefixime, TET = tetracycline, CHL = chloramphenicol, SXT = trimethoprim–sulfametoxazole CIP = ciprofloxacin, NAL = nalidixic acid, CST = colistin sulfate, GMI = gentamicin, TZP = piperacillin–tazobactam, PIP = piperacillin, NTM = netilmicin, TMN = tobramycin, R = resistant, C3G = 3rd generation cephalosporins, C4G = 4th generation cephalosporins, N = number of strains
Epidemiology of resistance phenotypes observed
The results of the study show that 9/31 strains had a wild-type β-lactam phenotype (29%), 26/31 had a wild-type aminoglycoside phenotype (83.9%), and 25/31 had a wild-type quinolone phenotype (80.6%). The most resistant phenotype represented was the ESBL phenotype (21/31 [67.7%]). Five strains had a carbapenemase phenotype (16.1%) (3 EAEC [9.6%], one EPEC [3.2%], and one EHEC [3.2%]) (Table 2).
Table 2.
Distribution of resistance phenotypes prevalence by E. coli pathovars
| Resistance phenotypes | Prevalence of resistance phenotypes N (%) | Total N (%) | ||||
|---|---|---|---|---|---|---|
| EPEC | EHEC | EIEC | EAEC | ETEC | ||
| PSβL | 4 (12.9) | 1 (3.2) | 0 (0) | 4 (12.9) | 0 (0) | 9(29) |
| PBN | 1 (3.2) | 0 (0) | 0 (0) | 3 (9.7) | 0 (0) | 4 (12.9) |
| PHN | 5 (16.1) | 2 (6.4) | 3 (9.7) | 8 (25.8) | 1 (3.2) | 19 (61.2) |
| ESBL | 2 (6.4) | 1 (3.2) | 4 (12.9) | 5 (16.1) | 0 (0) | 12 (38.7) |
| ESBL+ CASE | 0 (0) | 0 (0) | 0 (0) | 1 (3.2) | 1 (3.2) | 2 (6.4) |
| ESBL+ carbapenemase | 1 (3.2) | 1 (3.2) | 0 (0) | 1 (3.2) | 0 (0) | 3 (9.7) |
| ESBL+ carbapenemase + KTGNt | 0 (0) | 0 (0) | 0 (0) | 1 (3.2) | 0 (0) | 1 (3.2) |
| ESBL+ carbapenemase + KTGNt + RCQ | 0 (0) | 0 (0) | 0 (0) | 1 (3.2) | 0 (0) | 1 (3.2) |
| ESBL + RCQ | 1 (3.2) | 0 (0) | 0 (0) | 1 (3.2) | 0 (0) | 2 (6.4) |
| PSA | 6 (19.4) | 3 (9.7) | 4 (12.9) | 12 (38.7) | 1 (3.2) | 26 (83.9) |
| KTGNt | 1 (3.2) | 0 (0) | 0 (0) | 2 (6.4) | 0 (0) | 3 (9.7) |
| PSQ | 6 (19.4) | 2 (6.4) | 4 (12.9) | 12 (38.7) | 1 (3.2) | 25 (80.6) |
| RCQ | 1 (3.2) | 1 (3.2) | 0 (0) | 3 (9.7) | 0 (0) | 5 (16.1) |
Legend: PSβL = β-lactamins wild phenotype, PBN = low-level penicillinases, PHN = high-level penicillinases, CASE = cephalosporinases, ESBL = extended spectrum β-lactamases, PSA = aminoglycosides wild phenotype, KTGNt = cross-resistance phenotype with kanamycin–tobramicin–gentamicin–netilmicin, PSQ = quinolone wild phenotype, RCQ = cross-resistance phenotype to quinolones, EPEC = enteropathogenic E. coli, EHEC = enterohemorrhagic E. coli, EIEC = enteroinvasive E. coli, EAEC = enteroaggregative E. coli, ETEC = enterotoxigenic E. coli
Discussion
The β-lactams (penicillins, cephalosporins, monobactam, and carbapenems) constitute the family of antibiotic most used in antibiotic therapy. The misuse of these antibiotics soon led to the emergence of resistance against these antibiotics. As regards penicillin resistance, the present study showed high resistance levels to amoxicillin (77.4%) and amoxicillin–clavulanic acid (77.4%). A similar level of resistance to amoxicillin–clavulanic acid (73.3%) was observed in a recent study in Burkina Faso by Dembélé et al. [8]. However, another previous study carried out in 2011 in Dar es Salaam, Tanzania, noted low resistance of E. coli to amoxicillin–clavulanic acid (7.8%) [15]. This shows an increase in the resistance rate to penicillins and the geographical disparity of resistances.
