Abstract
The role of Notch signaling in human innate lymphoid cell (ILC) differentiation is unclear, although IL-7 and IL-15 promote differentiation of natural cytotoxicity receptor (NCR) NKp44+ group 3 ILCs (NCR+ILC3s) and conventional NK (cNK) cells from CD34+ hematopoietic progenitor cells (HPCs) ex vivo. In this study, we analyzed the functions of Notch in the differentiation of NCR+ILC3s and cNK cells from human HPC subpopulations circulating in peripheral blood by limiting dilution and clonal assays using high-throughput flow cytometry. We demonstrated that Notch signaling in combination with IL-7 induced NCR+ILC3 differentiation, but conversely suppressed IL-15–dependent cNK cell generation in CD45RA+Flt-3−c-Kitlow, a novel innate lymphocyte-committed HPC subpopulation. In contrast, Notch signaling induced CD45RA−Flt-3+c-Kithigh multipotent HPCs to generate CD34+CD7+CD62Lhigh, the earliest thymic progenitor–like cells, which preserved high cNK/T cell potential, but lost NCR+ILC3 potential. These findings implicate the countervailing functions of Notch signaling in the fate decision between NCR+ILC3 and cNK cell lineages at different maturational stages of human HPCs. Inhibition of Notch functions by Abs specific for either the Notch1 or Notch2 negative regulatory region suggested that both Notch1 and Notch2 signals were involved in the fate decision of innate lymphocyte-committed HPCs and in the generation of earliest thymic progenitor–like cells from multipotent HPCs. Furthermore, the synergistic interaction between Notch and IL-7 in NCR+ILC3 commitment was primarily explicable by the induction of IL-7 receptor expression in the innate lymphocyte–committed HPCs by Notch stimulation, suggesting the pivotal role of Notch in the transcriptional control required for human NCR+ILC3 commitment.
Introduction
Innate lymphoid cells (ILCs) are innate immune cells of lymphoid origin that lack expression of Ag receptors, but retain pivotal effector and regulatory functions for innate immunity and tissue homeostasis (1). In addition to conventional NK (cNK) cells, ILCs are classified into three additional groups based on their discrete patterns of cytokine production in response to particular stimulation signals and their distinct requirements of transcription factors (TFs) for their development and functional maintenance (2–4). Group 3 ILCs (ILC3s) are characterized by the expression of the retinoic acid receptor–related orphan receptor (ROR)γt TF and the production of IL-17 and/or IL-22 (5, 6). Furthermore, the aryl hydrocarbon receptor (AHR) is required for the postnatal differentiation of mouse ILC3s (7–9). Human ILC3s can be further divided into three subsets: lymphoid tissue inducer cells, natural cytotoxicity receptor (NCR) NKp44+ ILC3s, and NKp44− ILC3s. Human RORγt+NCR+ ILC3s, as well as cNK cells, differentiate simultaneously from cord blood (CB) CD34+ hematopoietic progenitor cells (HPCs) in the presence of IL-7 and IL-15 (10). RORγt+CD34dim cells residing in human tonsils (To) and intestinal lamina propria (LP) can specifically differentiate into ILC3s, whereas RORγt−CD34+ cells have the potential to differentiate into both ILC3 and cNK cells (11). Although these studies demonstrated that ILC3 potential is preserved in human HPCs, the transcriptional regulation involved in the differentiation of ILC3s from HPCs, particularly lineage divarication from cNK cells, is largely unknown.
The developmental program of ILCs has been studied extensively in mice (3). The presence of common innate lymphoid progenitors at the downstream stage of common lymphoid progenitors was predicted by the finding that mice lacking expression of the transcriptional regulatory inhibitor of DNA binding 2 (Id2) have a loss of all ILC lineages, whereas T and B cell development is largely unaffected (12–14). In Id2 reporter mice, a single Id2+ cell with the CD127+Flt-3−CD25−integrin α4β7+ phenotype gave rise to all ILC lineages, but not to cNK cells (15), suggesting that this Id2+ progenitor cell functions as a common helper-like ILC progenitor (CHILP) (3) without cNK cell potential. Furthermore, the promyelocytic leukemia zinc finger TF is partly involved in the development of CHILP (15, 16).
In addition to Id2 and its potential target gene promyelocytic leukemia zinc finger TFs, Notch signals are a major common denominator in ILC lineage differentiation in mice. CHILP generates progeny of all ILC lineages following coculture with OP9 stroma cells that express Notch ligand delta-like 1 (OP9-DL1) (15). ILC1s and NCR+ILC3s are greatly decreased in mice lacking the Notch signaling adaptor RBP/J (17). In vivo and in vitro development of ILC2s requires Notch signals (18, 19). ILC3 differentiation also requires Notch signals, although the requirement is different in fetal and adult progenitors (14, 20). Acquisition of T-box expressed in T cells (T-bet) expression, which is required for NCR+ILC3 differentiation, also requires Notch stimulation by OP9-DL1 stroma cells (21). Recent studies have shown that Notch signaling is required for differentiation of NCR+ILC3 from NCR−ILC3 and for maintenance of the NCR+ILC3 population (22, 23). In humans, Notch signals enhance cNK cell differentiation from HPCs in culture (24–27). However, although only one report noted that ILC2 lineage cells can be induced from postnatal thymic CD34+CD1a− cells by Notch signals using OP9-DL1 cells (28), the significance of Notch signaling in the commitment of human ILCs is largely unknown.
In the present study, we clarified the functions of Notch in the differentiation of human NCR+ILC3s and cNK cells from human HPCs circulating in peripheral blood (PB). A recent report using parabiotic mice demonstrated that ILCs in both lymphoid and nonlymphoid organs were locally renewed and expanded in response to acute infection (29). However, although the homeostasis and lifespan of human ILCs in peripheral organs are totally unknown, we assume that ILC precursors migrate from bone marrow (BM) to peripheral organs through the bloodstream in adults, particularly during severe epithelial damage in association with an extensive shrinkage of the ILC pool. We previously developed a cell sorting–based limiting dilution assay (LDA) and clonal analyses using coculture with OP9-DL1 cells in 384-well microplates for quantification and characterization of various progenitors among CD34+ HPC populations circulating in PB of adult humans (26, 30). LDA and clonal assays are thought to be essential for accurate quantification of lineage commitment in a specific progenitor population, because extensive generation of progeny that originate from a limited number of highly proliferative progenitor clones skews experimental output. In this study, we further improved the LDA and clonal assay with high-throughput flow cytometry (HTFC) (31) to efficiently detect and count the progeny that differentiated from HPCs distributed in a large number of culture wells. Using this method, we identified CD45RA+Flt-3−c-Kitlow, novel innate lymphocyte–committed progenitor cells in the human circulating HPC population. We demonstrated that Notch signaling induced NCR+ILC3 differentiation in the presence of IL-7, but conversely suppressed IL-15–dependent cNK cell generation from this HPC subpopulation. In contrast, Notch stimulated the CD45RA−Flt-3+c-Kithigh multipotent HPC subpopulation to generate CD34+CD7+CD62Lhigh cells, which retained the high cNK/T cell potential but reduced the NCR+ILC3 potential. These results suggest opposing roles for Notch signaling in the fate decision between NCR+ILC3 and cNK cell lineages at different maturational stages of human HPCs. Because it was reported that DL1 molecules stimulate both Notch1 and Notch2 receptor (32), we evaluated whether Notch1 or Notch2 signals function in the fate decision by blocking experiments with Abs specific for the receptor negative regulatory region (NRR) of either Notch1 or Notch2 (33).
Materials and Methods
Cytokines and Abs
Recombinant human Kit ligand (KL), Flt-3 ligand (FL), IL-1β, and IL-7 were purchased from PeproTech. IL-12p70 and IL-15 were purchased from Tonbo Biosciences. IL-18 and IL-23 were purchased from MBL International and eBioscience, respectively.
Anti-CD3 (UCHT1), CD7 (CD7-6B7), CD11a (TS2/4 and HI111), CD11c (Bu15), CD14 (M5E2), CD15 (H198 and W6D3), CD16 (3G8), CD19 (HIB19), CD20 (2H7), CD34 (581), CD56 (HCD56), CD62L (DREG-56), CD117 (104D2), CD135 (BV10A4H2), CD161 (HP-3G10), CD235a (HI235), CD294 (BM16), NKp44 (P44-8), NKp46 (9E2), IL-8 (E8N1), and IL-17A (BL168) Abs and human IgG1 isotype control (ET901) were purchased from BioLegend. Anti-CD3 (SK7), CD5 (UCHT2), CD7 (M-T701), CD16 (3G8), CD235a (GA-R2), HLA-DR (L243), and IFN-γ (25723.11) Abs were purchased from BD Biosciences. Anti-CD16a (3G8), CD56 (N901), CD159a (Z199), and CD314 (ON72) Abs were purchased from Beckman Coulter. Anti-CD10 (CB-CALLA), CD14 (61D3), CD25 (BC96), IL-22 (22URTI), GM-CSF (GM2F3), RORγt (AFKJS-9), AHR (FF3399), T-bet (eBio4B10), and eomesodermin (Eomes; WD1928) Abs were obtained from eBioscience. Anti-CD25 (BC96), CD45RA (HI100), and CD127 (R34-34) Abs were purchased from Tonbo Biosciences. Anti-CD14 (TÜK4) and granzyme B (GB12) Abs were purchased from Invitrogen. Anti-Notch1 NRR (NRR1) and anti-Notch2 NRR (NRR2) Abs were provided by C.W. Siebel (Genentech, South San Francisco, CA). Development and characterization of anti-NRR1 and anti-NRR2 Abs were as described elsewhere (33).
Preparation of PB HPCs
Human PB samples were collected from 11 healthy in-house volunteer donors (Japanese) with informed consent (5 males and 6 females; age range, 38–61 y), following the guidance of the Institutional Review Board (Human Investigation Committee of the Radiation Effects Research Foundation), which approved this study. PBMCs were separated from 10-ml PB samples by Ficoll density gradient centrifugation (lymphocyte separation medium 1077; Wako Pure Chemical Industries). For sorting CD34+lineage (Lin)−CD7− HPCs, PBMCs were stained with allophycocyanin-labeled anti-CD34, PerCP-Cy5.5–labeled anti-CD7, and PE-labeled anti-lineage marker (CD3, CD14, CD15, CD16, CD19, CD20, CD56, and CD235a) Abs for 30 min on ice, and dead cells were excluded by 1 μg/ml DAPI (Invitrogen). CD34+Lin−CD7− HPCs were sorted by FACSAria II (BD Biosciences). For sorting HPC subpopulations, PBMCs were stained with allophycocyanin-Cy7–conjugated anti-CD34, allophycocyanin-conjugated anti–c-Kit, PE-conjugated anti–Flt-3, PerCP-Cy5.5–conjugated anti-CD45RA (RA), FITC-conjugated anti-lineage markers, and FITC-conjugated anti-CD7 Abs. HPC subpopulations with RA−Flt-3+c-Kithigh, RA+Flt-3+c-Kitlow, and RA+Flt-3−c-Kitlow phenotypes were sorted.
