Abstract
Pituitary adenylate cyclase activating polypetide (PACAP) constitutes a neuropeptide that is widely distributed in the host exerting essential cytoprotective properties, whereas PACAP–/– mice display increased susceptibility to distinct immunopathological conditions. The orchestrated interplay between the gut microbiota and the host is pivotal in immune homeostasis and resistance to disease. Potential pertubations of the intestinal microbiota in PACAP–/– mice, however, have not been addressed so far. For the first time, we performed a comprehensive survey of the intestinal microbiota composition in PACAP–/– and wildtype (WT) mice starting 2 weeks postpartum until 18 months of age applying quantitative culture-independent techniques. Fecal enterobacteria and enterococci were lower in PACAP–/– than WT mice aged 1 month and ≥6 months, respectively. Whereas Mouse Intestinal Bacteroides were slightly higher in PACAP–/– versus WT mice aged 1 and 6 months, this later in life held true for Bacteroides/Prevotella spp. (≥12 months) and lactobacilli (>15 months of age). Strikingly, health-beneficial bifidobacteria were virtually absent in the intestines of PACAP–/– mice, even when still breastfed. In conclusion, PACAP deficiency is accompanied by distinct changes in fecal microbiota composition with virtually absent bifidobacteria as a major hallmark that might be linked to increased susceptibility to disease.
Keywords: intestinal microbiota dynamics, pituitary adenylate cyclase activating polypeptide (PACAP), bifidobacteria, intestinal ecology, gut–brain axis, predisposition to immunopathology, probiotic immunomodulation
Introduction
Pituitary adenylate cyclase activating polypeptide (PACAP) was initially identified as a neuropeptide in the hypothalamus stimulating adenylate cyclase activity in the pituitary gland [1]. Beyond the nervous system, PACAP is widely expressed in peripheral organs of the endocrine, respiratory, reproductive, and digestive system as well as in lymphoid organs including immune cells [2]. PACAP can bind to VPAC1, VPAC2, and PAC1 receptors that are present on innate immune cell subsets such as lymphocytes and macrophages, for instance [3–5]. In the context of its virtual ubiquitous distribution, PACAP exerts a broad mode of biological actions with cytoprotective including anti-inflammatory and anti-apoptotic properties in particular [5, 6]. Hence, PACAP-deficient (PACAP–/–) mice display several morphological, biochemical, and behavioral abnormalities that are characterized by decreased fertility but higher mortalitity rates due to compromised respiratory and metabolic functions, in additon to hyperactivity, decreased anxiety, depression-like behavior, and abnormal stress responses as reviewed by Reglodi and colleagues [6]. PACAP–/– mice have been shown to be more vulnerable to harmful stimuli to the central nervous system, but also peripheral organs including peripheral nerves, kidney, and the intestinal tract [6]. For instance, applying the acute dextran sodium sulfate (DSS)-induced colitis model, PACAP–/– mice displayed 50% higher mortality rates as compared to wildtype (WT) mice [7]. In addition, PACAP–/– mice were suffering from more severe chronic DSS-induced colitis, whereas 60% of diseased animals additionally developed colorectal tumors with aggressive phenotype [8]. Interestingly, naive PACAP–/– mice did not display histopathological changes in their intestinal mucosa. Hence, lack of endogeneous PACAP resulted in increased susceptibility of the murine host to intestinal inflammation and inflammation-induced colonic cancer development.
There is compelling evidence that the host microbiota constitutes a key factor in health and disease of vertebrates. In fact, it has been shown that the gut microbiota is essentially involved in a multitude of physiological processes including food digestion [9], fat metabolism [10], vitamin synthesis [11], intestinal angiogenesis [12], enteric nerve function [13], and protection from pathogens [14, 15] as well as immune cell development [16]. Conversely, pertubations of this complex intestinal ecosystem termed dysbiosis are associated with increased susceptibility of the host to distinct intestinal (e.g., inflammatory bowel disease, irritable bowel syndrome, coeliac disease) as well as to extra-intestinal immunopathological conditions (e.g., multiple sclerosis, autism, depression, allergy, asthma, cardiovascular morbidities) [17]. Hence, the fine-tuned orchestrated interplay between the intestinal commensal microbiota and the host plays a pivotal role in immune homeostasis, host cell physiology, and resistance to disease [17].
To date, however, no data are available regarding the intestinal microbiota composition of PACAP–/– mice. In the present study, we therefore addressed whether PACAP deficiency was associated with qualitative and/or quantitative differences in the gut microbiota repertoire that in turn may be linked to the phenotypes and increased susceptibility to immunopatholocal conditions observed in PACAP gene-deficient mice. To accomplish this, we performed a comprehensive kinetic survey of the intestinal microbiota composition in PACAP–/– and corresponding WT mice starting 2 weeks postpartum until 18 months of age applying culture-independent molecular techniques.
Methods
Mice
PACAP gene-deficient (PACAP–/–) mice and corresponding WT counterparts were raised and maintained in open cages within an experimental semi-barrier (accessible only with laboratory coat, overshoes, caps, and sterile gloves) under standard conditions (22–24 °C room temperature, 55 ± 15% humidity, 12 h light/12 dark cycle) in the animal facilities of the University of Pecs (Pecs, Hungary). Mice had unlimited access to autoclaved tap water and standard chow. Comparable numbers of male and female age-matched PACAP–/– and WT mice were included in the study at defined time points postpartum. Genotypes of mice were confirmed by polymerase chain reaction (PCR) as described elsewhere [18].