Another important observation in this study with the DEC isolates was that they were highly resistant to third generation cephalosporins used (particularly in children below 2 years). These resistances varied between 41.9% and 48.4%. Similar high prevalence of third generation cephalosporins antimicrobial resistance was been reported from younger children in Lusaka, Zambia [16]. However, low resistance to cefotaxime (4.7%) was observed in a previous study in Dar es Salaam, Tanzania [15]. Indeed, resistance to C3G/C4G from Enterobacteriaceae remains a major public health challenge with further increasing resistance rates. This is particularly true because treatment of severe gastroenteritis is mainly based on two antibiotics (C3G and fluoroquinolones) [17]. In Burkina Faso, the treatment of severe gastroenteritis in children below 2 years is based on β-lactams, particularly third generation cephalosporins (generally ceftriaxone). These increases in resistance may be attributed to the widespread misuse of these drugs. However, this resistance could be explained by the production of extended-spectrum β-lactamases and, to a lesser extent, plasmid cephalosporinases (AmpC) [17]. Generally, when children reach 6 months of age, their mothers begin to give them several foods in addition to breast-feeding. Otherwise, between 6 and 23 months of age, children are sent to the nursery (susceptibility to lack of hygiene). Previous studies have shown that milk [18] and meat [19] were common carriers of the main cephalosporins resistant E. coli. This beginning of environmental exposure and increased introduction of contaminated foods in younger children whose immune system is still developing could also explain this high resistance. Nevertheless, the low resistance rate of E. coli in older children (>2 years) could be associated with the development of immunity. Older children have a higher chance than children below 2 years to get diarrhea from hand-contamination, especially while playing on the ground, playing with their toys or other objects, and unknowingly putting their dirty fingers into their mouths. In addition, the risk of ingesting contaminated materials is high, especially in unhygienic environments.
Monobactams (aztreonam) are β-lactams reserved for severe hospital infections. Resistance to these antibiotics has begun to be observed in recent years. In the present study, E. coli strains were resistant to aztreonam (45.2%). These results confirm those of Nordmann and Carre [20] who also noted aztreonam-resistant phenotypes. Indeed, strains that had this aztreonam-resistant phenotype possessed both the Klebsiella pneumoniae Carbapenemsa (KPC) gene [20]. Resistance to this antibiotic could be explained by genetic mutations [21].
The present study unraveled 5 multiresistant E. coli isolates that exhibited ESBL and/or kanamycin–tobramicin–gentamicin–netilmicin (KTGNt), “Résistance croisée aux quinolones” (RCQ) phenotype in addition to their carbapenemase phenotype. Previous studies have shown that most of the E. coli strains that produce carbapenemase (KPC) also express other β-lactamases, including many types of ESBL (Temoniera [TEM], Suplhydryl Variable [SHV], Cefotaximase-Munich [CTX-M]) and possess a certain degree of resistance by impermeability [22]. Resistance to carbapenems in enterobacteria is essentially due to two mechanisms, both of which involve β-lactamases. The first mechanism associates the production of a chromosomal (AmpC)/plasmid cephalosporinase or ESBL with a quantitative or qualitative decrease in the expression of transmembrane proteins, such as porins [20]. The occurrence of the ESBL phenotype of E. coli strains is thus an important problem requiring special attention.
The phenotypic analysis of E. coli was in favor of a production of ESBL, i.e., 21/31 (67.7%). This rate of occurrence of ESBL isolates in children less than 5 years of age is comparable to those reported in Guinea-Bissau (32.6%) by Isendahl et al. [23] and Burkina Faso (38.3%) by Dembélé et al. [8]. The emergence and subsequent spread of ESBLs, the first mechanism involved in the resistance of enterobacteria to β-lactams, are strictly linked to their sequence of introduction into the therapeutic arsenal and to the consumption of the various β-lactams.
