Abstract
Purpose
The 2015 American College of Medical Genetics and Genomics–Association for Molecular Pathology (ACMG–AMP) guidelines were a major step toward establishing a common framework for variant classification. In practice, however, several aspects of the guidelines lack specificity, are subject to varied interpretations, or fail to capture relevant aspects of clinical molecular genetics. A simple implementation of the guidelines in their current form is insufficient for consistent and comprehensive variant classification.
Methods
We undertook an iterative process of refining the ACMG–AMP guidelines. We used the guidelines to classify more than 40,000 clinically observed variants, assessed the outcome, and refined the classification criteria to capture exceptions and edge cases. During this process, the criteria evolved through eight major and minor revisions.
Results
Our implementation: (i) separated ambiguous ACMG–AMP criteria into a set of discrete but related rules with refined weights; (ii) grouped certain criteria to protect against the overcounting of conceptually related evidence; and (iii) replaced the “clinical criteria” style of the guidelines with additive, semiquantitative criteria.
Conclusion
Sherloc builds on the strong framework of 33 rules established by the ACMG–AMP guidelines and introduces 108 detailed refinements, which support a more consistent and transparent approach to variant classification.
Keywords: ACMG laboratory guideline, clinical genetic testing, clinical interpretation, variant classification, variant interpretation
Introduction
Variant classification is the cornerstone of clinical molecular genetic testing. The validity and utility of genetic testing require that variant classifications be evidence-based, objective, and systematic.1, 2, 3 Clinical and medical geneticists must be able to distinguish between established facts and reasonable hypotheses and must understand the evidence and logic underlying variant classifications.4 Pathogenicity evaluations must be reproducible and protected from the personal and professional biases that can be present in research laboratories, the investigative settings of diagnostic laboratories, and clinicians’ urgent desire to make a diagnosis.
The 2015 American College of Medical Genetics and Genomics–Association for Molecular Pathology (ACMG–AMP) guidelines for the interpretation of sequence variants were a major step toward establishing a shared framework for variant classification.5 However, during the process of applying the ACMG–AMP guidelines to the classification of thousands of variants, we and other groups6 identified several areas in which the guidelines lacked specificity or were subject to ambiguous or contradictory interpretations. To address this, we developed and validated Sherloc (semiquantitative, hierarchical evidence-based rules for locus interpretation), a variant classification framework that is an effective refinement of the ACMG–AMP criteria. Sherloc addresses several key issues.
Certain ACMG–AMP rules conflate concepts that should be considered separately. Sherloc expands overly encumbered criteria into a set of discrete but related rules and weights these rules separately.
Certain pairings of ACMG–AMP rules capture types of evidence that contribute to the same basic argument, which creates a “double counting” effect in which an argument is overvalued by invoking the same basic observation more than once. Sherloc groups evidence types into broader lines of argument to prevent this inadvertent error.
The “clinical criteria” style of the ACMG–AMP guidelines introduces difficulty in intuitively understanding the cumulative strength of the evidence, and in appreciating how much additional evidence is required to move to a confident conclusion. Sherloc substitutes a categorical framework and numerically weighted criteria to support a more intuitive understanding of variant classification. Consistent with the ACMG–AMP guidelines, Sherloc is a system for the evaluation of constitutional variants within a Mendelian disease framework.
Materials and methods
Sherloc was developed through an iterative process using the ACMG–AMP guidelines as a starting point. The ACMG–AMP draft guidelines were released for member comment in August 2013 and adopted for internal use at Invitae. A working group was formed comprising American Board of Medical Genetics and Genomics–certified laboratory directors, doctoral-level scientists, and American Board of Genetic Counseling–certified genetic counselors with experience in many clinical areas of diagnostic genetic testing, including hereditary cancer, cardiology, neurology, and pediatric genetics.
The working group interpreted variants observed during diagnostic testing by using the implemented framework, and identified variants for which (i) strict adherence to the framework led to classifications at odds with the established understanding of clinical significance, or (ii) uncertainty or disagreement arose about the correct application of the rule set. The working group met weekly to discuss these cases, identify the underlying genetic issues, and refine the rules and their valuations. This iterative process continued for more than two years and through more than 40,000 unique variants identified during clinical laboratory testing across more than 500 genes and conditions. The framework has developed through many major and minor iterations. The rule set described herein is Sherloc version 4.2. All interpreted variants are routinely deposited into ClinVar.7
Results
Our experience using the ACMG–AMP criteria was mixed. The guidelines presented a logical framework for categorizing and valuing evidence that generally matched the perspective of our clinical staff, many of whom had participated in ACMG–AMP surveys and discussions during guideline development. However, the criteria left many aspects of clinical molecular genetics undescribed and subject to personal interpretation. We routinely encountered variants that caused uncertainty about the appropriate rule usage, which led to classification inconsistencies and debate. Generally, discrepancies were due either to uncertainty about how to categorize evidence that did not fit neatly into the available rules, or subjectivity about when to count evidence as strong or moderate.
To address these questions, we set out to describe every use-case with explicit evidence criteria. When ambiguity arose, we developed more granular rules to capture the necessary complexity. For example, the ACMG–AMP guidelines contain one caveat-laden rule (PVS1) capturing premature termination codon (PTC) variants and no alternative criteria for PTC variants that fail to fulfill all of the requirements, even though a PTC that does not meet every usage note criteria can have a predictably disruptive effect on a gene product. To address this shortcoming, we established a set of variably weighted criteria for PTCs (see “Variant type and the expected consequence for gene products” below), in which the value is modulated based on the location of the stop codon relative to the pre–messenger RNA (mRNA) structure (5′ truncations that lead to nonsense-mediated decay versus 3′ PTCs that yield translated, truncated proteins8) and the molecular mechanism of disease for the gene (confirmed versus unconfirmed loss-of-function (LOF) mechanism).
We recognized that the full complexity of clinical genetics was unlikely to be captured prospectively, and expected that regular iterations to Sherloc would be necessary. We therefore designed Sherloc to support refinements that could maintain backward compatibility. Over time, this approach expanded the original set of 33 ACMG–AMP criteria to the 108 criteria contained within Sherloc version 4.2. The iterative process continues and is an essential part of laboratory process quality improvement.
Evidence required for confident classifications
The ACMG–AMP framework assigns a strength level to each evidence criterion and requires various combinations of strong, moderate, and supporting evidence for a confident classification. However, we regularly identified variants that could be formally classified as likely pathogenic but seemed insufficiently supported or, conversely, variants that could formally be classified as variants of uncertain significance despite persuasive evidence that was not handled well by the ACMG–AMP framework.
For example, CDH1 c.1118C>T (p.Pro373Leu) is a variant in a gene associated with hereditary diffuse gastric cancer and lobular breast cancer.9 It is absent from the Exome Aggregation Consortium (ExAC) database and is supported by strong functional studies: in vitro functional characterization shows that p.Pro373Leu impairs cell–cell adhesion and leads to increased cellular motility and activation of EGFR, mitogen-activated protein kinase, and Src kinase.10, 11 Computational predictors recapitulate this conclusion. Clinical observations, however, are inconclusive: the variant has been found in affected and unaffected individuals in the same family.12 A strict application of the ACMG–AMP rules should yield a likely pathogenic classification: PS3 (well-established functional studies) +PP3 (predicted deleterious) +PM2 (absent from population). However, without supporting clinical observations, this conclusion seems premature, particularly because PS3 and PP3 redundantly describe the functional argument that the protein is disrupted.