Molecular analysis of fecal microbiota
Fresh fecal pellets were collected from corresponding mice at 2 weeks, 1 month, 3 months, 6 months, 12 months, and >15 months (i.e., between 15 and 18 months) of age, immediately snap-frozen in liquid nitrogen and stored at –80 °C until further processing. DNA was extracted from fecal samples as described previously [19, 20]. In brief, DNA was quantified by using Quant-iT PicoGreen reagent (Invitrogen, UK) and adjusted to 1 ng/µl. Then, the main bacterial groups abundant in the murine intestinal microbiota including enterobacteria, enterococci, lactobacilli, bifidobacteria, Bacteroides/Prevotella spp., Mouse Intestinal Bacteroides, Clostridium coccoides group, and Clostridium leptum group as well as total eubacterial loads were assessed by quantitative real-time PCR (qRT-PCR) with species-, genera-, or group-specific 16S rRNA gene primers (Tib MolBiol, Germany) as described previously [15, 21–23], and numbers of 16S rRNA gene copies per nanogram DNA of each sample were determined.
Statistical analysis
Medians and levels of significance were determined by Mann–Whitney U test (GraphPad Prism v5, La Jolla, CA, USA) as indicated. Two-sided probability (p) values of ≤0.05 were considered significant.
Ethics statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments (BA02/2000-15024/2011).
Results
Comprehensive culture-independent analysis of the fecal microbiota in PACAP–/– and WT mice over time
In order to provide a comprehensive kinetic survey of the main bacterial groups abundant in the intestinal tract of PACAP–/– and WT mice, we collected fecal samples of age- and sex-matched mice starting 2 weeks postpartum until the age of 18 months. Notably, due to limited numbers in progressively aged and surviving mice lacking the PACAP gene, results derived from mice aged 15 to 18 months of either genotype were pooled (indicated as >15 months age group) and subjected to statistical analyses.
Total eubacterial gene numbers were highest in fecal samples derived from both PACAP–/– and WT mice at the beginning of the observation period as early as 2 weeks postpartum and higher as compared to any time points analyzed thereafter (i.e., >15 months; p < 0.001; Fig. 1). Furthermore, irrespective of the genotype, total bacterial loads were lower in fecal samples taken from mice ≥6 months as compared to 1-month-old animals (p < 0.05–0.01; Fig. 1). By the age of ≥15 months, WT but not PACAP–/– mice displayed lower eubacterial gene numbers in their intestines as compared to corresponding mice ≤6 months of age (p < 0.05–0.001; Fig. 1A). It is important to note, however, that the observed differences were rather minor (approximately 0.5 order of magnitude).
Fig. 1.
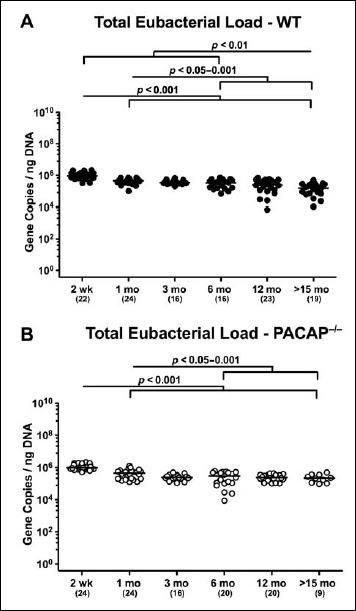
Total eubacterial gene numbers in fecal samples derived from PACAP-deficient and wildtype mice over time. Individual intestinal total eubacterial loads were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
When assessing gene numbers of commensal enterobacteria including Escherichia coli, 2-week-old and 1-month-old WT mice harbored up to four orders of magnitude more enterobacteria in their intestines as compared to WT mice aged ≥3 months (p < 0.001; Fig. 2A). In fact, fecal enterobacteria burdens were lower in mice of the oldest cohort as compared to any other time point (p < 0.05–0.001; Fig. 2A). In 2-week-old PACAP–/– mice, fecal enterobacteria loads were higher as compared to animals aged ≥3 months (p < 0.001; Fig. 2B). Furthermore, PACAP–/– mice over 12 months of age exhibited less enterobacteria in their feces than 1-month-old and 3-month-old animals (p < 0.001; Fig. 2B). The overall decrease in fecal enterobacteria burdens observed during the entire survey period was approximately 2.5 orders of magnitude in PACAP–/– animals (Fig. 2B).
Fig. 2.