In the present study, resistances to quinolones (nalidixic acid [19.4%]) were observed. Extensive multidrug resistance to nalidixic acid for E. coli has been reported in many countries around the world [8, 24]. However, the geographical distribution of these resistances is very heterogeneous [3]. With respect to fluoroquinolones, the study of antibiotic resistance found no pathogenic E. coli strain resistant to ciprofloxacin. This confirms the results of the study in children in Dar es Salaam, Tanzania by Moyo et al. [15]. However, another study on isolated E. coli strains in adults (15–65 years) showed that ciprofloxacin resistance was increasing over time (5.51%, 6%, 7.3%, 8.1%, 9.4%, 10.3%, and 9.9% in 2004, 2005, 2006, 2007, 2008, 2009, and 2010), respectively, compared to 0% in this study [25]. The efficacy of fluoroquinolones may be explained by the fact that ciprofloxacin is not recommended in children. However, in the case of multidrug resistance, their prescription in the short term, seeing as a single dose is possible in a pediatric setting. Indeed, there is a need for continuous monitoring to the potential development of resistance to this antibiotic family.
High prevalence of resistance to tetracycline, colistin sulfate, trimethoprim/sulfamethoxazole, and chloramphenicol was also found in children less than 2 years of age compared to older ages (Table 1). A similar study, conducted in Northern Ghana, showed resistance to tetracycline (98%), trimethoprim/sulfametoxazole (90%), and chloramphenicol (51%) more frequently among isolates obtained from infants (<1 year) when compared with older children (1–4 years) [26]. Resistance to colistin sulfate antibiotics was also observed in China among E. coli isolates obtained from infants when compared with older children [27]. However, in the pediatric patients, colistin was effective for the treatment of life threatening gram-negative infections caused by bacteria that were resistant to multiple antimicrobial agents in Turkey [28]. Resistance to polymyxins (colistin sulfate) mainly depends on modification of lipopolysaccharide (LPS), which is often chromosomally mediated [29]. The high proportions of resistance to tetracycline (which is generally not used in children) in samples from children (<2 years) indicate the acquisition of resistant bacteria by the children rather than resistance induced through antimicrobial treatment.
Aminoglycosides are among the antibiotics used in the management of E. coli gastroenteritis. The present study found resistance to tobramycin (22.6%), gentamicin (16.1%), and netilmicin (12.9%). A previous study in Spain in children less than 5 years of age revealed low resistance to tobramycin (2.4%) and gentamicin (2.4%) [30]. In addition, low levels of resistance to gentamicin (3.8%) were also found in 2011 [25]. This increase in resistance to aminoglycosides over the years in children could be explained by the selection pressure by antibiotics and cross-transmission, two factors that fuel bacterial resistance.
From this study, high prevalence of multidrug-resistant (MDR) E. coli was found in children. Similarly, high proportions of MDR E. coli isolates in children were reported in Nigeria [31], Zimbabwe [32], Ethiopia [33, 34], Kenya [35], South India [36], and Ghana [37]. The prevalence of resistance to antibiotics was very high in children less than 2 years of age compared to older ages groups of children. The high rate of resistant bacteria (no resistance to ciprofloxacin, which is generally not used in children) in samples from this age groups of children indicate that the resistance was induced through antimicrobial treatment. It has been previously suggested that resistance levels are likely to be higher in those communities with a higher proportion of young children because of their high consumption of antibiotics [38].