Conversely, TTC8 c.459G>A (p.Thr153=) is a very rare silent change (0.02% in ExAC) in a gene that can cause Bardet–Biedl syndrome. Although not in the consensus +1/+2 splice site, it is located at the last nucleotide of the exon and is predicted to disrupt normal splicing. It has been observed in the homozygous state in three affected siblings in a single family.13 A strict application of the ACMG–AMP rules yields a variant of uncertain significance classification of PP1 (cosegregation with disease) +PP3 (computational evidence). In our assessment, however, the rules fail to capture relevant sequence context and undervalue the clinical observations. This variant has been observed in our laboratory in the homozygous state in an unrelated affected individual and is now classified as pathogenic.
Such examples suggested that the ACMG–AMP criteria were not capturing certain qualitative considerations. Therefore, we first posed a normative question: “What kind of evidence, and how much, should be required for a pathogenic classification?” We first recognized that there are two general types of evidence: clinical and functional. Clinical evidence describes the correlation of the variant with disease (or absence of disease) in human populations, and includes observations in affected and unaffected individuals and families. Functional evidence describes the molecular consequence of a variant on various gene products and includes the results of molecular and cellular experiments, and predictions about functional effects based on variant type or complex computational algorithms. Clearly, clinical and functional evidence are both important: a variant is pathogenic if it disrupts a gene product in a way that leads to human disease, and is benign if it has an effect that does not lead to disease in humans. Although both clinical and functional evidence are relevant, they have a hierarchical relationship. Clinical data describe human disease directly, whereas functional data are relevant to disease only to the extent to which the measured property correlates with disease physiology. Therefore, when a discrepancy or conflict arises between clinical and functional observations, the clinical observations should be considered more persuasive. Broadly speaking, a variant should not be considered pathogenic if it is present in a large percentage of healthy individuals (clinical data), even if a measurable effect on protein function has been observed in an experimental assay (functional data). Conversely, a variant should be considered pathogenic if it is present in many affected individuals and has not been observed in healthy individuals (clinical data), even if it is predicted to be nondeleterious and has been demonstrated to have no effect on a measured protein property (functional data).
Examples of these kinds of conflicts include CDKN2A c.9_32dup24 (p.Ala4_Pro11dup) and SCN5A c.3578G4A (p.Arg1193Gln). CDKN2A c.9_32dup24 is an in-frame duplication predicted to have no effect on protein function and demonstrated not to affect CDK4 or CDK6 binding.14, 15, 16 However, the variant has been identified in several individuals affected with melanoma15, 17, 18 and has been shown to segregate with disease (incomplete penetrance) in several melanoma families.19, 20, 21, 22 The abundance of positive clinical evidence trumps the negative functional evidence. It is possible that the effect on binding was mismeasured or that CDK4/6 binding efficiency is not the relevant molecular consequence of this variant. Conversely, SCN5A c.3578G>A (p.Arg1193Gln) is a missense change in the voltage-gated cardiac sodium channel. Pathogenicity seems to be supported by functional evidence: the variant was demonstrated to destabilize inactivation gating and to lead to a persistent current in vitro.23 However, a glycine is present at the equivalent position in the horse ortholog, and the variant is present in more than 7% of the East Asian population, with 17 homozygotes reported in ExAC. The abundance of negative clinical evidence outweighs the positive functional evidence.
This principle of the primacy of clinical evidence establishes a framework for evidentiary thresholds: a rare variant supported by nothing but functional evidence should be classified as a variant of uncertain significance in the absence of supporting clinical data linking the molecular dysfunction to a clinical phenotype.
Assigning points to evidence types
In the ACMG–AMP system, the overall strength of the total evidence set is evaluated in a manner analogous to the familiar style of diagnostic clinical criteria. Evidence types are roughly binned into one of four levels, and different combinations of evidence from the bins suffice for a confident classification. In practice, however, we have found that this style of assessing an argument introduced obstacles to the accuracy and flexibility of an evolving system. In many cases, we needed to introduce more subtle gradations to the evidence valuation than the four levels could support. We also found that the combinatorial logic of the ACMG–AMP criteria made it very difficult to predict the consequence of introducing new criteria or changing the valuation of criteria. Because there are different paths to a threshold, it was difficult to understand intuitively how much more evidence might be required for a confident classification.
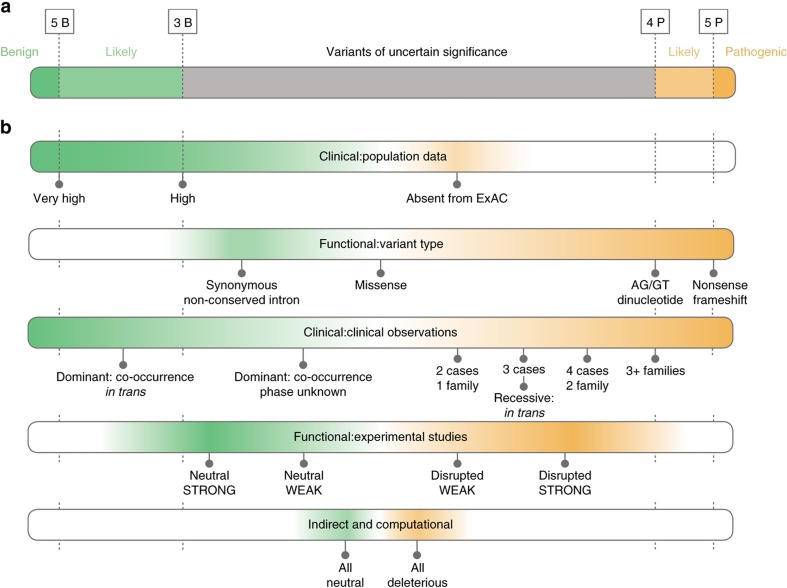
We knew Sherloc would evolve, so we needed a weighting system that provided more precision and flexibility. We therefore established a semiquantitative system in which each criterion is awarded a preset number of points on orthogonal benign or pathogenic scales (i.e., 1B-5B or 1P-5P), which reflect the value of the data type toward the overall classification argument (Figure 1). Accumulated benign and pathogenic evidence types are summed separately and compared against preset thresholds. When substantial evidence supports both pathogenic and benign conclusions, that dichotomy may indicate low-penetrance variants, genetic or environmental modifiers, or other ambiguity within the Mendelian framework. For most variants supported by evidence toward both poles, however, the clinical/functional hierarchical framework described previously and the practical approach described in Figure 5 provide guidance for evaluating these apparent contradictions. Point thresholds for likely benign and likely pathogenic classifications are asymmetric (3B versus 4P), and reflect the fact that neutral genetic variation is abundant and pathogenic variants are rare. The burden of proof to reach a benign classification is therefore lower.
Figure 1.
Classification scoring thresholds and evidence categories. (a) Point score thresholds for pathogenic (P), likely pathogenic, variant of uncertain significance, likely benign, and benign (B) classifications. Pathogenic and benign evidence is scored separately. Evidence in both directions can suggest a non-Mendelian variant. (b) Five evidence categories in the order in which they are evaluated, and with the point value of select criteria indicated. Clinical criteria include population data and clinical findings. Functional criteria include sequence observations, molecular studies, and indirect and computational data. ExAC, Exome Aggregation Consortium.