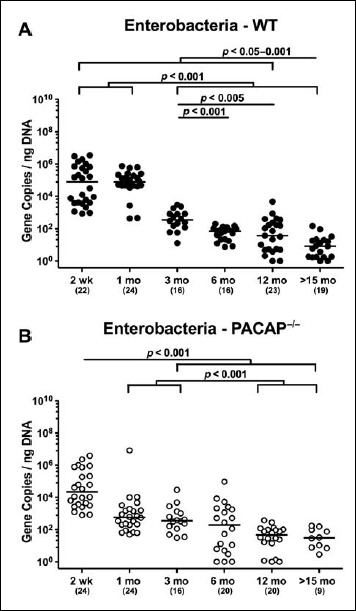
Fecal gene numbers of enterobacteria in PACAP-deficient and wildtype mice over time. Individual intestinal loads of enterobacteria were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
In mice of either genotype, not only fecal enterococci gene numbers were rather low but also kinetic changes were minor. Three-month-old WT mice displayed 1.0–1.5 orders of magnitude lower enterococci burdens as compared to younger counterparts (p < 0.005; Fig. 3A). In ≤1-month-old PACAP–/– mice, enterococci gene numbers were higher than those detected in feces of ≥3-month-old animals (p < 0.005; Fig. 3B). Notably, enterococci loads in gene-deficient mice older than 3 months were close to the detection limit (Fig. 3B).
Fig. 3.
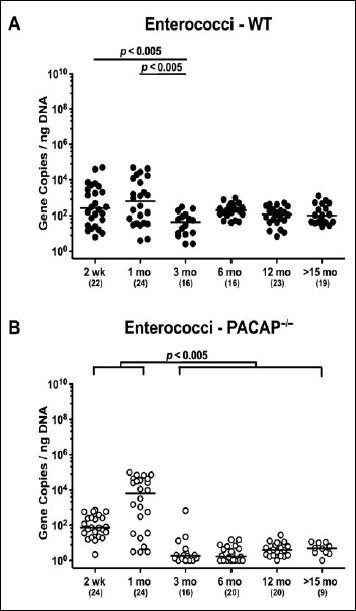
Fecal gene numbers of enterococci in PACAP-deficient and wildtype mice over time. Individual intestinal loads of enterococci were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Irrespective of the genotype, intestinal lactobacilli loads were approximately two orders of magnitude higher in ≤3-month-old mice than in older counterparts (p < 0.05–0.001; Fig. 4). In addition, WT mice in the youngest cohort displayed slightly higher fecal burdens of lactobacilli as compared to 1-month-old animals (p < 0.001; Fig. 4A).
Fig. 4.
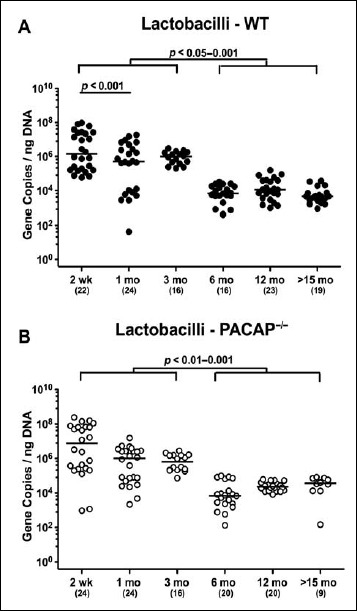
Fecal gene numbers of lactobacilli in PACAP-deficient and wildtype mice over time. Individual intestinal loads of lactobacilli were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Two-week-old WT mice harbored higher bifidobacteria gene numbers in their feces as compared to younger (except for 3-month-old) mice (p < 0.001; Fig. 5A). Even though 6-month-old WT animals exhibited very low intestinal bifidobacteria numbers in their intestines, the fecal loads further declined to the detection limit thereafter (p < 0.05–0.01; Fig. 5A). Remarkably, even in the youngest cohort, bifidobacteria were (if at all) abundant only in very low numbers (i.e., close to the detection limit) in the intestinal tract of PACAP–/– mice (Fig. 5B).
Fig. 5.
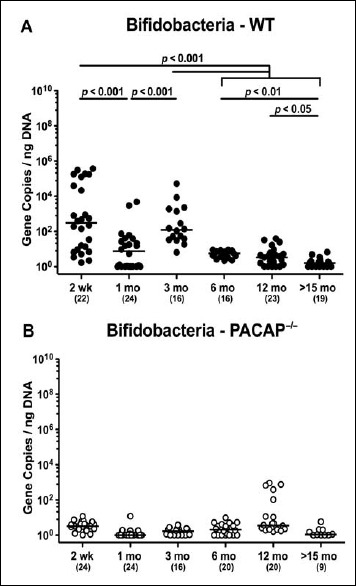
Fecal gene numbers of bifidobacteria in PACAP-deficient and wildtype mice over time. Individual intestinal loads of bifidobacteria were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
When analyzing obligate anaerobic gram-negative commensals, fecal loads of Bacteroides/Prevotella spp. were highest in the youngest and lowest in the oldest cohort of WT mice (p < 0.05–0.001; Fig. 6A). In PACAP–/– mice, however, fecal gene numbers of Bacteroides/Prevotella spp. slightly decreased until 3 months of age (p < 0.001) but were higher thereafter (p < 0.05–0.005) and comparable to loads observed in the youngest cohort (Fig. 6B).
Fig. 6.
