Abstract
We compared the analytical and clinical performance of cobas® CT/NG for use on the Cobas® 6800/8800 Systems with the Cobas® 4800 CT/NG Test from urogenital and extragenital specimens in over 12,000 specimens from both male and female subjects in Germany and the United States. The analytical sensitivity was ≤40 EB/ml for Chlamydia trachomatis (CT) and ≤1 CFU/ml for Neisseria gonorrhoeae (NG). Using clinical specimens, the overall percent agreement with the Cobas® 4800 CT/NG Test was >98.5%. Across urogenital specimens, there were 93 discrepant specimens; 76 (93.8%) of 81 CT discrepant specimens were 6800+/4800– and 10 (83.3%) of 12 NG discrepant specimens were 6800+/4800–. Sequencing verified CT results for 45 (61.6%) of 73 samples positive by 6800 and 1 (20%) of 5 positive by 4800. Similarly, 7 (70.0%) of 10 NG samples positive by 6800 and 1 of 2 positive by 4800 were confirmed by sequencing. Among discrepant extragenital specimens (all 6800+/4800–), 7 (50%) of 14 oropharyngeal and 23 (76.7%) of 30 anorectal CT discordant samples were confirmed as CT positive by sequencing; all 8 anorectal and 20 (90.9%) of 22 oropharyngeal NG discordant results were also confirmed as NG positive. In conclusion, Cobas® CT/NG for use on the Cobas® 6800/8800 Systems provides high-throughput automated solutions for sexually transmitted infection (STI) screening programs.
Keywords: cobas®CT/NG, Chlamydia trachomatis, Neisseria gonorrhoeae, molecular diagnostics, PCR, extragenital infection, genital infection
Introduction
Infections with Chlamydia trachomatis (CT) and Neisseria gonorrhoeae (NG) are the leading bacterial causes of sexually transmitted infections (STIs) worldwide [1, 2]. Prevalence is highest in persons aged 15–24 years [3]. Left untreated, these infections can cause significant sequelae to sexual health. Infections with these organisms have been strongly associated with development of pelvic inflammatory disease, adverse pregnancy outcomes, and an increased risk of human immunodeficiency virus (HIV) acquisition [3–7]. Many infections remain asymptomatic, with high numbers of infected patients who may not seek care and thus remaining a reservoir for infection. Many countries have screening or testing recommendations for CT and/or NG, particularly aimed at young women among whom the disease burden and the consequences of untreated infection are highest [8].
While control strategies for CT and NG focus on urogenital testing during the past decade, various studies from the United States of America (USA) and Australia have highlighted the importance of testing oropharyngeal and anorectal specimens in addition to urogenital tests in men who have sex with men (MSM) [9–14]. One study in San Francisco showed that 53% of C. trachomatis and 64% of N. gonorrhoeae infections in MSM involved non-urethral sites and would be missed if screening was done only for urethral infection [9]. In addition, the prevalence of anorectal CT in women attending STI clinics has been shown to be similar to that in MSM (up to 15% in women vs. 14% in MSM) [15–17]. Furthermore, recent studies in the USA looking at STI rates after the implementation of HIV preexposure prophylaxis (PrEP) with antiretroviral therapy have shown that no new HIV infections were noted with increasing use of HIV PrEP, despite high rates of STIs and decreased condom use. Thus, as PrEP use expands, STI screening will play an important role in patient management [18].
The diagnosis of infection with CT and NG is established with sensitive and specific NAATs. There are many commercially available choices for STI NAATs. The Cobas® 4800 CT/NG Test addresses the needs for low to mid-throughput testing for CT/NG [19]. The performance of the test has been confirmed in large clinical trials [20–22]. In many countries, centralized testing in large reference laboratories using high-throughput, automated NAAT systems has been the trend for large STI screening programs [23–25]. Therefore, Cobas® CT/NG for use on the Cobas® 6800/8800 Systems was recently launched addressing the needs of high-throughput laboratories with up to 960 samples per 8-h shift.
In the present study, the analytical and clinical performance of Cobas® CT/NG for use on the Cobas® 6800/8800 Systems was demonstrated using a variety of urogenital, oropharyngeal, and anorectal specimens from female and male individuals.
Methods
cobas® CT/NG for use on the cobas® 6800 and cobas® 8800 Systems
cobas® CT/NG is a qualitative nucleic acid test performed on the cobas® 6800 System and cobas® 8800 System. The test received CE-IVD mark in 2016. The system features, e.g., the instrument’s high-throughput capacity (384 samples per 8-h shift for the cobas® 6800 System and 960 samples per 8-h shift on the cobas® 8800 System), have recently been reviewed in detail [23]. Figure 1 shows the key features of the cobas® 6800 System and cobas® 8800 System. cobas® CT/NG enables the detection of CT/NG DNA from vaginal (clinician collected and self-collected with clinician instruction) and endocervical swabs, and female and male urine, as well as detection from female and male oropharyngeal and anorectal swabs collected and stabilized with cobas® PCR media. Cervical specimens collected in PreservCyt® solution were also evaluated. The volumes of sample required for the test are 400 µl for all swab samples and for PreservCyt® samples, and 850 µl for urine samples. Test duration is <3.5 h to the first set of 96 results.
Fig. 1.
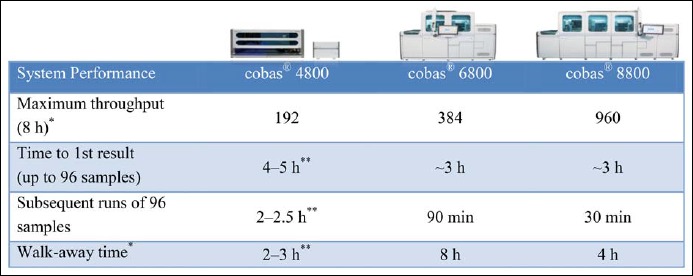
System performance characteristics of the cobas® 4800, cobas® 6800, and cobas® 8800 Systems. The cobas® 8800 System is identical to the cobas® 6800 System but designed for a higher throughput and contains 2 instead of 1 processing module (for sample extraction) and 4 instead of 1 thermocyclers (analytical module). Results presented include samples and controls. Times depicted do not include pre-analytic/post-analytic processing steps. *May vary based on workflow demands. **Varies based on assay and sample type
cobas® CT/NG is based on fully automated sample preparation (nucleic acid extraction and purification) followed by PCR amplification and detection. The cobas® 6800/8800 Systems consist of the sample supply module, the transfer module, the processing module, and the analytic module. Automated data management is performed by the cobas® 6800/8800 Systems software which assigns test results for all tests as positive, negative, or invalid. Results can be reviewed directly on the system screen, exported, or printed as a report.
