Abstract
The World Health Organization has rated multidrug-resistant (MDR) Pseudomonas aeruginosa as a critical threat to human health. In the present study, we performed a survey of intestinal colonization, and local and systemic immune responses following peroral association of secondary abiotic mice with either a clinical MDR P. aeruginosa or a commensal murine Escherichia coli isolate. Depletion of the intestinal microbiota following antibiotic treatment facilitated stable intestinal colonization of both P. aeruginosa and E. coli that were neither associated with relevant clinical nor histopathological sequelae. Either stable bacterial colonization, however, resulted in distinct innate and adaptive immune cell responses in the intestines, whereas a pronounced increase in macrophages and monocytes could be observed in the small as well as large intestines upon P. aeruginosa challenge only, which also applied to colonic T lymphocytes. In addition, TNF secretion was exclusively elevated in large intestines of P. aeruginosa-colonized mice. Strikingly, association of secondary abiotic mice with MDR P. aeruginosa, but not commensal E. coli, resulted in pronounced systemic pro-inflammatory responses, whereas anti-inflammatory responses were dampened. Hence, intestinal carriage of MDR P. aeruginosa as compared to a mere commensal Gram-negative strain in otherwise healthy individuals results in distinct local and systemic pro-inflammatory sequelae.
Keywords: Pseudomonas aeruginosa, multi-drug resistant Gram-negative bacteria, susceptibility to infection, intestinal microbiota, colonization resistance, broad-spectrum antibiotic treatment, secondary abiotic (gnotobiotic) mice, pro-inflammatory immune responses, systemic sequelae of infection, commensal Escherichia coli
Introduction
The strictly aerobic, non-fermenting Gram-negative bacterium Pseudomonas aeruginosa can be ubiquitously found and particularly thrives in moist environments. Whereas a single flagellum and multiple cell surface pili contribute to bacterial motility and adherence to surfaces, biofilm formation, alginate secretion, quorum-sensing, intrinsic antibiotic resistance, and an elaborated secretion system constitute further virulence factors of P. aeruginosa facilitating immune escape and establishment in the host ecosystem [1, 2]. In the hospital setting, water bottles, respiratory equipment, and sinks are typical environmental reservoirs [3]. P. aeruginosa is considered an opportunistic pathogen and one of the most frequent causative agents of acute nosocomial infections, particularly affecting immune-compromised (such as neutropenic) individuals or patients admitted to the intensive care unit (ICU) [4, 5]. P. aeruginosa-induced infections including ventilator-associated pneumonia, burn wound, and surgical site infections, as well as urinary tract infections are associated with very high (i.e., >30%) mortality rates [4]. In addition, P. aeruginosa drives chronic respiratory infections in patients suffering from cystic fibrosis, chronic obstructive lung disease, or bronchiectasis [5–7]. In recent years, emerging infections with strains that were resistant to a plethora of antibiotic classes due to β-lactamases, 16S rRNA methylases, and carbapenemases have been associated with significant morbidity and increasing mortality [5, 7, 8]. This fatal scenario has prompted the World Health Organization (WHO) in the beginning of 2017 to rate multidrug-resistant (MDR) Gram-negative species including P. aeruginosa as serious threats for human health, further emphasizing the urgent need for novel treatment approaches [9]. In addition to colonized patients, contaminated surfaces, and objects in the hospital setting as potential external sources for P. aeruginosa acquisition, the human gastrointestinal tract might be considered as important internal source for P. aeruginosa infection [10, 11]. Although P. aeruginosa is not considered part of the human commensal gut microbiota, intestinal colonization usually precedes P. aeruginosa infection. This might particularly be the case upon antibiotic treatment compromising the intestinal microbiota integrity and, hence, physiological colonization resistance subsequently facilitating establishment of the opportunistic pathogen in the intestinal ecosystem [5, 12]. In fact, a recent study revealed that prior rectal colonization was a predictor of subsequent development of P. aeruginosa infection of ICU patients [13].
To date, however, no data are available whether intestinal P. aeruginosa carriage by otherwise healthy individuals that are treated with antibiotic compounds is accompanied with host immune responses. This prompted us in the present study to treat mice with broad-spectrum antibiotics and to challenge them with either a clinical MDR P. aeruginosa or a murine commensal Escherichia coli strain perorally. We determined intestinal colonization capacities of respective strains either under continuous antibiotic treatment or after antibiotic withdrawal as compared to uncompromised gut microbiota conditions and further assessed intestinal as well as systemic pro-and anti-inflammatory responses in otherwise healthy bacterial carriers.