Stroma cells
Generation of mouse OP9-DL1 stroma cells from OP9 cells (34) engineered to express the GFP and mouse DL1 has been described previously (35). OP9-DL1 and the OP9 stroma cells were maintained by culturing in αMEM (Life Technologies) supplemented with 20% FBS (Hyclone), 4 × 10−6 M 2-ME, and penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Coculture of HPCs with stroma cells
Details of the methods of HPC coculture with stroma cells were as described previously (26). Briefly, OP9-DL1 or OP9 stroma cells were seeded in the wells of a 24-well plate or 384-well flat-bottom black plate (BD Biosciences), which were coated with gelatin. Briefly, at least 4 h prior to HPC sorting, culture medium in each well was replaced with 50 μl of phenol red–free αMEM containing 20% knockout serum replacement (Life Technologies), 10−4 M monothioglycerol (Sigma-Aldrich), 50 μg/ml gentamicin (Sigma-Aldrich), KL and FL in combination with IL-7 and/or IL-15 (10 ng/ml). For bulk culture of HPCs, HPCs and their subpopulation cells were cocultured with stroma cells in a 24-well plate at a density of ≥500 HPCs per well for 2–5 wk.
Surface phenotyping of ILCs and cNK cells
For surface phenotyping of progeny harvested from coculture of HPCs with stroma cells, cells were stained with PE-Cy7–labeled anti-CD56, allophycocyanin-Cy7–labeled CD16, allophycocyanin-labeled or PE-labeled CD11a, and PerCP-Cy5.5–labeled or PE-labeled CD7 Abs, in combination with PerCP-Cy5.5–labeled CD161, PE-labeled anti-NKG2A, PE-labeled anti-NKp44, PerCP-Cy5.5–labeled anti-NKp46, allophycocyanin-labeled anti–c-Kit, allophycocyanin-labeled anti-CD25, and allophycocyanin-labeled anti-CD127 Abs. For detection of CD127 expression, progeny were precultured in the absence of IL-7 for 5 d.
Cytoplasmic and nuclear immunostaining
For detection of intracellular cytokines, ILC3 (CD56+CD11a−) and cNK cells (CD56+CD11a+) sorted from coculture of HPCs with stroma cells as described above were stimulated with either IL-1β and IL-23 or IL-12 and IL-18 for 12 h in the absence of stroma cells in U-bottom 96-well plates (BD Biosciences). After cytokine stimulation, cells were treated with brefeldin (eBioscience) for 4 h to inhibit the protein transporter, and cells were harvested and stained with a Zombie Green fixable viability kit (BioLegend) to gate out dead cells by flow cytometry. Cells were stained with allophycocyanin-conjugated anti-CD11a, PE-Cy7–conjugated anti-CD56, and PerCP-Cy5.5–conjugated anti-CD7 Abs for 30 min on ice and washed with PBS. Fixation and permeabilization of cells were performed using an intracellular fixation and permeabilization buffer set (eBio). Fixed and permeabilized cells were stained with PE-conjugated anti–IL-8, IL-17, IL-22, GM-CSF, and IFN-γ Abs.
For detecting expression of TFs, sorted ILC3 and cNK cells were stained with Zombie Green and stained with anti-CD7, CD56, and CD11a Abs, as described above. Fixation and permeabilization of cells were performed using a Foxp3 TF staining buffer set (eBioscience). Fixed and permeabilized cells were stained with PE-conjugated anti-RORγt, AHR, T-bet, and Eomes Abs.
Limiting dilution and single-cell analyses by HTFC
For the LDA of total HPCs and the RA−Flt-3+c-Kithigh HPC subpopulation, HPCs were sorted into 80 wells of a 384-well plate at 20, 15, 10, or 5 cells per well (20 wells for each cell number). For LDA of the RA+Flt-3+c-Kitlow and RA+Flt-3−c-Kitlow HPC subpopulations, HPCs were sorted at six, four, two, and one cell per well. LDA culture was maintained at 37°C in a humidified atmosphere of 5% CO2, and half the culture medium (25 μl) was changed every week. After 3–5 wk of culture, all cells grown in wells of the 384-well plate were harvested by vigorous pipetting and transferred to a 96-well plate. For detection of NCR+ILC3 and cNK cell lineage progeny, harvested progeny cells were stained with PE-Cy7–labeled anti-CD56, PerCP-Cy5.5–labeled anti-CD7, PE-labeled anti-CD11a, allophycocyanin-labeled anti-NKp44, and allophycocyanin-Cy7–labeled CD16 Abs in PBS containing 2 mM EDTA, 0.01% NaN3, and 1% FBS (washing buffer [WB]). For detection of T and myeloid cells, progeny were stained with PE-labeled anti-CD7, allophycocyanin-labeled anti-CD5, PE-Cy7–labeled anti-CD56, allophycocyanin–Alexa Fluor 750-labeled CD14, and PerCP-Cy5.5–labeled anti-CD15 Abs. When cell samples were assayed for both ILC3/cNK and T/myeloid cells, cell suspensions were split into two 96-well plates for staining with each Ab set. After washing cells with WB, all stained cells suspended in 30 μl of WB containing DAPI were aspirated and transferred to a CyAn (Beckman Coulter) flow cytometer by HyperCyt (IntelliCyt) (31). The tubing line was washed with Triton X-100 (2%) and WB after each sampling of cells from wells to avoid cross contamination of cell samples between wells. A well exhibiting six or more positive events was designated as a positive well for LDA. When cell samples were assayed for both ILC3/cNK and T/myeloid cells, a well exhibiting three or more positive events was considered positive. Responding progenitor frequencies (PFs) of ILC3, cNK, and T cells were calculated by online analysis using ELDA software (36), available on the home page of Walter and Eliza Hall Institute Bioinformatics Division (http://bioinf.wehi.edu.au/software/elda/index.html). The 95% confidence interval of each PF and the p values of the differences in PFs between two different culture conditions were obtained by this software.
For clonal HPC assays, total HPCs and their subpopulation cells were sorted into 160 wells of a 384-well plate at one cell per well. The methods of OP9-DL1 coculture and flow cytometry of progeny for clonal HPC culture were the same as for LDA as described above.
Inhibition of Notch functions by anti-NRR Abs
Anti-NRR1, anti-NRR2, or human isotype control IgG1 was added to the coculture of HPC subpopulations with OP9-DL1 stroma cells at the initiation of culture. The optimum concentration of Abs (10 μg/ml) was determined by preliminary dose response experiments (data not shown). Effects of Abs on generation of NCR+ILC3, cNK cell, and T cell progenies from HPCs were evaluated by LDA, as described above.
Results
Differentiation of NCR+ILC3s and cNK cells from circulating HPCs in the presence or absence of Notch signaling
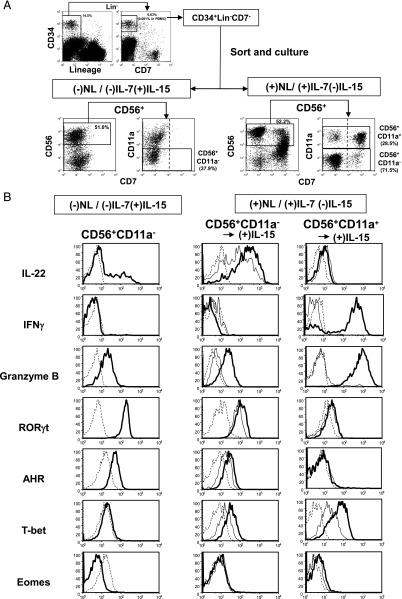
Notch function in the generation of NCR+ILC3 and cNK cell progeny from CD34+Lin−CD7− cells (HPCs) circulating in PB was evaluated by coculture with OP9 control stroma cells and OP9-DL1 stroma cells expressing the Notch ligand (NL), Delta1, in combination with IL-7 and/or IL-15, in the presence of KL and FL (Fig. 1A). As reported previously (10), we used CD11a− and CD11a+ phenotypes as markers for discriminating NCR+ILC3 and cNK cells in the CD56+ cell population, respectively. We sorted the CD7− fraction of HPCs to examine whether Notch signaling can induce CD7 expression along with ILC and cNK cell differentiation (Fig. 1A), as CD7 is one of the first surface markers expressed during the course of cNK cell differentiation (37). HPCs gave rise to CD56+CD11a− and CD56+CD11a+ cells, which were mainly detected in the CD7− and CD7+ fractions, respectively, in coculture with OP9-DL1 stroma cells in the presence of IL-7 and in the absence of IL-15 conditioning, abbreviated as (+)NL/(+)IL-7(−)IL-15. In contrast, neither CD56+CD11a− nor CD56+CD11a+ cells expressed CD7 in coculture with OP9 stroma cells in the absence of IL-7 and in the presence of IL-15 conditioning [(−)NL/(−)IL-7(+)IL-15]. Upon (−)NL/(+)IL-7(−)IL-15 conditioning, CD56+CD11a− and CD56+CD11a+ cells were detected in the CD7− fraction, but their generation was found to be very low (data not shown).
FIGURE 1.
Expression of intracellular ILC3 and cNK cell markers in progeny generated from total HPCs in the presence or absence of Notch signaling. (A) CD34+Lin−CD7− cells (HPCs) among PBMCs were sorted and cultured under either (−)NL/(−)IL-7(+)IL-15 or (+)NL/(+)IL-7(−)IL-15 conditions in the presence of KL and FL for 4.5 wk. CD56+CD11a− and CD56+CD11a+ progeny were sorted for intracellular immunofluorescence. (B) Representative flow histograms of cytokine (IL-22 and IFN-γ), granzyme B, and TF (RORγt, AHR, T-bet, and Eomes) expression in sorted CD56+CD11a− and CD56+CD11a+ cells. Detection of IL-22– and IFN-γ–producing cells was conducted by stimulation of sorted cells with IL-1β and IL-23 and with IL-12 and IL-18, respectively, for 12 h in the absence of stroma cells. CD56+CD11a− and CD56+CD11a+ cell progeny generated under (+)Notch/(+)IL-7(−)IL-15 conditions were further cultured for 1 wk in the presence (thick line) or absence of IL-15 (thin line) without stroma cells (middle and right panels). Dotted lines represent IgG isotype control. Similar results were obtained for the other two donors.
