Abstract
Rhizoma Anemarrhenae, a famous traditional Chinese medicine (TCM), is the dried rhizome of Anemarrhena asphodeloides Bge. (Anemarrhena Bunge of Liliaceae). The medicine presents anti-inflammatory, antipyretic, sedative, and diuretic effects. The chemical constituents of Rhizoma Anemarrhenae are complex and diverse, mainly including steroidal saponins, flavonoids, phenylpropanoids, benzophenones, and alkaloids. In this study, UPLC-Q-TOF/MS was used in combination with data postprocessing techniques, including characteristic fragments filter and neutral loss filter, to rapidly classify and identify the five types of substances in Rhizoma Anemarrhenae. On the basis of numerous literature reviews and according to the corresponding characteristic fragments produced by different types of compounds in combination with neutral loss filtering, we summarized the fragmentation patterns of the main five types of compounds and successfully screened and identified 32 chemical constituents in Rhizoma Anemarrhenae. The components included 18 steroidal saponins, 6 flavonoids, 4 phenylpropanoids, 2 alkaloids, and 2 benzophenones. The method established in this study provided necessary data for the study on the pharmacological effects of Rhizoma Anemarrhenae and also provided the basis for the chemical analysis and quality control of TCMs to promote the development of a method for chemical research on TCMs.
1. Introduction
Rhizoma Anemarrhenae is one of the traditional Chinese medicines (TCMs) that are commonly used for clearing heat and purging fire, nourishing yin, moistening dryness, reducing anxiety and thirst, and promoting excretion. This TCM is the dried rhizome of the Liliaceae Anemarrhena genus and has been widely used for clinical treatment of febrile disease, high fever and thirst, cough and asthma, osteopyrexia and fever, and other diseases [1, 2]. The pharmacologically active components of Rhizoma Anemarrhenae include steroidal saponins, flavonoids, phenylpropanoids, alkaloids, and benzophenones, with saponins being the major constituents [3–5]. At present, the analytical methods of TCM ingredients mainly include thin layer chromatography (TLC), gas chromatography (GC), high-performance liquid chromatography (HPLC), and ultraperformance liquid chromatography/time-of-flight mass spectrometry (UPLC-Q-TOF/MS), among others. As a comprehensive analysis technique, UPLC-Q-TOF/MS has been increasingly applied for the qualitative and quantitative analysis of components in TCMs and compound preparations [6–11]. However, this method displays several disadvantages, including complicated and time-consuming data processing. Therefore, a newly developed method to rapidly identify components in TCMs is urgently needed to improve the efficiency and accuracy of the qualitative analysis and to promote the development of modernization and internationalization of TCMs.
With the rapid development of modern technology, data postprocessing techniques, mainly including characteristic fragments filter (CFF) and neutral loss filter (NLF), play increasingly important roles in TCM research [12–17]. CFF is based on compounds with the same or similar mother nucleus structures that can produce characteristic fragments to determine a certain type of compound under the same mass spectrometric conditions. NLF is based on a certain type of compounds that frequently lose the same neutral fragments by energy collision splitting. According to the loss of these neutral fragments, substances with a class of characteristic substituted groups can be briefly classified [18–22]. CFF is favorable for the rapid screening of similar compounds, whereas NLF helps further confirmation of compounds. Thus, the combination of both techniques markedly reduces qualitative analysis difficulty and was gradually applied in the field of pharmaceutical analysis.
In recent years, UPLC-Q-TOF/MS is widely used to analyze constituents in Rhizoma Anemarrhenae [23, 24]. In this study, Rhizoma Anemarrhenae was selected as an example to establish an efficient approach for the rapid screening of chemical components by using UPLC-Q-TOF/MS in combination with data postprocessing techniques. First, through reading and integration of abundant information from the literature, we summarized the fragmentation patterns of five kinds of compounds (steroidal saponins, flavonoids, phenylpropanoids, alkaloids, and benzophenones) in Rhizoma Anemarrhenae. Secondly, a full spectrum scan was performed by UPLC-Q-TOF/MS. Finally, the compounds were characterized based on the CFF and NLF rule. Compared with traditional methods, the study could rapidly classify and identify a certain type of compounds. The method of UPLC-Q-TOF/MS coupled with data postprocessing techniques not only partly overcame the difficulty of rapid classification and identification of complex components of TCM but also laid the foundation for the rapid development of composition identification and provided an analytical method for the further research and development of TCMs.