Materials and methods
Mice and broad-spectrum antibiotic treatment
C57BL/6j wildtype mice were reared and maintained under specific pathogen-free (SPF) conditions in the Forschungseinrichtungen für Experimentelle Medizin (FEM, Charité – University Medicine Berlin). At the age of 8 to 10 weeks, female mice were subjected to broad-spectrum antibiotic treatment. In brief, mice were transferred to sterile cages and treated with a quintuple antibiotic cocktail consisting of ampicillin plus sulbactam (1 g/l; Ratiopharm, Ulm, Germany), vancomycin (500 mg/l; Cell Pharm, Hannover, Germany), ciprofloxacin (200 mg/l; Bayer Vital, Leverkusen, Germany), imipenem (250 mg/l; MSD, Haar, Germany), and metronidazole (1 g/l; Fresenius, Bad Homburg, Germany) via the drinking water ad libitum for 8 weeks [14, 15]. Cultural and culture-independent (i.e., 16S rRNA based molecular) quality control measures revealed virtual absence of bacteria in fecal samples as described earlier [15, 16]. In one group of thus generated secondary abiotic (i.e., gnotobiotic) mice, the antibiotic cocktail was replaced by sterile water three days before P. aeruginosa infection, whereas in another group, antibiotic treatment was continued until the end of the experiment. Sex- and age-matched conventionally colonized mice served as antibiotics-untreated control group.
Bacterial strains
The MDR P. aeruginosa isolate was initially cultured from respiratory material of a patient with nosocomial pneumonia and kindly provided by Prof. Dr. Bastian Opitz (Charité – University Medicine Berlin, Berlin, Germany). Notably, the bacterial strain displayed antimicrobial sensitivity to fosfomycin and colistin only [17].
The commensal E. coli strain had been isolated from a conventional wildtype mouse recently [14]. Of note, the genome of the applied E. coli strain did not contain known virulence genes of pathogenic E. coli such as stx 1 and 2, hlyA, cspA, catA, astA, and katP as confirmed by PCR analysis in a reference laboratory [18].
Prior infection, the P. aeruginosa and E. coli strains were grown on cetrimid agar (Oxoid) for 48 h and MacConkey agar (Oxoid) for 24 h (both from Oxoid), respectively, in an aerobic atmosphere at 37 °C.
Bacterial challenge
On two consecutive days (i.e., days 0 and 1), mice were perorally challenged either with 109 colony forming units (CFU) of the MDR P. aeruginosa or the commensal E. coli strain by gavage in a total volume of 0.3 ml PBS as reported earlier [14, 17, 18].
Cultural analysis of P. aeruginosa and E. coli
Fecal samples were homogenized in sterile PBS. Serial dilutions were then streaked onto Columbia agar supplemented with 5% sheep blood (Oxoid, Germany) and cetrimid or MacConkey agar and incubated in an aerobic atmosphere at 37 °C for 48 and 24 h in order to assess intestinal P. aeruginosa and E. coli loads, respectively, as described previously [14, 17, 18].
Clinical conditions
Macroscopic and/or microscopic abundance of fecal blood was assessed in individual mice on a daily basis by the Guajac method using Haemoccult (Beckman Coulter/PCD, Germany) as described earlier [18–20].
Sampling procedures
Mice were sacrificed 4 weeks (28 days) postinfection (p.i.) by isoflurane treatment (Abott, Germany). Cardiac blood (for serum) and tissue samples from spleen, mesenteric lymph nodes (MLN), ileum, and colon were removed under sterile conditions. Intestinal samples were collected from each mouse in parallel for microbiological, immunological, and immunohistochemical analyses.
Immunohistochemistry
Five-micrometer thin paraffin sections of colonic and ileal ex vivo biopsies were used for in situ immunohistochemical analysis as reported previously [19–21]. In brief, primary antibodies against cleaved caspase-3 (Asp175, Cell Signaling, Beverly, MA, USA, 1:200), Ki67 (TEC3, Dako, Glostrup, Denmark, 1:100), F4/80 (no. 14-4801, clone BM8, eBioscience, 1:50), CD3 (no. N1580, Dako, 1:10), FOXP3 (FJK-16s, eBioscience, San Diego, CA, USA, 1:100), and B220 (eBioscience, 1:200) were used to assess apoptotic cells, proliferating cells, macrophages/monocytes, T lymphocytes, regulatory T cells (Treg), and B lymphocytes, respectively. The average numbers of positively stained cells within at least six high power fields (HPF, 0.287 mm2; 400× magnification) were determined by an independent blinded investigator.
Cytokine detection
Ex vivo biopsies (approximately 1 cm2) derived from colon (cut longitudinally and washed in PBS), MLN, and spleen were placed in 24-well flat-bottom culture plates (Falcon, Germany) containing 500 ml serum-free RPMI 1640 medium (Gibco, life technologies) supplemented with penicillin (100 U/ml, Biochrom, Germany) and streptomycin (100 µg/ml; Biochrom). After 18 h at 37 °C, culture supernatants and serum samples were tested for TNF, IFN-γ, and IL-10 by the Mouse Inflammation Cytometric Bead Assay (CBA; BD Bioscience) on a BD FACSCanto II flow cytometer (BD Bioscience).