The original translation of the ACMG–AMP guidelines into a point system aimed simply to recapitulate conclusions reached via the ACMG–AMP combinatorial scoring method. The value of criteria has changed over time as the rule set was expanded and refined. The current rules and weights are described throughout this paper and in Supplementary Table S1 online. Although the criteria in Sherloc version 1 had a 1:1 mapping to the ACMG–AMP criteria, subsequent versions diverged from that strict correlation. The derivation history or closest mapping of Sherloc evidence criteria (EVs) to ACMG–AMP criteria is presented in Supplementary Table S1.
This approach (exhaustive criteria set, fixed point values, and a consistent evaluation protocol) promotes consistency, reproducibility, and efficiency among users. The same steps are performed, the same weight is granted to each element of an argument, and users are guided toward the correct application of evidence types.
Clinical criteria
Clinical data include population frequency information and observations of variants in well-characterized affected and unaffected individuals and families. Sherloc contains sets of evidence types to capture these data. Detailed knowledge about the symptoms and phenotypes associated with each condition, the penetrance and age at onset of features, and the percentage of clinical cases accounted for by pathogenic variants in known genes are essential prerequisites for using these data effectively. The sections that follow describe the original ACMG–AMP criteria, the derived Sherloc criteria, and the evaluation process for each clinical data type.
Population data
Variant frequencies from large population data sets can provide strong evidence that a variant is benign, or can reveal that one is sufficiently rare to be considered a candidate for pathogenicity. Most variants encountered during testing have previously been observed at a frequency inconsistent with the incidence of monogenic disease. With appropriate safeguards, beginning variant evaluation with population data maximizes accuracy and efficiency. The full set of frequency criteria are shown in Figure 2.
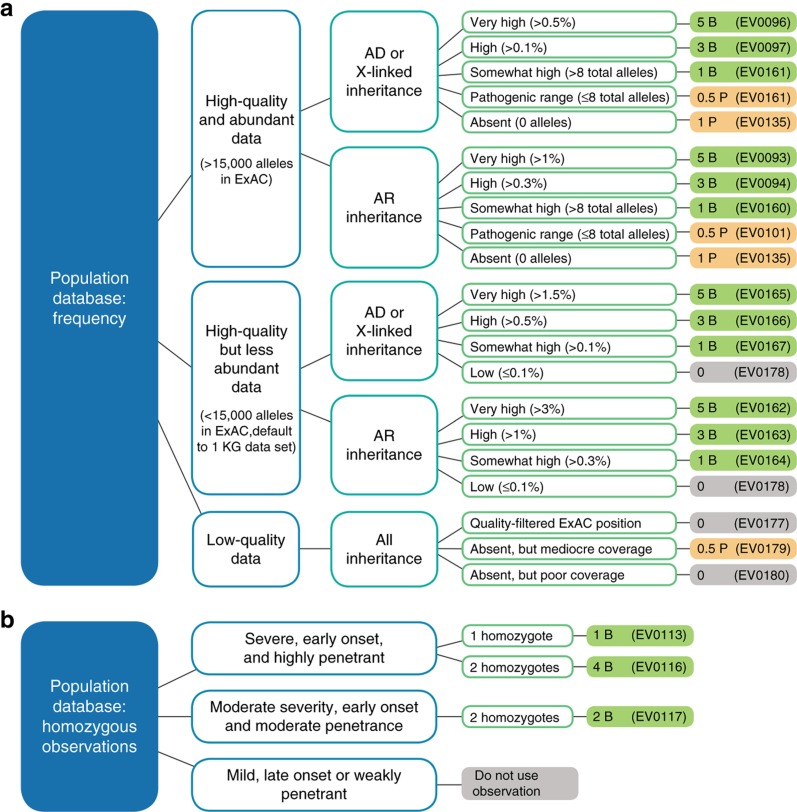
Figure 2.
Population data: Sherloc criteria and decision tree. (a) A single evidence type criterion from the frequency set of criteria is chosen for each variant. This decision tree guides users to the correct criterion based on the quality and abundance of the Exome Aggregation Consortium (ExAC) data at the locus in question, the mode of inheritance of the gene, and the frequency of the variant in ExAC. Points and directionality (pathogenic versus benign) are indicated in the far right column. (b) Decision tree for using observations of homozygotes in the ExAC database depending on the severity, onset, and penetrance of the biallelic phenotype, and the number of homozygotes present. Loci flagged with data quality issues are excluded. Solid orange color corresponds to pathogenic evidence, solid green corresponds to benign evidence, and solid grey corresponds to neutrally weighted evidence. AD, autosomal dominant; AR, autosomal recessive.
The ACMG–AMP guidelines contain two benign rules and two pathogenic rules to capture the impact of population frequency on variant classification: BA1 (allele frequency >5%), BS1 (allele frequency > expected), PM2 (absent from controls), and PS4 (higher prevalence in affected individuals versus controls). We encountered a number of limitations to this approach related to the effective placement of frequency thresholds, the known presence of pathogenic variants in population data sets, and the quality and abundance of population data.
While it is clear that a variant present in 5% of the population cannot be a cause of a rare, monogenic disease, this is also true for variants at much lower frequencies. The ACMG–AMP framework suggests that frequency thresholds be set based on a synthesis of disease prevalence, penetrance, and percent attribution.5, 24 This guidance is impractical, primarily because accurate prevalence, penetrance, and gene attribution numbers have not been established for most disorders and can vary two- to tenfold even for well-studied disorders, depending on the subpopulation and the total number of unique pathogenic variants. Moreover, frequency data are inherently quantitative. All other things being equal, the likelihood that a variant is benign increases as its observed frequency increases. A single threshold does not adequately capture this variable likelihood of pathogenicity.
To address these issues, Sherloc captures five frequency levels:
Absent in ExAC (1P)
“Within pathogenic range” (0.5P): low frequency, but consistent with previously well-characterized pathogenic variants
“Somewhat high” (1B): low frequency and inconsistent with previously well-characterized pathogenic variants
“High” (3B): sufficiently common for a likely benign classification without additional corroborating evidence
“Very high” (5B): sufficiently common for a benign classification without additional corroborating evidence
To define the bottom four tiers quantitatively, we developed an empirical approach to establish the frequency spectrum of pathogenic variants in the ExAC database.25 ExAC contains pathogenic variants, and their frequencies can be characterized. Such analysis revealed that in a set of 79 disease genes (39 dominant, 40 recessive, 1508 total variants), 97.3% of pathogenic variants had an allele frequency of less than 0.01%, and 94% were present at eight alleles or fewer.
These observations are used cautiously to support frequency thresholds that, although much more aggressive than the ACMG–AMP recommended 5%, are still safely conservative. For dominant genes, Sherloc incorporates thresholds of 0.5%, 0.1%, and >8 alleles for the “very high”, “high” and “somewhat high” levels, respectively. For recessive genes, it incorporates thresholds of 1%, 0.3%, and >8 alleles, respectively. These cutoffs support the use of ethnic subpopulation allele frequencies, which can be less precise due to smaller sample sizes but are critical for identifying ethnicity-enriched polymorphisms. When a gene is associated with more than one inheritance pattern, an a priori gene-level decision is made to use either the higher or the lower frequency thresholds based on the severity and age at onset of the monoallelic phenotype.