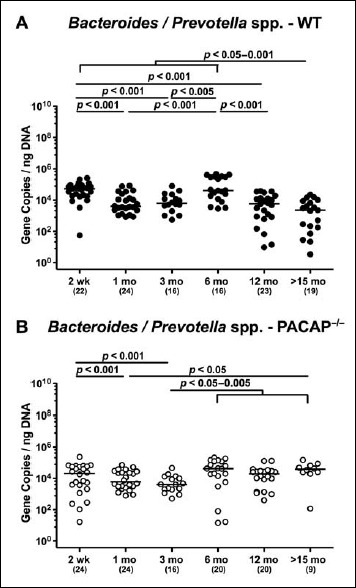
Fecal gene numbers of Bacteroides/Prevotella spp. in PACAP-deficient and wildtype mice over time. Individual intestinal loads of Bacteroides/Prevotella spp. were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
In both PACAP–/– and WT mice, mouse intestinal Bacteroides were up to 1.5 orders of magnitude higher in the fecal samples derived from animals aged ≥3 months as compared to 2-week-old and 1-month-old counterparts (p < 0.05–0.001; Fig. 7). Conversely, fecal gene numbers of Clostridium coccoides were slightly lower in >15-month-old as compared to ≤3-month-old WT mice (p < 0.05–0.005; Fig. 8A). In >6-month-old PACAP–/– mice, C. coccoides loads were up to 1.5 orders lower in the feces as compared to those detected in younger animals (p < 0.05–0.001; Fig. 8B). Fecal gene numbers of the C. leptum group increased after 2 weeks of age in WT animals by approximately one order of magnitude and remained stable until the end of the observation period (p < 0.05–0.001; Fig. 9A). Interestingly, intestinal C. leptum loads were virtually comparable over time in PACAP–/– mice (n.s.; Fig. 9B).
Fig. 7.
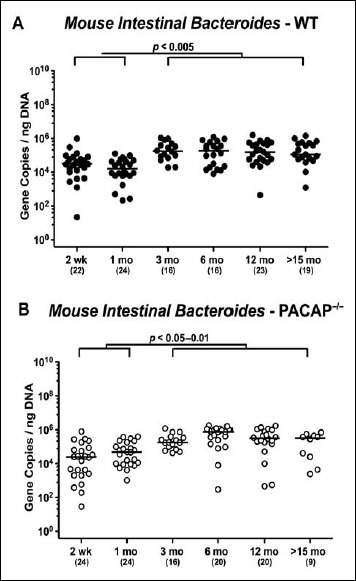
Fecal gene numbers of Mouse Intestinal Bacteroides in PACAP-deficient and wildtype mice over time. Individual intestinal loads of Mouse Intestinal Bacteroides were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Fig. 8.
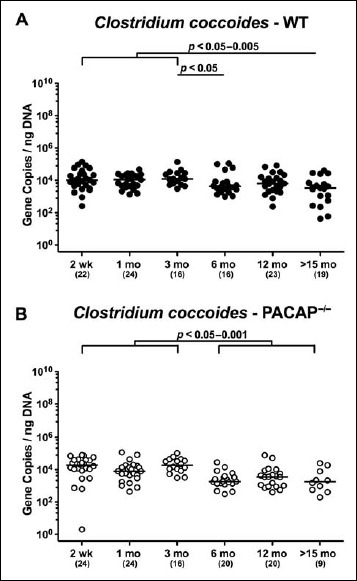
Fecal gene numbers of Clostridium coccoides in PACAP-deficient and wildtype mice over time. Individual intestinal loads of Clostridium coccoides were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Fig. 9.
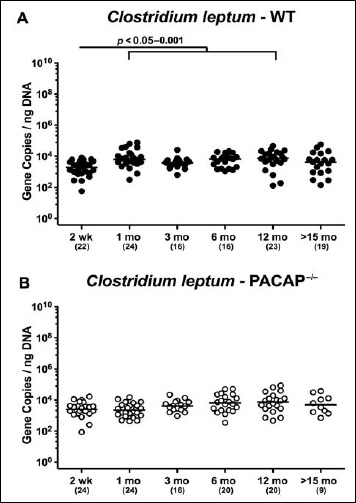
Fecal gene numbers of Clostridium leptum in PACAP-deficient and wildtype mice over time. Individual intestinal loads of Clostridium leptum were quantitatively assessed in fecal samples derived from (A) wildtype (WT, closed circles) and (B) pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses) and significance levels (p values), determined by Mann–Whitney U test are indicated
Genotype-dependent differences in fecal microbiota of PACAP-/- and WT mice at defined time points postpartum
When comparing total eubacterial gene numbers in fecal samples derived from WT and PACAP–/– mice at defined time points postpartum, no differences could be observed (n.s.; Fig. 10). Fecal enterobacteria loads, however, were two orders of magnitude lower in 1 month old PACAP–/– as compared to age-matched WT mice (p < 0.001; Fig. 11 A), whereas, conversely, respective loads were slightly higher in the former versus the latter when older than 15 months (p < 0.05; Fig. 11 A). PACAP–/– mice harbored more enterococci in their intestines as compared to WT counterparts, whereas the opposite was true for ≥6-month-old mice (p < 0.001; Fig. 11B). Whereas lactobacilli were higher in fecal samples derived from PACAP–/– as compared to WT mice of the respective oldest cohorts (i.e., >15 months of age, p < 0.005; Fig. 12A), intestinal bifidobacteria were virtually absent in PACAP–/– mice and up to two orders of magnitude lower until the age of 6 months (p < 0.001; Fig. 12B). Obligate anaerobic gram-negative commensals such as Bacteroides/Prevotella spp. were up to 1.5 order of magnitude higher in ≥12-month-old PACAP–/– as compared to age-matched WT control animals (p < 0.05–0.001; Fig. 13A), which also held true for Mouse Intestinal Bacteroides in respective 1-month-old and 6-month-old mice (p < 0.05; Fig. 13B). Interestingly, fecal loads of obligate gram-positive species such as C. coccoides and C. leptum groups did not differ in PACAP–/– and WT mice at either time point postpartum (n.s.; Fig. 14).