Nucleic acid from patient samples, external controls, and added internal control DNA (DNA-IC) molecules are simultaneously extracted. In summary, bacterial nucleic acid is released by addition of proteinase and lysis reagent to the sample. The released nucleic acid binds to the silica surface of the added magnetic glass particles. Unbound substances and impurities, such as denatured protein, cellular debris, and potential PCR inhibitors are removed with subsequent wash steps, and purified nucleic acid is eluted from the magnetic glass particles with elution buffer at elevated temperature.
Selective amplification of target nucleic acid from the sample is achieved by the use of target-specific forward and reverse primers which are selected from highly conserved plasmid and genomic regions of CT and NG. A region on the CT cryptic plasmid and the ompA gene (dual target) and two conserved sequences of the NG DR-9 region are amplified by cobas® CT/NG. Selective amplification of DNA IC is achieved by the use of sequence-specific forward and reverse primers which are selected to have no homology with either the CT or NG target regions. A thermostable DNA polymerase enzyme is used for PCR amplification. The target and DNA-IC sequences are amplified simultaneously utilizing a universal PCR amplification profile with predefined temperature steps and number of cycles. The master mix includes deoxyuridine triphosphate (dUTP), instead of deoxythimidine triphosphate (dTTP), which is incorporated into the newly synthesized DNA (amplicon). Any contaminating amplicon from previous PCR runs is eliminated by the AmpErase enzyme, which is included in the PCR master mix, during the first thermal cycling step. However, newly formed amplicons are not eliminated since the AmpErase enzyme is inactivated once exposed to temperatures above 55 °C.
The cobas® CT/NG master mix contains two detection probes specific for the CT target sequences, two detection probes specific for the NG target sequences, and one for the DNA-IC. The probes are labeled with target specific fluorescent reporter dyes allowing simultaneous detection of CT targets, NG targets, and DNA-IC in three different target channels. When not bound to the target sequence, the fluorescent signal of the intact probes is suppressed by a quencher dye. During the PCR amplification step, hybridization of the probes to the specific single-stranded DNA template results in cleavage of the probe by the 5′ to 3′ exonuclease activity of the DNA polymerase resulting in separation of the reporter and quencher dyes and the generation of a fluorescent signal. With each PCR cycle, increasing amounts of cleaved probes are generated and the cumulative signal of the reporter dye increases concomitantly. Real-time detection and discrimination of PCR products are accomplished by measuring the fluorescence of the released reporter dyes for the CT targets, NG targets, and DNA-IC, respectively.
The DNA-IC, used to monitor the entire sample preparation and PCR amplification process, is introduced into each reaction well, along with the specimen during sample processing. In addition, the test utilizes a low titer positive and a negative control.
Compared to the cobas® 4800 CT/NG Test, improvements have been made which resulted in an increase in analytical sensitivity. Specifically, continuous mixing of sample during the lysis incubation step allows for a higher rate of DNA recovery.
Limit of detection and precision studies
The limit of detection (LOD) was determined using negative background for each specimen type by pooling prescreened individual specimens that had tested negative for both CT and NG with cobas® CT/NG. Quantified stocks of C. trachomatis, serovars D and I, and the N. gonorrhoeae strains 2948 and 891 were used to produce panels in each specimen type consisting of five target positive levels and a negative level tested over three reagent lots.
In-house precision was examined using a panel composed of CT and NG cultures diluted into pools of negative endocervical swab specimen, negative urine, and negative cervical specimen matrices. Four levels (negative, very low: ≤0.7 EB/ml and ≤0.07 CFU/ml, low: ≤4 EB/ml and ≤0.4 CFU/ml, and medium: ≤12 EB/ml and ≤1.2 CFU/ml) were tested over the course of eight non-consecutive days (2 runs/day) using three lots of reagents on each of two instruments; each run (total 24 runs with 864 valid CT results and 864 valid NG results) contained three replicates of each sample for each specimen type. For each target and specimen type, the approximate 1 × LOD level hit rate was calculated confirming ~95% reactivity. For each target and specimen type, the approximate 3× LOD level hit rate was calculated confirming ≥99% reactivity.
Analytical specificity/cross-reactivity, exogenous and endogenous interfering substances
A panel of 193 microorganisms, consisting of 151 unique subspecies of bacteria, fungi, protozoa, and viruses, including those commonly found in the male and female urogenital tract, 21 representatives of non-gonorrhoeae Neisseria strains, and 130 other phylogenetically unrelated organisms, were tested with cobas® CT/NG to assess analytical specificity (see package insert for details). The organisms were spiked at concentrations of approximately 1 × 106 units (CFU, IFU or cells)/ml for bacteria and approximately 1 × 105 units/ml for viruses (TCID50 endpoint dilution assay) into pools of negative swab specimens. Testing was performed with each potentially interfering organism alone as well as with each organism mixed with CT and NG cultures at ≤12 EB/ml and ≤1.2 CFU/ml.
The effect of over-the-counter or prescription feminine products that may be present in urogenital specimens (n = 18 including antibacterial and antifungal cream, moisturizer, and contraceptive foam), over-the-counter oral hygiene products that may be present in oropharyngeal specimens (n = 8 including tooth paste, cough syrup, and mouthwash), and hygiene and prescription products that may be present in anorectal specimens (n = 11 including suppositories/hemorrhoidal treatment, rash ointment, and vaseline) were evaluated. Testing was done using pooled clinical specimens with spiking of potential interfering substances at levels expected from normal patient usage. Interferents were tested in CT/NG negative specimen pools as well as in specimen pools with CT/NG at ≤120 EB/ml and ≤1.2 CFU/ml, depending on the specimen type tested. CT serovars D and I and NG strains 2948 and 891 were used.
Endogenous substances potentially present in urogenital, oropharyngeal, or anorectal specimens including albumin, glucose, white blood cells, saliva, stool, whole blood, and mucus were tested for interference. Testing was done using pooled clinical specimens with spiking of potential endogenous interferents. Interferents were tested in CT/NG negative specimen pools as well as in the presence of CT/NG at ≤120 EB/ml and ≤1.2 CFU/ml, depending on the specimen type tested. CT serovars D and I and NG strains 2948 and 891 were used in this study.