Conclusion
In the present study, the five strains of carbapenemase phenotype had both the ESBL phenotype and/or KTGNt, RCQ favoring the risks of therapeutic deadlock. Antibiotic resistance is a public health issue and of particular concern when it affects carbapenems, quinolones, aminoglycosides, and molecules of choice for the treatment of multidrug-resistant bacterial infections. Unlike other antibiotics, no strain of pathogenic E. coli resistant to ciprofloxacin was found during the study. Ciprofloxacin is an antibiotic not recommended to children, which confirms that bacterial resistance is due, on the one hand, to the selection pressure exerted by the antibiotics and on the other hand to the cross-transmission of these multidrug resistant strains. Antibiotic resistance is therefore a factor of persistence of E. coli responsible for gastroenteritis in Burkina Faso. Antimicrobial resistance therefore needs the dynamism of research in Burkina Faso and a reinforcement of collective and multidisciplinary actions for a better efficiency. If the activity of existing antibiotics is to be safeguarded, the spread of E. coli resistant must be tackled by improving individual, collective, and hospital hygiene. Phenotypic methods such as the antibiogram are insufficient to study the antibiotics resistance mechanisms of bacteria; the spread of antibiotic-resistant bacteria and their corresponding genes must imperatively be two of the major challenges of the coming decades in developing countries, particularly in Burkina Faso by genotype methods. Given that genotypic methods are very costly and out of the ordinary routine for Sub-Saharan Africa, we propose that the Burkina Faso authorities focus on raising population awareness of the rules of good practice and good use of antibiotics. To ensure compliance with antibiotics prescription standards within health facilities and establish an epidemiological surveillance system of antibiotic resistance (national observatory) is therefore imperative.
Acknowledgements
The authors gratefully thank “Campus France and REMENTA/PARRAF” for financial support. The authors thank the authorities of “Institut Pasteur de Côte d’Ivoire” for technical support. We also thank the parents and guardians of children as well as the authorities of the CMA Paul VI and CMA of Schiphra for their frank cooperation.
Footnotes
Funding sources
“Campus France” and “Réseau de Recherche sur les Maladies Entériques à potentiel épidémique en Afrique de l’Ouest (REMENTA)/Programme d’Appui à la Recherche en Réseau en Afrique (PARRAF)” financed this study. The funders had no role in study design, data collection and analysis, preparation of the article, or decision to publish.
Authors’ contributions
N.B., A.S.T., N.K.G., R.D., and A.K. contributed to study concept and design. A.K., R.D., F.K.K., and I.K.K. participated in the laboratory analysis and interpretation of data. A.K. drafted the primary article and performed statistical data analyses. A.K. is recipient of a thesis grant from Campus France. N.B. and N.K.G. contributed to study supervision. All authors contributed to the preparation and revision of the article. All authors read and approved the final article for publication.
Conflict of interest
The authors declare that they have no conflict interests.
References
- 1.Liu L, Johnson HL, Cousens S: Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. The Lancet 379, 2151–2161 (2012) [DOI] [PubMed] [Google Scholar]
- 2.Nitiema LW, Nordgren J, Ouermi D: Burden of Rotavirus and other enteropathogens among children with diarrhea in Burkina Faso. Int J Infect Dis 15, 646–652 (2011) [DOI] [PubMed] [Google Scholar]
- 3.Imbert P: Prise en charge des diarrhées aiguës de l’enfant en milieu tropical. Med Trop 61, 226–230 (2001) [PubMed] [Google Scholar]
- 4.Organisation Mondiale de la Santé (OMS) (2014): Antimicrobial Resistance: Global Report on Surveillance 2014 [Google Scholar]