Fig. 10.
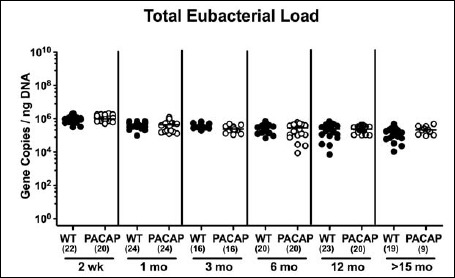
Fecal total eubacterial gene numbers in PACAP-deficient as compared to wildtype mice over time. Individual intestinal total eubacterial loads were quantitatively assessed in fecal samples derived from wildtype (WT, closed circles) and pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians and numbers of analyzed animals (in parentheses) are indicated
Fig. 11.
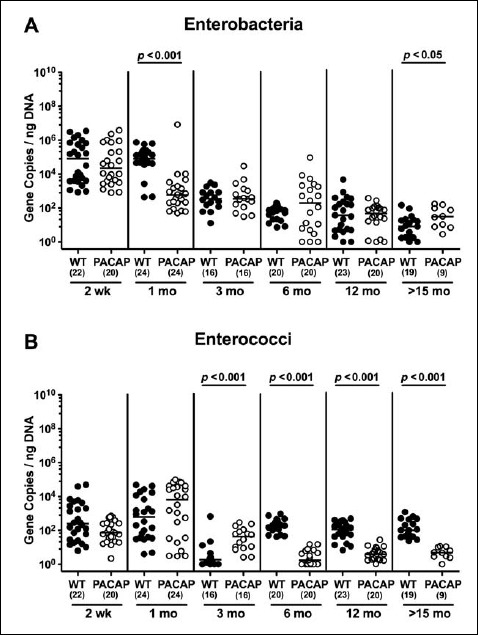
Fecal gene numbers of enterobacteria and enterococci in PACAP-deficient as compared to wildtype mice over time. Individual loads of (A) enterobacteria and (B) enterococci were quantitatively assessed in fecal samples derived from wildtype (WT, closed circles) and pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Fig. 12.
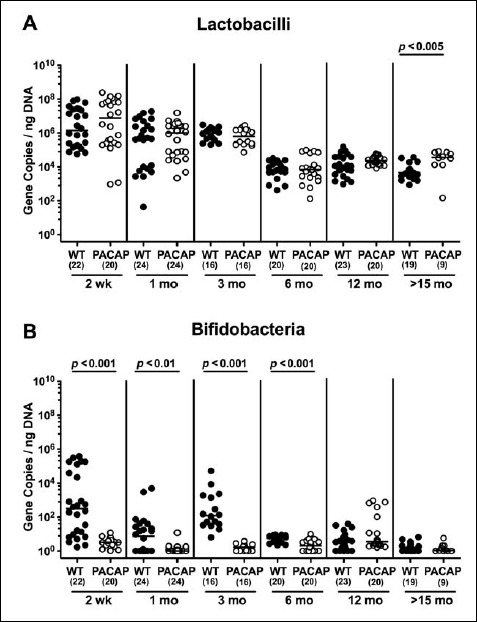
Fecal gene numbers of lactobacilli and bifidobacteria in PACAP-deficient as compared to wildtype mice over time. Individual loads of (A) lactobacilli and (B) bifidobacteria were quantitatively assessed in fecal samples derived from wildtype (WT, closed circles) and pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Fig. 13.
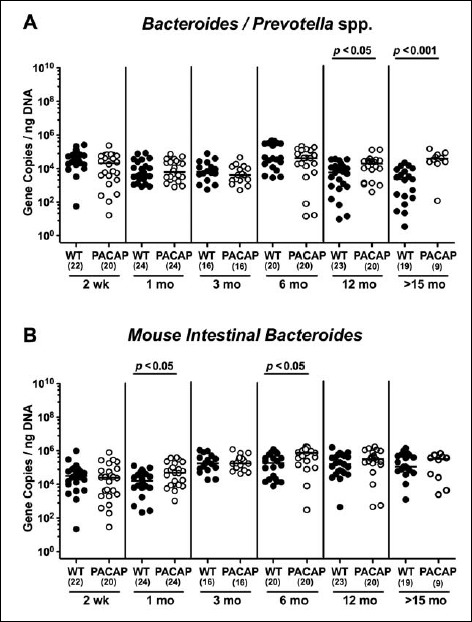
Fecal gene numbers of Bacteroides/Prevotella spp. and Mouse Intestinal Bacteroides in PACAP-deficient as compared to wildtype mice over time. Individual loads of (A) Bacteroides/Prevotella spp. and (B) Mouse Intestinal Bacteroides were quantitatively assessed in fecal samples derived from wildtype (WT, closed circles) and pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians, numbers of analyzed animals (in parentheses), and significance levels (p values) determined by Mann–Whitney U test are indicated
Fig. 14.