Statistical analysis
Medians and levels of significance were determined using Mann–Whitney U test. Two-sided probability (p) values of ≤0.05 were considered significant. Experiments were reproduced twice.
Ethics statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments headed by the “Landesamt für Gesundheit und Soziales” (LaGeSo, Berlin; registration numbers G0039/15). Animal welfare was monitored twice daily by assessment of clinical conditions and weight loss of mice.
Results
Broad-spectrum antibiotic treatment facilitates intestinal colonization of mice with MDR P. aeruginosa
We first addressed how efficient an MDR P. aeruginosa strain was able to colonize the intestinal tract of clinically uncompromised wildtype mice with a conventional gut microbiota. The first 3 days following peroral challenge with 109 CFU P. aeruginosa, fecal loads between 103 and 105 per gram could be assessed on two consecutive days (p < 0.001 versus uninfected mice; Fig. 1A) but were progressively declining thereafter. In fact, only 15.8% of conventional mice harbored P. aeruginosa in their intestines between days 4 and 6 postinfection (p.i.) (with median loads of 0 CFU per gram; n.s. versus uninfected mice; Fig. 1A), whereas P. aeruginosa could be isolated from fecal samples in single cases only later on.
Fig. 1.
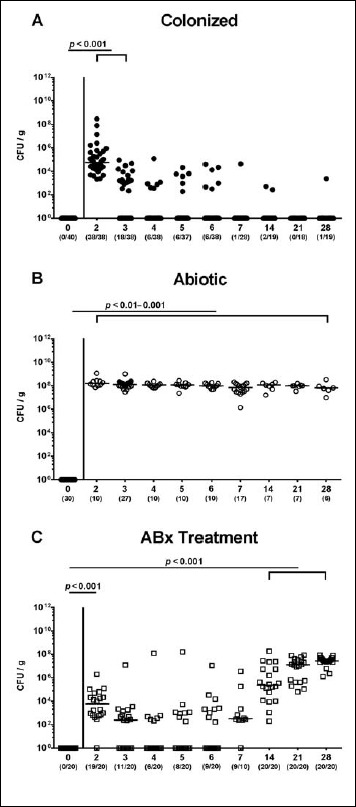
Fecal P. aeruginosa loads over time following peroral challenge of mice. (A) Conventionally colonized wildtype mice (black circles), (B) secondary abiotic mice generated by broad-spectrum antibiotic (ABx) treatment as described in methods (ABx treatment withdrawn 3 days prior infection; white circles), and (C) secondary abiotic mice under continuous ABx treatment (white squares) were perorally associated with a multidrug resistant P. aeruginosa strain on day (d) 0. Intestinal colonization densities were determined in fecal samples until day 28 postinfection by culture and expressed as colony forming units per gram (CFU/g). Numbers of mice harboring P. aeruginosa out of the total number of analyzed mice (in parentheses), medians (black bars), and significance levels (p values) determined by Mann–Whitney U test are indicated. Data shown were pooled from at least three independent experiments
Given that the commensal intestinal microbiota is known to provide the host with a physiological colonization resistance thereby preventing from (opportunistic) pathogenic infection [15, 22], we next generated secondary abiotic mice following broad-spectrum antibiotic treatment. Three days following an antibiotic withdrawal, mice with a virtually depleted gut microbiota were perorally challenged with 109 CFU P. aeruginosa on days 0 and 1. Within 24 h p.i., P. aeruginosa could establish within the intestinal tract of secondary abiotic mice at high median loads of 108 CFU per gram feces that remained stable until day 28 p.i. (p < 0.01–0.001; Fig. 1B).
We further perorally associated secondary abiotic mice under continuous antibiotic treatment with 109 CFU P. aeruginosa, but this time without antibiotic withdrawal and washout. Whereas within 24 h p.i. viable bacteria could be isolated from almost all fecal samples with median loads of 104 CFU per gram (p < 0.001 versus day 0; Fig. 1C), P. aeruginosa detection rates progressively declined until day 6 p.i. After this nadir, however, P. aeruginosa could again be isolated from fecal samples in 90% of mice at day 7 p.i., and median bacterial loads further increased up to 108 CFU per gram until day 28 p.i. (Fig. 1C) which had been observed in secondary abiotic mice (Fig. 1B). Notably, under continuous antibiotic treatment, also small colony variants of P. aeruginosa could be detected, particularly during the nadir between days 4 and 6 p.i., but also thereafter.
Hence, depletion of the intestinal microbiota following antibiotic treatment facilitated stable MDR P. aeruginosa colonization of the intestinal tract.