We confirmed that CD56+CD11a− cells, which were generated under (−)NL/(−)IL-7(+)IL-15 conditions, produced IL-22 following stimulation with IL-1β and IL-23, but did not produce IFN-γ following stimulation with IL-12 and IL-18 (Fig. 1B). CD56+CD11a− cells in either the CD7+ or CD7− fraction that were generated under (+)NL/(+)IL-7(−)IL-15 conditions also produced IL-22, which was enhanced by additional culture with IL-15. These CD56+CD11a− cells produced a high level of IL-8 and a low level of IL-17, but no GM-CSF (Supplemental Fig. 1A). Alternatively, CD56+CD11a+ cells from the (+)NL/(+)IL-7(−)IL-15 culture did not produce IL-22, but did produce IFN-γ following additional culture with IL-15 (Fig. 1B). Additionally, IL-15 extensively enhanced granzyme B expression in CD56+CD11a+ cells, but induced only very low levels of expression in CD56+CD11a− cells. Furthermore, CD56+CD11a− cells generated under both the (+)NL and (−)NL conditions expressed a high level of RORγt and significant levels of AHR and T-bet, but not Eomes TFs. The expression of RORγt, AHR, and T-bet in CD56+CD11a− cells under (+)NL/(+)IL-7(−)IL-15 conditions was significantly elevated by additional stimulation with IL-15. Surprisingly, only a low level of Eomes expression was detected in Notch-induced CD56+CD11a+ cells, even in the presence of IL-15. Because CD56+CD11a+ cells under (−)NL/(−)IL-7(+)IL-15 conditions expressed a significant level of Eomes (Supplemental Fig. 1B), Notch signaling may have suppressed Eomes expression. These results demonstrated that CD56+CD11a− cells were generated from HPCs under either (−)NL/(−)IL-7(+)IL-15 or (+)NL/(+)IL-7(−)IL-15 conditions being typical RORγt+ ILC3s with a capacity for IL-22 production. Alternatively, CD56+CD11a+ cells were cNK cells with IFN-γ/granzyme B–producing ability.
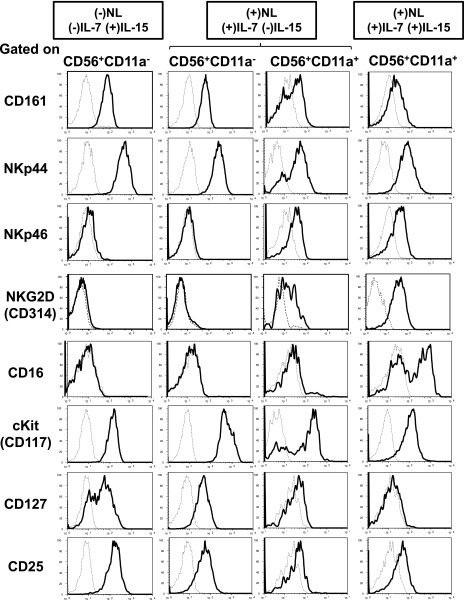
We further analyzed the surface phenotype of ILC3 and cNK cell progeny generated under the different conditions. As shown in Fig. 2, CD56+CD11a− cells generated under both the (−)NL/(−)IL-7(+)IL-15 and (+)NL/(+)IL-7(−)IL-15 conditions expressed high levels of c-Kit and NKp44 and significant levels of CD161, CD127, and CD25, but no CD314 (NKG2D) or CD16. In contrast, CD56+CD11a+ progeny generated in the presence of IL-15 exhibited significant levels of CD314 and CD16 but no CD127 expression (Fig. 2, Supplemental Fig. 1B), whereas CD56+CD11a+ progeny generated under (+)NL/(+)IL-7/(−)IL-15 conditions exhibited immature phenotypes such as a significant level of CD127, but only low levels of CD314 and CD16 expression (Fig. 2). We found that immature CD56+CD11a+ cells generated under (+)NL/(+)IL-7(−)IL-15 conditions expressed mature cNK cell markers, such as CD314 and CD159a (NKG2A), after 2 wk of culture with IL-15 in the absence of stromal cells (data not shown).
FIGURE 2.
Surface phenotype of progeny generated from total HPCs in the presence or absence of Notch signaling. Representative flow histograms of ILC3 and cNK cell surface markers in progeny generated from PB total HPCs under (−)NL/(−)IL-7(+)IL-15, (+)NL/(+)IL-7(−)IL-15, and (+)NL/(+)IL-7(+)IL-15 conditions for 4.5 wk, as shown in Fig. 1A. IL-15 was added to the culture from the beginning under (+)NL/(+)IL-7(+)IL-15 conditions. Each histogram was obtained by gating for CD56+CD11a− and CD56+CD11a+ fractions of the progeny population. Similar results were obtained for the other two donors.
Taken together, we demonstrated that Notch signaling in combination with IL-7 induced CD56+CD11a− cells, representing mature NCR+ILC3s from human adult circulating HPCs, as observed previously in CD56+CD11a− cells generated from CB HPCs under (−)NL/(+)IL-7(+)IL-15 conditions (10). Circulating HPCs may also give rise to CD56+CD11a+ cells, representing either immature cNK cells induced by Notch signaling in the presence of IL-7 or more mature cNK cells generated in the presence of IL-15.
Role of Notch signaling in NCR+ILC3 and cNK cell commitments analyzed by LDA using HTFC
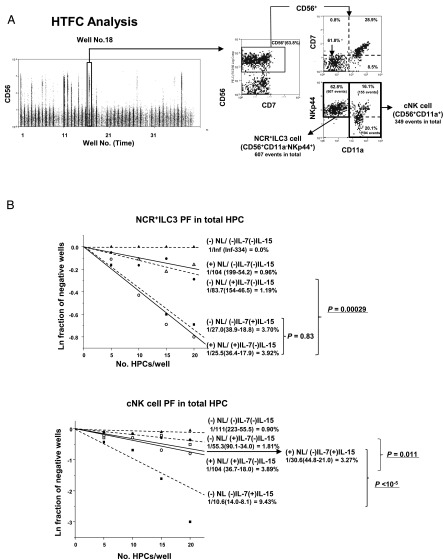
To quantitatively evaluate the role of Notch signaling in NCR+ILC3 and cNK cell commitment, we determined NCR+ILC3 and cNK cell PFs of HPCs with cell sorting–based LDA (Fig. 3). For this assay, the generation of NCR+ILC3 and cNK cell progeny was determined by the presence of CD56+NKp44+CD11a− and CD56+NKp44+ or lowCD11a+ cells, respectively, both of which were detected by HTFC (Fig. 3A). Again, most NKp44+ ILC3 and cNK cell progeny were CD7− and CD7+, respectively. Notch signaling in combination with IL-7 stimulated ILC3 commitment of total HPCs, even in the absence of IL-15 (p = 0.00029), whereas this signal was not required for ILC3 differentiation in the presence of IL-15 (Fig. 3B, upper), as observed in the bulk culture (Fig. 1). Moreover, Notch stimulated IL-7–induced cNK cell differentiation (p = 0.011), as reported by our previous studies (26, 30). In contrast, Notch signaling significantly suppressed cNK cell commitment induced by IL-15 (p < 10−5) (Fig. 3B, lower). The differing actions of Notch signaling on NCR+ILC3 and cNK cell commitments observed in the total HPC population were thought to be attributable to the differences in Notch functions in different HPC subpopulations.
FIGURE 3.
NCR+ILC3 and cNK cell commitments of total HPC population analyzed by LDA using HTFC. One thousand HPCs were sorted into 80 wells of 384-well plates at different cell numbers (20 wells for each HPC dose) and cultured in the presence or absence of Notch signaling with IL-7 or IL-15 for 5 wk. (A) The whole population of progeny cells in each well was analyzed for expression of CD56, CD7, CD11a, and NKp44 by HTFC. Time plots of HTFC were obtained without any gating procedures. Events in each well were discriminated in the time plot and analyzed for their expression of surface markers. The numbers of CD56+CD11a− and CD56+CD11a+ cell events per well for ILC3 and cNK cells, respectively, were obtained for calculation of PF by LDA. Representative flow cytograms of NKp44+CD56+CD11a− and NKp44+ or low CD56+CD11a+ cells, which were generated under (+)NL/(+)IL-7(−)IL-15 conditions, are shown. (B) Representative LDA plots for NCR+ILC3 PFs (upper) and cNK PFs (lower) under different culture conditions of Notch signaling and cytokines: (+)NL/(+)IL-7(−)IL-15 (○); (+)NL/(−)IL-7(+)IL-15 (⬜); (+)NL/(−)IL-7(−)IL-15 (△) (−)NL/(+)IL-7(−)IL-15 (●); (−)NL/(−)IL-7(+)IL-15 (■); (−)NL/(-)IL-7(−)IL-15 (▴). Presence of NCR+ILC3 and cNK cell progeny was analyzed for each culture condition. The natural log of negative well fractions was plotted against the number of HPCs per well. The PFs and their 95% confidence intervals (parentheses) obtained by online analysis using ELDA software (36) are shown in each LDA plot. The p values of the differences in PFs between two different culture conditions also were calculated using ELDA software. The PFs were converted to a percentage in each HPC population. Similar results were obtained for the other two donors.
Different roles of Notch signaling in NCR+ILC3 and cNK cell commitments of HPC subpopulations
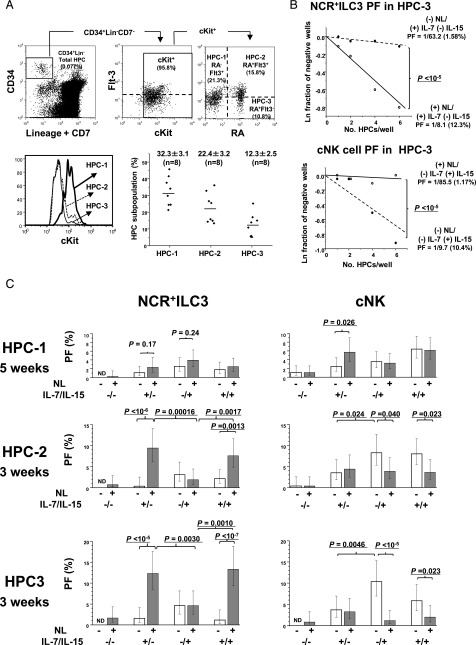
We examined whether the role of Notch signaling in the commitments of NCR+ILC3 and cNK cells differs among HPC subpopulations, including CD45RA (RA)−Flt-3+c-Kithi (HPC-1), RA+Flt-3+c-Kitlo (HPC-2), and RA+Flt-3−c-Kitlo (HPC-3) fractions (Fig. 4A). The CD38 marker for fractionation of PB HPCs was not used, because the major lymphoid-restricted HPCs are present in the RA+ fraction of both CD38− and CD38+ adult BM HPC populations (38). The RA−Flt-3− HPC population was excluded from the present analysis, because these cells mainly constitute erythroid and megakaryocyte progenitors (38). Thus, the HPC-1 subpopulation was considered to comprise hematopoietic stem cells (HSCs), multipotent progenitors (MPPs), and common myeloid progenitors. HPC-2 included lymphoid-primed MPPs (LMPPs), common lymphoid progenitors, and granulocyte-monocyte progenitors. HPC-3 cells with the CD34+Lin−CD7−c-KitlowFlt-3− phenotype have not been reported in human CB or BM (38). Thus, HPC-3 progenitors may be a novel HPC subpopulation detected only in adult PB, although the population size was relatively small (Fig. 4A).
FIGURE 4.