Cross-contamination studies
Potential cross-contamination causing false-positive results was investigated by alternating very high positive and negative samples of the three media types used with cobas® CT/NG for use on the cobas® 6800/8800 Systems (swabs collected in and urine stabilized with cobas® PCR media, cervical specimens collected in PreservCyt® Solution) over multiple runs. Sample-to-sample cross-contamination was evaluated for each media type by processing 144 CT/NG negative samples and 138 high positive CT/NG samples in a checkerboard configuration across three runs. Run-to-run cross-contamination was evaluated for each media type by testing a full run of 94 CT/NG negative samples following the three checkerboard runs. High positive samples in the study generated Ct values that exceeded 95% or more of signal obtained from specimens of infected patients in the intended use population.
Method correlation studies using clinical specimens
Clinical specimens were collected in the US and in Europe. The collection of specimens in the U.S. included endocervical swabs, vaginal swabs (clinician collected and self-collected with clinician instruction), oropharyngeal swabs, anorectal swabs, urine specimens collected and stabilized in cobas® PCR Media, and cervical specimens collected in PreservCyt® Solution which were tested using the cobas® 4800 CT/NG Test and cobas® CT/NG for use on the cobas® 6800/8800 Systems at Roche Molecular Systems in Pleasanton. All clinician-collected and self-collected vaginal specimens were collected with the woven swab contained in the cobas® PCR Female Swab Sample Kit; endocervical swabs were collected using the woven swab contained in the cobas® PCR Female Swab Sample Kit (for the cobas® 4800 CT/NG Test) and a new flocked swab contained in the cobas® PCR Media Dual Swab Sample Kit (for cobas® CT/NG for use on the cobas® 6800/8800 Systems). Many of the sites for collection of specimens in the US were the same geographically diverse sites used for the “VENUS” trial, the US clinical trial conducted to obtain Food and Drug Administration (FDA) clearance for the cobas® CT/NG Test for use on the cobas® 4800 System (for details see Refs. [20–22]). Anorectal and oropharyngeal swab specimens were collected from a distinct cohort of subjects in the U.S.
An additional sample set including vaginal swabs, and female and male urines were collected from subjects tested at two laboratories in Germany (Bioscientia, Ingelheim and Amedes, Hannover); these laboratories also provided oropharyngeal and anorectal swabs from male and female subjects.
Samples were tested to determine positive percent agreement (PPA), negative percent agreement (NPA), and overall percent agreement (OPA) with 95% confidence intervals (CIs) for cobas® CT/NG for use on the cobas® 6800/8800 Systems compared to the cobas® 4800 CT/NG Test. Testing was conducted from May 2016 through September 2016. Specimens, for which a result was not available for one or both platforms, whether due to failed processing or invalid results, were excluded from the study. Sanger sequencing using a unique set of CT and NG primers was performed on all specimens with discrepant results for CT or NG, as well as to confirm all concordant NG positive oropharyngeal and anorectal specimens; the sequencing process was validated for sensitivity, specificity, and reproducibility using all collected specimen types. Specimens were extracted on the cobas® 6800/8800 Systems and tested at either Roche Molecular Systems in Pleasanton, CA or GENEWIZ (South Plainfield, NJ).
Statistical analysis
To determine precision (reproducibility), standard deviation and percent coefficient of variation (CV) of the Ct values were calculated. PPA, NPA, and OPA of cobas® CT/NG for use on the cobas® 6800/8800 Systems were calculated separately for detection of CT and NG compared to the Cobas® 4800 CT/NG Test. The corresponding two-sided 95% score (Wilson) CI is reported.
Results
Limit of detection and precision
The CT analytical sensitivity for Cobas® CT/NG for use on the Cobas® 6800/8800 Systems was determined to be 40 elementary bodies (EBs) per milliliter for all serovars (A, B, Ba, C, D, E, F, G, H, I, J, K, L1, L2, L3) as well as for the Swedish variant (nvCT), in all claimed specimen types. Some serovars tested positive below 40 EB/ml. In urine, the analytical sensitivity was found to be 20 EB/ml. The NG analytical sensitivity for the assay was determined to be 1.0 colony forming units (CFU) per milliliter (45 NG strains tested) in all claimed specimen types including extragenital specimens. Some gonorrhoeae strains tested positive below 1.0 CFU/ml. Urine revealed an analytical sensitivity of 0.5 CFU/ml.
In regard to precision, all CT/NG samples for each specimen type tested negative in the negative panel, had a hit rate between 20% and 80% for very low positive panel members, had a ~95% hit rate in the low positive panel members (~1× LOD), and had a 100% hit rate in medium positive panel members (~3× LOD). Intra-laboratory precision of Cobas® CT/NG for use on the Cobas® 6800/8800 Systems was found to be very high. Negative panel members tested negative throughout the study. Analysis of standard deviation and percent CV of the Ct values from valid tests performed on positive panel yielded overall CV (%) ranges from 1.62% to 4.05% for CT and from 1.17% to 3.55% for NG. All panels tested above the LOD showed a detection rate of 100%.
Analytical specificity/cross-reactivity, endogenous and exogenous interfering substances
Results of analytical specificity/cross-reactivity indicated that none of the bacteria, fungi, and viruses, including non-gonorrhoeae Neisseria strains and other phylogenetically unrelated organisms tested, interfered with the detection of CT and NG or produced false-positive results in the CT/NG negative matrices. Of the over-the-counter (OTC) feminine hygiene and prescription products tested in urogenital specimens, metronidazole gel, Replens, RepHresh Odor Eliminating Vaginal Gel, and RepHresh Clean Balance produced false-negative or invalid results at levels that may potentially be present in clinical specimens. None of the OTC oral hygiene products tested in oropharyngeal swabs or the OTC anorectal hygiene and prescription products tested in anorectal swabs produced interference to the test when examined at concentrations expected through typical product use.
Interference was noted with whole blood at 10% for urine and PreservCyt® specimens, with stool at 0.4% in anorectal specimens and with cervical mucus at 1% in endocervical specimens.