Comparative colonization properties of MDR P. aeruginosa and commensal E. coli in secondary abiotic mice
We next addressed whether the intestinal colonization properties of the applied MDR P. aeruginosa strain differed from those of a commensal murine Gram-negative strain such as E. coli that had been isolated from feces of a conventionally colonized wildtype mouse previously. Therefore, after a 3-day antibiotic washout period, secondary abiotic mice were either perorally associated with 109 CFU MDR P. aeruginosa or commensal E. coli by gavage and the bacterial loads were determined in fecal samples thereafter. Whereas until day 7 p.i. intestinal E. coli loads were approximately one order of magnitude higher as compared to P. aeruginosa (p < 0.01–0.001; Fig. 2), respective fecal bacterial burdens were comparable thereafter until the end of the experiment (i.e., 108 CFU per gram at day 28 p.i.; n.s., Fig. 2).
Fig. 2.
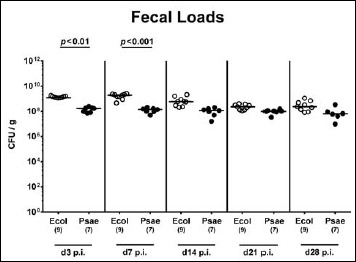
Fecal bacterial loads over time following peroral MDR P. aeruginosa association of secondary abiotic mice. Secondary abiotic mice (antibiotic treatment withdrawn 3 days prior infection) were perorally associated with a murine commensal E. coli (Ecol, white circles) or a multidrug resistant P. aeruginosa strain (Psae, black circles) on day (d) 0. Intestinal colonization densities were determined in fecal samples over time until day 28 postinfection (p.i.) by culture and expressed as colony forming units per gram (CFU/g). Numbers of mice, medians (black bars), and significance levels (p values) determined by Mann–Whitney U test are indicated. Data shown were pooled from two independent experiments
Hence, MDR P. aeruginosa displayed virtually comparable intestinal colonization properties like a commensal Gram-negative strain such as E. coli.
Macroscopic and microscopic inflammatory sequelae following MDR P. aeruginosa colonization of secondary abiotic mice
We further assessed whether potential inflammatory sequelae of MDR P. aeruginosa colonization of secondary abiotic mice could be observed on macroscopic (i.e., clinical) or microscopic (i.e., histological) level. Notably, neither following MDR P. aeruginosa nor upon commensal E. coli challenge, secondary abiotic mice were clinically compromised and exhibited symptoms such as weight loss, wasting, diarrhea, or macroscopic/microscopic abundance of fecal blood (data not shown). We next quantitatively assessed apoptotic cell numbers in intestinal epithelia applying in situ immunohistochemistry. Neither in the colon nor in the ileum of MDR P. aeruginosa-associated secondary abiotic mice increased apoptotic epithelial cell numbers could be observed (n.s., Fig. 3), whereas caspase3+ cells were even higher in the large intestinal epithelia of mice harboring commensal E. coli as compared to naive controls (p < 0.001; Fig. 3A).
Fig. 3.
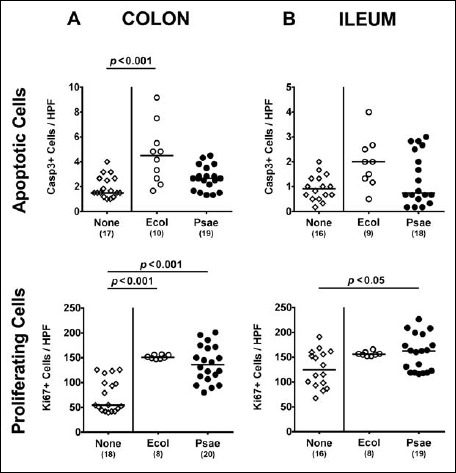
Apoptotic and proliferating intestinal epithelial cells following peroral MDR P. aeruginosa association of secondary abiotic mice. Secondary abiotic mice were perorally associated with a murine commensal E. coli (Ecol, white circles) or a multidrug resistant P. aeruginosa strain (Psae, black circles) on day (d) 0. Four weeks thereafter (i.e., on day 28 postinfection), the average numbers of epithelial apoptotic (positive for caspase 3, Casp3; upper panel) and proliferating cells (positive for Ki67; lower panel) were determined in ex vivo biopsies derived from the (A) colon (left panel) and (B) ileum (right panel) in six high power fields (HPF, 400× magnification) per animal in immunohistochemically stained intestinal paraffin sections. Uninfected mice (none; open diamonds) served as negative controls. Numbers of mice, medians (black bars), and significance levels (p values) determined by the Mann–Whitney U test are indicated. Data shown were pooled from three independent experiments
We further determined numbers of Ki67+ cells in intestinal epithelia indicative for cell proliferation and regeneration counteracting potential bacteria-induced intestinal inflammatory responses. Remarkably, Ki67+ counts were multifold increased in large intestinal epithelia at day 28 following association of secondary abiotic mice with either strain (p < 0.001; Fig. 3A), whereas in the ileal epithelia numbers of proliferating cell were elevated upon MDR P. aeruginosa challenge only (p < 0.05; Fig. 3B).