2. Experimental
2.1. Sample Preparation
Prepared Rhizoma Anemarrhenae was accurately weighed (5 g) and extracted twice with 50 and 40 mL of 75% methanol, respectively, and each reflux time was 1 h. The extracts were filtered, merged, and then concentrated to 0.01 g/mL. The concentrated solution was filtered using a 0.22 μm syringe filter, with 2 μL injected for UPLC-Q-TOF/MS analysis.
2.2. UPLC-Q-TOF/MS Conditions
Chromatographic separation was conducted on a Waters Acquity UPLC Class I series, which consisted of a quatpump, an autosampler, a DAD detector, and a column compartment. All separation was carried out on a Waters ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm particle size) with a set column temperature of 35°C. The solvent flow rate was 0.3 mL/min, and 2 μL of sample solution was injected in each run. The mobile phase was composed of water (solvent A) and acetonitrile (solvent B). The gradient elution program employed was as follows: 0–0.5 min, 2%–5% B; 0.5–3 min, 5%–20% B; 3–5 min, 20%–22% B; 5-6 min, 22%-22% B; 6–10 min, 22%–38% B; 10–14 min, 38%–50% B; 14-15 min, 50%-50% B; 15–18 min, 50%–100% B; 18–20 min, 100%-100% B; 20-21 min, 100%–2% B; 21–23 min, 2%-2% B.
UPLC was coupled to a Q-TOF-MS system (Waters, USA) equipped with an electrospray ionization source (ESI) for scanning samples in negative ion modes. Ultrahigh purity helium (He) was used as the collision gas, and high-purity nitrogen (N2) was used as nebulizing gas. The conditions of ESI source were as follows: drying gas temperature: 325°C; drying gas flow rate: 10 mL min−1; desolvation gas flow: 600 L/h; capillary voltage: 2.0 kV; collision energy: 20–40 eV; scan spectra from m/z 50–1500; nebulizing gas pressure: 350 psi. Leu-enkephalin ions at m/z 556.2771 and 554.2615 were used to ensure accuracy in spectral acquisition.
2.3. Data Analysis
Original data were analyzed using Masslynx (Waters, USA) software 4.1. to detect and align the peaks. Data were processed and converted to an Excel format containing complete information on mass, retention time, and peak area of the samples. Target compounds were obtained by output data processing.
3. Results and Discussion
3.1. Method Development
The chemical constituents of Rhizoma Anemarrhenae are complex and diverse, but similar kinds of compounds exhibit certain similarities in chemical structures. We summarized the fragmentation patterns of compounds in Rhizoma Anemarrhenae according to the mass spectrometric behavior; that is, the same type of compounds with the same or similar public skeleton usually show the same fracture mode to produce the same CFs. The method that uses CFs to determine a certain type of compounds is called CFF. For example, phenylpropanoid compounds can produce characteristic fragment ions at m/z 107 [C7H7O]− and 91 [C7H7]− in negative ion mode. The ions could be screened from the total ion current chromatograms according to their CFs and then identified according to specific fragments and molecular ion peaks. In MS collision-induced dissociation, the differences between molecular ion peaks and high mass-to-charge ratio fragment peaks also play an important role in component identification. The method that uses NLs to screen compounds is called NLF. Mangiferin could easily lose neutral fragments at m/z 90 (C3H6O3) and 120 (C4H8O4) and could be filtered according to the two characteristic neutral fragments. CFF and NLF perform an increasingly important role in TCM research. Therefore, the complex components in Rhizoma Anemarrhenae can be rapidly and accurately classified and identified by combining UPLC-Q-TOF/MS with data postprocessing techniques (CFs and NLs). Finally, Based on the results of our study and those described in relevant and previous experiments, the rules of CFF and NLF regarding the five categories were established (shown in Table 1).
Table 1.