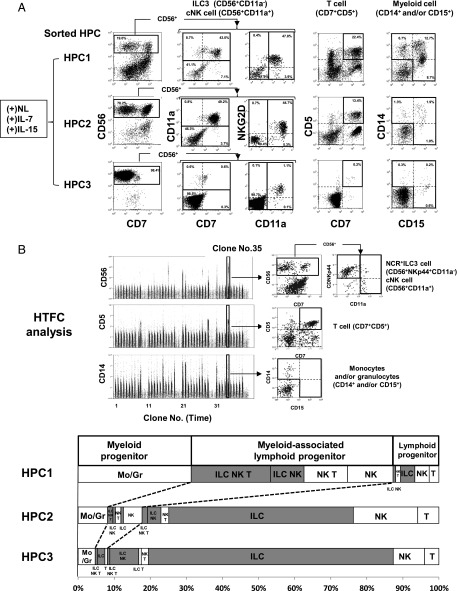
NCR+ILC3 and cNK cell commitments of three different HPC subpopulations. (A) Gating strategy of three HPC subpopulations and their c-Kit expression levels. CD34+Lin−CD7−c-Kit+ PB HPCs were gated into three subpopulations based on the expression levels of Flt-3 and CD45RA (RA): RA−Flt-3+ (HPC-1), RA+Flt-3+ (HPC-2), and RA+Flt-3− (HPC-3) (upper). The expression levels of c-Kit in each subpopulation are represented in a histogram (lower left). The frequencies (%) of the three HPC subpopulations in total HPCs are shown for eight donors (lower right). (B) Representative LDA plots for NCR+ILC3 PFs (upper) and cNK PFs (lower) of HPC-3 subpopulation. Different numbers of HPC-3 cells were sorted into the wells of 384-well plates and cultured in the presence or absence of Notch signaling with IL-7 and/or IL-15 for 3 wk. PFs and percentages are shown, as described in the legend for Fig. 3B. The p values of the differences in PFs between (+)NL (○) and (−)NL (●) conditions were calculated by online analysis using ELDA software (36). (C) NCR+ILC3 and cNK PFs of different HPC subpopulations under different culture conditions of Notch signaling and cytokines (3 or 5 wk culture). The PFs for the HPC-1, HPC-2, and HPC-3 subpopulations were obtained by LDA. The bars represent the 95% confidence intervals of PFs. The p values of the differences in PFs between the two different culture conditions are presented. Similar results were obtained for the other three donors. ND, not detectable due to no positive wells in LDA.
NCR+ILC3 and cNK cell PFs of each subpopulation were determined by LDA using HTFC (Fig. 4B, 4C). The opposite effects of Notch signaling on IL-7–induced NCR+ILC3 versus IL-15–induced cNK cell commitments observed in the total HPCs (Fig. 3B) were most prominent in the HPC-3 subpopulation (p < 10−5) (Fig. 4B, 4C). In this subpopulation, IL-7 was more effective than IL-15 for Notch-dependent NCR+ILC3 differentiation (p = 0.0030), whereas IL-15 was more effective for Notch-independent cNK cell induction (p = 0.0046). Thus, the opposite effect of Notch on NCR+ILC3 and cNK cell commitment was observed, even in the presence of both IL-7 and IL-15. The HPC-2 subpopulation exhibited a similar characteristic as the HPC-3 subpopulations, but the suppressive effects of Notch on IL-15–induced cNK commitment were weaker in HPC-2 than in HPC-3 (Fig. 4C) Thus, the opposite effects of Notch on NCR+ILC3 and cNK cell differentiation observed in the total HPCs can largely be attributed to the HPC-3 subpopulation. In contrast to HPC-2 and HPC-3 cells, Notch signaling significantly enhanced IL-7–induced NK cell generation in the HPC-1 subpopulation (p = 0.026), whereas the enhancement of IL-7–induced NCR+ILC3 generation by Notch stimulation was not significant in this subpopulation (p = 0.17) (Fig. 4C) Hence, the enhancement of IL-7–induced cNK cell commitment observed in the total HPCs (Fig. 3B, lower) was attributable to the HPC-1 subpopulation. These findings suggest that Notch signaling works on the NCR+ILC3 and cNK cell commitment of HPC-1 cells in a different manner from the case of HPC-2 and HPC-3 commitments. We speculate that the different actions of Notch signaling in the HPC subpopulations are associated with Notch-dependent commitment to other lineages linked with NCR+ILC3 and/or cNK cell lineage commitment.
Clonal analyses of differentiation potential revealed by HPC subpopulations in the presence of Notch signaling
To elucidate the lineage commitment relationship between NCR+ILC3/cNK cell and other lineages, the differentiation potential toward T and myeloid cells in addition to NCR+ILC3 and cNK cells was evaluated for the three HPC subpopulations under (+)NL/(+)IL-7(+)IL-15 conditions by bulk (Fig. 5A) and single-cell (Fig. 5B, 5C) culture. In bulk culture, the HPC-1 population possessed high NCR+ILC3 (CD56+CD11a−), NK cell (CD56+CD11a+), T cell (CD7+CD5+), and myeloid cell (CD14+ and/or CD15+) potential. HPC-3 cells predominantly generated NCR+ILC3s along with a small population of cNK cells, as expected from LDA results. The potential of HPC-2 cells was intermediate between HPC-1 and HPC-3 cells. Although data are not shown, we confirmed that most CD56+CD11a− and CD56+CD11a+ cells generated from these HPC subpopulations under NL(+)/IL7(+)IL-15(+) conditions were RORγt+EOMES− and RORγt−EOMESlo populations, respectively. Almost no T cell progeny were detected in the three HPC subpopulations in the absence of Notch signaling (data not shown).
FIGURE 5.
Differentiation potential of HPC subpopulations analyzed with bulk and single-cell cultures. (A) Five hundred HPC-1, -2, and -3 cells were cultured with OP9-DL1 stroma cells in the presence of IL-7 and IL-15 [(+)NL/(+)IL-7(+)IL-15 condition] for 5 wk. Progeny from each HPC subpopulation were analyzed for their surface phenotype of ILC3 (CD56+CD11a−), cNK cells (CD56+CD11a+), T cells (CD7+CD5+), and myeloid cells (CD14+ monocytes and/or CD15+ granulocytes). CD7 and NKG2D expression in ILC3 and cNK cells is also presented. (B) NCR+ILC3, cNK cell, T cell, and myeloid cell progeny derived from a single HPC in each subpopulation were analyzed by HTFC. Time plots and flow cytograms show the representative HTFC analysis of HPC-1–derived clones (upper). Samples from three human donors were examined for each HPC subpopulation. Numbers of clones producing different lineage combinations of progeny from the donors were combined and are listed in Supplemental Table I. Progenitor clones retaining lymphoid potential were classified as lymphoid precursors with or without myeloid potential (lower). Lineage classification of lymphoid potential is represented by the combination of NCR+ILC3s (ILC), cNK cells (NK), and T cells (T) in bar graphs. Classification is shown as the percentage of clones retaining a given lineage combination of the total lymphoid progenitor clones. Gray bars represent the percentage of lymphoid progenitor clones retaining ILC potential.
The linkage of lineage commitment was evaluated by clonal analyses of the HPC subpopulations. For these analyses, we detected NCR+ILC3, cNK cell, T cell, and myeloid cell progeny generated from a single HPC using HTFC (Fig. 5B, upper) and classified the lineage of the progeny (Fig. 5B, lower; Supplemental Table I). Based on the combination of lineages, lymphoid potentials were classified into ILC3/cNK/T triple, ILC3/cNK dual, cNK/T dual, ILC3/T dual, ILC3 single, cNK single, and T single lineages with or without myeloid cell potential. Because ∼60% of HPC-1–derived cells were lymphoid progenitors associated with myeloid potential, most HPC-1 clones were classified as MPP or MPP-derived LMPP. Most of these MPP/LMPP clones had ILC3/NK/T triple lymphoid lineage potential, and the minority had ILC3/cNK dual, cNK/T dual, and cNK single lineage potentials. Lymphoid progenitor clones lacking myeloid potential mainly retained ILC3, NK, and T single lineage potential. Of note, almost no clones exhibited ILC3/T dual lineage potential, suggesting early dichotomy between ILC3 and T cell commitment in the downstream differentiation pathway from HPC-1. In contrast to HPC-1, HPC-3 clones had ILC3 and/or NK cell potential but very low T and myeloid potential (Fig. 5B, lower), indicating that this subpopulation consists of lymphoid progenitors mainly committed to innate lymphocyte lineages. Furthermore, we confirmed by LDA that the HPC-3 subpopulation had very low B cell potential. This is because even total PB HPCs retained only low B cell PFs in comparison with CB and adult BM HPCs (Supplemental Fig. 2). As expected, HPC-2 clones showed intermediate characteristics between HPC-1–derived and HPC-3–derived clones.
Induction of CD34+Lin−CD7+ and CD34low/−Lin−CD7+CD127+ cell generation by Notch signaling in HPC subpopulations
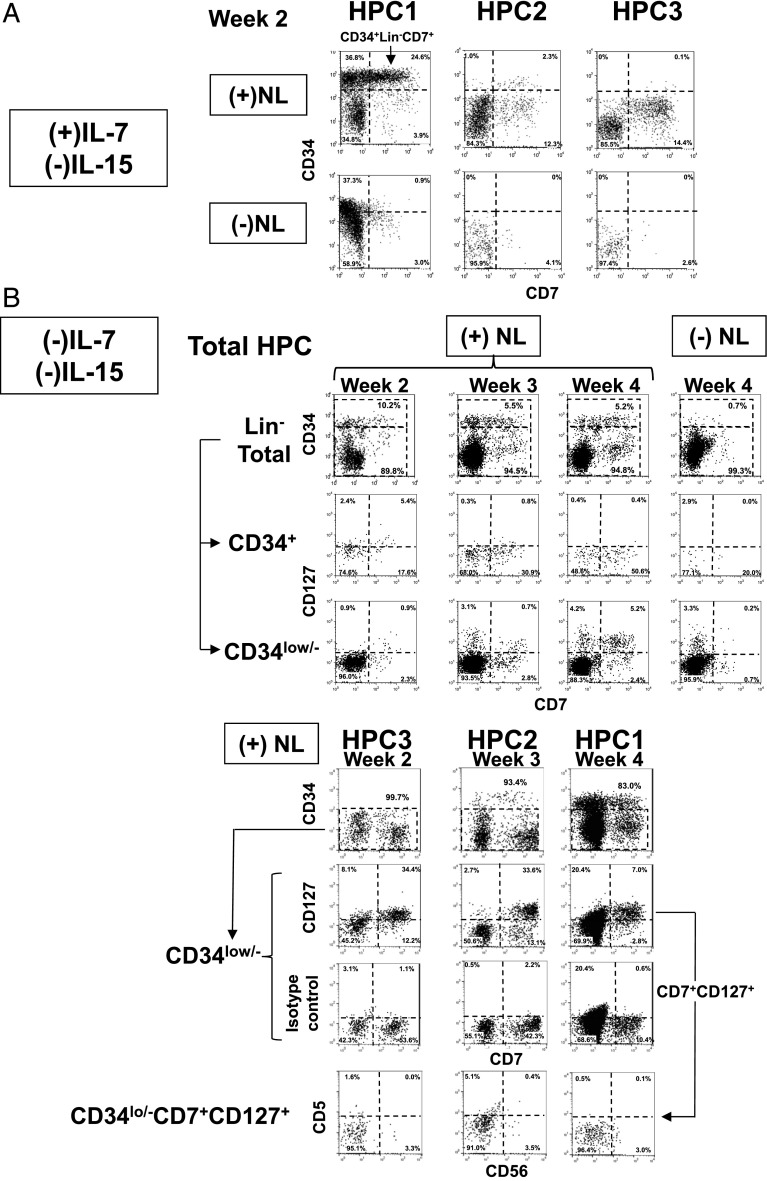
As mentioned above, because HPC-1 clones substantially exhibited either cNK/T or NCR+ILC3/cNK dual lineages, but almost no NCR+ILC3/T dual lineages (Fig. 5B, lower; Supplemental Table I), we expected that Notch signaling would induce HPC-1 cells to generate downstream progenitors that preferentially gave rise to T and cNK cell lineage progeny independent of NCR+ILC3 commitment. Because T/cNK cell common progenitors express CD34 in association with CD7 (39, 40), we examined the induction of CD7 expression in intermediate precursors generated from each HPC subpopulation in the presence or absence of Notch signaling (Fig. 6). Notch signaling induced CD34+Lin−CD7+ cells mainly from HPC-1 cells, only partly from HPC-2 cells, and not at all from HPC-3 cells in 2-wk culture with IL-7 (Fig. 6A). Alternatively, HPC-2 and HPC-3 cells mainly generated CD7+ cells lacking CD34 expression following Notch stimulation. Under this culture condition, most CD34low/−Lin−CD7+ cells in HPC-2 and HPC-3 were found to be CD56+ cells (data not shown). Moreover, the generation of both CD34+Lin−CD7+ and CD34low/−Lin−CD7+ cells induced by Notch was found to be independent of IL-7 or IL-15 (Fig. 6B). Importantly, along with CD7 expression Notch induced CD127 expression mainly in CD34low/−Lin− cells in the total HPCs and three HPC subpopulations (Fig. 6B). HPC-3 cells were found to most promptly express CD127 (2 wk) among the three subpopulations. More than 90% of these CD34low/−Lin−CD7+CD127+ cells under the (−)IL-7(−)IL-15 condition were found to be a CD5−CD56− immature phenotype. Furthermore, most CD34+Lin−CD7+ cells expressed high levels of CD62L and only low levels of CD10, whereas CD34low/−Lin−CD7+ cells expressed normal levels of CD62L and CD10. Both populations were RA+ phenotype (Fig. 7).