Variant frequencies can be elevated for founder mutations and within mutational hotspots. As such, high-frequency variants should not be classified as likely benign or benign without a literature review. (For literature review, our laboratory uses custom software, which combines direct searches using Google and SETH/PubTator with indirect reference reviews from the Human Gene Mutation Database, Online Mendelian Inheritance in Man database, and ClinVar. Manual search protocols could provide similar security.) The frequency thresholds used for these criteria may change as public data sets grow in size, but the concept and weighting will probably remain the same.
Data quality
There are technical considerations for using aggregate population data. For example, KCNQ1 c.1795-11803A>C is absent from the ExAC exome data, which has weak coverage of intronic regions, but is present at 32.3% in 1000 Genomes. As this example illustrates, the absence of a variant from ExAC does not always indicate that it is rare. At some loci, smaller data sets, such as 1000 Genomes, may have more relevant information, although the frequency may be less reliable. Sherloc includes a second set of frequency criteria for observations supported by fewer alleles. Certain loci in ExAC are also troubled by data quality issues. An example is ATM c.566G>A. This variant is present in ExAC at a frequency sufficient to justify a likely benign classification; however, the data are flagged as not passing the variant quality score recalibration filter. Incidentally, the Exome Sequencing Project and 1000 Genomes data do not report a variant at this position. Frequency information at quality-flagged loci should be used cautiously, if at all; Sherloc contains criteria to formally capture these cases. Likewise, ExAC contains loci with high-quality data but low total allele count. Low-frequency observations may be unreliable at these loci due to small sample sizes.
For all variants, a single “population” rule is selected based on a simple decision tree that reflects these considerations (Figure 2a).
Zygosity
Finally, we considered the zygosity of ExAC observations. For example, RAD50 c.280A>C (p.Ile94Leu) has been observed at a frequency of 0.7% in the south Asian population in ExAC, a cohort that includes two homozygous observations. For genes such as RAD50, in which biallelic pathogenic variants are expected to be lethal or severe (Nijmegen breakage syndrome26), observations of homozygous variants are strong evidence that the variant is benign. Therefore, Sherloc contains three rules of varying strength relating to observations of homozygotes in databases (Figure 2b).
Observations in well-characterized individuals
Individuals who are well characterized both phenotypically and genotypically can support inferences that are more powerful than those that can be drawn from discrete entries in general databases. The ACMG–AMP criteria contain five rules for capturing observations in well-characterized individuals: BS4 (nonsegregation with disease), BP2 (cis'trans with a pathogenic variant), BP5 (case with an alternative cause), PP1 (cosegregation with disease), and PP4 (individual’s phenotype or family history is highly specific). Other ACMG–AMP criteria, such as PM2 and PS6 (de novo criteria) and many of the variant type criteria, depend implicitly on an observation in an individual with a relevant phenotype.
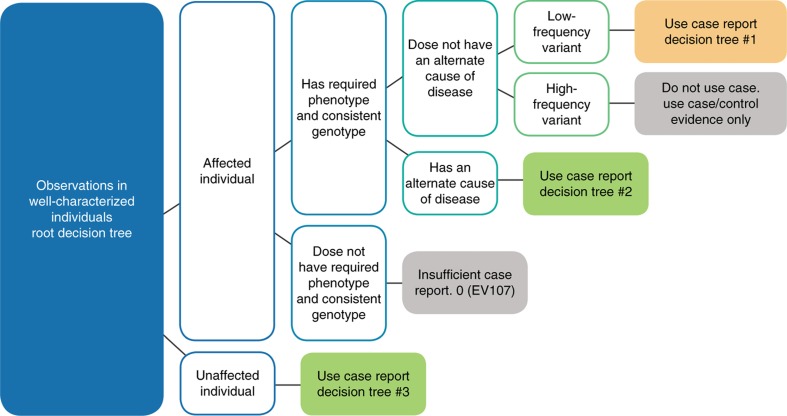
We identified three distinct classes of clinical observations that should be considered separately: (i) variants in unaffected individuals (suggests benign), (ii) variants in affected individuals with an alternate cause of disease (also suggests benign), and (iii) variants in affected individuals without an alternate cause of disease (suggests pathogenic). Case report criteria for each are described below. In general, these criteria are additive: each unrelated observation is considered an independent data point that further contributes to the argument. The three classes of observations are depicted in the case report root decision tree (Figure 3).
Figure 3.
Root decision tree for clinical case report criteria. Case reports are divided into one of three types based on the affected status of the proband, the relevance of the phenotype to the gene in question, and the presence of a known disease etiology. This root decision tree guides the user to the correct detailed decision tree (Supplementary Figures S2–S4) based on these considerations. The variant frequency is an essential lens through which to understand the relevance of case reports. The more frequent a variant is, the more likely it becomes that case reports are simply coincidental.
Observations of variants in affected individuals.
Using case report data accurately requires rigor regarding the appreciation of variant frequency, the distinctiveness of the phenotype, the relevance of the phenotype to the gene in question, and the diagnostic yield of the genetic test in patients with the observed phenotype (Supplementary Figure S1).
Variant frequency is a primary concern in evaluations of the relevance of positive case reports, and must be a preliminary evaluation. Individual case reports should be considered as possible evidence only if the variant is rare (absent from ExAC or present within the “pathogenic range”). If the variant is not rare, spurious case reports should be expected; segregation or case-control data are required to confirm the relevance of any single observation.
Using the clinical phenotype of affected individuals consistently and accurately proved challenging with the ACMG–AMP criteria. The genes BRCA1, JAG1, and MYH7 demonstrate how observations of a rare variant in an affected individual might contribute to an argument of variant causality to different extents.
Pathogenic variants in BRCA1 strongly predispose to breast cancer. However, isolated breast cancer is common, and approximately 90% of cases have nongenetic etiologies.27, 28 Most observations of BRCA1 variants in affected individuals, therefore, must be coincidental, not causal, so the observation of a variant in an isolated affected individual is not strong evidence that the variant is pathogenic unless it is first established that the individual’s disease is hereditary. Contrast BRCA1 with JAG1, which causes Alagille syndrome, a rare and clinically distinctive condition without known nongenetic etiologies. For classically affected individuals who meet clinical diagnostic criteria,29 the diagnostic yield for the molecular test is greater than 95%.30 It is therefore substantially more likely that a novel JAG1 variant in a clinically distinct individual is causal. JAG1/Alagille syndrome observations are therefore stronger evidence than BRCA1/breast cancer observations. Between these two extremes lies MYH7 and hypertrophic cardiomyopathy (HCM). Pathogenic variants in MYH7 can lead to hypertrophic or dilated cardiomyopathy, phenotypes that can have genetic origins but can also have substantial nongenetic etiologies (up to 70%).31, 32, 33 Case reports of rare MYH7 variants in individuals with HCM are cautiously considered evidence of pathogenicity, as a substantial likelihood remains that the combination is coincidental. A cohort of affected individuals with the same rare variant, however, becomes persuasive.
These examples demonstrate that the value of a clinical observation should vary based on both the specificity of the clinical phenotype and the diagnostic yield of the molecular test for that phenotype. The higher the pretest probability that a proband with a particular phenotype will have a particular gene disruption, the greater the likelihood that an observed variant in that gene or set of genes is the explanation for disease. Conversely, the lower the yield of a test, the greater the likelihood that the true etiology lies elsewhere and the higher the likelihood that observed variants are coincidental.