Hence, MDR P. aeruginosa did neither induce macroscopic (i.e., clinical) nor microscopic (i.e., apoptotic) sequelae upon stable intestinal colonization of secondary abiotic mice.
Intestinal immune cell responses following MDR P. aeruginosa colonization of secondary abiotic mice
We next determined whether association of secondary abiotic mice with MDR P. aeruginosa was accompanied by an increased influx of pro-inflammatory immune cell populations into the intestinal mucosa and lamina propria. To address this, we applied quantitative in situ immunohistochemistry of small and large intestinal ex vivo biopsies that had been stained with antibodies against distinct innate and adaptive immune cell subsets. Following association with MDR P. aeruginosa, but not commensal E. coli, numbers of innate immune cells such as macrophages and monocytes increased more than two-fold in both colon and ileum of secondary abiotic mice at day 28 p.i. (p < 0.001; Fig. 4). Whereas increased T lymphocyte numbers could be determined in the colon of P. aeruginosa associated mice only (p < 0.001; Fig. 4A), this held true for increased ileal CD3+ counts at day 28 upon E. coli challenge (p < 0.001; Fig. 4B). Following association with either bacterial strain, colonic numbers of Treg and B lymphocytes were higher as compared to naive secondary abiotic mice (p < 0.01–0.001; Fig. 4A), whereas this held true for Treg counts in the ileal mucosa and lamina propria at day 28 p.i. (p < 0.001; Fig. 4B).
Fig. 4.
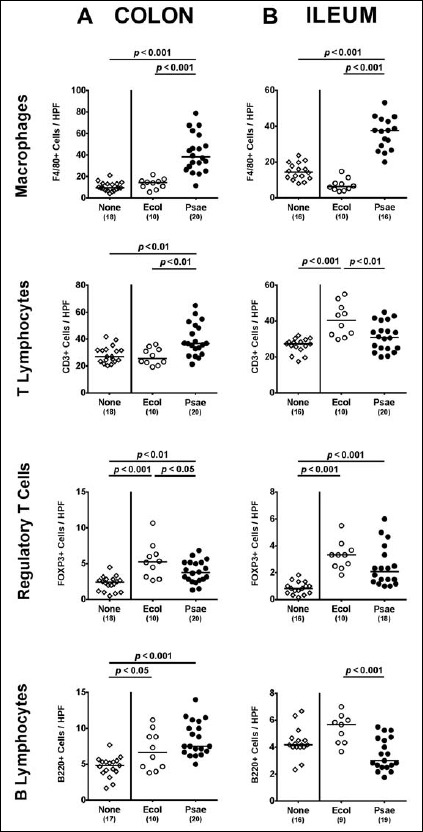
Intestinal immune cell responses following peroral MDR P. aeruginosa association of secondary abiotic mice. Secondary abiotic mice were perorally associated with a murine commensal E. coli (Ecol, white circles) or a multidrug resistant P. aeruginosa strain (Psae, black circles) on day (d) 0. Four weeks thereafter (i.e., on day 28 postinfection), the average numbers of macrophages and monocytes (positive for F4/80), colonic T lymphocytes (positive for CD3), regulatory T cells (positive for FOXP3), and B lymphocytes (positive for B220) were determined in ex vivo biopsies derived from the (A) colon (left panel) and (B) ileum (right panel) in six high power fields (HPF, 400× magnification) per animal in immunohistochemically stained intestinal paraffin sections. Uninfected mice (none; open diamonds) served as negative controls. Numbers of mice, medians (black bars), and significance levels (p values) determined by the Mann–Whitney U test are indicated. Data shown were pooled from three independent experiments
Hence, both MDR P. aeruginosa and commensal E. coli association resulted in distinct innate and adaptive immune cell responses in the intestines of secondary abiotic mice, whereas a pronounced increase in macrophages and monocytes could be observed in the small as well as large intestines upon MDR P. aeruginosa challenge only, which also applied to colonic T lymphocytes.
Intestinal cytokine secretion following peroral MDR P. aeruginosa association of secondary abiotic mice
Next, we addressed whether the observed intestinal immune cell responses upon bacterial challenge were accompanied by pronounced local secretion of pro- and anti-inflammatory cytokines. At day 28 following MDR P. aeruginosa, but not commensal E. coli association of secondary abiotic mice, colonic TNF concentrations were more than two-fold higher as compared to uninfected controls (p < 0.05; Fig. 5A), whereas higher TNF secretion could be determined in MLN of E. coli associated versus non-associated mice (p < 0.001, Fig. 5B). At day 28 following challenge with either E. coli or P. aeruginosa, increased IFN-γ secretion could be measured in MLN as compared to naive controls (p < 0.05 and p < 0.001, respectively; Fig. 5B) with even higher concentrations in the former as compared to the latter associated mice (p < 0.05; Fig. 5B). Notably, secretion of the anti-inflammatory cytokine IL-10 was virtually unchanged in the colon and MLN at day 28 following either bacterial association.