Characteristic fragments and neutral loss information of chemical substances in the Rhizoma Anemarrhenae.
| Compound classification | Subclass | Characteristic fragments | Neutral loss |
|---|---|---|---|
| Steroidal saponins | |||
| Spirostanol saponins [M-H]− |
Type I saponins | 273 [C19H29O]−, 255 [C19H27]−, 301 [C21H33O]−, 283 [C21H31]− | 162 (C6H10O5), 132 (C5H8O4) 18 (H2O) 142 (C8H14O2) |
| Type II and type III saponins | 289 [C19H29O2]−, 317 [C21H33O2]− |
||
| Type IV saponins | 271 [C19H27O]−, 253 [C19H25]−, 299 [C21H31O]−, 281 [C21H29]− |
||
| Furostan saponins [M-H-OH]− [M-H-OCH3]− |
Type V saponins | 269 [C19H25O]−, 251 [C19H23]− | |
| Flavonoids | Mangiferin-type flavonoids | 90 (C3H6O3), 120 (C4H8O4) |
|
| Chalcone-type flavonoids | 119 [C8H7O]− or 146 [C9H6O2]− | ||
| Flavanones | 119 [C8H7O]−, 165 [C8H5O4]− |
||
| Homoisoflavonoid | 133 [C9H9O]− | ||
| Icariin-type flavonoids | 367 [C21H19O6]− | 162 (C6H10O5), 132 (C5H8O4) | |
| Phenylpropanoids | coumarins | 107 [C7H7O]−, 91 [C7H7]− |
|
| Benzophenones | 94 (C6H6O) 44 (CO2) |
||
| Alkaloids | Amide alkaloids | 119 (C8H7O) 15 (CH3) |
A full scan was conducted in both positive and negative modes to analyze the Rhizoma Anemarrhenae extract, and results show that the ESI negative ion mode was simpler, more stable, and easier to interpret compared with the positive ion mode, and the characteristic ions obtained in the negative ion mode could be evidently observed to distinguish the two types of steroidal saponins. Thus, the negative ion mode was used in this study. The full scan MS chromatograms of substances in Rhizoma Anemarrhenae extracts are shown in Figure 1. The fragmentation patterns of the five types of main constituents in Rhizoma Anemarrhenae were summarized, and 32 compounds were identified (shown in Table 2).
Figure 1.
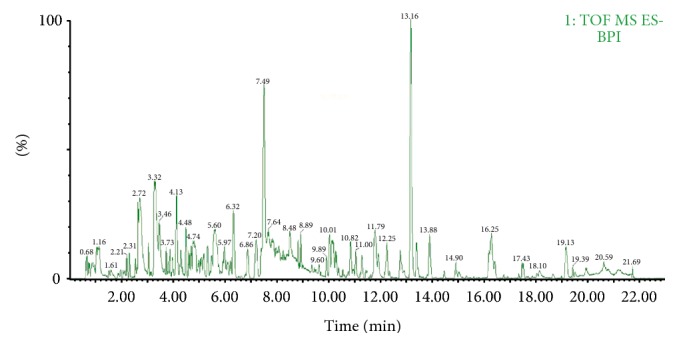
Base peak ion (BPI) chromatograms of substances in the extract of the Rhizoma Anemarrhenae herb, under negative ion mode.
Table 2.
Identification of the chemical constituents of Rhizoma Anemarrhenae extract by using UPLC-Q-TOF/MS in negative ion mode.
