Abstract
The rising incidences of infections with multidrug-resistant (MDR) Gram-negative bacteria including Pseudomonas aeruginosa (PA) have gained increasing attention in medicine, but also in the general public and global health politics. The mechanisms underlying opportunistic pathogen–host interactions are unclear, however. To address this, we challenged secondary abiotic IL10–/– mice deficient for Toll-like receptor-4 (TLR4–/– × IL10–/–), the main receptor of the Gram-negative cell wall constituent lipopolysaccharide, with a clinical MDR PA isolate. Despite higher intestinal colonization densities, apoptotic colonic epithelial cell numbers were lower in TLR4–/– × IL10–/– mice as compared to IL10–/– controls at day 14 postinfection (p.i.), whereas proliferating/regenerating cells had increased in the latter only. Furthermore, PA-colonized TLR4–/– × IL10–/– mice displayed less distinct innate and adaptive immune cell responses in the colon as compared to IL10–/– counterparts that were accompanied by lower nitric oxide concentrations in mesenteric lymph nodes in the former at day 14 p.i. Conversely, splenic NO levels were higher in both naive and PA-colonized TLR4-deficient IL10–/– mice versus IL10–/– controls. Remarkably, intestinal MDR PA was able to translocate to extra-intestinal including systemic compartments of TLR4–/– × IL10–/– mice only. Hence, MDR PA-induced intestinal and systemic immune responses observed in secondary abiotic IL10–/– mice are TLR4-dependent.
Keywords: Pseudomonas aeruginosa, multidrug-resistant Gram-negative bacteria, colonization resistance, susceptibility to infection, gut microbiota shifts, secondary abiotic (gnotobiotic) IL10-deficient mice, pro-inflammatory immune responses, bacterial translocation, Toll-like receptor-4, lipopolysaccharide
Introduction
The fine-tuned interactions of immune cells, pattern recognition receptors such as Toll-like receptors (TLR), and evolving signaling pathways are pivotal for preventing the host from invading (opportunistic) pathogens and for bacterial clearance [1, 2]. TLR4 is the receptor for sensing cell wall constituents such as lipopetide and lipopolysaccharide (LPS) derived from Gram-negative bacteria including Pseudonomas aeruginosa (PA) [2–4]. Besides LPS, this aerobic opportunistic pathogen harbors a diverse arsenal of virulence factors facilitating adhesion and invasion, further immune escape, establishment, and persistence within the host [5, 6]. Particularly in hospitalized patients with deprived immune functions, PA is one of the most common nosocomial pathogens responsible for severe infections with significant mortality [7]. Acute PA-related morbidities include ventilator-associated pneumonia, infections of surgical sites, burn wounds, and of the urinary tract [8]. Furthermore, PA is a frequent cause for chronic pulmonary infections in patients suffering from chronic obstructive lung disease, bronchiectasis, or cystic fibrosis, for instance [9, 10]. Given its preferential growth in moist environments, water bottles, sinks, and respiratory equipment constitute typical reservoirs in the health-care-associated setting, whereas due to its adhesive properties, PA might be acquired from any contaminated surfaces, instruments, and objects on wards [7, 11]. The human gastrointestinal tract, however, needs to be considered as internal source for subsequent infections, even though PA is not regarded as part of the commensal intestinal microbiota [11, 12]. Particularly antimicrobial treatment, however, compromising the colonization resistance exerted by the complex intestinal microbiota and physiologically preventing the host from (opportunistic) pathogenic infection [13], facilitates human PA colonization. An earlier study revealed that intestinal PA colonization rates of patients increased with duration of hospitalization [11]. Remarkably, an intestinal PA carriage before admission to an intensive care unit (ICU) has been shown to be associated with an up to 15-fold increased risk for subsequent PA infection as compared to non-colonized patients [14]. Particularly the rise of emerging multidrug-resistant (MDR) PA strains expressing extended spectrum, including metallo-β-lactamases, carbapenemases, or 16S rRNA methylases during the past years [10, 15, 16], has prompted the World Health Organization (WHO) in the beginning of 2017 to rate MDR Gram-negative bacteria including PA as serious threat for human health with an urgent need for novel treatment options [17].
Recent surveys revealed that PA could be more frequently detected in the intestinal tract of patients with underlying intestinal inflammatory diseases such as irritable bowel syndrome [18] or ulcerative colitis [19]. Even though the pathogenic properties of PA are well known, there is a large gap in knowledge regarding the crosstalk between mere intestinal carriage of MDR PA and innate immunity. In the present study, we therefore assessed whether 1) intestinal colonization, 2) macroscopic and microscopic sequelae, 3) intestinal and systemic pro-inflammatory immune responses, and 4) bacterial translocation to extra-intestinal including systemic compartments following peroral MDR PA challenge of secondary abiotic IL10–/– mice occurred TLR4-dependently
Materials and methods
Mice and broad-spectrum antibiotic treatment
Female and male IL10–/– mice and IL10–/– mice lacking TLR4 (TLR4–/– × IL10–/–) (all in C57BL/10ScSn background) were bred and housed within the same specific pathogen-free unit of the Forschungseinrichtungen für Experimentelle Medizin (FEM, Charité – University Medicine Berlin). Immediately after weaning (i.e., at the age of 3 weeks), sex-matched mice of either genotype were subjected to a broad-spectrum antibiotic treatment as described earlier [13, 20]. In brief, mice were transferred to sterile cages and treated with a quintuple antibiotic cocktail consisting of ampicillin plus sulbactam (1 g/l; Ratiopharm, Ulm, Germany), vancomycin (500 mg/l; Cell Pharm, Hannover, Germany), ciprofloxacin (200 mg/l; Bayer Vital, Leverkusen, Germany), imipenem (250 mg/l; MSD, Haar, Germany), and metronidazole (1 g/l; Fresenius, Bad Homburg, Germany) via the drinking water ad libitum for 4 months. Cultural and culture-independent (i.e., 16S rRNA based molecular) quality control measures revealed virtual absence of bacteria in fecal samples as described previously [13, 21].