Fig. 5.
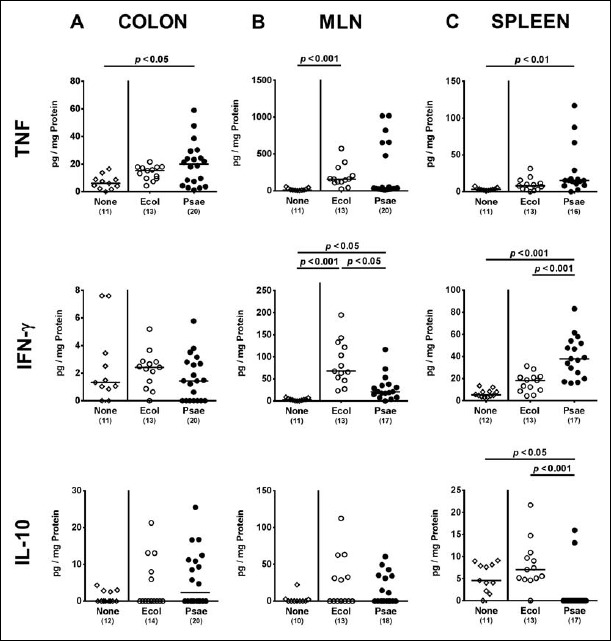
Intestinal cytokine responses following peroral MDR P. aeruginosa association of secondary abiotic mice. Secondary abiotic mice were perorally associated with a murine commensal E. coli (Ecol, white circles) or a multidrug resistant P. aeruginosa strain (Psae, black circles) on day (d) 0. Four weeks thereafter (i.e., on day 28 postinfection), proinflammatory cytokines including TNF (upper panel) and IFN-γ (middle panel) as well as the anti-inflammatory mediator IL-10 (lower panel) were measured in supernatants of ex vivo biopsies derived from the (A) colon (left panel), (B) mesenteric lymph nodes (MLN; middle panel), and (C) spleen (right panel). Uninfected mice (none; open diamonds) served as negative controls. Numbers of mice, medians (black bars), and significance levels (p values) determined by the Mann–Whitney U test are indicated. Data shown were pooled from three independent experiments
Hence, TNF secretion was exclusively increased in large intestines of MDR P. aeruginosa associated secondary abiotic mice.
Systemic cytokine secretion following peroral MDR P. aeruginosa association of secondary abiotic mice
We finally addressed systemic pro- and anti-inflammatory sequelae upon bacterial association of secondary abiotic mice. Remarkably, P. aeruginosa but not E. coli challenge induced a multifold increased secretion of pro-inflammatory cytokines such as TNF and IFN-γ in the spleen at day 28 (p < 0.01 and p < 0.001 versus naive, respectively; Fig. 5C), whereas anti-inflammatory IL-10 concentrations were downregulated in spleens of P. aeruginosa associated mice far below baseline levels (p < 0.001 versus naive; Fig. 5C).
Hence, association of secondary abiotic mice with MDR P. aeruginosa but not commensal E. coli resulted in pronounced systemic pro-inflammatory sequelae, whereas anti-inflammatory responses were dampened.
Discussion
Whereas infections with MDR Gram-negative bacteria including P. aeruginosa have become a serious threat, particularly in hospitalized patients under antibiotic therapy [2, 11], it is to date unclear, however, whether mere carriage of an MDR P. aeruginosa strain in the gastrointestinal tract of otherwise healthy individuals provokes pro-inflammatory immune responses. In the present study, we therefore treated mice with broad-spectrum antibiotic compounds and challenged them perorally with a clinical MDR P. aeruginosa strain – either during continuous antibiotic treatment or after antibiotic withdrawal to assure antibiotic wash-out. Upon cessation of antibiotics, mice could be stably colonized by the opportunistic pathogenic strain as early as 24 h p.i. and displayed continuously high bacterial burdens in their feces until the end of the observation period (i.e., day 28 p.i.). These results are indicative for successful disruption of the colonization resistance (that is physiologically provided by the complex and diverse commensal intestinal microbiota) which in turn facilitates establishment and invasion of opportunistic or obligate pathogens in mice and men [15, 22–24]. The vast majority of mice with an uncompromised intestinal microbiota, however, had expelled the clinical isolate within 4 days p.i.; after 7 days p.i., P. aeruginosa could be cultured from fecal samples in single cases and at very low loads only. Our results are well in line with an earlier study where mice had been treated with five different antibiotics and exhibited distinct intestinal P. aeruginosa loads at day 7 following peroral infection depending on the degree of the compromised colonization resistance due to respective antimicrobial spectra of the applied compound [23]. Furthermore, ingested P. aeruginosa could be cultured from fecal samples of healthy volunteers with an intact gut microbiota up to 6 days following peroral bacterial challenge [25]. In subjects who had a 4-day course of oral ampicillin treatment, however, P. aeruginosa persisted for 14 days.