FIGURE 6.
Expression of CD7 and CD127 induced in HPC subpopulations by Notch signaling. (A) Generation of CD34+Lin−CD7+ and CD34low/−Lin−CD7+ cells from HPC-1, HPC-2, and HPC-3 cells in culture with IL-7 in the presence and absence of Notch signaling for 2 wk. In this experiment, the CD7− fraction of each HPC subpopulation was sorted to exclude the possible expansion of endogenous CD34+CD7+ cells. T, B, and myeloid lineage cells were gated out by using CD3, CD11c, CD14, CD15, CD19, and CD20 Abs for detection of CD34+Lin−CD7+ cells and CD34low/−Lin−CD7+ cells. (B) Time course for the generation of CD34+Lin−CD7+ and CD34low/−Lin−CD7+ cells in association with CD127 expression from the total HPC population (upper) and HPC subpopulation (lower) under (+) or (−)NL/(−)IL-7(−)IL-15 conditions. Six color analyses using CD5, CD7, CD34, CD56, CD127, and lineage (CD3, CD11c, CD14, CD15, CD19, and CD20) were conducted. CD127 expression in the CD34low/− cell populations from the three HPC subpopulations is shown at the week when each subpopulation started to express CD127 (lower). CD5 and CD56 expressions in CD34low/−Lin−CD7+CD127+cells were analyzed for HPC subpopulations. Similar results were obtained from two experiments for the other two donors.
FIGURE 7.
Surface phenotype of CD34+Lin−CD7+ and CD34low/−Lin−CD7+cells generated from HPC-1 cells. Expression of CD62L, CD10, and CD45RA in CD34+Lin−CD7+ and CD34low/−Lin−CD7+ cells generated from HPC-1 cells by 3 wk culture under NL(+)/IL-7(−)IL-15(−) conditions was analyzed by four-color flow cytometry using CD7, CD34, and lineage (CD3, CD11c, CD14, CD15, CD19 and CD20) in combination with CD62L, CD10, CD45RA, or isotype control. CD34+ (left) and CD34low/− (right) populations were gated from the Lin− fraction (upper) for the analyses of CD62L, CD10, and CD45RA expression (lower). Similar results were obtained from two experiments for the other two donors.
Based on these findings, we hypothesized that CD34+Lin−CD7+ cells generated from the HPC-1 subpopulation preserve T and cNK cell potentials but lose NCR+ILC3 potential, whereas the CD34low/−Lin−CD7+CD127+ cell population contains NCR+ILC3 precursors in addition to the cNK/T cell precursors derived from CD34+Lin−CD7+ cells.
Low NCR+ILC3 potential in Notch-induced CD34+Lin−CD7+cells in which high cNK and T cell potentials are preserved
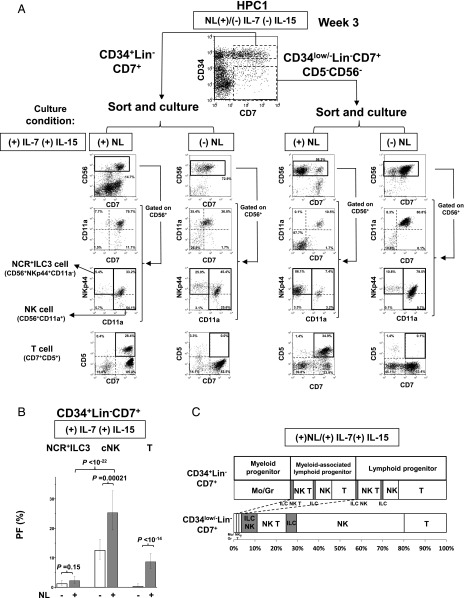
To examine the differentiation potentials of CD34+Lin−CD7+ and CD34low/−Lin−CD7+cells, we sorted these cell populations generated from HPC-1 cells under (+)NL/(−)IL-7(−)IL-15 conditions (Fig. 8A) and evaluated their lymphoid and myeloid potential under (+)NL/(+)IL-7(+)IL-15 or (−)NL/(+)IL-7(+)IL-15 conditions by bulk cell cultures (Fig. 8A) and LDA (Fig. 8B) along with clonal analysis (Fig. 8C).
FIGURE 8.
Differentiation potential of CD34+Lin−CD7+ cells generated from HPC-1 cells. (A) Differentiation of NCR+ILC3s, cNK cells, and T cells from HPC-1–derived CD34+Lin−CD7+ and CD34low/−Lin−CD7+ cells in culture with IL-7 and IL-15 in the presence or absence of Notch signaling. CD34+Lin−CD7+ and CD34low/−Lin−CD7+ cells were induced by culture of HPC-1 cells under (+)NL/(−)IL-7(−)IL-15 conditions for 3 wk, as shown in Fig. 6B. (B) LDA for determination of NCR+ILC3, cNK cell, and T cell PFs in HPC-1–derived CD34+Lin−CD7+ cells in the presence or absence of Notch signaling with both IL-7 and IL-15. The p values of the differences in PFs between two different culture conditions were calculated by online analysis using ELDA software (36). The bars represent the 95% confidence intervals of PFs. (C) Clonal analysis of the differentiation potential of CD34+Lin−CD7+ (upper) and CD34low/−Lin−CD7+ (lower) cells. Lineage classification of the lymphoid potential was conducted according to Fig. 4B. Samples from three human donors were examined for each HPC subpopulation. The numbers of clones producing different lineage combinations of progeny from the donors were combined and listed in Supplemental Table II.
In the bulk culture, CD34+Lin−CD7+ cells exhibited high cNK and T cell potential, but very low NCR+ILC3 potential in the presence of Notch signaling (Fig. 8A, left). In the absence of Notch signal, CD34+Lin−CD7+ cells did not give rise to any T cells at all, but exhibited high cNK cell and minor NCR+ILC3 potential. Thus, CD34+Lin−CD7+ cells may have largely lost their NCR+ILC3 potential in the process of divarication from HPC-1 cells. LDA confirmed the observations obtained in bulk cell culture (Fig. 8B). Notably, in contrast to the suppressive effect of Notch signaling on cNK cell commitment in HPC-2 and HPC-3 subpopulations (Fig. 4C), a stimulatory action of Notch on cNK cell differentiation was observed for CD34+Lin−CD7+ cells (Fig. 8B). Clonal assays confirmed that CD34+Lin−CD7+ cells retained NK and/or T cell potential with or without myeloid potential but showed very low NCR+ILC3 potential (Fig. 8C, Supplemental Table II). We also confirmed that CD7+CD5+ cells generated from HPC-1–derived CD34+Lin−CD7+ cells could develop into CD3+CD4+CD8+ triple-positive cells by prolonged culture for 8 wk under (+)NL/(+)IL-7(−)IL-15 conditions (data not shown).
In contrast to the CD34+Lin−CD7+ cells, the CD34low/−Lin−CD7+ population retained significant NCR+ILC3 potential (Fig. 8A). Clonal analyses demonstrated that CD34low/−Lin−CD7+ cells were able to give rise to NCR+ILC3 progeny in addition to cNK and T cell progeny, but almost no myeloid progeny. NCR+ILC3 progeny were classified into NCR+ILC3 single and NCR+ILC3/cNK dual lineage but not NCR+ILC3/T dual lineage (Fig. 8C, Supplemental Table II), supporting the notion that the differentiation pathway for NCR+ILC3 is independent of the pathway for T cells. We suspected that T cell precursors in the CD34low/−Lin−CD7+ population were generated from CD34+Lin−CD7+ cells by concurrent downregulation of CD34 expression. Furthermore, we confirmed that CD34low/−Lin−CD7+ cells generated from either HPC-2 or HPC-3 cells under (+)NL/(−)IL-7(−)IL-15 conditions showed high NCR+ILC3, but very low cNK cell potential in bulk culture under (+)NL/(+)IL-7(+)IL-15 conditions (data not shown).
Effects of Notch Abs on lineage commitments in HPC subpopulations
Finally, we evaluated the effects of anti-NRR1 and anti-NRR2 Abs on the generation of NCR+ILC3s, cNK cell and T cell progenies from HPC subpopulations to determine whether Noch1 or Notch2 signals were required for the commitment of each lineage (Fig. 9).
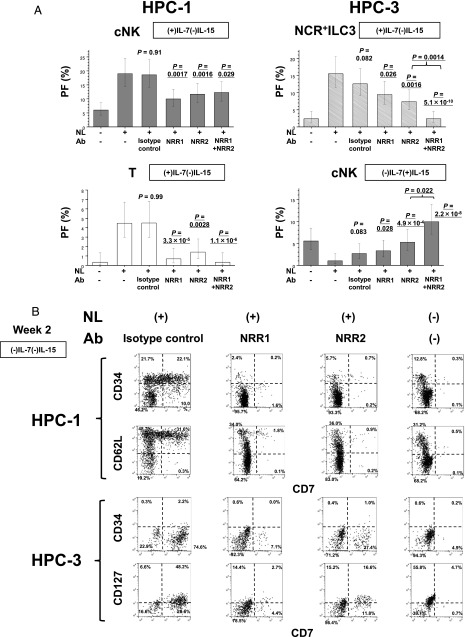
FIGURE 9.