Bacterial strain and murine challenge
The MDR P. aeruginosa isolate was initially isolated from respiratory material of a patient with nosocomial pneumonia and kindly provided by Prof. Dr. Bastian Opitz (Charité – University Medicine Berlin, Berlin, Germany). Notably, the bacterial strain exhibited exclusive antimicrobial sensitivity to fosfomycin and colistin [22]. Before peroral challenge, the P. aeruginosa isolate was grown on Cetrimide agar (Oxoid) for 48 h in an aerobic atmosphere at 37 °C. Three days before peroral bacterial association, the antibiotic cocktail was withdrawn and replaced by sterile water (ad libitum) to assure antibiotic washout [20, 23]. Mice were perorally challenged with 109 colony forming units (CFU) of the MDR P. aeruginosa strain by gavage in a total volume of 0.3 mL PBS as reported earlier [22].
Cultural analysis of P. aeruginosa
To assess intestinal colonization properties of P. aeruginosa, fecal samples were homogenized in sterile PBS. Serial dilutions were then streaked onto Columbia agar supplemented with 5% sheep blood and Cetrimide agar (both Oxoid, Germany) and incubated in an aerobic atmosphere at 37 °C for 48 h in order to assess intestinal P. aeruginosa loads, as described previously [22].
Clinical conditions
Macroscopic and/or microscopic abundance of fecal blood was assessed in individual mice on a daily basis by the Guajac method using Haemoccult (Beckman Coulter/PCD, Germany) as reported earlier [23–25].
Sampling procedures
Mice were sacrificed at day 14 postinfection (p.i.) by isoflurane treatment (Abott, Germany). Tissue samples from spleen, liver, kidney, mesenteric lymph nodes (MLNs), ileum, and colon were removed under sterile conditions. Intestinal samples were collected from each mouse in parallel for microbiological, immunological, and immunohistochemical analyses.
Bacterial translocation
At day of necropsy, P. aeruginosa loads were determined in homogenates of whole tissue ex vivo biopsies derived from MLN, liver (approximately 1 cm3), kidney, and spleen. Serial dilutions (dissolved in sterile PBS) were cultured on Columbia agar supplemented with 5% sheep blood and Cetrimide agar (both Oxoid) for 2 days at 37 °C under aerobic conditions.
The respective weights of feces or tissue samples were determined by the difference of the sample weights before and after asservation. The detection limit of viable bacteria was ≈ 100 CFU per gram.
Immunohistochemistry
Five-micrometer-thin paraffin sections of colonic ex vivo biopsies were used for in situ immunohistochemical analysis as reported previously [24, 26]. In brief, primary antibodies against cleaved caspase-3 (Asp175, Cell Signaling, Beverly, MA, USA, 1:200), Ki67 (TEC3, Dako, Glostrup, Denmark, 1:100), F4/80 (no. 14-4801, clone BM8, eBioscience, 1:50), CD3 (no. N1580, Dako, 1:10), FOXP3 (FJK-16s, eBioscience, San Diego, CA, USA, 1:100), and B220 (eBioscience, 1:200) were used to detect apoptotic cells, proliferating cells, macrophages/monocytes, T lymphocytes, regulatory T cells (Treg), and B lymphocytes, respectively. The average numbers of positively stained cells within at least six high power fields (HPF, 0.287 mm2; 400× magnification) were determined by an independent blinded investigator.
Nitric oxide detection
Ex vivo biopsies (approximately 1 cm2) derived from colon and ileum (cut longitudinally and washed in PBS), as well as from MLN and spleen were placed in 24-well flat-bottom culture plates (Falcon, Germany) containing 500 mL serum-free RPMI 1640 medium (Gibco, Life Technologies) supplemented with penicillin (100 U/ml, Biochrom, Germany) and streptomycin (100 µg/ml; Biochrom). After 18 h at 37 °C, culture supernatants were tested for nitric oxide (NO) secretion by the Griess reaction as described earlier [20].
Statistical analysis
Mean values, medians, and levels of significance were determined using Mann–Whitney U test or Wilcoxon test. Two-sided probability (p) values <0.05 were considered significant. Experiments were reproduced twice.
Ethics statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments headed by the “Landesamt für Gesundheit und Soziales” (LaGeSo, Berlin; registration numbers G0097/12 and G0039/15). Animal welfare was monitored twice daily by assessment of clinical conditions.