Interestingly, under continuous broad-spectrum antibiosis, P. aeruginosa could stably establish within the intestinal tract rather late (i.e., after 2 to 3 weeks) p.i., but finally reached comparably high fecal loads as compared to mice in which the antibiotic cocktail had been withdrawn before bacterial challenge. Even though tested resistant against each antibiotic compound in our in vitro antimicrobial susceptibility assays, the nadir in fecal P. aeruginosa loads between day 3 and day 14 p.i. in addition to the abundance of small colony variants point toward synergistic antimicrobial properties of the components within the quintuple antibiotic cocktail. Remarkably, neither antimicrobial treatment nor respective bacterial colonization was associated with any clinical symptoms in challenged mice. This is in line with an earlier study by Hentges et al. who did not report any clinical symptoms in mice upon treatment with five different antibiotic compounds and subsequent P. aeruginosa infection [23], which also held true for healthy volunteers subjected to P. aeruginosa with or without preceding ampicillin treatment [25]. Furthermore, following peroral application of the clinical isolate or the commensal E. coli strain in our study, secondary abiotic mice (with withdrawn antibiosis) harbored comparable fecal bacterial loads indicating that the clinical P. aeruginosa isolate exhibited intestinal colonization like a murine commensal Gram-negative strain.
In line with lacking macroscopic (i.e., clinical) sequelae, P. aeruginosa colonization was neither associated with histopathological nor apoptotic responses affecting the intestinal mucosa and epithelial cells in particular. Interestingly, either bacterial association, however, resulted in increased proliferating/regenerating epithelial cell numbers in both small and large intestines thereby counteracting potential cell damage. Remarkably, MDR P. aeruginosa colonization induced pronounced innate as well as adaptive immune cell responses as indicated by elevated numbers of macrophages/monocytes as well as T lymphocytes, Treg and B cells, respectively, in intestinal mucosa and lamina propria at day 28 p.i., that were accompanied by multifold increased pro-inflammatory cytokine concentrations such as TNF and IFN-γ in colon and MLN, respectively. Strikingly, P. aeruginosa carriage induced not only immune responses in the intestinal tract but also systemically as indicated by increased splenic TNF and IFN-γ secretion at day 28 p.i., whereas anti-inflammatory IL-10 concentration was virtually undetectable by then.
Preceding antibiotic treatment facilitates intestinal P. aeruginosa colonization, which in turn bears a 15-fold increased risk for a nosocomial infection on ICU [13]. Whereas the pathogenic potential of P. aeruginosa is well known, its potential in initiation and perpetuation of immunopathologies, particularly within the intestinal tract, is only incompletely understood. In a very recent study, we were able to show that acute ileitis facilitated infection with MDR P. aeruginosa in mice harboring a human gut microbiota, but the inflammation model was too acute to decipher additional inflammatory sequelae induced by the MDR bacterial strain [17].
We conclude that upon preceding broad-spectrum antibiotic treatment a clinical MDR P. aeruginosa isolate is capable of colonizing the intestinal tract like a commensal Gram-negative strain such as murine E. coli. Already mere MDR P. aeruginosa carriage, however, is associated with both intestinal and systemic immune responses that might be potentiated in concert with other comorbidities. Further studies need to unravel the orchestrated interplay of MDR Gram-negative strains including P. aeruginosa, host microbiota and immunity, particularly under inflammatory conditions.
Acknowledgements
We thank Michaela Wattrodt, Ursula Rüschendorf, Alexandra Bittroff-Leben, Ines Puschendorf, Gernot Reifenberger, Ulrike Fiebiger, and the staff of the animal research facility at Charité – University Medicine Berlin for excellent technical assistance and animal breeding.
Footnotes
Funding sources
This work was supported by grants from the German Research Foundation (DFG) to E.V.K. (SFB633, Immuco), S.B. (SFB633, TP A7), and M.M.H. (SFB633, TP B06 and SFB TR84, TP A05).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Conflict of interest
Stefan Bereswill and Markus M. Heimesaat are Editorial Board members.