Effects of anti-NRR1 and anti-NRR2 Abs on the generation of NCR+ILC3s, cNK cells, and T cells from HPC subpopulations. (A) Effects of anti-NRR1 and anti-NRR2 Abs on the generation of cNK and T cells from HPC-1 cells under (+)IL-7(−)IL-15 culture conditions for 2 wk (left) and NCR+ILC3 and cNK cells from HPC-3 cells under (−)IL-7(+)IL-15 conditions for 4 wk (right). Anti-NRR1 Ab, anti-NRR2 Ab, anti-NRR1 plus anti-NRR2 Abs, or human isotype control IgG1 was added to the coculture of HPC subpopulations with OP9-DL1 stroma cells at the initiation of culture. The PFs of cNK cells, T cells, and NCR+ILC3 were obtained by LDA using ELDA software (36). The bars represent the 95% confidence intervals of PFs. The p values of the differences in PFs between (+)NL(−) Ab (control) and (+)NL (+) NRR1 Ab, (+)NL (+) NRR2 Ab, or (+)NL (+) NRR1 plus NRR2 Abs are presented. The p values of the differences PFs in HPC-3 between (+)NL(+)NRR2 Ab and (+)NL(+) NRR1 plus NRR2 Abs are also presented (right). (B) Effects of anti-NRR1 Ab, anti-NRR2 Ab, and isotype control on the Notch-dependent generation of CD34+Lin−CD7+CD62Lhigh cells (top) and CD34low/−Lin−CD7+ CD127+ cells (bottom) from HPC-1 and HPC-3 subpopulations, respectively, under (−)IL-7(−)IL-15 conditions for 2 wk. Similar results were obtained from two experiments for the other two donors.
More than 60% of Notch-dependent generation of cNK cell progenies from the HPC-1 subpopulation was inhibited by either anti-NRR1 or anti-NRR2 Ab (Fig. 9A). Each Ab also blocked >80% of Notch-dependent generation of T cells from HPC-1 cells. No significant additive effects between anti-NRR1 and anti-NRR2 Abs on the generation of both cNK and T cells were observed. This finding suggested that simultaneous signaling through both Nocth1 and Notch2 may be required for the commitment of cNK and T cell lineages in HPC-1 subpopulation. Similarly, nearly half of Notch-dependent NCR+ILC3 differentiation from HPC-3 cells was inhibited by either anti-NRR1 or anti-NRR2 Ab (Fig. 9A). Conversely, Notch-dependent suppression of cNK cell generation from HPC-3 was found to be reverted by either Notch1 or Notch2 blockade. Interestingly, significant additive effects between anti-NRR1 and anti-NRR2 Abs were observed on both ILC3 generation and cNK cell suppression in HPC-3. This observation suggested that Notch1 and Notch2 signals may independently operate in the commitment in different subpopulations of HPC-3, which expressed either Notch1 or Notch2.
Because Notch signaling induced HPC-1 and HPC-3 cells to generate intermediate-stage cells, CD34+Lin−CD7+CD62Lhigh and CD34low/−Lin−CD7+ CD127+ cells, respectively, as described above (Figs. 6, 7, 8), we tested the effect of anti-NRR1 and anti-NRR2 Abs on the generation of intermediate cells from HPC-1and HPC-3 cells under (+)NL/(−)IL-7(−)IL-15 conditions (Fig. 9B). Both Abs were found to almost completely inhibit the generation of Notch-dependent CD34+Lin−CD7+CD62Lhigh cells from HPC-1 cells. Furthermore, both Abs partly suppressed Notch-dependent generation of CD34low/−Lin−CD7+CD127+ from HPC-3 cells (Fig. 9B). Thus, the inhibitory effects of Notch Abs on the differentiation of ILC3s, cNK cells, and T cells can be attributed to the suppression of intermediate cell formation from HPCs (Fig. 10).
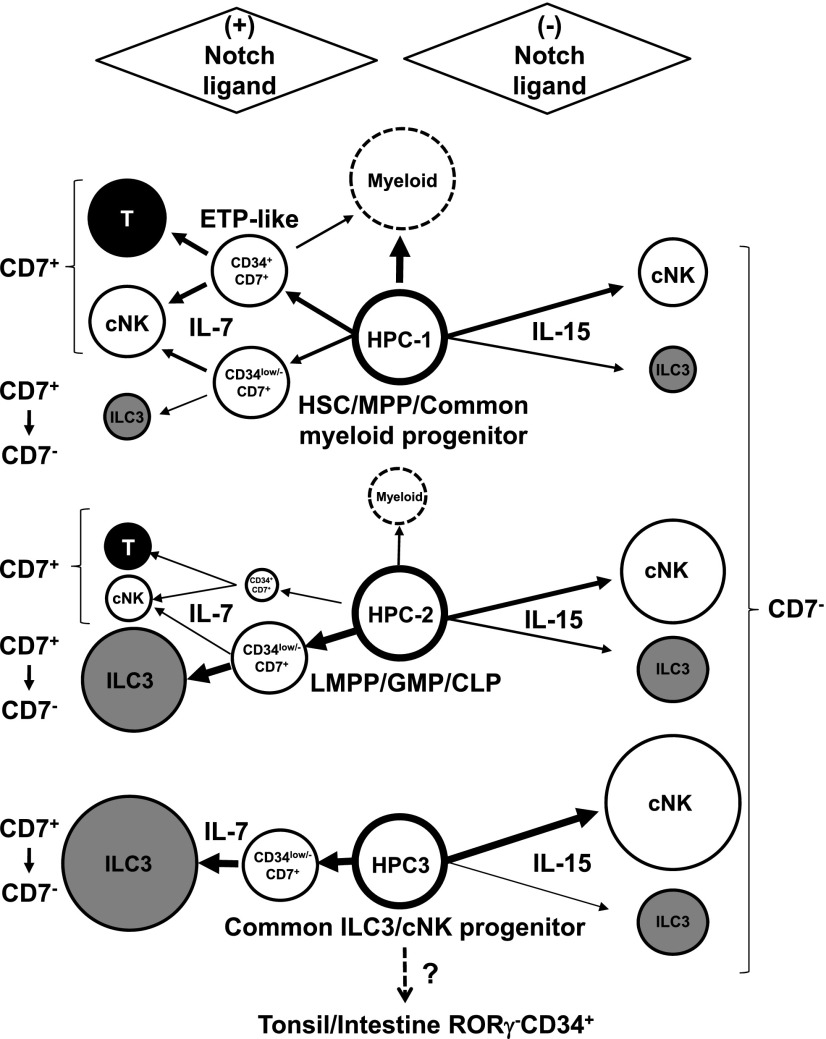
FIGURE 10.
Proposed model for differentiation pathways and commitments of NCR+ILC3 and cNK cell lineages from HPC subpopulations. Red, orange, green, and brown lines indicate NCR+ILC3, cNK cell, T cell, and myeloid cell commitment, respectively.
Discussion
In the present study, we demonstrated the crucial role of Notch signaling in the commitment of human adult circulating HPCs to the NCR+ILC3 or cNK cell lineage ex vivo. Notch functions had countervailing properties that stimulated IL-7–dependent NCR+ILC3 differentiation, but in turn suppressed IL-15–induced cNK cell differentiation in HPCs at large. These characteristics of Notch were most marked in the HPC-3 subpopulation, the differentiation potential of which was considerably inclined toward the NCR+ILC3 and cNK cell lineages as observed with LDA and clonal analysis. Thus, Notch signaling is thought to drive the innate lymphocyte progenitor toward the NCR+ILC3 commitment. Because the HPC-3 population had relatively high ILC3 and NK potential among HPC subpopulations, the countervailing functions of Notch observed in the total HPC population can mainly be attributed to those in the HPC-3 subpopulation. In contrast to Notch function in HPC-3 cells, Notch signaling enhanced IL-7–induced cNK cell but not NCR+ILC3 differentiation from the HPC-1 subpopulation. This enhancement of cNK cell differentiation was attributed to Notch dependency in the generation of CD34+Lin−CD7+ cells that retained high cNK and T cell potentials but only minor NCR+ILC3 potential. Further stimulation of CD34+Lin−CD7+ cells through Notch signaling promoted cNK cell differentiation, but not NCR+ILC3 differentiation. The HPC-2 subpopulation retained intermediate levels of ILC3 and cNK cell potential between those of HPC-1 and HPC-3 cells and exhibited similar Notch functions to HPC-3, although Notch-dependent suppression of cNK cell commitment from HPC-2 was found to be weaker than that from HPC-3. These results suggest that Notch signaling plays pivotal roles in the fate decision of human HPC commitment to NCR+ILC3 or cNK cell lineages through the opposite functions at different stages of HPC differentiation, although the molecular mechanisms underlying the countervailing aspect of Notch signaling are unknown.
Notch signaling acted synergistically with IL-7 for cNK cell differentiation from the HPC-1 subpopulation and NCR+ILC3 cell differentiation from HPC-2 and HPC-3 subpopulations. In this context, Notch signaling is required for the expression of CD127 (IL-7R α-chain) in human early T cell progenitors (41, 42), and IL-7R α-chain transduces IL-7 signals to promote cell survival as well as differentiation of CD4−CD8− double-negative thymocytes (43). Analogous to T cell development, we presume that Notch signaling also induces IL-7R expression in cNK/ILC3 precursor cells. In fact, we found that Notch stimulation induced CD127 expression along with acquisition of CD7 and loss of CD34 expression in the HPC-1, HPC-2, and HPC-3 subpopulations. The induction of CD127 and CD7 expression in these cells was found to be independent of IL-7. Thus, these findings serve to primarily explain the synergistic interaction between Notch and IL-7 in NCR+ILC3 differentiation from HPC-2 and HPC-3. IL-7 signaling may promote cell survival and differentiation of NCR+ILC3 precursors. However, an additional, yet unanswered question is whether IL-7 signaling is sufficient for further ILC3 differentiation. Other Notch target genes should be investigated.
The interaction between Notch and IL-15 signals involved in NCR+ILC3 and cNK cell differentiation is thought to be different from the case of Notch and IL-7 in NCR+ILC3 differentiation. Although the presence of IL-15 produced an analogous trophic effect on ILC3 generation in all the HPC subpopulations in the absence of Notch signaling, this trophic effect of IL-15 was not significantly augmented by Notch signaling. Rather, Notch markedly suppressed IL-15–dependent differentiation of cNK cells from HPC-3 cells. Therefore, we primarily propose that Notch signaling suppresses the expression of the IL-15R required for cNK cell differentiation from HPC-3 cells. This hypothesis needs to be tested by quantification of IL-15R mRNA produced in these cells, as Abs with sufficient reactivity and specificity for detecting IL-15R in HPCs are not available at present. An alternative hypothesis is that Notch target genes may include TFs that suppress cNK cell differentiation. Because Notch signaling was shown to suppress Eomes expression in CD56+CD11a+ cells by the present study, a putative Notch-induced cNK cell repressor TF may suppress Eomes expression. In this context, it was reported that Notch signaling suppressed the expression of the Id2 NK cell program activator through the activation of the Bcl11b NK repressor at the double-negative stage 2 during early T cell development in mice (44).