- 5.United Nations International Children’s Emergency Fund (UNICEF) (2007): La Situation des Enfants dans le Monde 2008. UNICEF, New York, pp. 1–154 [Google Scholar]
- 6.Sanou I, Kam KL, Tougouma A, Sangare L, Nikiema JHP, Sanou I, Koueta F, Dao L, Sawadogo SA, Soudre RB: Diarrhées aiguës de l’enfant: aspects épidémiologiques, cliniques et évolutifs en milieu hospitalier pédiatrique à Ouagadougou. Méd Afr Noire 46, 21–26 (1999) [Google Scholar]
- 7.Bonkoungou IJO, Lienemann T, Martikainen O, Dembélé R, Sanou I, Traoré AS, Siitonen A, Barro N, Haukka K: Detection of diarrhoeagenic Escherichia coli by 16-plex PCR from young children in urban and rural Burkina Faso. Clin Microbiol Infect 18, 901–906 (2012) [DOI] [PubMed] [Google Scholar]
- 8.Dembélé R, Bonkoungou IJO, Konaté A, Bsadjo Tchamba G, Ibrahim Bawa H, Bako E, Bagré TS, Kagambèga A, Zongo C, Traoré AS, Barro N: Serotyping and antibiotic resistance of enteropathogenic Escherichia coli and E. coli O157 isolated from diarrheal children in rural area of Burkina Faso. Afr J Microbiol Res 9, 1053–1059 (2015) [Google Scholar]
- 9.Antikainen J, Tarkka E, Haukka K, Siitonen A, Vaara M, Kirveskari J: New 16 plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. European J Clin Microbiol Infect Dis 28, 899–908 (2009) [DOI] [PubMed] [Google Scholar]
- 10.European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2017): Recommendation 2017, Éd. V.1.0 Mars, pp. 1–127 [Google Scholar]
- 11.Ye X, Wang X, Fan Y, Peng Y, Li L, Li S, Huang J, Yao Z, Chen S: Genotypic 1 and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl Environ Microbiol. 82(13), 3892–3899 (2016), DOI: 10.1128/AEM.00091-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guessennd N, Kacou-N’Douba A, Gbonon V, Yapi D, Ekaza E, Dosso M, Courvalin P. Prévalence et profil de résistance des entérobactéries productrices de β-lactamases à spectre élargi (BLSE) à Abidjan côte d’ivoire de 2005 à 2006. J Sci Pharm Biol 9, 63–70 (2008) [DOI] [PubMed] [Google Scholar]
- 13.Philippon A, Arlet G. Entérobactéries et β-lactamines: phénotypes de résistance naturelle. Pathol Biol 60, 112–126 (2012) [DOI] [PubMed] [Google Scholar]
- 14.Kamga HG, Nzengang R, Toukam M, Sando Z, Shiro SK: Phénotypes de resistance des souches d’Escherichia coli responsables des infections urinaires communautaires dans la ville de Yaoundé (Cameroun). Afr J Pathol Microbiol 3, 1–4 (2014) [Google Scholar]
- 15.Moyo SJ, Gro N, Matee MI, Kitundu J, Myrmel H, Mylvaganam H, Maselle SY, Langeland N: Age specific aetiological agents of diarrhoea in hospitalized children aged less than five years in Dar es Salaam, Tanzania. BMC Pediatr 11, 19 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiyangi H, Muma JB, Malama S, Manyahi J, Abade A, Kwenda G, Matee MI: Identification and antimicrobial resistance patterns of bacterial enteropathogens from children aged 0–59 months at the University Teaching Hospital, Lusaka, Zambia: a prospective cross sectional study. BMC Infect Dis 17, 117 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madec JY, Haenni M, Jouy E, Granier S, Weill FX, Le Hello S: Les entérobactéries résistantes aux céphalosporines de dernières générations: de l’animal à l’Homme. Bull Epidémiol, Santé Animale et Alimentation 53, 37–40 (2010) [Google Scholar]
- 18.Bagré TS, Kagambèga A, Bawa HI, Bsadjo Tchamba G, Dembélé R, Zongo C, Savadogo A, Aggad H, Traorél AS, Barro N: Antibiotic susceptibility of Escherichia coli and Salmonella strains isolated from raw and curds milk consumed in Ouagadougou and Ziniaré, Burkina Faso. Afr J Microbiol Res 8, 1012–1016 (2014) [Google Scholar]
- 19.Bawa IH, Bsadjo Tchamba G, Bagré TS, Bouda SC, Konaté A, Bako E, Kagambèga A, Zongo C, Somda M, Savadogo A, Traoré AS, Barro N: Antimicrobial susceptibility of Salmonella enterica strains isolated from raw beef, mutton and intestines sold in Ouagadougou, Burkina Faso. J Applied Biosci 95, 8966–8972 (2015) [Google Scholar]