Cross-contamination studies
Cross-contamination was assessed as an important risk characteristic for high-throughput molecular systems. No cross-contamination events were observed in checkerboard runs with contrived swab or PreservCyt® samples. All results from the CT/NG negative sample runs following the checkerboard runs of each media type were negative for both CT and NG. Two CT/NG negative samples from the urine checkerboard runs were positive for both CT and NG leading to an overall cross-contamination rate of 0.5% (2/432 samples). Importantly, the high positive urine samples used in this study ranged from a mean Ct value of 19.8 to 21.0, which exceeded 95% or more of signal obtained from specimens of infected patients. Since samples with extremely high bacterial concentrations were used in these studies we calculated that sample to sample cross-contamination rates in routine use of cobas® CT/NG would likely be less than 0.5% × 5% × CT (or NG) prevalence in the testing population. Applying a theoretical maximum prevalence of 100%, the cross-contamination rate would be 0.5% × 5% × 100% = 0.025%. Run to run cross-contamination was not observed in this study.
Method correlation study with clinical specimens
To determine the accuracy of cobas® CT/NG for use on the cobas® 6800/8800 Systems, more than 800 samples were tested for each specimen type and compared to results obtained using the cobas® 4800 CT/NG Test. The accuracy of the cobas® 4800 CT/NG test has been studied extensively in prospective clinical trials [20–22]. There were at least 87 CT positive results per sample type except for oropharyngeal swabs (n = 37), and there were at least 18 NG positive results. Correlation analysis for CT and NG results between cobas® CT/NG for use on the cobas® 6800/8800 Systems and the cobas® 4800 CT/NG Test in all samples is presented in Table 1. Overall, there were 120 samples that tested positive for CT on the Cobas® 6800/8800 Systems but negative on the Cobas® 4800 System but only five samples that tested positive for CT on the Cobas® 4800 System and negative on the Cobas® 6800/8800 Systems. Similarly, 40 samples tested NG positive on the Cobas® 6800/8800 Systems but negative on the Cobas® 4800 System whereas only two samples tested NG positive on the Cobas® 4800 System and NG negative on the Cobas® 6800/8800 Systems.
Table 1.
Numbers of positive and negative results in urogential and extragenital samples using cobas® CT/NG for use on the cobas® 6800/8800 Systems compared to the cobas® 4800 CT/NG Test
| Specimen type | Chlamydia trachomatis | Neisseria gonorrhoeae | ||||||
|---|---|---|---|---|---|---|---|---|
| Concordant + | Concordant – | 6800+/4800– | 6800–/4800+ | Concordant + | Concordant – | 6800+/4800– | 6800–/4800+ | |
| Endocervical swab | 114 | 1778 | 15 | 0 | 22 | 1883 | 1 | 1 |
| CC* vaginal swab | 87 | 1040 | 15 | 0 | 20 | 1111 | 1 | 0 |
| SC** vaginal swab | 90 | 1028 | 14 | 0 | 18 | 1100 | 3 | 0 |
| Female urine | 272 | 2083 | 18 | 0 | 23 | 2340 | 4 | 0 |
| Male urine | 114 | 717 | 3 | 0 | 30 | 803 | 0 | 1 |
| PreservCyt† | 157 | 1905 | 11 | 5 | 25 | 2049 | 1 | 0 |
| Oropharyngeal swab | 37 | 1915 | 14 | 0 | 74 | 1864 | 22 | 0 |
| Anorectal swab | 100 | 1871 | 30 | 0 | 71 | 1923 | 8 | 0 |
| All specimens combined | 971 | 12,337 | 120 | 5 | 283 | 13,073 | 40 | 2 |
*CC = clinician-collected
**SC = self-collected
†PreservCyt = cervical sample collected in liquid-based cytology medium
Table 2 shows positive, negative, and overall percent agreement between the two systems. PPAs were greater than 98.4% for CT and greater than 95% for NG in all specimen types with the majority of specimen types having a PPA of 100% for CT and NG. Negative and overall percent agreements were greater than 98.6% for both CT and NG in all specimen types. These results indicate high percent agreement between the CT/NG tests on the cobas® 4800 versus the cobas® 6800/8800 Systems. The vast majority of discordant results were positive with cobas® CT/NG for use on the cobas® 6800/8800 Systems but negative with the cobas® 4800 CT/NG Test pointing towards a higher sensitivity of the former compared to the latter test.
Table 2.
Positive, negative, and overall percent agreement of cobas® CT/NG for use on the cobas® 6800/8800 Systems with the cobas® 4800 CT/NG Test in urogenital and extragential samples
| Specimen type | Chlamydia trachomatis | Neisseria gonorrhoeae | ||||
|---|---|---|---|---|---|---|
| Result (%) | 95% CI (%) | Result | 95% CI (%) | |||
| Endocervical swab | PPA* | 100 | 96.8–100 | PPA | 95.7 | 78.1–99.9 |
| NPA** | 99.2 | 98.6–99.5 | NPA | 99.9 | 99.7–100 | |
| OPA† | 99.2 | 98.7–99.6 | OPA | 99.9 | 99.6–100 | |
| CC‡ vaginal swab | PPA | 100 | 95.8–100 | PPA | 100 | 83.2–100 |
| NPA | 98.6 | 97.7–99.2 | NPA | 99.9 | 99.5–100 | |
| OPA | 98.7 | 97.8–99.3 | OPA | 99.9 | 99.5–100 | |
| SC§ vaginal swab | PPA | 100 | 96.0–100 | PPA | 100 | 81.5–100 |
| NPA | 98.7 | 97.8–99.3 | NPA | 99.7 | 99.2–99.9 | |
| OPA | 98.8 | 97.9–99.3 | OPA | 99.7 | 99.2–99.9 | |
| NPA | 99.1 | 98.6–99.5 | NPA | 99.8 | 99.6–100 | |
| OPA | 99.2 | 98.8–99.5 | OPA | 99.8 | 99.6–100 | |
| Male urine | PPA | 100 | 96.8–100 | PPA | 96.8 | 83.3–99.9 |
| NPA | 99.6 | 98.8–99.9 | NPA | 100 | 99.5–100 | |
| OPA | 99.6 | 99.0–99.9 | OPA | 99.9 | 99.3–100 | |
| PreservCyt|| | PPA | 96.9 | 92.9–99.0 | PPA | 100 | 86.3–100 |
| NPA | 99.4 | 99.0–99.7 | NPA | 99.9 | 99.7–100 | |
| OPA | 99.2 | 98.8–99.6 | OPA | 99.9 | 99.7–100 | |
| Oropharyngeal swab | PPA | 100 | 90.5–100 | PPA | 100 | 95.1–100 |
| NPA | 99.3 | 98.8–99.6 | NPA | 98.8 | 98.2–99.3 | |
| OPA | 99.3 | 98.8–99.6 | OPA | 98.9 | 98.3–99.3 | |
| Anorectal swab | PPA | 100 | 96.4–100 | PPA | 100 | 94.9–100 |
| NPA | 98.4 | 97.8–98.9 | NPA | 99.6 | 99.2–99.8 | |
| OPA | 98.5 | 97.9–99.0 | OPA | 99.6 | 99.2–99.8 | |
| All specimens combined | PPA | 99.5 | 98.8–99.8 | PPA | 99.3 | 97.5–99.9 |
| NPA | 99.0 | 98.8–99.2 | NPA | 99.7 | 99.6–99.8 | |
| OPA | 99.1 | 98.9–99.2 | OPA | 99.7 | 99.6–99.8 | |
*PPA = positive percent agreement
**NPA = negative percent agreement
†OPA = overall percent agreement
‡CC = clinician-collected
§SC = self-collected
||PreservCyt = cervical sample collected in liquid-based cytology medium
The numbers of negative and positive results for CT and NG in oropharyngeal swabs comparing cobas® CT/NG for use on the cobas® 6800/8800 Systems with the cobas® 4800 CT/NG Test are depicted in Table 1. High concordance was found between the two platforms on a large number of oropharyngeal and anorectal swabs. All discordant samples showed positive results with cobas® CT/NG for use on the cobas® 6800/8800 Systems but negative results with the cobas® 4800 CT/NG Test suggesting a higher sensitivity of cobas® CT/NG for use on the cobas® 6800/8800 Systems compared to the cobas® 4800 CT/NG Test.