In contrast to the suppressive function of Notch in IL-15–induced cNK cell differentiation from HPC-3 cells, Notch signaling augmented IL-7–induced cNK cell differentiation from HPC-1 cells, because a substantial proportion of the cNK cell population is generated from CD34+Lin−CD7+ cells, which are induced from HPC-1 cells by Notch signaling. Similarly, Notch signaling was reported to induce cNK cell differentiation from human HPCs in culture (24, 26, 30, 45), although only the total CD34+ cells were used for cNK cell induction. Moreover, because cNK cells generated from CD34+Lin−CD7+ cells expressed CD127 but were devoid of CD16 expression (data not shown), these cNK cells appeared to correspond to CD127+ thymic NK cells or an immature stage (III or IV) of cNK cells (46). These findings suggest that CD34+Lin−CD7+ cells induced by Notch signaling correspond to the earliest thymic progenitors (ETPs) (40). CD62Lhigh, CD10−, and CD127− phenotypes of the CD34+Lin−CD7+ cells also supported the similarity to BM and thymus ETPs (47). It is known that Notch signaling is essential for T cell commitment in ETPs (32, 48). In contrast, Notch signaling is dispensable for thymic NK cell differentiation in mice (49, 50). In fact, our LDA results demonstrated high levels of cNK cell differentiation from CD34+Lin−CD7+ cells, even in the absence of Notch signaling. Furthermore, ETPs have myeloid potential but lack erythroid potential (51, 52), as observed for CD34+Lin−CD7+ cells in our previous (30) and present studies, although in vivo differentiation of myeloid cells in the thymus is controversial (53). As we found that CD34+Lin−CD7+ cells had only low NCR+ILC3 potential, CD34+Lin−CD7+ cells appeared to be committed to mainly generate T and cNK cells but lost their ILC3 potential in the presence of Notch signal. Based on the findings regarding the phenotypic similarity between CD34+Lin−CD7+ cells and ETPs, we anticipate that ETPs will not significantly give rise to NCR+ILC3 cells in the human thymus in vivo.
Notch signaling induced CD7 expression in a portion of both CD34+ and CD34− immature precursor populations, which were derived from HPC-1, -2, and -3 subpopulations. As observed in cNK cell differentiation from CD34+Lin−CD7+ cells, once the cNK precursor cells began to express CD7, most cNK cells maintained CD7 expression during further differentiation, regardless of the presence of Notch signaling. Alternatively, most NCR+ILC3 cells lost CD7 expression during the differentiation from the CD7+ precursors, even after Notch stimulation. We observed that maintenance of CD7 expression in ILC3 cells required a continuous supply of fresh OP9-DL1 cells providing strong Notch signaling (data not shown). Neither ILC3 nor cNK cells began to express CD7 during differentiation in the absence of Notch signaling. Thus, CD7 expression in cNK cells may be a marker for a history of receiving Notch signaling, although no significant differences in functions or surface phenotypes were observed between CD7+ and CD7− cell fractions (data not shown). In this case, because the vast majority of PB cNK cells express CD7, we infer that these PB cNK cells developed in the presence of Notch signaling.
Ab-mediated blocking of Notch functions showed that cosignaling through both Notch1 and Notch2 is involved in the generation of CD34+Lin−CD7+CD62Lhigh cells, ETP-like cells, which preserve high cNK and T cell potential. Our finding is considered to contradict mouse studies showing that Notch1 is critical for intrathymic T cell development at the earliest stage, whereas Notch2 is dispensable (32, 48). Conversely, Notch2 can promote early T cell commitment of Notch1-deficient HSCs in coculture with OP9-DL1 stroma cells and possibly in vivo in the spleen after BM transplantation (32). Thus, Notch2 compensates for the loss of Notch1 during mouse ETP generation in OP9-DL1 coculture, whereas Ab blocking of Notch1 signaling was found to almost completely inhibit human ETP generation in the present study. Such contradiction of ex vivo observation between mice and humans may reflect the difference in HSC characteristics in vivo between the two species. However, owing to the lack of human in vivo experimental systems in a physiological setting, a definitive conclusion is thought to be currently unobtainable. Similarly, both Notch1 and Notch2 signals are involved in promotion of IL-7–dependent ILC3 commitment and in the suppression of IL-15–dependent cNK cell commitment of HPC-3 cells, although Notch1 and Notch2 signals may independently function in the commitment of different subpopulations of HPC-3. In mice, although Notch2 signaling was reported to be required for in vivo differentiation of intestinal NCR+ILC3 from NCR−ILC3 and for maintenance of the NCR+ILC3 population (22, 23), the development of intestinal NCR+ILC3 was not dependent on Notch signaling (22, 23). We showed that human NCR+ILC3s were induced to some extent from the total HPC population or any HPC subpopulations by IL-15 in the absence of Notch signaling. It was also reported that differentiation of NCR+ILC3 from human To and intestinal LP CD34+ cells could be induced by a cytokine mixture containing IL-15 in the absence of Notch signaling ex vivo (11). Thus, if this finding is also true for mice, it is presumed that the commitment of intestinal NCR+ILC3 in mice may be Notch-independent, but IL-15–dependent.
Considering our current findings, we propose a model for the differentiation pathways of NCR+ILC3 and cNK cells from human PB HPC subpopulations in the presence or absence of Notch signaling (Fig. 10). HPC-1 is considered to contain an MPP population that retains at least ILC3, cNK, T, and myeloid potential, although the ILC3 potential was much lower than cNK and T cell potential even under (+)NL conditions. This may be because Notch signaling mainly induced cNK cells along with T cells via CD34+Lin−CD7+ cells, which have almost no ILC3 potential. Therefore, in the presence of Notch signaling, most ILC3s are generated from HPC-1 cells via a pathway that does not contain CD34+Lin−CD7+ cells, but rather CD34low/−Lin−CD7+ cells. Furthermore, our finding on the presence of the NCR+ILC3/cNK dual lineage progeny generated from CD34low/−Lin−CD7+ cells indicated that at least some fraction of cNK cells is differentiated from HPC-1 via CD34low/−Lin−CD7+ cells. In the absence of Notch signaling, HPC-1 cells are able to generate mature cNK cells and ILC3s in the presence of IL-15. In contrast, HPC-3 cells have almost no T cell, B cell, or myeloid potential, but retain NCR+ILC3 and cNK cell potentials. Whether these ILC3 and cNK cell progeny are derived from a bipotential or distinctive monopotential HPC-3 clone is unclear, because most ILC3 and cNK cell progeny are exclusively generated under (+)NL or (−)NL culture conditions, respectively. Nevertheless, as under (+)NL culture conditions, a significant fraction of HPC-3 clones (∼10%) gave rise to both ILC3 and cNK cell progeny, being indicated that a portion of the HPC-3 population is likely bipotential. This HPC subpopulation may therefore be defined as common ILC3/cNK cell progenitors.
The HPC-3 subpopulation had similar phenotypes to To and intestinal LP CD34+ cells, which were reported to express CD45RA and differentiate to ILC3 and cNK cells (11), although it was unknown whether To/LP CD34+ cells expressed Flt-3. The To/LP CD34+ cell population comprised RORγt−CD34bright, RORγt−CD34dim, and RORγt+CD34dim subpopulations. RORγt−CD34bright cells were able to differentiate into either ILC3 or cNK cells, whereas RORγt+CD34dim cells selectively differentiated into ILC3 cells. As HPC-3 cells expressed neither RORγt nor CD161 (data not shown), HPC-3 cells were thought to differ from RORγt+CD34dim cells, which expressed CD161. Thus, the HPC-3 subpopulation may be a direct precursor for To/LP RORγt−CD34bright cells (Fig. 10). If this is the case, ILC3/cNK cell precursors can be continuously supplied through PB from BM to the peripheral lymphoid organs in human adults. Further studies on the human To/LP microenvironment, including the distribution of NL-expressing cells, is needed to clarify whether Notch signaling is required for human ILC3 commitment in vivo.
In conclusion, we demonstrated that signaling functions of Notch play a crucial role in the fate decision of human circulating HPC subpopulations between ILC3 and cNK cell lineage commitments.
Supplementary Material
Acknowledgments
We thank C. Siebel for supplying anti-NRR Abs, M. Yamaoka and A. Nishikiori for assistance with FACS analyses, and E. Douple and R. Ullrich for valuable suggestions.
This work was supported by National Institute of Allergy and Infectious Diseases Contract HHSN272200900059C. This study was based on Radiation Effects Research Foundation Research Protocol RP 5-09. The Radiation Effects Research Foundation, Hiroshima and Nagasaki, Japan, is a private, nonprofit foundation funded by the Japanese Ministry of Health, Labor and Welfare and the U.S. Department of Energy, the latter in part through U.S. Department of Energy Award DE-HS0000031 to the National Academy of Sciences. The views of the authors do not necessarily reflect those of the two governments.
The online version of this article contains supplemental material.