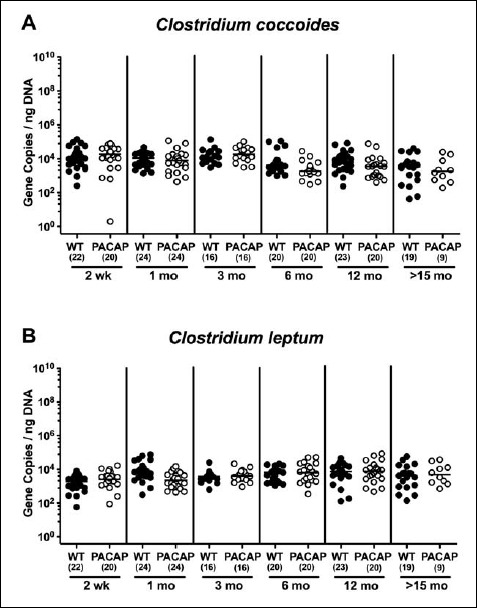
Fecal gene numbers of Clostridium coccoides and Clostridum leptum in PACAP-deficient as compared to wildtype mice over time. Individual loads of (A) Clostridium coccoides and (B) Clostridum leptum were quantitatively assessed in fecal samples derived from wildtype (WT, closed circles) and pituitary adenylate cyclase activating polypeptide-deficient (PACAP–/–, open circles) mice at defined time points postpartum (as indicated on the x axis; wk: weeks; mo: months) by qRT-PCR and expressed as 16S rRNA gene numbers per nanogram DNA. Medians and numbers of analyzed animals (in parentheses) are indicated
Discussion
During the past years, the commensal microbiota has gained increasing attention concerning their impact in health and disease [24]. Microbial colonization and host immune development and maturation run in parallel and are essentially involved in regulation of intestinal (patho) physiology in mice and men [25, 26]. Given the pivotal interactions between the intestinal microbiota and host physiology, pertubations of this fine-tuned, orchestrated interplay result in increased vulnerability of the host to external (harmful) stimuli and increased susceptibility to disease not only of the intestinal tract [17, 27], in fact, phenomena that are shared by PACAP gene-deficient mice [6]. This prompted us to present a comprehensive kinetic survey of the intestinal microbiota composition and their changes “during lifetime” of PACAP–/– and corresponding WT mice for the first time.
The microbial colonization of the intestinal tract starts immediately after birth with facultative anaerobic commensal species such as enterobacteria, enterococci, and lactobacilli as the first colonizers [28], as we could confirm with highest respective fecal loads in the youngest murine cohort (i.e., 2 weeks postpartum) in our study. Irrespective of the genotype, fecal gene numbers of respective commensals declined with progressive aging, particularly after 3 months of age. Conversely, obligate anaerobic bacterial species including Bacteroides and Clostridium species, but also bifidobacteria, usually establish gradually in mice and men [29, 30]. Our results are well in line, given that Mouse Intestinal Bacteroides increased in either mice after 1 month of age, for instance, whereas Bacteroides/Prevotella spp. were lowest in feces of >15-month-old WT but not PACAP–/– mice. Strikingly, bifidobacteria were virtually absent in the intestinal tract of PACAP–/– mice, even in still breastfed 2-week-old infants. In WT mice, however, bifidobacterial loads were highest in 2-week-old mice and subsequently declined after 3 months of age.
Bifidobacteria belong to the phylum Actinobacteria and constitute the predominant microbial group in healthy breastfed babies. It remains relatively stable during adulthood and tend to decrease upon senescence [28]. Whereas bifidobacteria are considered important for intestinal homeostasis, a delayed bifidobacterial colonization of the intestinal tract during infancy results in increased susceptibility to diseases in childhood and later in life [31]. For instance, dysbiosis with decreased bifidobacterial numbers have been associated with distinct immunopathologies such as inflammatory bowel disease, irritable bowel disease, celiac disease, and atopic disease [28]. In our own studies, conventionally colonized mice gene-deficient for nucleotide-binding oligomerization domain (NOD) 2 or Toll-like receptor (TLR) 9 were virtually deprived from intestinal bifidobacteria but suffered from more distinct Toxoplasma gondii-induced inflammatory sequelae in intestinal and extra-intestinal compartments including the brain [20, 32].
Genome-based analyses revealed a number of factors that might explain host-beneficial mechanisms exerted by bifidobacteria as reviewed by Klijn et al. [33]: production of 1) bacteriocins, further of 2) lactic acid and 3) acetic acid, thereby lowering the intraluminal pH, 4) exerting functional oxidative stress response mechanisms, and thus supporting the host phalanx angainst invading (entero) pathogens as well as providing metabolic features such as 5) production of vitamins, 6) hydrolysis of bile salts, 7) production of conjugated linoleic acid, and 8) degradation of oxalate. 9) Importantly, a very recent report provided a direct link between bifidobacteria and neuromodulatory activities, given that a particular Bifidobacterium dentium strain was able to modulate visceral sensitivity in the intestinal tract by production of γ-aminobutyric acid (GABA), the primary inhibitory neurotransmitter in the central nervous system of mammals [34]. This report together with our study further underlines the (patho)physiological importance of the gut–brain axis.