Results
Intestinal colonization properties of MDR P. aeruginosa in secondary abiotic IL10–/– mice lacking TLR4
We first generated secondary abiotic TLR4-deficient IL10–/– mice and IL10–/– counterparts to assure stable intestinal MDR PA colonization. In order to prevent mice from potential colitogenic stimuli derived from the commensal intestinal microbiota, mice were subjected to a 4-month course of broad-spectrum antibiotic treatment (via the drinking water) immediately after weaning (i.e., 3 weeks post partum). To assure antibiotic washout, the antibiotic cocktail was withdrawn 3 days prior bacterial challenge. At day 0, mice were perorally subjected to 109 viable PA (a clinical MDR isolate) by gavage and bacterial colonization properties were followed up in fecal samples by culture. As early as 24 h p.i., both TLR4–/– × IL10–/– mice and IL10–/– control animals harbored high median PA loads of approximately 108 CFU per gram feces (Fig. 1). At days 7, 10, and 14 p.i., respective fecal PA counts in IL10–/– mice were slightly lower as compared to day 1 p.i. (p < 0.01; Fig. 1A), whereas this held true for TLR4–/– × IL10–/– mice at the end of the observation period (i.e., day 14 p.i.; p < 0.001; Fig. 1B). Interestingly, at day 7 p.i. and later on, fecal PA numbers were higher in TLR4–/– × IL10–/– mice as compared to IL10–/– counterparts at respective time points (p < 0.01–0.001; Fig. 1). At day 14 p.i., mean PA loads were up to two orders of magnitude higher in both ileum and colon of TLR4–/– × IL10–/– versus IL10–/– mice (p < 0.001; Fig. 2). Hence, MDR PA could stably establish in the intestinal tract of mice irrespective of their genotype, but with higher colonization densities in TLR4-deficient IL10–/– mice.
Fig. 1.
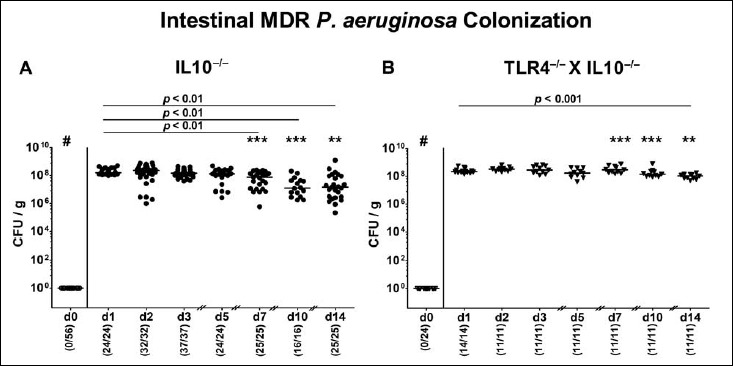
Kinetic analysis of intestinal multidrug-resistant P. aeruginosa colonization properties following peroral association of secondary abiotic IL10–/– mice lacking TLR4. Secondary abiotic (A) IL10-deficient (IL10–/–; closed circles) and (B) TLR4-deficient IL10–/– mice (TLR4–/– × IL10–/–; closed triangles) were generated by broad-spectrum antibiotic treatment as described in Materials and methods section. Following peroral challenge with a clinical multidrug-resistant P. aeruginosa strain at day (d) 0, intestinal colonization densities were determined in fecal samples until d14 postinfection by culture and expressed as colony forming units per gram (CFU/g). Numbers of mice harboring P. aeruginosa out of the total number of analyzed mice (in parentheses), medians (black bars), and significance levels (p values) determined by Wilcoxon and Mann–Whitney U test are indicated. Asterisks illustrate significant differences between genotypes at defined time points (*p < 0.05; **p < 0.01; ***p < 0.001), # indicates significant differences (p < 0.001) between d0 and respective time points thereafter. Data shown were pooled from at least three independent experiments
Fig. 2.
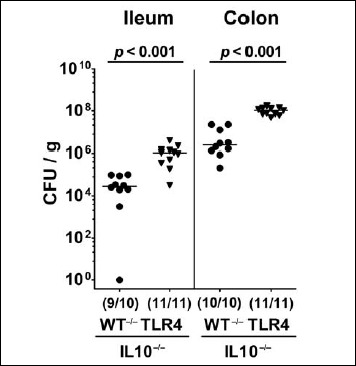
Intestinal loads of multidrug-resistant P. aeruginosa following peroral challenge of secondary abiotic IL10–/– mice lacking TLR4. Secondary abiotic IL10-deficient (WT IL10–/–; closed circles) and TLR4-deficient IL10–/– mice (TLR4–/– IL10–/–; closed triangles) were perorally challenged with a clinical multidrug-resistant P. aeruginosa strain. Two weeks thereafter, the intestinal bacterial loads were quantitatively assessed in luminal samples taken from the ileum and colon by culture and expressed as colony forming units per gram (CFU/g). Numbers of mice harboring P. aeruginosa out of the total number of analyzed mice (in parentheses), medians (black bars), and significance levels (p values) determined by Mann–Whitney U test are indicated. Data shown were pooled from three independent experiments
Macroscopic and microscopic inflammatory sequelae following MDR P. aeruginosa colonization of secondary abiotic IL10–/– mice lacking TLR4
Stable intestinal colonization of secondary abiotic IL10–/– mice with high loads of MDR PA did not lead to overt macroscopic (i.e., clinical) sequelae, given that neither the mice did display any symptoms such as abundance of fecal blood, diarrhea, or wasting nor could a shrinkage of intestinal lengths (as a macroscopic parameter for intestinal inflammation [20, 23]) be observed upon necropsy (not shown).
We therefore assessed potential inflammatory responses upon MDR PA colonization on microscopic level. Given that apoptosis constitutes a well-established parameter for histopathological grading of intestinal inflammation [13], we quantitatively determined apoptotic cells numbers in large intestinal epithelia applying in situ immunohistochemistry Whereas PA colonization did not induce apoptosis in mice of either genotype, apoptotic colonic epithelial cell numbers were lower in PA-colonized TLR4–/– × IL10–/– mice as compared to IL10–/– counterparts at day 14 p.i. (p < 0.01; Fig. 3A). We further determined Ki67+ cell numbers in colonic epithelia indicative for cell proliferation and regeneration counteracting potential MDR PA-induced intestinal inflammatory responses. Interestingly, Ki67+ counts slightly increased within 14 days post MDR PA challenge in IL10–/– (p < 0.05; Fig. 3B), but not in TLR4-deficient IL10–/– mice (n.s.; Fig. 3B). Hence, despite higher intestinal colonization densities, colonic apoptotic cells numbers were lower in TLR4–/– × IL10–/– mice as compared to IL10–/– controls at day 14 p.i., whereas proliferating/regenerating cells had increased in the latter only.