Discrepancy analysis using Sanger sequencing
All discordant samples as well as all concordant NG positive oropharyngeal and anorectal specimens were Sanger sequenced. Overall, there were 125 discrepant specimens for CT (120 positive on the Cobas® 6800/8800 Systems, five positive on the Cobas® 4800 System), and there were 42 discrepant specimens for NG (40 positive on the Cobas® 6800/8800 Systems, two positive on the Cobas® 4800 System) across all specimen types. The sequencing results for 122 of the 125 discrepant CT specimens (three endocervical swab specimens were unavailable for sequencing) are shown in Table 3. Of 117 discordant samples (CT positive on the Cobas® 6800/8800 Systems but negative on the Cobas® 4800 System), 75 were verified as true positives for CT by Sanger sequencing. Of the five discrepant specimens that were CT positive on the Cobas® 4800 System but CT negative on the Cobas® 6800/8800 System, one was verified as a true positive for CT by Sanger sequencing. The majority of discordant samples with for PCR positive/sequencing negative results showed Ct values of >37.5, suggesting target titers below the sensitivity of Sanger sequencing.
Table 3.
Sanger sequencing results for CT positive discrepant specimens
| Specimen type | Chlamydia trachomatis | |||||
|---|---|---|---|---|---|---|
| 6800+/4800– | Sanger* + | Sanger– | 6800–/4800+ | Sanger + | Sanger – | |
| Endocervical swab** | 15 | 11 | 1 | 0 | N/A | N/A |
| CC† vaginal swab | 15 | 11 | 4 | 0 | N/A | N/A |
| SC‡ vaginal swab | 14 | 7 | 7 | 0 | N/A | N/A |
| Female urine | 18 | 7 | 11 | 0 | N/A | N/A |
| Male urine | 3 | 3 | 0 | 0 | N/A | N/A |
| PreservCyt§ | 11 | 6 | 5 | 5 | 1 | 4 |
| Oropharyngeal swab | 14 | 7 | 7 | 0 | N/A | N/A |
| Anorectal swab | 30 | 23 | 7 | 0 | N/A | N/A |
| All specimens combined | 120* | 75 | 42 | 5 | 1 | 4 |
*Sanger = Sanger sequencing
**3 Endocervical swab samples were missing and not available for sequencing
†CC = clinician-collected
‡SC = self-collected
§PreservCyt = cervical sample collected in liquid-based cytology medium
Sequencing results for all discrepant NG specimens are shown in Table 4. Of the urogenital specimens, 8 of 12 NG discordant samples were confirmed by sequencing. Twenty of 22 discrepant oropharyngeal specimens (NG positive on the Cobas® 6800/8800 Systems but negative on the cobas® 4800 System) were confirmed NG positive by Sanger sequencing, and all 8 anorectal specimens that were NG discrepant (cobas® 6800/8800 Test positive) were confirmed NG positive. In addition, 71 of 74 concordant NG positive oropharyngeal specimens were confirmed positive for NG by Sanger sequencing; similarly, 64 of 71 concordant NG positive anorectal specimens were confirmed NG positive by Sanger sequencing. The majority of PCR positive/sequencing negative results showed Ct values at or below the sensitivity of Sanger sequencing.
Table 4.
Sanger sequencing results for NG discrepant and NG concordant positive specimens
| Specimen type | Neisseria gonorrhoeae | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Concordant + | Sanger* + | Sanger – | 6800+/4800– | Sanger + | Sanger– | 6800–/4800+ | Sanger + | Sanger – | |
| Endocervical swab | 22 | N/A | N/A | 1 | 1 | 0 | 1 | 0 | 1 |
| CC** vaginal swab | 20 | N/A | N/A | 1 | 1 | 0 | 0 | N/A | N/A |
| SC† vaginal swab | 18 | N/A | N/A | 3 | 1 | 2 | 0 | N/A | N/A |
| Female urine | 23 | N/A | N/A | 4 | 3 | 1 | 0 | N/A | N/A |
| Male urine | 30 | N/A | N/A | 0 | N/A | N/A | 1 | 1 | 0 |
| PreservCyt‡ | 25 | N/A | N/A | 1 | 1 | 0 | 0 | N/A | N/A |
| Oropharyngeal swab | 74 | 71 | 3 | 22 | 20 | 2 | 0 | N/A | N/A |
| Anorectal swab | 71 | 64 | 7 | 8 | 8 | 0 | 0 | N/A | N/A |
| All specimens combined | 283 | 135 | 10 | 40 | 35 | 5 | 2 | 1 | 1 |
*Sanger = Sanger sequencing
**CC = clinician-collected
†SC = self-collected
‡PreservCyt = cervical sample collected in liquid-based cytology medium
Discussion
Results presented in this study demonstrate the excellent performance of the cobas® 6800/8800 Systems for high-throughput CT/NG testing. Using these systems, centralized laboratories are able to handle mid to high volume CT/NG testing using both a large variety of established urogenital specimens as well as extragenital specimens (oropharyngeal and anorectal swabs). cobas® CT/NG for use on the cobas® 6800/8800 Systems is the first CE-IVD test that has been validated for use with extragenital specimens in addition to urogenital specimens. Studies have demonstrated the importance of testing extragenital specimens in certain high-risk populations and that >50% of infections may be missed in MSMs when only testing with traditional urogenital specimens [9–14]. In this regard, a recent study in a cohort of female patients attending a Dutch STD clinic revealed that routine universal anorectal screening for CT in women who do not report anal sex or symptoms is feasible, highly acceptable, and superior to the current selective testing on indication since 80% of anorectal CT infections would not have been tested for [17]. In contrast, selective testing appeared to be an appropriate control strategy for anorectal NG infection in women.