To develop a consistent approach to determining the value of clinical observations, we divided genes based on their yield for particular phenotypes. If the diagnostic yield is high (>75%), isolated case reports may be considered very significant, and finding a rare variant in a relatively small number of classically affected probands will be sufficient for a pathogenic classification. However, if the test accounts for less than 75% of cases, case reports are relevant only to the extent that the disease is first established to be hereditary based on the nature of the phenotype or a pedigree analysis. Furthermore, at least two unrelated case reports are required to begin counting the observations as relevant data to protect against circular reasoning in diagnostic testing. A cohort of similarly affected individuals is generally required for a pathogenic classification.
If the index proband is not a confirmed hereditary case, an isolated case is not used as evidence, and segregation data are required. Sherloc has three levels of segregation data to capture the fact that additional families and higher LOD scores quantitatively substantiate a classification argument (see Supplementary Information, Supplement 4: “Quantifying segregation”). When asserting a correlation between a positive genotype and a positive phenotype, the genomic linkage of the observed variant to the causative variant is a concern. Therefore, the value of a single family, no matter how large, is capped to protect against this possibility; other types of supporting evidence (functional data or observations in unrelated, affected individuals or families) are required for a confident classification.
It is important to predefine the distinct clinical features required to count an observation as a relevant case. Accepted clinical criteria, if they exist, should be used. Summary or simplified condition information, such as that often deposited in ClinVar or included on test requisition forms, may be insufficient for case-based conclusions, and additional communication may be required to capture necessary detail. Sherloc contains a 0-point criterion, “Observed in patient with nonspecific phenotype or insufficient genotype” (EV0107), to acknowledge reports of poorly characterized individuals or well-characterized individuals with nonspecific phenotypes.
Finally, the full genotype and variant inheritance can provide additional support to a pathogenicity argument. Two rare variants in trans in a recessive gene, or a hemizygous variant in an affected male, can provide an additional level of certainty that the case report is valuable. The observation that a variant has arisen de novo in a relevant and established disease gene is counted as strong evidence. Sherloc contains a less heavily weighted criteria for capturing de novo variants in candidate genes, an important consideration for exome analysis, in which multiple de novo events are common.
Observations in affected individuals with an alternate explanation for disease
Most Mendelian conditions are rare and explained by a single genetic etiology. Therefore, when an affected individual has an identified genetic etiology, additional variants in the same or a different gene are less likely to be pathogenic. In certain cases, co-occurrence observations of this sort can be used as evidence that the additional variants are benign. Sherloc contains four evidence types capturing these scenarios (Supplementary Figure S2). Within the same gene, co-occurrence applies only to variants in trans with pathogenic variants; once an allele is disrupted, there is no selective pressure preventing that allele from acquiring additional variants that would be pathogenic in isolation. It also applies only to variants causing dominantly inherited disease (or in X-linked genes in male probands). Recessive carrier status is no more or less likely in affected individuals.
Finally, this logic does not apply to relatively common conditions with locus heterogeneity—that is, when there are known case reports of affected individuals who inherit pathogenic variants in related genes from both parents. For example, up to 5% of individuals with a pathogenic HCM variant also have a second pathogenic HCM variant in a different gene34. The second variant cannot be presumed to be benign, and co-occurrence evidence types are not applied for HCM cases.
Observations in unaffected individuals
The observation of a variant in an unaffected individual, which in a genetic context should lead to disease if it were pathogenic, suggests that the variant may be benign (Supplementary Figure S3). For example, CHARGE syndrome is a dominant, highly penetrant, congenital disease caused by pathogenic variants in CHD7.35 A CHD7 variant in an unaffected adult is therefore strong evidence that the variant is benign. Likewise, biallelic pathogenic variants in BRCA2 cause Fanconi anemia,36 and there is no difference in the mutation spectrums of pathogenic variants that lead to BRCA2-mediated Fanconi anemia versus hereditary breast and ovarian cancer.37 A variant in trans with a pathogenic BRCA2 variant in an individual without Fanconi anemia must therefore be benign.
The strength of these assertions depends on the penetrance and expressivity of a condition. Pathogenic variants in JAG1, as described previously, cause Alagille syndrome, which has highly variable expressivity and severity; some individuals reach childbearing age without recognizing that they are affected.38 The observation of a JAG1 variant in an “unaffected” individual is evidence only to the extent that the individual has been phenotyped thoroughly and found to be devoid of subtle features.
To address examples like these, Sherloc modulates “unaffected case report” rules on inheritance, zygosity, and the penetrance and age at onset of associated diseases. For genes that lead to highly penetrant, early-onset diseases, even a single observation in a well-characterized unaffected individual can be strong evidence that a variant is benign; however, for genes in which we might expect to see unaffected, genotype-positive individuals, additional observations are required.
The use of these evidence types must be based on the confirmed absence of a particular phenotype. The fact that a phenotype has not been mentioned in a test requisition form or publication is insufficient evidence that it is absent. Furthermore, the term “unaffected” is relative; these rules also apply when the individual is affected by a disease but is not affected by the disease associated with the gene in question.
The ACMG–AMP criterion BS4 (lack of segregation) is challenging to parse, as nonsegregation can refer two separate phenomena that should be considered separately, and remain challenging to quantify. Within the context of a family with multiple individuals affected by a hereditary condition, the variant is (i) present in an unaffected individual (genotype-positive/phenotype-negative), or (ii) absent in an affected individual (genotype-negative/phenotype-positive). Observations of genotype-positive/phenotype-negative individuals, even in the context of a complex pedigree, are treated simply as independent data points using the criteria described in Supplementary Figure S3. Linkage to a causative variant is not a concern in the negative correlation, although the concerns about penetrance and expressivity described above are relevant. On the other hand, the absence of a variant in an affected individual (genotype-negative/phenotype-positive) is currently awarded 1 to 2 “miscellaneous benign points” based on the specificity and rarity of the condition. EV criteria do not yet exist and will be addressed more formally in future versions; a method for objectively establishing symptom specificity and phenocopy rates, and weighing this evidence against other data is still being developed.
Functional criteria
Variant type and the expected consequence for gene products
A variant is more likely to be pathogenic if it has a consequence for a gene product (a transcript or protein, or both) that is consistent with the disease mechanism of the gene. The ACMG–AMP guidelines contain six rules related to variant type: BP1 (missense variant in a gene with an LOF mechanism), BP3 (in-frame indels in a repetitive region), BP7 (silent change with no predicted impact on splicing), PVS1 (null variant in an LOF gene), PM4 (in-frame indel in a nonrepetitive region), and PP2 (missense variant in a gene in which missense changes are rare). Sherloc expands these rules to 27 criteria that address a more comprehensive set of variant types, specify differences based on variant location in the mRNA exonic structure, and incorporate assertions about molecular mechanism (Supplementary Figure S5, Supplementary Information: Supplement 1, “Variant type”). A single variant type rule is applied to each variant. In some cases (truncations, indels, and missense variants), additional dependent rules incorporate information about other nearby pathogenic variants. Missense changes and in-frame deletions/insertions are given a weight of 0 points. Variants that do not reliably affect protein sequence or abundance (such as silent or some intronic variants) are presumed more likely to be tolerated, and variants that exert a more dramatic effect on protein sequence or abundance (such as premature stops and splice junctions) are presumed more likely to be deleterious.
The relevance of a null variant depends on the disease mechanism of the gene, and ACMG–AMP and Sherloc both give special weight to null mutations in LOF genes. An objective approach to determining whether a disease mechanism has been established as LOF is described in the Supplementary Information (Supplement 2, “Establishing loss of function as a mechanism”), and a cohort of three unrelated affected individuals with null variants is typically required to support this conclusion. Variant effect rules are grouped and nonadditive.