Fig. 3.
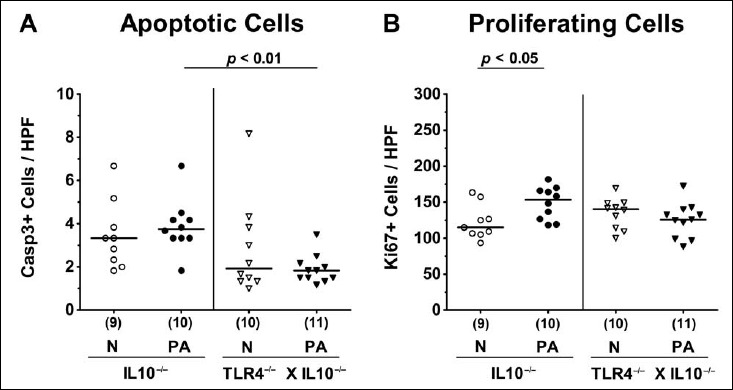
Apoptotic and proliferating intestinal epithelial cells following peroral multidrug-resistant P. aeruginosa colonization of secondary abiotic IL10–/– mice lacking TLR4. Secondary abiotic IL10-deficient (IL10–/–; circles) and TLR4-deficient IL10–/– mice (TLR4–/– × IL10–/–; triangles) were perorally challenged with a clinical multidrug-resistant P. aeruginosa strain (PA; closed symbols). Two weeks thereafter, the average numbers of colonic epithelial (A) apoptotic (positive for caspase 3, Casp3+) and (B) proliferating cells (positive for Ki67; Ki67+) were determined in six high power fields (HPF, 400× magnification) per animal in immunohistochemically stained large intestinal paraffin sections. Non-infected mice (N; open symbols) served as negative controls. Numbers of mice (in parentheses), means (black bars), and significance levels (p values) determined by the Mann–Whitney U test are indicated. Data shown were pooled from two independent experiments
Colonic innate and adaptive immune cell responses following MDR P. aeruginosa colonization of secondary abiotic IL10–/– mice lacking TLR4
We next addressed whether stable colonization of secondary abiotic TLR4-deficient IL10–/– mice with MDR P. aeruginosa was accompanied by an increased influx of innate and adaptive immune cell subsets into the large intestinal mucosa and lamina propria applying quantitative in situ immunohistochemistry of colonic ex vivo biopsies. Within 14 days following PA colonization, numbers of innate immune cells including macrophages and monocytes had increased in the colonic mucosa and lamina propria of both IL10–/– and TLR4–/– × IL10–/– mice (p < 0.001 and p < 0.05, respectively; Fig. 4A). These increases, however, were far less pronounced in TLR4-deficient IL10–/– mice as compared to IL10–/– controls (p < 0.001; Fig. 4A). In fact, TLR4–/– × IL10–/– mice displayed only one third of F4/80+ colonic cell numbers that had been observed in IL10–/– mice at day 14 p.i. (Fig. 4A). Intestinal PA colonization further resulted in elevated numbers of adaptive immune cell subsets such as T lymphocytes and Tregs in the large intestines of IL10–/–, but not TLR4–/– × IL10–/– mice (p < 0.001 and p < 0.05, respectively; Fig. 4B,C). At day 14 p.i., colonic numbers of T lymphocytes, Tregs, and B lymphocytes were lower in TLR4-deficient IL10–/– mice as compared to IL10–/– controls (p < 0.001, p < 0.05, p < 0.01, respectively; Fig. 4B-D). Of note, already in the basal state, colonic Treg counts were lower in the former than the latter (p < 0.05; Fig. 4C). Hence, TLR4 deficiency of PA-colonized IL10–/– mice is associated with less distinct innate as well as adaptive immune cell responses in the large intestines.
Fig. 4.