The excellent analytical performance of cobas® CT/NG for use on the cobas® 6800/8800 Systems thus provides vastly improved laboratory efficiency for STI screening programs, i.e., all sample types seen in routine practice are validated and demonstrate excellent performance. Improved laboratory efficiencies may also be driven by increased screening in MSMs using PrEP. A recent meta-analysis reported that MSM using PrEP were 25.3 times more likely to acquire NG infection, 11.2 times more likely to acquire CT infection, and 44.6 times more likely to acquire syphilis versus MSM not using PrEP [26]. However, using a modeling approach, Jenness et al. [27] predicted an overall decrease in CT and NG infections in MSMs using PrEP based on adherence to CDC guidelines for MSMs using PrEP [28] with more frequent STD screening despite reduced condom use [27].
Analytical sensitivity of the Cobas® 4800 CT/NG Test was found to be around 40 EB/ml for CT and 1 CFU/ml for NG in most specimen types and was as low as 20 EB/ml and 0.5 CFU/ml for urine samples. Clinical sensitivity expressed as percent agreement with the Cobas® 4800 CT/NG Test was very high ranging from a PPA of ≥98.4% for CT and ≥95% for NG in all specimen types to an NPA and OPA of ≥98.6% for both CT and NG in all specimen types. Sanger sequencing confirmed the validity of the vast majority of 6800/8800 positive and 4800 negative results, thus demonstrating the superior sensitivity of Cobas® CT/NG for use on the Cobas® 6800/8800 Systems. Such high sensitivity is desirable as many STIs are asymptomatic but can readily be transmitted to partners.
Importantly, cross-reactivity was not observed with other non-gonorrheal Neisseria strains. When testing mucosal sites, i.e., genital and especially oropharyngeal specimens, high specificity for NG and lack of cross-reactivity with non-gonorrheal Neisseria spp. is critical [29, 30]. In this regard, during development of Cobas® CT/NG for use on the Cobas® 6800/8800 Systems, algorithm parameters were optimized to decrease the likelihood of reporting a false-positive result. In the present study, exclusivity studies with a large number of non-gonorrheal Neisseria strains as well as results of discordant analysis using sequencing with a validated and distinct target region for clinical specimens confirmed the absence of cross-reactivity; this finding is further strengthened by the lack of cross-reactivity observed when sequencing extragenital samples that gave concordant results.
Lastly, cross-contamination as determined using very high titer samples was very low (0.5%). In the screening population, we determined a theoretical cross-contamination rate of approximately 0.025%. This rate is noteworthy in light of the high-throughput capabilities of the systems allowing approximately 1000 samples to be run in an 8-h shift (see below and Fig. 1). The value of supporting large screening programs has been demonstrated through the identification of infected individuals and the initiation of treatment for the individual and partner(s). Altogether, these measures will support public health efforts to reduce the incidence of CT and NG infections [8].
Correlation analysis between cobas® CT/NG for use on the cobas® 6800/8800 Systems and the cobas® 4800 CT/NG Test found that the overall percent agreement was >95% for both CT and NG in all specimen types. Discordant analysis further demonstrated the improved sensitivity of cobas® CT/NG for use on the cobas® 6800/8800 Systems compared to the cobas® 4800 CT/NG Test at continued high specificity. For the PCR+/sequencing– results, the majority of Ct values were ≥37.5, suggesting target titers below the sensitivity of Sanger sequencing. The increased sensitivity of cobas® CT/NG for use on the cobas® 6800/8800 Systems is likely caused by changes that have been introduced to the assay. While primer and probe design are the same for the assays for use on the cobas® 4800 and the cobas® 6800/8800 Systems, continuous mixing of the sample during the lysis incubation step was introduced and allows for a higher rate of DNA recovery. In addition, a lower elution volume provides a more concentrated extract, also contributing to higher sensitivity.
cobas® CT/NG for use on the cobas® 6800/8800 Systems offers simplicity and flexibility for mid to high volume laboratories. The cobas® 6800/8800 Systems currently offer the highest throughput solution for molecular CT/NG testing, achieving up to 384 CT/NG results in an 8-h shift on the cobas® 6800 System and up to 960 CT/NG results in an 8-h shift on the cobas® 8800 System. In addition, based on the large onboard storage capacity, enough CT/NG reagents, and positive and negative controls can be loaded to process more than 4600 results. With this large reagent capacity and an open stability for reagent cassettes of 90 days, the system offers a true walk-away solution. The operator interaction required for performance of the test is minimal and consists of loading of samples and consumables (at the start of a shift and as needed during shifts, depending on test mix and volume), clearing the waste containers from time to time, and managing the assays via onboard touch screen interface. Unlike other solutions for CT/NG testing, cobas® CT/NG offers ready to load reagent cartridges, with no reagent reconstitution, no pouring, no mixing, no thawing required. With a walk-away time of 8 h (cobas® 6800) or 4 h (cobas® 8800), staff can efficiently be used to perform other duties while samples are being processed.
In a German laboratory performing high-throughput screening for CT [31], several mid-throughput platforms were retired after the introduction of a single cobas® 8800 System. Figure 2 shows performance characteristics for the cobas® 4800, cobas® 6800, and cobas® 8800 systems when run in batch mode (1 batch = 94 samples) at this site. The implementation of the high-throughput system allowed for an increased number of batches per day to be performed with a theoretical maximum of 16 batches (1504 samples) in one working day (two shifts, total working hours 7 AM to 6.30 PM). At this time point, due to the very recent implementation of the cobas® 8800, only up to 12 batches are performed in a single working day. Thus, one Cobas® 8800 alone can handle the same number of batches as five cobas® 4800 systems during the aforementioned laboratory hours; an even higher throughput could be achieved using the walk-away ability of the cobas® 8800 system, i.e., initiating runs at the end of the working day and releasing results the next morning.