Given their health-promoting features, viable bifidobacterial strains have been introduced into probiotic formulations such as VSL#3 and have been shown efficient in ameliorating symptoms and maintaining remission of patients suffering from inflammatory bowel disease [28]. Very recently, we were able to demonstrate the extensive impact of VSL#3 treatment on mucosal, peripheral, and systemic innate and adaptive immunity exerting beneficial anti-inflammatory effects in intestinal as well as systemic compartments of mice that had been subjected to broad-spectrum antibiotic treatment [27]. In addition, VSL#3 treatment was sufficient to attenuate intestinal, but also extra-intestinal including systemic pro-inflammatory responses upon murine infection with the enteropathogen Campylobacter jejuni [35].
It is, hence, highly likely that the lack of intestinal bifidobacteria observed in PACAP–/– mice might be one key factor explaining their vulnerability to cell-toxic stimuli and increased susceptibility to immunopathological conditions in both intestinal and extra-intestinal compartments.
In addition, we were able to show that facultative anaerobic commensals such as enterobacteria (e.g., E. coli) were lower in the intestines of 1-month-old PACAP–/– as compared to WT mice, which also held true for enterococci later in murine life (i.e., ≥6 months of age). Whereas gene numbers of obligate anaerobic gram-negative commensals such as Mouse Intestinal Bacteroides were slightly higher in the feces of PACAP–/– versus WT mice aged 1 and 6 months, this held true for Bacteroides/Prevotella spp. in the intestines later in life (i.e., ≥12 months of age).
Conclusion
We conclude that the described genotype-dependent differences in gut microbiota composition and their development over time with the virtually absent intestinal bifidobacteria as hallmark might explain the distinct phenotypes observed in PACAP–/– mice upon cell-toxic (i.e., oxidative, traumatic, inflammatory) challenges further supporting the major (patho)physiological impact of the gut–brain axis.
Footnotes
Funding sources
This work was supported by grants from the German Research Foundation (DFG) to M.M.H. (HE3040/3-1) and from the National Research, Development and Innovation Fund K119759, 115874 (A.T.); GINOP-2.3.2-15-2016-00050 “Pepsys” (D.R. and A.T.), PTE AOK KA Research grant 2017-17 (G.H.); National Brain Research program (KTIA_13_NAP-A-III/5) (D.R. and A.T.); TAMOP 4.2.4.A/2-11-1-2012-0001 National Excellence Program; UNKP-16-4-IV New National Excellence Program of the Ministry of Human Capacities (A.T.); MTA TKI 14016 Program (D.R.), Bolyai Scholarship (GH. and A.T.); EFOP-3.6.3-VEKOP-16-2017-00008 project (co-financed by the European Union); and the European Social Fund (D.R., V.V., G.H., A.T., B.F.). The present scientific contribution is dedicated to the 650th anniversary of the foundation of the University of Pécs, Hungary.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Conflict of interest
Stefan Bereswill and Markus M. Heimesaat are Editorial Board members.
References
- 1.Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, Culler MD, Coy DH: Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun 164, 567–574 (1989) [DOI] [PubMed] [Google Scholar]
- 2.Vaudry D, Gonzalez BJ, Basille M, Yon L, Fournier A, Vaudry H: Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol Rev 52, 269–324 (2000) [PubMed] [Google Scholar]
- 3.Gomariz RP, Juarranz Y, Abad C, Arranz A, Leceta J, Martinez C: VIP-PACAP system in immunity: new insights for multitarget therapy. Ann N Y Acad Sci 1070, 51–74 (2006) [DOI] [PubMed] [Google Scholar]
- 4.Abad C, Gomariz RP, Waschek JA: Neuropeptide mimetics and antagonists in the treatment of inflammatory disease: focus on VIP and PACAP. Curr Top Med Chem 6, 151–163 (2006) [DOI] [PubMed] [Google Scholar]
- 5.Vaudry D, Falluel-Morel A, Bourgault S, Basille M, Burel D, Wurtz O, Fournier A, Chow BK, Hashimoto H, Galas L, Vaudry H: Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol Rev 61, 283–357 (2009) [DOI] [PubMed] [Google Scholar]
- 6.Reglodi D, Kiss P, Szabadfi K, Atlasz T, Gabriel R, Horvath G, Szakaly P, Sandor B, Lubics A, Laszlo E, Farkas J, Matkovits A, Brubel R, Hashimoto H, Ferencz A, Vincze A, Helyes Z, Welke L, Lakatos A, Tamas A: PACAP is an endogenous protective factor-insights from PACAP-deficient mice. J Mol Neurosci 48, 482–492 (2012) [DOI] [PubMed] [Google Scholar]
- 7.Azuma YT, Hagi K, Shintani N, Kuwamura M, Nakajima H, Hashimoto H, Baba A, Takeuchi T: PACAP provides colonic protection against dextran sodium sulfate induced colitis. J Cell Physiol 216, 111–119 (2008) [DOI] [PubMed] [Google Scholar]
- 8.Nemetz N, Abad C, Lawson G, Nobuta H, Chhith S, Duong L, Tse G, Braun J, Waschek JA: Induction of colitis and rapid development of colorectal tumors in mice deficient in the neuropeptide PACAP. Int J Cancer 122, 1803–1809 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper LV, Midtvedt T, Gordon JI: How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22, 283–307 (2002) [DOI] [PubMed] [Google Scholar]
- 10.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. : The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101, 15718–15723 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M: Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 24, 160–168 (2013) [DOI] [PubMed] [Google Scholar]