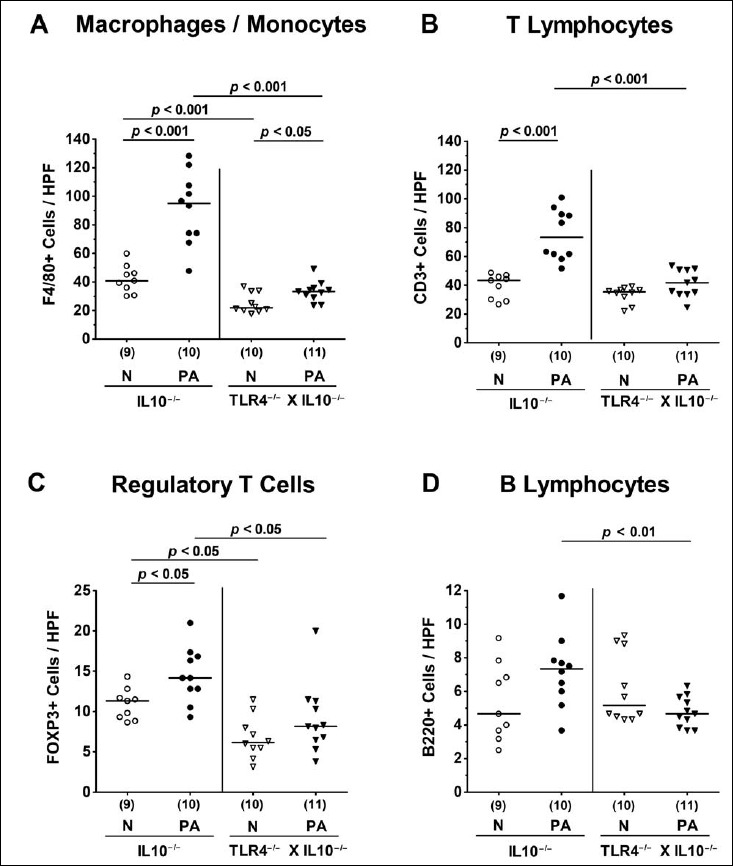
Colonic innate and adaptive immune cell responses following peroral multidrug-resistant P. aeruginosa association of secondary abiotic IL10–/– mice lacking TLR4. Secondary abiotic IL10-deficient (IL10–/–; circles) and TLR4-deficient IL10–/– mice (TLR4–/– × IL10–/–; triangles) were perorally challenged with a clinical multidrug-resistant P. aeruginosa strain (PA; closed symbols). Two weeks thereafter, the average numbers of colonic macrophages and monocytes (positive for F4/80), T lymphocytes (positive for CD3), regulatory T cells (positive for FOXP3), and B lymphocytes (positive for B220) were determined in six high power fields (HPF, 400× magnification) per animal in immunohistochemically stained large intestinal paraffin sections. Non-infected mice (N; open symbols) served as negative controls. Numbers of mice, means (black bars), and significance levels (p values) determined by the Mann–Whitney U test are indicated. Data shown were pooled from two independent experiments
Intestinal and systemic nitric oxide secretion following peroral MDR P. aeruginosa association of secondary abiotic IL10–/– mice lacking TLR4
Next, we addressed whether the observed intestinal immune cell responses upon MDR PA challenge of secondary abiotic IL10–/– mice lacking TLR4 were accompanied by local or even systemic secretion of the pro-inflammatory mediator NO. At day 14 p.i., NO concentrations were increased in the colon of TLR4-deficient IL10–/– mice only (p < 0.05; Fig. 5A), whereas in IL10–/– counterparts at least a trend towards elevated colonic NO levels could be observed (n.s. due to high standard deviation; Fig. 5A). In the ileum, however, the opposite was true, given that increased NO concentrations could be measured in PA-colonized IL10–/– mice only (p < 0.05; Fig. 5B). Furthermore, In MLN, NO secretion was less pronounced in IL10–/– mice lacking TLR4 as compared to IL10–/– controls mice at day 14 p.i. (p < 0.01; Fig. 5C).
Fig. 5.
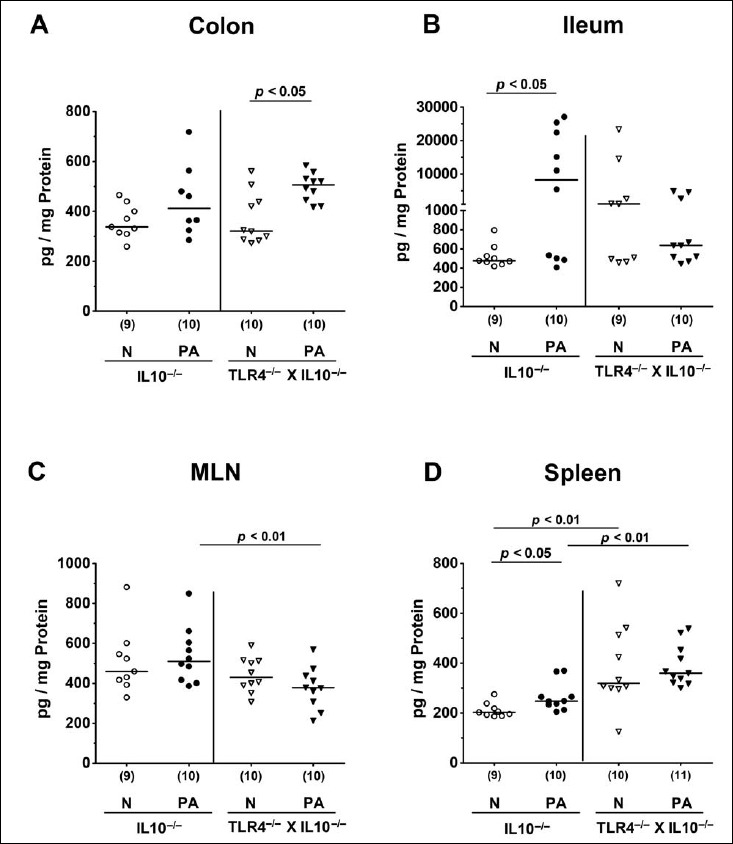
Intestinal and systemic nitric oxide secretion following peroral multidrug-resistant P. aeruginosa association of secondary abiotic IL10–/– mice lacking TLR4. Secondary abiotic IL10-deficient (IL10–/–; circles) and TLR4-deficient IL10–/– mice (TLR4–/– × IL10–/–; triangles) were perorally challenged with a clinical multidrug-resistant P. aeruginosa strain (PA; closed symbols). Two weeks thereafter, nitric oxide concentrations were measured in supernatants of ex vivo biopsies derived from (A) colon, (B) ileum, (C) mesenteric lymph nodes (MLN), and (D) spleen. Non-infected mice (N; open symbols) served as negative controls. Numbers of mice (in parentheses), means (black bars), and significance levels (p values) determined by the Mann–Whitney U test are indicated. Data shown were pooled from two independent experiments
Remarkably, MDR PA-induced pro-inflammatory immune response could be observed also systemically, given that NO secretion was increased in splenic ex vivo biopsies derived from IL10–/– mice (p < 0.05; Fig. 5D), but not TLR4-deficient IL10–/– animals at day 14 following PA colonization (n.s.; Fig. 5D). Interestingly, splenic NO concentrations were approximately 30% higher in both naive and PA-colonized TLR4–/– × IL10–/– mice as compared to respective IL10–/– counterparts (p < 0.01; Fig. 5D). Hence, MDR PA colonization induced less distinct nitric oxide secretion in MLN of TLR4-deficient IL10–/– as compared to IL10–/– controls, whereas, conversely, systemic NO levels were higher in both naive and PA-colonized TLR4-deficient IL10–/– mice.