Fig. 2.
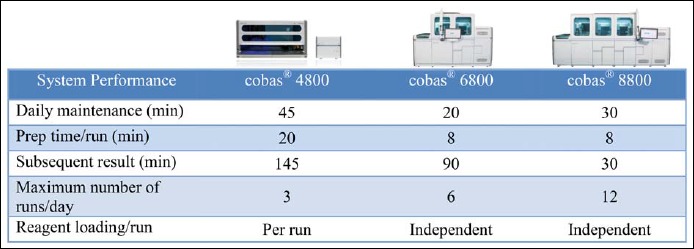
Comparison of system performances between the cobas® 4800, cobas® 6800, and cobas® 8800 Systems (1 run = 96 results, including controls) at a high-throughput customer site
While the cobas® 6800/8800 Systems allow for test consolidation, it also allows for capacity to accommodate growing testing needs. For example, high-throughput testing is a hallmark of screening for cervical cancer. cobas® CT/NG for use on the cobas® 6800/8800 Systems has also been validated for detection of CT/NG in cervical specimens collected in PreservCyt® solution (commonly performed for cervical cancer screening via molecular tests and/or Pap smears). On the cobas® 6800 and 8800 Systems, CT/NG and HPV can be processed from the same cervical sample (aliquot from PreservCyt® solution) during the same run. In addition, all sample types for cobas® CT/NG can be processed during the same run as samples being processed using cobas® HPV.
Furthermore, as testing menus expand on the cobas® 6800 and 8800 Systems, the ability to consolidate testing will become a key to a laboratories workflow and testing expansion. The cobas® 6800 and 8800 Systems are able to leverage common reagents, consumables, and processing profiles, where technically feasible. Using this approach will allow laboratories to process CT/NG and TV/MG (currently in development) from one patient sample during the same run with no sorting or reloading of patient samples, reducing the turn-around-time for delivering results back to a clinician. Since CT/NG and TV/MG are both combination tests, there would be enough patient sample available for cases where a repeat test or additional testing is required. Other reproductive health tests (i.e., Ureaplasma urealyticum) could be developed and run as laboratory-developed tests (LDTs) using the cobas® omni utility channel on the cobas® 6800/8800 Systems. The cobas® omni utility channel allows laboratories to develop and run LDTs on the same system (processed during different runs) as their IVDs reducing instrument footprint and staff training requirements in the laboratory. The flexibility and ability to accommodate additional testing can provide laboratories with a system that allows for growth, efficiency, and administrative planning.
Conclusion
cobas® CT/NG for use on the cobas® 6800 and 8800 Systems exhibited high sensitivity, specificity, PPV, and NPV for the detection of CT and NG in urogenital (endocervical swabs, urine specimens, and vaginal swabs collected in cobas® PCR Media and cervical specimens collected in PreservCyt® Solution) as well as extragenital (oropharyngeal and anorectal swabs) specimens to support screening of at risk populations. The unprecedented capacity for high-throughput testing of up to 960 samples in an 8-h shift makes the cobas® 6800 and 8800 Systems an attractive platform for molecular laboratories.
Acknowledgements
We are grateful to Pari Hemyari, Enrique Marino, and Jesse Canchola for expert statistical support. We are also grateful to Kate Suileabhain and Michael Lewinski for critical review of the article.
Footnotes
Funding sources
This study was funded by Roche Molecular Systems, Pleasanton, CA.
Conflict of interest
E.M., D.H., M.K., R.A., T.S., B.S., and O.L. are employees of Roche Molecular Diagnostics.
References
- 1.Detels R, Green AM, Klausner JD, Katzenstein D, Gaydos C, Handsfield H, Pequegnat W, Mayer K, Hartwell TD, Quinn TC: The incidence and correlates of symptomatic and asymptomatic Chlamydia trachomatis and Neisseria gonorrhoeae infections in selected populations in five countries. Sex Transm Dis 38, 503–509 (2011) [PMC free article] [PubMed] [Google Scholar]
- 2.Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, Low N, Stevens G, Gottlieb S, Kiarie J, Temmerman M: Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10, e0143304 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price MJ, Ades AE, De Angelis D, Welton NJ, Macleod J, Soldan K, Simms I, Turner K, Horner PJ: Risk of pelvic inflammatory disease following Chlamydia trachomatis infection: analysis of prospective studies with a multistate model. Am J Epidemiol 178, 484–492 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb SL, Xu F, Brunham RC: Screening and treating Chlamydia trachomatis genital infection to prevent pelvic inflammatory disease: interpretation of findings from randomized controlled trials. Sex Transm Dis 40, 97–102 (2013) [DOI] [PubMed] [Google Scholar]
- 5.Rekart ML, Gilbert M, Meza R, Kim PH, Chang M, Money DM, Brunham RC: Chlamydia public health programs and the epidemiology of pelvic inflammatory disease and ec-topic pregnancy. J Infect Dis 207, 30–38 (2013) [DOI] [PubMed] [Google Scholar]
- 6.Romoren M, Hussein F, Steen TW, Velauthapillai M, Sundby J, Hjortdahl P, Kristiansen IS: Costs and health consequences of chlamydia management strategies among pregnant women in sub-Saharan Africa. Sex Transm Infect 83, 558–566 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sexton J, Garnett G, Rottingen JA: Metaanalysis and metaregression in interpreting study variability in the impact of sexually transmitted diseases on susceptibility to HIV infection. Sex Transm Dis 32, 351–357 (2005) [DOI] [PubMed] [Google Scholar]
- 8.Workowski KA, Bolan GACenters for Disease C, Prevention: Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep 64, 1–137 (2015) [PMC free article] [PubMed] [Google Scholar]
- 9.Kent CK, Chaw JK, Wong W, Liska S, Gibson S, Hubbard G, Klausner JD: Prevalence of rectal, urethral, and pharyngeal chlamydia and gonorrhea detected in 2 clinical settings among men who have sex with men: San Francisco, California, 2003. Clin Infect Dis 41, 67–74 (2005) [DOI] [PubMed] [Google Scholar]