Molecular, cellular, and animal experimental data
Experimental data can demonstrate an effect on certain aspects of protein or RNA function, localization and abundance but speak only indirectly to the question of pathogenicity. Experimental data are persuasive to the extent that the measured property is recapitulated in vivo and is relevant to the disease mechanism of the gene. The ACMG–AMP guidelines contain two rules capturing functional studies: BS3 (well-established assay, no deleterious effect) and PS3 (well-established assay, deleterious effect). However, we found it challenging to address the evidence provided by less-well-established functional assays, and found that the value awarded to even well-established functional assays was excessive, as we routinely encountered examples in which experimental evidence was later overturned by contradictory experimental evidence or by new population data. For example, ACTN2 c.26A>G (p.Gln9Arg) is an actinin variant observed in a number of patients with dilated cardiomyopathy or HCM, or both.39, 40 p.Gln9Arg expression in a skeletal muscle cell line was significantly different from the equivalent expression of a wild-type construct and failed to support normal cellular morphological changes and protein localization.39 However, the frequency of this variant in ExAC is greater than 0.1%, which is inconsistent with pathogenicity and high enough to cast substantial doubt on the relevance of case reports.
Likewise, MLH1 c.794G>A (p.Arg265His) is a rare variant observed in many individuals and families affected with Lynch syndrome.41, 42, 43 At least two experimental studies have reported cellular or molecular phenotypes attributed to this variant: a mutator phenotype in S288c and SK1 cells,44 and altered splicing in an ex vivo splicing assay.41 However, subsequent functional studies demonstrated repeatedly that this variant was mismatch-repair competent when transfected into HEK-293 cells,45, 46 did not alter β-galactosidase activities in a yeast two-hybrid assay,47 and did not lead to a yeast mutator phenotype.48 Furthermore, the variant has been observed to co-occur with pathogenic MLH1 variants in multiple families. The evidence justifies a likely benign classification in both of these cases, despite initial functional evidence demonstrating a deleterious effect.
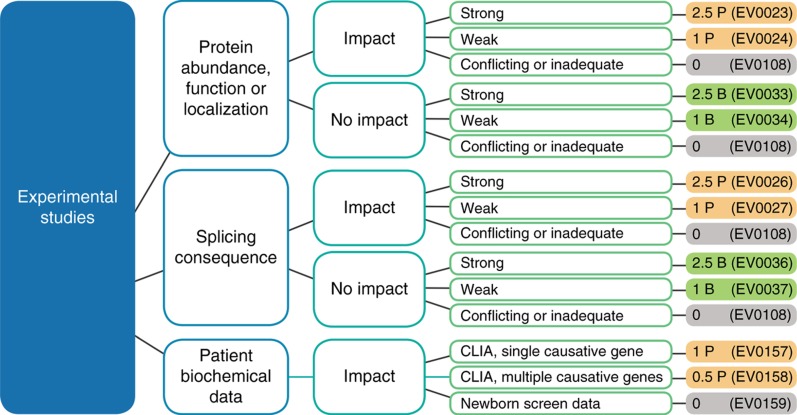
Disagreement about the value of functional data is a major source of classification discrepancies among genetics professionals. Clearly, the value of functional data depends on the relevance of the measured property to the disease biology, the quality of the experiment, the reproducibility of the result, and the amount of measured change, although a consistent evaluation of these considerations is challenging. To address this complexity, Sherloc contains rules capturing varying degrees of confidence, distinguishing splicing and protein effect experiments, and capturing and discounting poorly performed or inconclusive studies (Figure 4). Detailed guidelines for distinguishing between strong and weak categorizations are included in the Supplementary Information (Supplement 3, “Experimental evidence”). Functional evidence types are grouped and nonadditive. When multiple criteria are appropriate, only the strongest is counted.
Figure 4.
Functional data: Sherloc criteria and decision tree. Functional evidence is evaluated based on the type of experiment performed and the relevance and validity of the assay. Clinical Laboratory Improvement Amendments–generated biochemical data from affected individuals are also considered a type of in vivo functional experiment, although this evidence type is usually used to augment the value of a case report.
Patient biochemical data
A well-established, clinically validated assay that measures enzyme activity or analyte abundance in a patient-derived sample is a special data type that straddles the boundary between clinical phenotype and functional data. It is captured here as functional data in recognition of the essential quantitative objectivity of the data. These evidence types augment the value of a case report when appropriate. Newborn screen data are captured but not valued in recognition of the high false-positive rate.
Computational predictors and conservation
Many computational tools exist to predict the effect of missense changes on gene products. Although these tools are useful in prioritizing variant lists in gene discovery exercises, their clinical validity as predictors of disease is not well established.49, 50 Sherloc contains a series of weakly weighted criteria to capture these observations (Supplementary Figure S5). Splicing predictors, although also weakly weighted, are used to indicate which silent or intronic variants should be considered further.
The presence of an equivalent missense change in a mammalian species is granted a special weight reflecting the assumption that variation within a mammalian clade speaks more directly to questions of mammalian physiology and is therefore more relevant to human disease. Computational predictor evidence types are nonadditive. They are also grouped with, and superseded by, functional evidence criteria. When multiple evidence types from this large group are used, only the strongest is counted.
Discussion
Sherloc is an implementation and refinement of the ACMG–AMP variant classification guidelines and a robust method for the consistent valuation of classification-related evidence. Sherloc is a collection of specific, interdependent, and consistently weighted evidence types supported by a set of hierarchical decision trees.
Sherloc is built on a number of basic principles:
Variant classification should be reproducible and auditable. When confronted with the same evidence, different people should come to the same conclusions. Conclusions should be derived directly from the consistent use of evidence.
Evidence classification should be specific. A single evidence type should capture a single, well-defined use-case, and every use-case should be captured by an available evidence criterion. Ambiguities should be addressed by iterative refinements to the system.
Evidence should not be counted twice. Certain types of evidence contribute to the same basic argument. The rule set should contain dependencies to correct for double counting.
Some observations are additive. Certain arguments become more persuasive when supported by additional observations. Some evidence types can be invoked more than once and are additive when invoked multiple times.
Data types can be interrelated. Certain data types meaningfully affect the significance of other data types. The frequency of a variant, for example, changes our expectations for and use of case reports and segregation data.
Clinical genetics can be more complicated than Mendelian inheritance. Contradictory evidence may exist, and the evidence supporting a variant classification of benign or pathogenic should be considered separately. When substantial evidence supports both conclusions, that dichotomy may reflect complex, non-Mendelian mechanisms, exceptionally low-penetrance variants, or genetic modifiers or environmental effects, or may otherwise be ambiguous within the Mendelian framework.
Efficient application of a complex rule set
Molecular genetics is complicated, and that complexity is reflected in the Sherloc rule set. In practice, however, a simple hierarchical approach to evidence types yields accurate and thorough results quickly.