- 12.Stappenbeck TS, Hooper LV, Gordon JI: Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A 99, 15451–15455 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husebye E, Hellstrom PM, Midtvedt T: Intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of migrating myoelectric complex. Dig Dis Sci 39, 946–956 (1994) [DOI] [PubMed] [Google Scholar]
- 14.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB: Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76, 4726–4736 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Dasti JI, Zautner AE, Muñoz M, Loddenkemper C, Gross U, Göbel UB, Heimesaat MM: Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6, e20953 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cebra JJ: Influences of microbiota on intestinal immune system development. Am J Clin Nutr 69, 1046S–1051S (1999) [DOI] [PubMed] [Google Scholar]
- 17.Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, Scheffold A, Kühl AA, Bereswill S, Heimesaat MM: Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front Immunol 8, 397 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farkas J, Sandor B, Tamas A, Kiss P, Hashimoto H, Nagy AD, Fulop BD, Juhasz T, Manavalan S, Reglodi D: Early neurobehavioral development of mice lacking endogenous PACAP. J Mol Neurosci 61, 468–478 (2017) [DOI] [PubMed] [Google Scholar]
- 19.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, Jahn HK, Dunay IR, Moter A, Gescher DM, Schumann RR, Göbel UB, Liesenfeld O: Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol 177, 8785–8795 (2006) [DOI] [PubMed] [Google Scholar]
- 20.Bereswill S, Kuhl AA, Alutis M, Fischer A, Mohle L, Struck D, Liesenfeld O, Göbel UB, Dunay IR, Heimesaat MM: The impact of Toll-like-receptor-9 on intestinal microbiota composition and extra-intestinal sequelae in experimental Toxoplasma gondii induced ileitis. Gut Pathogens 6, 19 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C, Steinhoff U, Tchaptchet S, Thiel E, Freudenberg MA, Göbel UB, Uharek L: MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 59, 1079–1087 (2010) [DOI] [PubMed] [Google Scholar]
- 22.Rausch S, Held J, Fischer A, Heimesaat MM, Kühl AA, Bereswill S, Hartmann S: Small intestinal nematode infection of mice is associated with increased enterobacterial loads alongside the intestinal tract. PLoS One 8, e74026 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heimesaat MM, Boelke S, Fischer A, Haag LM, Loddenkemper C, Kühl AA, Göbel UB, Bereswill S: Comprehensive postmortem analyses of intestinal microbiota changes and bacterial translocation in human flora associated mice. PLoS One 7, e40758 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Klitzing E, Oz F, Ekmekciu I, Escher U, Bereswill S, Heimesaat MM: Comprehensive survey of intestinal microbiota changes in offspring of human microbiota-associated mice. Eur J Microbiol Immunol (Bp) 7, 65–75 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, Marchesi JR, Collado MC: The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26, 26050 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI: The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1, 6ra14 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekmekciu I, von Klitzing E, Fiebiger U, Neumann C, Bacher P, Scheffold A, Bereswill S1, Heimesaat MM: The Probiotic compound VSL#3 modulates mucosal, peripheral, and systemic immunity following murine broad-spectrum antibiotic treatment. Front Cell Infect Microbiol 7, 167 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tojo R, Suarez A, Clemente MG, de los Reyes-Gavilan CG, Margolles A, Gueimonde M, Ruas-Madiedo P: Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol 20, 15163–15176 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arboleya S, Solis G, Fernandez N, de los Reyes-Gavilan CG, Gueimonde M: Facultative to strict anaerobes ratio in the preterm infant microbiota: a target for intervention? Gut Microbes 3, 583–588 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langille MG, Meehan CJ, Koenig JE, Dhanani AS, Rose RA, Howlett SE, Beiko RG: Microbial shifts in the aging mouse gut. Microbiome 2, 50 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar A, Mandal S: Bifidobacteria-Insight into clinical outcomes and mechanisms of its probiotic action. Microbiol Res 192, 159–171 (2016) [DOI] [PubMed] [Google Scholar]
- 32.Heimesaat MM, Dunay IR, Alutis M, Fischer A, Möhle L, Göbel UB, Kühl AA, Bereswill S: Nucleotide-oligomerization-domain-2 affects commensal gut microbiota composition and intracerebral immunopathology in acute Toxoplasma gondii induced murine ileitis. PLoS One 9, e105120 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klijn A, Mercenier A, Arigoni F: Lessons from the genomes of bifidobacteria. FEMS Microbiol Rev 29, 491–509 (2005) [DOI] [PubMed] [Google Scholar]
- 34.Pokusaeva K, Johnson C, Luk B, Uribe G, Fu Y, Oezguen N, Matsunami RK, Lugo M, Major A, Mori-Akiyama Y, Hollister EB, Dann SM, Shi XZ, Engler DA, Savidge T, Versalovic J: GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol Motil 29 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekmekciu I, Fiebiger U, Stingl K, Bereswill S, Heimesaat MM: Amelioration of intestinal and systemic sequelae of murine Campylobacter jejuni infection by probiotic VSL#3 treatment. Gut Pathog 9, 17 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]