References
- 1.Driscoll JA, Brody SL, Kollef MH: The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368 (2007) [DOI] [PubMed] [Google Scholar]
- 2.Gellatly SL, Hancock RE: Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67, 159–173 (2013) [DOI] [PubMed] [Google Scholar]
- 3.Stratton CW: Pseudomonas aeruginosa revisited. Infect Control Hosp Epidemiol 11, 101–104 (1990) [DOI] [PubMed] [Google Scholar]
- 4.Vincent JL: Nosocomial infections in adult intensive-care units. Lancet 361, 2068–2077 (2003) [DOI] [PubMed] [Google Scholar]
- 5.Oliver A, Mulet X, Lopez-Causape C, Juan C: The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updates 21–22, 41–59 (2015) [DOI] [PubMed] [Google Scholar]
- 6.Lyczak JB, Cannon CL, Pier GB: Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2, 1051–1060 (2000) [DOI] [PubMed] [Google Scholar]
- 7.Potron A, Poirel L, Nordmann P: Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45, 568–585 (2015) [DOI] [PubMed] [Google Scholar]
- 8.Livermore DM: Has the era of untreatable infections arrived? J Antimicrob Chemother 64 Suppl 1, 29–36 (2009) [DOI] [PubMed] [Google Scholar]
- 9.Tacconelli E, Magrini N: Global priority list of antibiotic-resistant bacteria to guide research discovery and development of new antibiotics. World Health Organization (2017) [Google Scholar]
- 10.Shooter RA, Walker KA, Williams VR, Horgan GM, Parker MT, Asheshov EH, Bullimore JF: Faecal carriage of Pseudomonas aeruginosa in hospital patients. Possible spread from patient to patient. Lancet 2, 1331–1334 (1966) [DOI] [PubMed] [Google Scholar]
- 11.Cohen R, Babushkin F, Cohen S, Afraimov M, Shapiro M, Uda M, Khabra E, Adler A, Ben Ami R, Paikin S: A prospective survey of Pseudomonas aeruginosa colonization and infection in the intensive care unit. Antimicrob Resist Infect Control 6, 7 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohara T, Itoh K: Significance of Pseudomonas aeruginosa colonization of the gastrointestinal tract. Intern Med 42, 1072–1076 (2003) [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Zorrilla S, Camoez M, Tubau F, Canizares R, Periche E, Dominguez MA, Ariza J, Pena C: Prospective observational study of prior rectal colonization status as a predictor for subsequent development of Pseudomonas aeruginosa clinical infections. Antimicrob Agents Chemother 59, 5213–5219 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, Jahn HK, Dunay IR, Moter A, Gescher DM, Schumann RR, Göbel UB, Liesenfeld O: Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol 177, 8785–8795 (2006) [DOI] [PubMed] [Google Scholar]
- 15.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Dasti JI, Zautner AE, Muñoz M, Loddenkemper C, Loddenkemper C, Gross U, Göbel UB, Heimesaat MM: Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6, e20953 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, Scheffold A, Kuhl AA, Bereswill S, Heimesaat MM: Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front Immunol 8, 397 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Klitzing E, Ekmekciu I, Bereswill S, Heimesaat MM: Acute ileitis facilitates infection with multidrug resistant Pseudomonas aeruginosa in human microbiota-associated mice. Gut Pathog 9, 4 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haag LM, Fischer A, Otto B, Plickert R, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM: Campylobacter jejuni induces acute enterocolitis in gnotobiotic IL-10–/– mice via Toll-like-receptor-2 and -4 signaling. PLoS One 7, e40761 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alutis ME, Grundmann U, Fischer A, Hagen U, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM: The role of gelatinases in Campylobacter jejuni infection of gnotobiotic mice. Eur J Microbiol Immunol (Bp) 5, 256–267 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alutis ME, Grundmann U, Hagen U, Fischer A, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM: Matrix metalloproteinase-2 mediates intestinal immunopathogenesis in Campylobacter jejuni-infected infant mice. Eur J Microbiol Immunol (Bp) 5, 188–198 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C, Steinhoff U, Tchaptchet S, Thiel E, Freudenberg MA, Göbel UB, Uharek L: MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 59, 1079–1087 (2010) [DOI] [PubMed] [Google Scholar]
- 22.Fiebiger U, Bereswill S, Heimesaat MM: Dissecting the interplay between intestinal microbiota and host immunity in health and disease: lessons learned from germfree and gnotobiotic animal models. Eur J Microbiol Immunol (Bp) 6, 253–271 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hentges DJ, Stein AJ, Casey SW, Que JU: Protective role of intestinal flora against infection with Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infect Immun 47, 118–122 (1985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall JC, Christou NV, Meakins JL: The gastrointestinal tract. The “undrained abscess” of multiple organ failure. Ann Surg 218, 111–119 (1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buck AC, Cooke EM: The fate of ingested Pseudomonas aeruginosa in normal persons. J Med Microbiol 2, 521–525 (1969) [DOI] [PubMed] [Google Scholar]