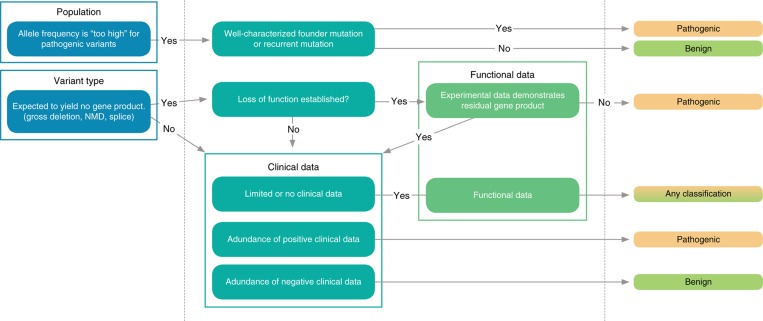
The most efficient process for variant classification moves from the most powerful and simplest information to the least powerful and most complex information. In practice, we adopt a four-step process, which usually leads to the invocation of three to five evidence types per variant. At each step, we usually choose the single most appropriate evidence type from a related collection of choices. The hierarchical steps are as follows (see Figure 5):
Evaluate population data. This step identifies variants that are too common to be causes of Mendelian disease and provides the lens through which clinical case reports must be evaluated.
Evaluate the expected effect of the variant on the gene product(s) (variant type). This step identifies variants strongly suspected to be pathogenic or benign and contextualizes the functional data.
Evaluate clinical case reports for substantial positive or negative evidence of enrichment in clinically affected individuals.
Evaluate functional experiments and predictive data. In most cases, functional data are consulted to confirm or refute the argument that has been established by the other data types.
Figure 5.
Hierarchical approach to efficient variant research. Because a hierarchical relationship exists between evidence types, an ordered approach to the evaluation of evidence can be very efficient. Evidence is evaluated starting with the simplest and potentially most powerful types and working toward the most complicated and subtle types (i.e., from population data and variant type toward clinical data and functional/prediction data). When sufficient evidence for a confident classification is identified, the remaining research can be focused on looking for contradictory evidence.
Once substantial evidence exists for a confident classification, the remaining steps can take the form of a scan for potentially contradictory evidence. Software can support the systematic efficiency of the evaluation process by providing a user interface that supports accurate rule usage and the automatic application of the discrete evidence types that depend on digitally available data. Population data and variant effect data are amenable to automatic classification, but the evaluation of case reports and functional studies are not. Novel variants unsupported by publications are highly amenable to automatic precategorization. Any system has limitations, however, and a purely software-generated classification cannot be considered a substitute for professional evaluation.
Should variant classification guidelines be disease specific?
Sherloc presents a general framework for the evaluation of evidence—an epistemological argument that simply addresses the questions, “When can we say we know the effect of a variant?,” “When do we merely suspect?,” and “How can we tell the difference?” The accurate use of this framework depends on specific knowledge of the molecular and clinical aspects of particular genes. A well-supported understanding of the disease mechanism associated with a gene should make rules that depend on the molecular mechanism applicable or inapplicable for variants in that gene. However, the weight granted to the general argument (that the variant is pathogenic because its mode of action is consistent with the mode of action associated with pathogenic variants in that gene) should be the same in all cases.
Specific knowledge about protein structure and function can lead to conclusions that a particular amino acid residue may be critical. This knowledge may be based on conservation, an understanding of protein domains, or knowledge about pairs of residues that bond to maintain a three-dimensional protein structure. However, the weight granted to the general argument—that a novel variant is pathogenic because it disrupts an amino acid residue suspected to be critical to the protein structure of a structural protein or the protein function of a channel or enzyme—should be the same in all cases.
Likewise, detailed knowledge about the phenotypes associated with pathogenic variants in a gene should make rules that depend on counting case reports relevant or irrelevant when considering observations of individuals with variants in that gene. However, the weight granted to the general argument—that a variant is more likely to be pathogenic when observed in an individual with a highly specific phenotype—should be the same in all cases.
ACMG-AMP framework
The 2015 ACMG–AMP guidelines for variant classification were a major step toward establishing the basic outlines of a shared framework for variant classifications. The conclusion of this paper is that the details of that framework can be further refined, and such refinements will improve the reproducibility and objectivity of variant classification across individuals and laboratories. However, the core value of the framework cannot be overstated: these guidelines help drive consensus by providing a shared framework for documenting the evidence considered in an evaluation, beginning the process of valuing certain evidence types, and turning professional disagreements about variant classifications into meaningful discussions about clinical and scientific data. A complex and nascent field, such as clinical genetics, will uncover cases about which reasonable professionals come to different conclusions. A shared language is the first requirement for achieving a common goal.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/gim
ADDITIONAL MEMBERS OF THE INVITAE CLINICAL GENOMICS GROUP
Sienna Aguilar, MS, Swaroop Aradhya, PhD, FACMG, Daniel Beltran, PhD, Brandon Bunker, PhD, Amy Daly, MS, Anne Deucher, MD, Tali Ekstein, MS, Ali Entezam, PhD, Karl Erhard, PhD, Ed Esplin MD, PhD, FACMG, FACP, Jennifer Fulbright, MS, Amy Fuller, MS, Kristen McDonald Gibson, PhD, FACMG, Tina Hambuch, PhD, FACMG, Rachel Harte, PhD, Christy Hartshorne, MS, Eden Haverfield, PhD, FACMG, Nastaran Heidari, PhD, Michelle Hogue, MS, Daniela Iacoboni, MS, Britt Johnson, PhD, FACMG, Hio Chung Kang, PhD, Rachel Lewis, PhD, Shiloh Martin, PhD, Sarah McCalmon, PhD, Scott Michalski, MS, Cindy Morgan, MS, Laura Murillo, PhD, Piper Nicolosi, PhD, Karen Ouyang, PhD, FACMG, Carolina Pardo, PhD, Rita Quintana, PhD, Marina Rabideau, MS, Darlene Riethmaier, MS, Amanda Stafford, PhD, Jackie Tahiliani, MS, Chris Tan, MS, S Paige Taylor, PhD, Shu-Huei Wang, PhD, Hannah White, MS, Ian Wilson, PhD, FACMG, Tom Winder, PhD, FACMG, and Michelle K Zeman, PhD.
All of the authors are employees and shareholders of Invitae, a commercial laboratory performing diagnostic genetic testing.
Supplementary Material
References
- National Institutes of Health, Secretary’s Advisory Committee on Genetic Testing. Enhancing the oversight of genetic tests: recommendations of the SACGT, 2000. http://osp.od.nih.gov/sites/default/files/oversight_report.pdf. Accessed 26 October 2016.