- 10.Annan NT, Sullivan AK, Nori A, Naydenova P, Alexander S, McKenna A, Azadian B, Mandalia S, Rossi M, Ward H, Nwokolo N: Rectal chlamydia – a reservoir of undiagnosed infection in men who have sex with men. Sex Transm Infect 85, 176–179 (2009) [DOI] [PubMed] [Google Scholar]
- 11.Gunn RA, O’Brien CJ, Lee MA, Gilchick RA: Gonorrhea screening among men who have sex with men: value of multiple anatomic site testing, San Diego, California, 1997–2003. Sex Transm Dis 35, 845–848 (2008) [DOI] [PubMed] [Google Scholar]
- 12.Ota KV, Fisman DN, Tamari IE, Smieja M, Ng LK, Jones KE, Diprima A, Richardson SE: Incidence and treatment outcomes of pharyngeal Neisseria gonorrhoeae and Chlamydia trachomatis infections in men who have sex with men: a 13-year retrospective cohort study. Clin Infect Dis 48, 1237–1243 (2009) [DOI] [PubMed] [Google Scholar]
- 13.Schachter J, Moncada J, Liska S, Shayevich C, Klausner JD: Nucleic acid amplification tests in the diagnosis of chlamydial and gonococcal infections of the oropharynx and rectum in men who have sex with men. Sex Transm Dis 35, 637–642 (2008) [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention: Recommendations for the laboratory-based detection of Chlamydia trachomatis and Neisseria gonorrhoeae – 2014. MMWR Recomm Rep 63, 1–19 (2014) [PMC free article] [PubMed] [Google Scholar]
- 15.Javanbakht M, Gorbach P, Stirland A, Chien M, Kerndt P, Guerry S: Prevalence and correlates of rectal chlamydia and gonorrhea among female clients at sexually transmitted disease clinics. Sex Transm Dis 39, 917–922 (2012) [DOI] [PubMed] [Google Scholar]
- 16.van Liere GA, Hoebe CJ, Wolffs PF, Dukers-Muijrers NH: High co-occurrence of anorectal chlamydia with urogenital chlamydia in women visiting an STI clinic revealed by routine universal testing in an observational study; a recommendation towards a better anorectal chlamydia control in women. BMC Infect Dis 14, 274 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Liere G, Dukers-Muijrers N, Levels L, Hoebe C: High proportion of anorectal Chlamydia trachomatis and Neisseria gonorrhoeae after routine universal urogenital and anorectal screening in women visiting the sexually transmitted infection clinic. Clin Infect Dis 64, 1705–1710 (2017) [DOI] [PubMed] [Google Scholar]
- 18.Volk JE, Marcus JL, Phengrasamy T, Blechinger D, Nguyen DP, Follansbee S, Hare CB: No new HIV infections with increasing use of HIV preexposure prophylaxis in a clinical practice setting. Clin Infect Dis 61, 1601–1603 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Der Pol B: Cobas(R) 4800: a fully automated system for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Expert Rev Mol Diagn 13, 131–140 (2013) [DOI] [PubMed] [Google Scholar]
- 20.Van Der Pol B, Liesenfeld O, Williams JA, Taylor SN, Lillis RA, Body BA, Nye M, Eisenhut C, Hook EW, 3rd: Performance of the cobas CT/NG test compared to the Aptima AC2 and Viper CTQ/GCQ assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J Clin Microbiol 50, 2244–2249 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Der Pol B, Taylor SN, Liesenfeld O, Williams JA, Hook EW, 3rd: Vaginal swabs are the optimal specimen for detection of genital Chlamydia trachomatis or Neisseria gonorrhoeae using the Cobas 4800 CT/NG test. Sex Transm Dis 40, 247–250 (2013) [DOI] [PubMed] [Google Scholar]
- 22.Taylor SN, Liesenfeld O, Lillis RA, Body BA, Nye M, Williams J, Eisenhut C, Hook EW, 3rd, Van Der Pol B: Evaluation of the Roche cobas(R) CT/NG test for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in male urine. Sex Transm Dis 39, 543–549 (2012) [DOI] [PubMed] [Google Scholar]
- 23.Cobb B, Simon CO, Stramer SL, Body B, Mitchell PS, Reisch N, Stevens W, Carmona S, Katz L, Will S, Liesenfeld O: The cobas(R) 6800/8800 System: a new era of automation in molecular diagnostics. Expert Rev Mol Diagn 17, 167–180 (2017) [DOI] [PubMed] [Google Scholar]
- 24.Zaninotto M, Plebani M: The “hospital central laboratory”: automation, integration and clinical usefulness. Clin Chem Lab Med 48, 911–917 (2010) [DOI] [PubMed] [Google Scholar]
- 25.Ledeboer NA, Dallas SD: The automated clinical microbiology laboratory: fact or fantasy? J Clin Microbiol 52, 3140–3146 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima N, Davey DJ, Klausner JD: Pre-exposure prophylaxis for HIV infection and new sexually transmitted infections among men who have sex with men. AIDS 30, 2251–2252 (2016) [DOI] [PubMed] [Google Scholar]
- 27.Jenness SM, Weiss KM, Goodreau SM, Gift T, Chesson H, Hoover KW, Smith DK, Liu AY, Sullivan PS, Rosenberg ES: Incidence of gonorrhea and chlamydia following HIV preexposure prophylaxis among men who have sex with men: a modeling study. Clin Infect Dis 65, 712–718 (2017), DOI: 10.1093/cid/cix439 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.US Public Health Service Preexposure prophylaxis for the prevention of HIV infection in the United States – 2014: a clinical practice guideline. https://wwwcdcgov/hiv/pdf/prepguideline s2014pdf (2014) [Google Scholar]
- 29.Tabrizi SN, Unemo M, Limnios AE, Hogan TR, Hjelmevoll SO, Garland SM, Tapsall J: Evaluation of six commercial nucleic acid amplification tests for detection of Neisseria gonorrhoeae and other Neisseria species. J Clin Microbiol 49, 3610–3615 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Upton A, Bromhead C, Whiley DM: Neisseria gonorrhoeae false-positive result obtained from a pharyngeal swab by using the Roche cobas 4800 CT/NG assay in New Zealand in 2012. J Clin Microbiol 51, 1609–1610 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohm I, Groning A, Sommer B, Muller HW, Krawczak M, Glaubitz R: A German Chlamydia trachomatis screening program employing semi-automated real-time PCR: results and perspectives. J Clin Virol 46 (Suppl 3), S27–S32 (2009) [DOI] [PubMed] [Google Scholar]