- 20.Nordmann P, Carre A: Les carbapénèmases des entérobactéries. Arch Pédiatr 17, 154–162 (2010) [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Walsh TR, Cuvillier V, Nordmann P: Multiplex PCR for detection of acquired carbapenemase genes. Diagnostic Microbiol Infect Dis 70, 119–123 (2011) [DOI] [PubMed] [Google Scholar]
- 22.Nordmann P, Cuzon G, Naas T: The real threat of Klebsiella pneumoniae carbapenemase producing bacteria. Lancet Infect Dis 9, 228–236 (2009) [DOI] [PubMed] [Google Scholar]
- 23.Isendahl J, Turlej-Rogacka A, Manjuba C, Rodrigues A, Giske CG, Naucler P: Fecal carriage of ESBL-producing E. coli and K. pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS One 7, e51981 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansouri Jamshidi N, Pakzad I, Tabaraei B, Hadadi A: Evaluating the frequency of ciprofloxacin resistance qnr genes in E. coli strains isolated from clinical samples of Imam Khomani and Milad Hospitals in Ilam and Tehran. Iran Sci J Ilam Univ Med Sci 21, 16–22 (2013) [Google Scholar]
- 25.Thibaut S, Caillon J, Grandjean G, Ballereau F: Réseau MedQual: Surveillance de l’évolution des résistances des souches d’Escherichia coli isolées en ville. Bull Epidémiol, Santé Animale et Alimentation 53, 21–25 (2011) [Google Scholar]
- 26.Djie-Maletz A, Reithe K, Danour S, Anyidoho L, Saad E, Danikuu F, Ziniel P, Weitzel T, Wagner J, Bienzle U, Stark K, Seidu-Korkor A, Mockenhaupt FP, Ignatius R: High rate of resistance to locally used antibiotics among entericbacteria from children in Northern Ghana. J Antimicrob Chemother 61, 1315–1318 (2008) [DOI] [PubMed] [Google Scholar]
- 27.Hu YY, Wang YL, Sun QL, Huang ZX, Wang HY, Zhang R, Chen GX: Colistin-resistance gene mcr-1 in children’s gut flora. Int J Antimicrob Agents (2017) [DOI] [PubMed] [Google Scholar]
- 28.Ozsurekci Y, Aykac K, Cengiz AB, Bayhan C, Sancak B, Oncel EK, Kara A, Ceyhan M: Is colistin effective in the treatment of infections caused by multidrug-resistant (MDR) or extremely drug-resistant (XDR) gram-negative microorganisms in children? Diagn Microbiol Infect Dis 85, 233–238 (2016), DOI: 10.1016/j.diagmicrobio.2016.02. 017 [DOI] [PubMed] [Google Scholar]
- 29.Olaitan AO, Morand S, Rolain JM: Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5, 643 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domínguez E, Zarazaga M, Sáenz Y, Briñas L, Torres C: Mechanisms of antibiotic resistance in Escherichia coli isolates obtained from healthy children in Spain. Microbial Drug Resistance 8, 321–327 (2002) [DOI] [PubMed] [Google Scholar]
- 31.Yah C, Chineye H, Eghafona N: Multi-antibiotics-resistance plasmid profile of enteric pathogens in pediatric patients from Nigeria. Biokemistri 19, 35–42 (2007) [Google Scholar]
- 32.Mbanga J, Dube S, Munyanduki H: Prevalence and drug resistance in bacteria of the urinary tract infections in Bulawayo province, Zimbabwe. East Afr J Public Health 7, 229–232 (2010) [DOI] [PubMed] [Google Scholar]
- 33.Beyene G, Tsegaye W: Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in Jimma University specialized hopsital, southwest Ethiopia. Ethiop J Health Sci 21, 141–146 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kibret M, Abera B: Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr Health Sci 11, 40–45 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang WK, Oundo V, Schnabel D: Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhoea in four provinces of Kenya. J Infect Dev Ctries 6, 572–578 (2012) [DOI] [PubMed] [Google Scholar]
- 36.Razak SK, Gurushantappa V: Bacteriology of urinary tract infection and antibiotic susceptibility pattern in a tertiary care hospital in South India. Int J Med Sci Public Heal 1, 109–112 (2012) [Google Scholar]
- 37.Hackman HK, Brown CA, Twum-Danso K: Antibiotic resistance profile of non-extended spectrum beta-lactamaseproducing Escherichia coli and Klebsiella pneumoniae in Accra, Ghana. J Biol Agric Health 4, 12–16 (2014) [Google Scholar]
- 38.Turnidge J, Christiansen K: Antibiotic use and resistance proving the obvious. The Lancet 365, 548–549 (2005) [DOI] [PubMed] [Google Scholar]