Bacterial translocation following MDR P. aeruginosa colonization of secondary abiotic IL10–/– mice lacking TLR4
We finally addressed whether systemic immune responses upon PA colonization of TLR4-deficient IL10–/– mice were associated with bacterial translocation from the intestinal tract to extra-intestinal including systemic compartments. In fact, at day 14 p.i., viable MDR PA could be cultured from MLN, liver, and kidney as well as from spleen of TLR4–/– × IL10–/– mice in 27.3%, 18.2%, 9.1%, and 18.2% of cases, respectively, but in none of challenged IL10–/– animals (Fig. 6). Hence, intestinal MDR PA was able to translocate to extra-intestinal including systemic compartments in TLR4-deficient IL10–/– mice only.
Fig. 6.
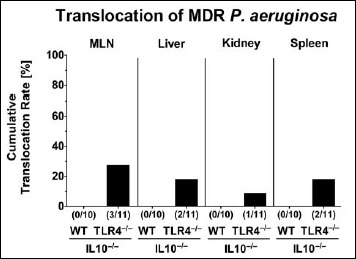
Bacterial translocation to extra-intestinal compartments in multidrug-resistant P. aeruginosa-colonized secondary abiotic IL10–/– mice lacking TLR4. Secondary abiotic IL10-deficient (WT IL10–/–; white bars) and TLR4-deficient IL10–/– mice (TLR4–/– × IL10–/–; black bars) were perorally challenged with a clinical multidrug-resistant P. aeruginosa strain (day 0). Two weeks thereafter, viable bacteria were detected in ex vivo biopsies derived from mesenteric lymph nodes (MLN), liver, kidney, and spleen by culture and the cumulative translocation rates out of three independent experiments indicated in %. Numbers of animals harboring the P. aeruginosa strain out of the total number of analyzed animals are given in parentheses
Discussion
Given a large gap in knowledge regarding the crosstalk between mere intestinal carriage of MDR Gram-negative strains including PA and innate immunity, we were able to show in the present study for the very first time that following peroral challenge with a clinical MDR PA strain, TLR4 deficiency of secondary abiotic IL10–/– mice was accompanied by 1) higher MDR PA colonization densities and bacterial translocation frequencies, but by 2) less distinct colonic epithelial apoptosis and 3) innate as well as adaptive immune responses in the large intestines, by 4) less NO secretion in MLN, but conversely, 5) higher systemic NO concentrations in both naive and colonized TLR4-deficient IL10–/– mice as compared to IL10–/– counterparts.
We here applied the secondary abiotic IL10–/– mouse model for the following reasons. First, preceding broad-spectrum antibiotic treatment not only mimics the clinical scenario posing the (mostly immune-compromised) patients at high risk for MDR PA acquisition and subsequent infection in the hospital setting, particularly when admitted to the ICU [12, 14], but also provides an important host immunity-depriving factor due to the gene deficiency of the important anti-inflammatory molecule IL10 [23]. Since the complex and diverse intestinal microbiota constitutes a very efficient barrier preventing the host from infection with (opportunistic) pathogens from outside [13, 27, 28], we accomplished stable colonization of the clinical MDR PA strain in the gastrointestinal tract of secondary abiotic mice. In addition, given that conventional IL10–/– mice usually develop chronic colitis due to TLR ligands derived from their own commensal gut microbiota [29], we had challenged IL10–/– mice with broad-spectrum antibiotic treatment starting immediately after weaning [23, 30]. This gave us furthermore the opportunity to observe potential large intestinal inflammatory responses upon MDR PA challenge that were not commensal microbiota-related. In fact, in previous studies, we were able to show that within 1 week following peroral infection with the enteropathogen Campylobacter jejuni, secondary abiotic IL10–/– mice developed severe non-selflimiting enterocolitis with bloody inflammatory diarrhea [23, 30]. This was, however, not the case when mice were associated with a commensal Escherichia coli strain that had been isolated from the murine gut before [23]. In line with these results, peroral challenge with PA in the present study did neither result in any overt macroscopic (i.e., clinical symptoms) nor in microscopic (i.e., histopathological) sequelae including apoptosis of colonic epithelia. TLR4-deficient IL10–/– mice, however, displayed even lower numbers of large intestinal apoptotic cells as compared to IL10–/– controls at day 14 p.i. This TLR4-dependent effect was accompanied by lower colonic numbers of both innate and adaptive immune cells in TLR4-deficient IL10–/– mice versus IL10–/– counterparts despite higher bacterial colonization densities in the small and large intestines of the former. In line with PA-induced effects observed here, Pseudomonas lipid A, a core moiety of Gram-negative bacterial LPS, has been shown to activate NFκB signaling via TLR4 resulting in pro-inflammatiry cytokine secretion [31]. Subsequently to the infection site recruited innate immune cells further accelerate against PA-directed host responses [6].