| RT | Formula | [M-H]− | Experimental m/z | ppm | Fragment ions | Chemical name | Chemical types | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.72 | C25H28O16 | 583.1299 | 583.1299 | 0.00 | 583 [M-H]−, 421 [M-H-C6H10O5]−, 331 [M-H-C6H10O5-C3H6O3]−, 301 [M-H-C6H10O5-C4H8O4]−, |
Neomangiferin | Mangiferin-type flavonoids | [5, 25] |
| 2 | 3.27 | C19H18O11 | 421.0771 | 421.0759 | 2.85 | 421 [M-H]−, 331 [M-H-C3H6O3]−, 301 [M-H-C4H8O4]−, |
Mangiferin | Mangiferin-type flavonoids | [5, 25] |
| 3 | 3.32 | C19H18O11 | 421.0771 | 421.0764 | 1.66 | 421 [M-H]−, 331 [M-H-C3H6O3]−, 301 [M-H-C4H8O4]−, |
Isomangiferin | Mangiferin-type flavonoids | [5, 25] |
| 4 | 4.48 | C13H10O5 | 245.0450 | 245.0454 | 1.63 | 245 [M-H]−, 151 [M-H-C6H6O]−, 107 [M-H-C6H6O-CO2]−, |
Iriflophenone | Benzophenones | [26] |
| 5 | 4.74 | C45H76O20 | 935.4852 | 935.4823 | 3.10 | 917 [M-H-H2O]−, 773 [M-H-C6H10O5]−, 289 [M-H-H2O-3C6H10O5-C8H14O2]−, 271 [M-H-2H2O-3C6H10O5-C8H14O2]−, 253 [M-H-3H2O-3C6H10O5-C8H14O2]−, 281 [M-H-3H2O-3C6H10O5-C6H10O2]−, |
Timosaponin E1 | Furostanol glycosides | [5, 27] |
| 6 | 4.79 | C45H76O20 | 935.4852 | 935.4827 | 2.67 | 935 [M-H]−, 773 [M-H-C6H10O5]−, 289 [M-H-H2O-3C6H10O5-C8H14O2]−, 271 [M-H-2H2O-3C6H10O5-C8H14O2]−, 253 [M-H-3H2O-3C6H10O5-C8H14O2]−, |
Timosaponin E | Furostanol glycosides | [5, 27] |
| 7 | 4.85 | C45H76O20 | 935.4852 | 935.4836 | 1.71 | 935 [M-H]−, 773 [M-H-C6H10O5]−, 289 [M-H-H2O-3C6H10O5-C8H14O2]−, |
Timosaponin N | Furostanol glycosides | [5, 27] |
| 8 | 5.09 | C56H94O29 | 1229.5803 | 1229.5806 | 0.24 | 1211 [M-H-H2O]−, 289 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]−, 271 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]−, 253 [M-H-3H2O-C5H8O4-4C6H10O5-C8H14O2]−, 281 [M-H-3H2O-C5H8O4-4C6H10O5-C6H10O2]−, |
Purpureagitoside | Furostanol glycosides | [5, 27] |
| 9 | 6.32 | C17H17NO3 | 282.1130 | 282.1132 | 0.71 | 282 [M-H]−, 163 [M-H-C8H7O]−, |
Coumaroyltyramine | Amide alkaloids | [28] |
| 10 | 6.86 | C18H19NO4 | 312.1236 | 312.1237 | 0.32 | 312 [M-H]−, 297 [M-H-CH3]−, 178 [M-H-CH3-C8H7O]−, |
N-trans-feruloyltyrami-ne or N-cis-feruloyltyramine | Amide alkaloids | [28] |
| 11 | 7.20 | C45H74O19 | 917.4746 | 917.4751 | 0.54 | 755 [M-H-C6H10O5]−, 289 [M-H-3C6H10O5-C8H14O2]−, 271 [M-H-3C6H10O5-C8H14O2-H2O]−, 253 [M-H-3C6H10O5-C8H14O2-2H2O]− 281 [M-H-3C6H10O5-C6H10O2-H2O]−, |
Timosaponin D | Furostanol glycosides | [5, 27] |
| 12 | 7.49 | C14H12O5 | 259.0607 | 259.0630 | 8.88 | 259 [M-H]−, 165 [M-H-C6H6O]−, 121 [M-H-C6H6O-CO2]−, |
2,6,4′-Trihydroxy-4-methoxyBenzophenone | Benzophenones | [26] |