- AHR
- aryl hydrocarbon receptor
- BM
- bone marrow
- CB
- cord blood
- CHILP
- common helper-like ILC progenitor
- cNK
- conventional NK
- DL1
- delta-like 1
- Eomes
- eomesodermin
- ETP
- earliest thymic progenitor
- FL
- Flt-3 ligand
- HPC
- hematopoietic progenitor cell
- HSC
- hematopoietic stem cell
- HTFC
- high-throughput flow cytometry
- Id2
- inhibitor of DNA binding 2
- ILC
- innate lymphoid cell
- ILC3
- group 3 ILC
- KL
- Kit ligand
- LDA
- limiting dilution assay
- LMPP
- lymphoid-primed MPP
- LP
- lamina propria
- MPP
- multipotent progenitor
- NCR
- natural cytotoxicity receptor
- NRR
- negative regulatory region
- NRR1
- Notch1 NRR
- NRR2
- Notch2 RRR
- PB
- peripheral blood
- PF
- responding progenitor frequency
- ROR
- retinoic acid receptor–related orphan receptor
- T-bet
- T-box expressed in T cells
- TF
- transcription factor
- To
- tonsil
- WB
- washing buffer.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.McKenzie A. N., Spits H., Eberl G. 2014. Innate lymphoid cells in inflammation and immunity. Immunity 41: 366–374. [DOI] [PubMed] [Google Scholar]
- 2.Spits H., Artis D., Colonna M., Diefenbach A., Di Santo J. P., Eberl G., Koyasu S., Locksley R. M., McKenzie A. N., Mebius R. E., et al. 2013. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13: 145–149. [DOI] [PubMed] [Google Scholar]
- 3.Diefenbach A., Colonna M., Koyasu S. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41: 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Immunological Genome Consortium 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montaldo E., Juelke K., Romagnani C. 2015. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur. J. Immunol. 45: 2171–2182. [DOI] [PubMed] [Google Scholar]
- 6.Hazenberg M. D., Spits H. 2014. Human innate lymphoid cells. Blood 124: 700–709. [DOI] [PubMed] [Google Scholar]
- 7.Kiss E. A., Vonarbourg C., Kopfmann S., Hobeika E., Finke D., Esser C., Diefenbach A. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334: 1561–1565. [DOI] [PubMed] [Google Scholar]
- 8.Klose C. S., Kiss E. A., Schwierzeck V., Ebert K., Hoyler T., d’Hargues Y., Göppert N., Croxford A. L., Waisman A., Tanriver Y., Diefenbach A. 2013. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature 494: 261–265. [DOI] [PubMed] [Google Scholar]
- 9.Lee J. S., Cella M., McDonald K. G., Garlanda C., Kennedy G. D., Nukaya M., Mantovani A., Kopan R., Bradfield C. A., Newberry R. D., Colonna M. 2011. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn Y. O., Blazar B. R., Miller J. S., Verneris M. R. 2013. Lineage relationships of human interleukin-22-producing CD56+ RORγt+ innate lymphoid cells and conventional natural killer cells. Blood 121: 2234–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montaldo E., Teixeira-Alves L. G., Glatzer T., Durek P., Stervbo U., Hamann W., Babic M., Paclik D., Stölzel K., Gröne J., et al. 2014. Human RORγt+CD34+ cells are lineage-specified progenitors of group 3 RORγt+ innate lymphoid cells. Immunity 41: 988–1000. [DOI] [PubMed] [Google Scholar]
- 12.Moro K., Yamada T., Tanabe M., Takeuchi T., Ikawa T., Kawamoto H., Furusawa J., Ohtani M., Fujii H., Koyasu S. 2010. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463: 540–544. [DOI] [PubMed] [Google Scholar]
- 13.Satoh-Takayama N., Lesjean-Pottier S., Vieira P., Sawa S., Eberl G., Vosshenrich C. A., Di Santo J. P. 2010. IL-7 and IL-15 independently program the differentiation of intestinal CD3−NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherrier M., Sawa S., Eberl G. 2012. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209: 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose C. S. N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157: 340–356. [DOI] [PubMed] [Google Scholar]
- 16.Constantinides M. G., McDonald B. D., Verhoef P. A., Bendelac A. 2014. A committed precursor to innate lymphoid cells. Nature 508: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J. S., Cella M., Colonna M. 2012. AHR and the transcriptional regulation of type-17/22 ILC. Front. Immunol. 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong S. H., Walker J. A., Jolin H. E., Drynan L. F., Hams E., Camelo A., Barlow J. L., Neill D. R., Panova V., Koch U., et al. 2012. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Q., Monticelli L. A., Saenz S. A., Chi A. W., Sonnenberg G. F., Tang J., De Obaldia M. E., Bailis W., Bryson J. L., Toscano K., et al. 2013. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38: 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Possot C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A., Golub R. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12: 949–958. [DOI] [PubMed] [Google Scholar]
- 21.Rankin L. C., Groom J. R., Chopin M., Herold M. J., Walker J. A., Mielke L. A., McKenzie A. N., Carotta S., Nutt S. L., Belz G. T. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14: 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chea S., Perchet T., Petit M., Verrier T., Guy-Grand D., Banchi E. G., Vosshenrich C. A., Di Santo J. P., Cumano A., Golub R. 2016. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci. Signal. 9: ra45. [DOI] [PubMed] [Google Scholar]
- 23.Viant C., Rankin L. C., Girard-Madoux M. J., Seillet C., Shi W., Smyth M. J., Bartholin L., Walzer T., Huntington N. D., Vivier E., Belz G. T. 2016. Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9: ra46. [DOI] [PubMed] [Google Scholar]
- 24.Haraguchi K., Suzuki T., Koyama N., Kumano K., Nakahara F., Matsumoto A., Yokoyama Y., Sakata-Yanagimoto M., Masuda S., Takahashi T., et al. 2009. Notch activation induces the generation of functional NK cells from human cord blood CD34-positive cells devoid of IL-15. J. Immunol. 182: 6168–6178. [DOI] [PubMed] [Google Scholar]
- 25.Benne C., Lelievre J. D., Balbo M., Henry A., Sakano S., Levy Y. 2009. Notch increases T/NK potential of human hematopoietic progenitors and inhibits B cell differentiation at a pro-B stage. Stem Cells 27: 1676–1685. [DOI] [PubMed] [Google Scholar]
- 26.Kyoizumi S., Kubo Y., Kajimura J., Yoshida K., Imai K., Hayashi T., Nakachi K., Young L. F., Moore M. A., van den Brink M. R., Kusunoki Y. 2013. Age-associated changes in the differentiation potentials of human circulating hematopoietic progenitors to T- or NK-lineage cells. J. Immunol. 190: 6164–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felices M., Ankarlo D. E., Lenvik T. R., Nelson H. H., Blazar B. R., Verneris M. R., Miller J. S. 2014. Notch signaling at later stages of NK cell development enhances KIR expression and functional maturation. J. Immunol. 193: 3344–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentek R., Munneke J. M., Helbig C., Blom B., Hazenberg M. D., Spits H., Amsen D. 2013. Modulation of signal strength switches Notch from an inducer of T cells to an inducer of ILC2. Front. Immunol. 4: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasteiger G., Fan X., Dikiy S., Lee S. Y., Rudensky A. Y. 2015. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350: 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyoizumi S., Kubo Y., Kajimura J., Yoshida K., Hayashi T., Nakachi K., Young L. F., Moore M. A., van den Brink M. R., Kusunoki Y. 2014. Linkage between dendritic and T cell commitments in human circulating hematopoietic progenitors. J. Immunol. 192: 5749–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez S., Aiken C. T., Andrzejewski B., Sklar L. A., Edwards B. S. 2003. High-throughput flow cytometry: validation in microvolume bioassays. Cytometry A 53: 55–65. [DOI] [PubMed] [Google Scholar]
- 32.Besseyrias V., Fiorini E., Strobl L. J., Zimber-Strobl U., Dumortier A., Koch U., Arcangeli M. L., Ezine S., Macdonald H. R., Radtke F. 2007. Hierarchy of Notch–Delta interactions promoting T cell lineage commitment and maturation. J. Exp. Med. 204: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Cain-Hom C., Choy L., Hagenbeek T. J., de Leon G. P., Chen Y., Finkle D., Venook R., Wu X., Ridgway J., et al. 2010. Therapeutic antibody targeting of individual Notch receptors. Nature 464: 1052–1057. [DOI] [PubMed] [Google Scholar]
- 34.Kodama H., Nose M., Niida S., Nishikawa S., Nishikawa S. 1994. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp. Hematol. 22: 979–984. [PubMed] [Google Scholar]
- 35.Schmitt T. M., Zúñiga-Pflücker J. C. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17: 749–756. [DOI] [PubMed] [Google Scholar]
- 36.Hu Y., Smyth G. K. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347: 70–78. [DOI] [PubMed] [Google Scholar]
- 37.Renoux V. M., Zriwil A., Peitzsch C., Michaëlsson J., Friberg D., Soneji S., Sitnicka E. 2015. Identification of a human natural killer cell lineage-restricted progenitor in fetal and adult tissues. Immunity 43: 394–407. [DOI] [PubMed] [Google Scholar]
- 38.Doulatov S., Notta F., Eppert K., Nguyen L. T., Ohashi P. S., Dick J. E. 2010. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11: 585–593. [DOI] [PubMed] [Google Scholar]
- 39.Haddad R., Guardiola P., Izac B., Thibault C., Radich J., Delezoide A. L., Baillou C., Lemoine F. M., Gluckman J. C., Pflumio F., Canque B. 2004. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood 104: 3918–3926. [DOI] [PubMed] [Google Scholar]
- 40.Hao Q. L., George A. A., Zhu J., Barsky L., Zielinska E., Wang X., Price M., Ge S., Crooks G. M. 2008. Human intrathymic lineage commitment is marked by differential CD7 expression: identification of CD7− lympho-myeloid thymic progenitors. Blood 111: 1318–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magri M., Yatim A., Benne C., Balbo M., Henry A., Serraf A., Sakano S., Gazzolo L., Lévy Y., Lelièvre J. D. 2009. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur. J. Immunol. 39: 1231–1240. [DOI] [PubMed] [Google Scholar]
- 42.González-García S., García-Peydró M., Martín-Gayo E., Ballestar E., Esteller M., Bornstein R., de la Pompa J. L., Ferrando A. A., Toribio M. L. 2009. CSL–MAML-dependent Notch1 signaling controls T lineage–specific IL-7Rα gene expression in early human thymopoiesis and leukemia. [Published erratum appears in 2009 J. Exp. Med. 206: 1633.] J. Exp. Med. 206: 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazzucchelli R., Durum S. K. 2007. Interleukin-7 receptor expression: intelligent design. Nat. Rev. Immunol. 7: 144–154. [DOI] [PubMed] [Google Scholar]
- 44.Rothenberg E. V. 2014. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 32: 283–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachanova V., McCullar V., Lenvik T., Wangen R., Peterson K. A., Ankarlo D. E., Panoskaltsis-Mortari A., Wagner J. E., Miller J. S. 2009. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol. Blood Marrow Transplant. 15: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freud A. G., Yokohama A., Becknell B., Lee M. T., Mao H. C., Ferketich A. K., Caligiuri M. A. 2006. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 203: 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohn L. A., Hao Q. L., Sasidharan R., Parekh C., Ge S., Zhu Y., Mikkola H. K., Crooks G. M. 2012. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat. Immunol. 13: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H. R., Aguet M. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10: 547–558. [DOI] [PubMed] [Google Scholar]
- 49.Ribeiro V. S., Hasan M., Wilson A., Boucontet L., Pereira P., Lesjean-Pottier S., Satoh-Takayama N., Di Santo J. P., Vosshenrich C. A. 2010. Cutting edge: thymic NK cells develop independently from T cell precursors. J. Immunol. 185: 4993–4997. [DOI] [PubMed] [Google Scholar]
- 50.Nozad Charoudeh H., Tang Y., Cheng M., Cilio C. M., Jacobsen S. E., Sitnicka E. 2010. Identification of an NK/T cell-restricted progenitor in adult bone marrow contributing to bone marrow- and thymic-dependent NK cells. Blood 116: 183–192. [DOI] [PubMed] [Google Scholar]
- 51.Wada H., Masuda K., Satoh R., Kakugawa K., Ikawa T., Katsura Y., Kawamoto H. 2008. Adult T-cell progenitors retain myeloid potential. Nature 452: 768–772. [DOI] [PubMed] [Google Scholar]
- 52.De Obaldia M. E., Bell J. J., Bhandoola A. 2013. Early T-cell progenitors are the major granulocyte precursors in the adult mouse thymus. Blood 121: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richie Ehrlich L. I., Serwold T., Weissman I. L. 2011. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood 117: 2618–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.