At the first sight, the NO data sets obtained in different parts of the intestinal tract are inconclusive, given that, in the colon, PA-induced NO secretion could be observed in TLR4-deficient IL10–/– mice, whereas, at day 14 p.i., elevated NO concentrations could by measured in the ileum of IL10–/– mice only. At the moment, however, we can only speculate about these results. It is well known that distinct molecules involved in pathogen recognition and innate immune functions, particularly cytokines of the IL10 cytokine family, can exert dichotomous mode of actions depending on the anatomical compartment, respective immune cell equipment, and surrounding cytokine milieu [32–35]. For instance, as shown by us and others, IL-22 (also being part of the IL10 cytokine family) exerts anti-inflammatory properties in the large intestines [33], whereas it has pro-inflammatory functions in the small intestinal tract [36–38]. Furthermore, lower NO levels could be measured in draining MLN of PA-colonized TLR4-deficient mice as compared to IL10–/– counterparts, whereas, conversely, NO secretion was more pronounced in the spleen of the former as compared to the latter at day 14 p.i. It is tempting to speculate that TLR4 deficiency was associated with less recruitment of leucocytes from the spleen to the lymph nodes draining the infection sites. More pronounced splenic NO secretion in TLR4-deficient IL10–/– mice, however, might also have been due to the observed higher translocation rates given that viable PA originating from the intestinal tract had gained access to extra-intestinal tissue sites including liver and kidney as well as to systemic compartments such as the spleen.
Conclusion
We conclude that TLR4-dependent signaling of PA LPS is important for intestinal and extra-intestinal including systemic host responses upon challenge with MDR PA. Furthermore, TLR4-mediated detection of PA LPS is essential for maintaining intestinal barrier function which subsequently prevents from extra-intestinal PA translocation with potentially fatal systemic spread. Future studies need to further unravel the molecular mechanisms underlying the orchestrated interplay between intestinal carriage of opportunistic pathogens such as MDR Gram-negative bacteria inclduing PA and host immunity in health and disease including intestinal inflammatory comorbidities.
Acknowledgements
We thank Michaela Wattrodt, Ursula Rüschendorf, Alexandra Bittroff-Leben, Ines Puschendorf, Gernot Reifenberger, Ulrike Fiebiger, and the staff of the animal research facility at Charité – University Medicine Berlin for excellent technical assistance and animal breeding.
Footnotes
Funding sources
This work was supported by grants from the German Research Foundation (DFG) to S.B. (SFB633, TP A7) and M.M.H. (SFB633, TP B06; SFB TR84, TP A05).
Conflicts of interest
Stefan Bereswill and Markus M. Heimesaat are Editorial Board members.
References
- 1.Uematsu S, Akira S: Innate immune recognition of viral infection. Uirusu 56, 1–8 (2006) [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O: Pathogen recognition and innate immunity. Cell 124, 783–801 (2006) [DOI] [PubMed] [Google Scholar]
- 3.King JD, Kocincova D, Westman EL, Lam JS: Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun 15, 261–312 (2009) [DOI] [PubMed] [Google Scholar]
- 4.Lam JS, Taylor VL, Islam ST, Hao Y, Kocincova D: Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2, 118 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driscoll JA, Brody SL, Kollef MH: The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368 (2007) [DOI] [PubMed] [Google Scholar]
- 6.Gellatly SL, Hancock RE: Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67, 159–173 (2013) [DOI] [PubMed] [Google Scholar]
- 7.Sievert DM Ricks P Edwards JR Schneider A Patel J Srinivasan A Kallen A Limbago B Fridkin S National Healthcare Safety Network T, Participating NF: Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009–2010. Infect Control Hosp Epidemiol 34, 1–14 (2013) [DOI] [PubMed] [Google Scholar]
- 8.Vincent JL: Nosocomial infections in adult intensive-care units. Lancet 361, 2068–2077 (2003) [DOI] [PubMed] [Google Scholar]
- 9.Lyczak JB, Cannon CL, Pier GB: Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2, 1051–1060 (2000) [DOI] [PubMed] [Google Scholar]
- 10.Potron A, Poirel L, Nordmann P: Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45, 568–585 (2015) [DOI] [PubMed] [Google Scholar]
- 11.Shooter RA, Walker KA, Williams VR, Horgan GM, Parker MT, Asheshov EH, Bullimore JF: Faecal carriage of Pseudomonas aeruginosa in hospital patients. Possible spread from patient to patient. Lancet 2, 1331–1334 (1966) [DOI] [PubMed] [Google Scholar]
- 12.Cohen R, Babushkin F, Cohen S, Afraimov M, Shapiro M, Uda M, Khabra E, Adler A, Ben Ami R, Paikin S: A prospective survey of Pseudomonas aeruginosa colonization and infection in the intensive care unit. Antimicrob Resist Infect Control 6, 7 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Dasti JI, Zautner AE, Munoz M, Loddenkemper C, Gross U, Göbel UB, Heimesaat MM: Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6, e20953 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Zorrilla S, Camoez M, Tubau F, Canizares R, Periche E, Dominguez MA, Ariza J, Pena C: Prospective observational study of prior rectal colonization status as a predictor for subsequent development of Pseudomonas aeruginosa clinical infections. Antimicrob Agents Chemother 59, 5213–5219 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livermore DM: Has the era of untreatable infections arrived? J Antimicrob Chemother 64(Suppl 1), i29–36 (2009) [DOI] [PubMed] [Google Scholar]