- Rehm HL, Berg JS, Brooks LD et al, ClinGen—the clinical genome resource. N Engl J Med 2015;372:2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Tahiliani J, Johnson NM et al, Clinical genetic testing for the cardiomyopathies and arrhythmias: a systematic framework for establishing clinical validity and addressing genotypic and phenotypic heterogeneity. Front Cardiovasc Med 2016;3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D. From evidence-based medicine to genomic medicine. Genomic Med 2007;1:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al, Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC et al, Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet 2016;98:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR et al, ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Brogna S. Nonsense-mediated mRNA decay. Biochem Soc Trans 2008;36:514–516. [DOI] [PubMed] [Google Scholar]
- Kaurah P, Huntsman DG. Hereditary diffuse gastric cancer. In: Pagon RA, Adam MP, Ardinger HH et al, (eds). GeneReviews. University of Washington: Seattle, WA, 2002. [Google Scholar]
- Corso G, Roviello F, Paredes J et al, Characterization of the P373L E-cadherin germline missense mutation and implication for clinical management. Eur J Surg Oncol 2007;33:1061–1067. [DOI] [PubMed] [Google Scholar]
- Mateus AR, Simões-Correia J, Figueiredo J et al, E-cadherin mutations and cell motility: a genotype-phenotype correlation. Exp Cell Res 2009;315:1393–1402. [DOI] [PubMed] [Google Scholar]
- Roviello F, Corso G, Pedrazzani C et al, Hereditary diffuse gastric cancer and E-cadherin: description of the first germline mutation in an Italian family. Eur J Surg Oncol 2007;33:448–451. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Faivre L et al, BBS8 is rarely mutated in a cohort of 128 Bardet-Biedl syndrome families. J Hum Genet 2006;51:81–84. [DOI] [PubMed] [Google Scholar]
- Parry D, Peters G. Temperature-sensitive mutants of p16CDKN2 associated with familial melanoma. Mol Cell Biol 1996;16:3844–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzon J, Liu L, Brill H et al, CDKN2A mutations in multiple primary melanomas. N Engl J Med 1998;338:879–887. [DOI] [PubMed] [Google Scholar]
- McNally EM, de Sá Moreira E, Duggan DJ et al, Caveolin-3 in muscular dystrophy. Hum Mol Genet 1998;7:871–877. [DOI] [PubMed] [Google Scholar]
- Jovanovic B, Egyhazi S, Eskandarpour M et al, Coexisting NRAS and BRAF mutations in primary familial melanomas with specific CDKN2A germline alterations. J Invest Dermatol 2010;130:618–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadt KA, Aoude LG, Krogh L et al, Molecular characterization of melanoma cases in Denmark suspected of genetic predisposition. PLoS One 2015;10:e0122662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland M, Meloni R, Gruis N et al, Germline mutations of the CDKN2 gene in UK melanoma families. Hum Mol Genet 1997;6:2061–2067. [DOI] [PubMed] [Google Scholar]
- Walker GJ, Hussussian CJ, Flores JF et al, Mutations of the CDKN2/p16INK4 gene in Australian melanoma kindreds. Hum Mol Genet 1995;4:1845–1852. [DOI] [PubMed] [Google Scholar]
- Eliason MJ, Larson AA, Florell SR et al, Population-based prevalence of CDKN2A mutations in Utah melanoma families. J Invest Dermatol 2006;126:660–666. [DOI] [PubMed] [Google Scholar]
- Flores JF, Pollock PM, Walker GJ et al, Analysis of the CDKN2A, CDKN2B and CDK4 genes in 48 Australian melanoma kindreds. Oncogene 1997;15:2999–3005. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chen S, Chen Q et al, The common SCN5A mutation R1193Q causes LQTS-type electrophysiological alterations of the cardiac sodium channel. J Med Genet 2004;41:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzkale H, Shen J, McLaughlin H et al, A systematic approach to assessing the clinical significance of genetic variants. Clin Genet 2013;84:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Yang S, Nykamp K et al, Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med 2017;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Demuth I, Digweed M. Nijmegen breakage syndrome. In: Pagon RA, Adam MP, Ardinger HH et al. (eds). GeneReviews. University of Washington: Seattle, WA, 1999. [Google Scholar]
- Campeau PM, Foulkes WD, Tischkowitz MD. Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet 2008;124:31–42. [DOI] [PubMed] [Google Scholar]
- Zimmerman BT. Understanding Breast Cancer Genetics. University Press of Mississippi: Jackson, MS, 2004. [Google Scholar]
- Saleh M, Kamath BM, Chitayat D. Alagille syndrome: clinical perspectives. Appl Clin Genet 2016;9:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn KJ, Yoon JK, Kim GB et al, Alagille syndrome and a JAG1 mutation: 41 cases of experience at a single center. Korean J Pediatr 2015;58:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles J, Sarina T, Yeates L et al, Clinical predictors of genetic testing outcomes in hypertrophic cardiomyopathy. Genet Med 2013;15:972–977. [DOI] [PubMed] [Google Scholar]
- Dunn KE, Caleshu C, Cirino AL et al, A clinical approach to inherited hypertrophy: the use of family history in diagnosis, risk assessment, and management. Circ Cardiovasc Genet 2013;6:118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls M. The 2014 ESC guidelines on the diagnosis and management of hypertrophic cardiomyopathy have been published. Eur Heart J 2014;35:2849–2850. [DOI] [PubMed] [Google Scholar]
- Bos JM, Will ML, Gersh BJ et al, Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc 2014;89:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Hefner MA, Belmont JW, Davenport SLH. CHARGE syndrome. In: Pagon RA, Adam MP, Ardinger HH et al. (eds). GeneReviews. University of Washington: Seattle, WA, 2006. [Google Scholar]
- Mehta PA, Tolar J. Fanconi anemia. In: Pagon RA, Adam MP, Ardinger HH et al. (eds). GeneReviews. University of Washington: Seattle, WA, 2002. [Google Scholar]
- Petrucelli N, Daly MB, Pal T. BRCA1 and BRCA2 hereditary breast and ovarian cancer. In: Pagon RA, Adam MP, Ardinger HH et al. (eds). GeneReviews. University of Washington: Seattle, WA, 1998. [Google Scholar]
- Jacquet A, Guiochon-Mantel A, Noël LH et al, Alagille syndrome in adult patients: it is never too late. Am J Kidney Dis 2007;49:705–709. [DOI] [PubMed] [Google Scholar]
- Mohapatra B, Jimenez S, Lin JH et al, Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab 2003;80:207–215. [DOI] [PubMed] [Google Scholar]
- Andreasen C, Nielsen JB, Refsgaard L et al, New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet 2013;21:918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier I, Vezain M, Martins A et al, A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat 2008;29:1412–1424. [DOI] [PubMed] [Google Scholar]
- Viel A, Genuardi M, Capozzi E et al, Characterization of MSH2 and MLH1 mutations in Italian families with hereditary nonpolyposis colorectal cancer. Genes Chromosomes Cancer 1997;18:8–18. [DOI] [PubMed] [Google Scholar]
- Genuardi M, Carrara S, Anti M et al, Assessment of pathogenicity criteria for constitutional missense mutations of the hereditary nonpolyposis colorectal cancer genes MLH1 and MSH2. Eur J Hum Genet 1999;7:778–782. [DOI] [PubMed] [Google Scholar]
- Wanat JJ, Singh N, Alani E. The effect of genetic background on the function of Saccharomyces cerevisiae mlh1 alleles that correspond to HNPCC missense mutations. Hum Mol Genet 2007;16:445–452. [DOI] [PubMed] [Google Scholar]
- Trojan J, Zeuzem S, Randolph A et al, Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology 2002;122:211–219. [DOI] [PubMed] [Google Scholar]
- Hinrichsen I, Brieger A, Trojan J et al, Expression defect size among unclassified MLH1 variants determines pathogenicity in Lynch syndrome diagnosis. Clin Cancer Res 2013;19:2432–2441. [DOI] [PubMed] [Google Scholar]
- Kondo E, Suzuki H, Horii A et al, A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res 2003;63:3302–3308. [PubMed] [Google Scholar]
- Shimodaira H, Filosi N, Shibata H et al, Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet 1998;19:384–389. [DOI] [PubMed] [Google Scholar]
- Tang H, Thomas PD. Tools for predicting the functional impact of nonsynonymous genetic variation. Genetics 2016;203:635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm DG, Azencott CA, Aicheler F et al, The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum Mutat 2015;36:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.