- 16.Oliver A, Mulet X, Lopez-Causape C, Juan C: The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22, 41–59 (2015) [DOI] [PubMed] [Google Scholar]
- 17.Tacconelli E, Magrini N: Global priority list of antibiotic-resistant bacteria to guide research discovery and development of new antibiotics. World Health Organisation (2017) [Google Scholar]
- 18.Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM: Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol 60, 236–245 (2011) [DOI] [PubMed] [Google Scholar]
- 19.Wang M, Molin G, Ahrne S, Adawi D, Jeppsson B: High proportions of proinflammatory bacteria on the colonic mucosa in a young patient with ulcerative colitis as revealed by cloning and sequencing of 16s rrna genes. Dig Dis Sci 52, 620–627 (2007) [DOI] [PubMed] [Google Scholar]
- 20.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, Jahn HK, Dunay IR, Moter A, Gescher DM, Schumann RR, Göbel UB, Liesenfeld O: Gram-negative bacteria aggravate murine small intestinal th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol 177, 8785–8795 (2006) [DOI] [PubMed] [Google Scholar]
- 21.Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, Scheffold A, Kühl AA, Bereswill S, Heimesaat MM: Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front Immunol 8, 397 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Klitzing E, Ekmekciu I, Bereswill S, Heimesaat MM: Acute ileitis facilitates infection with multidrug resistant Pseudomonas aeruginosa in human microbiota-associated mice. Gut Pathog 9, 4 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haag LM, Fischer A, Otto B, Plickert R, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM: Campylobacter jejuni induces acute enterocolitis in gnotobiotic IL-10–/– mice via toll-like-receptor-2 and -4 signaling. PLoS One 7, e40761 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alutis ME, Grundmann U, Fischer A, Hagen U, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM: The role of gelatinases in Campylobacter jejuni infection of gnotobiotic mice. Eur J Microbiol Immunol (Bp) 5, 256–267 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alutis ME, Grundmann U, Hagen U, Fischer A, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM: Matrix metalloproteinase-2 mediates intestinal immunopathogenesis in Campylobacter jejuni-infected infant mice. Eur J Microbiol Immunol (Bp) 5, 188–198 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C, Steinhoff U, Tchaptchet S, Thiel E, Freudenberg MA, Göbel UB, Uharek L: Myd88/tlr9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 59, 1079–1087 (2010) [DOI] [PubMed] [Google Scholar]
- 27.Masanta WO, Heimesaat MM, Bereswill S, Tareen AM, Lugert R, Gross U, Zautner AE: Modification of intestinal microbiota and its consequences for innate immune response in the pathogenesis of campylobacteriosis. Clin Dev Immunol 2013, 526860 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiebiger U, Bereswill S, Heimesaat MM: Dissecting the interplay between intestinal microbiota and host immunity in health and disease: Lessons learned from germfree and gnotobiotic animal models. Eur J Microbiol Immunol (Bp) 6, 253–271 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlgemuth S, Keller S, Kertscher R, Stadion M, Haller D, Kisling S, Jahreis G, Blaut M, Loh G: Intestinal steroid profiles and microbiota composition in colitic mice. Gut Microbes 2, 159–166 (2011) [DOI] [PubMed] [Google Scholar]
- 30.Heimesaat MM, Grundmann U, Alutis ME, Fischer A, Bereswill S: Absence of nucleotide-oligomerizationdomain-2 is associated with less distinct disease in Campylobacter jejuni infected secondary abiotic IL-10 deficient mice. Front Cell Infect Microbiol 7, 322 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korneev KV, Arbatsky NP, Molinaro A, Palmigiano A, Shaikhutdinova RZ, Shneider MM, Pier GB, Kondakova AN, Sviriaeva EN, Sturiale L, Garozzo D, Kruglov AA, Nedospasov SA, Drutskaya MS, Knirel YA, Kuprash DV: Structural relationship of the lipid a acyl groups to activation of murine toll-like receptor 4 by lipopolysaccharides from pathogenic strains of Burkholderia mallei, Acinetobacter baumannii, and Pseudomonas aeruginosa. Front Immunol 6, 595 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG: Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 29, 71–109 (2011) [DOI] [PubMed] [Google Scholar]
- 33.Eidenschenk C, Rutz S, Liesenfeld O, Ouyang W: Role of IL-22 in microbial host defense. Curr Top Microbiol Immunol 380, 213–236 (2014) [DOI] [PubMed] [Google Scholar]
- 34.Heimesaat MM, Grundmann U, Alutis ME, Fischer A, Göbel UB, Bereswill S: The IL-23/IL-22/IL-18 axis in murine Campylobacter jejuni infection. Gut Pathog 8, 21 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bereswill S, Grundmann U, Alutis ME, Fischer A, Kühl AA, Heimesaat MM: Immune responses upon Campylobacter jejuni infection of secondary abiotic mice lacking nucleotide-oligomerization-domain-2. Gut Pathog 9, 33 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munoz M, Heimesaat MM, Danker K, Struck D, Lohmann U, Plickert R, Bereswill S, Fischer A, Dunay IR, Wolk K, Loddenkemper C, Krell HW, Libert C, Lund LR, Frey O, Holscher C, Iwakura Y, Ghilardi N, Ouyang W, Kamradt T, Sabat R, Liesenfeld O: Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of Il-17. J Exp Med 206, 3047–3059 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz M, Liesenfeld O, Heimesaat MM: Immunology of Toxoplasma gondii. Immunol Rev 240, 269–285 (2011) [DOI] [PubMed] [Google Scholar]
- 38.Munoz M, Eidenschenk C, Ota N, Wong K, Lohmann U, Kühl AA, Wang X, Manzanillo P, Li Y, Rutz S, Zheng Y, Diehl L, Kayagaki N, van Lookeren-Campagne M, Liesenfeld O, Heimesaat M, Ouyang W: Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity 42, 321–331 (2015) [DOI] [PubMed] [Google Scholar]