| 13 | 8.12 | C56H92O28 | 1211.5697 | 1211.5642 | 4.54 | 1211 [M-H]−, 1193 [M-H-H2O]−, 271 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]−, 253 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]−, |
Timosaponin H1 | Furostanol glycosides | [5, 27] |
| 14 | 8.26 | C56H94O28 | 1213.5854 | 1213.5803 | 4.20 | 1213 [M-H]−, 273 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]−, 255 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]− |
Timosaponin I1/D1 | Furostanol glycosides | [5, 27] |
| 15 | 8.48 | C45H76O19 | 919.4903 | 919.4882 | 2.28 | 919 [M-H]−, 901 [M-H-H2O]−, 301 [M-H-H2O-3C6H10O5-C6H10O2]−, 273 [M-H-H2O-3C6H10O5-C8H14O2]−, 255 [M-H-2H2O-3C6H10O5-C8H14O2]−, |
Timosaponin B II or Timosaponin L or (25S)-Officinalisnin-I | Furostanol glycosides | [5, 27] |
| 16 | 9.60 | C39H64O15 | 771.4167 | 771.4174 | 0.91 | 771 [M-H]−, 609 [M-H-C6H10O5]−, 289 [M-H-2C6H10O5-C8H14O3]−, 271 [M-H-2C6H10O5-C8H14O3-H2O]−, 253 [M-H-2C6H10O5-C8H14O3-2H2O]−, |
Timosaponin F (C39) | Spirostanol glycosides | [27] |
| 17 | 10.01 | C45H74O18 | 901.4797 | 901.4773 | 2.66 | 901 [M-H]−, 739 [M-H-C6H10O5]−, 577 [M-H-2C6H10O5]−, 273 [M-H-3C6H10O5-C8H14O2]−, 255 [M-H-3C6H10O5-C8H14O2-H2O]−, |
Xilingsaponin B | Spirostanol glycosides | [27] |
| 18 | 10.14 | C45H74O18 | 901.4797 | 901.4768 | 3.22 | 739 [M-H-C6H10O5]−, 577 [M-H-2C6H10O5]−, 273 [M-H-3C6H10O5-C8H14O2]−, 255 [M-H-3C6H10O5-C8H14O2-H2O]−, |
Timosaponin B III | Furostanol glycosides | [5, 27] |
| 19 | 10.23 | C45H74O18 | 901.4797 | 901.4807 | 1.11 | 739 [M-H-C6H10O5]−, 577 [M-H-2C6H10O5]−, 273 [M-H-3C6H10O5-C8H14O2]−, 255 [M-H-3C6H10O5-C8H14O2-H2O]−, |
Macrostemonoside F or Timosaponin C | Furostanol glycosides | [5, 27] |
| 20 | 10.28 | C51H84O23 | 1063.5325 | 1063.5352 | 2.54 | 1063 [M-H]−, 273 [M-H-4C6H10O5-C8H14O2]−, 255 [M-H-4C6H10O5-C8H14O2-H2O]−, |
Timosaponin B IV (C51) | Furostanol glycosides | [5, 27] |
| 21 | 10.60 | C45H74O18 | 901.4797 | 901.4824 | 3.00 | 739 [M-H-C6H10O5]−, 577 [M-H-2C6H10O5]−, 273 [M-H-3C6H10O5-C8H14O2]−, 255 [M-H-3C6H10O5-C8H14O2-H2O]−, |
Macrostemonoside F or Timosaponin C | Furostanol glycosides | [5, 27] |
| 22 | 10.69 | C15H12O4 | 255.0658 | 255.0666 | 3.14 | 255 [M-H]−, 135 [M-H-C7H4O2]−, 119 [M-H-C7H4O3]−, |
2′,4′,4-Trihydroxychalcone | Chalcone-type flavonoids | [29] |
| 23 | 10.73 | C17H16O3 | 267.1021 | 267.1036 | 5.62 | 267 [M-H]−, 119 [M-H-C9H8O2]−, 107 [M-H-C10H8O2]−, 91 [M-H-C10H8O3]−, |
Oxy- hinokiresinol | Coumarins | [5] |
| 24 | 11.00 | C16H14O5 | 285.0763 | 285.0777 | 4.91 | 285 [M-H]−, 165 [M-H-C7H4O2]−, 119 [M-H-C8H6O4]−, |
(2S)-7,4′-Dihydroxy-5-methoxyflavone | Flavanones | [25] |
| 25 | 11.23 | C16H16O5 | 287.0920 | 287.0931 | 3.83 | 287 [M-H]−, 269 [M-H-H2O]−, 259 [M-H-CO]−, |
2′-O-Methylphloretin | Chalcone-type flavonoids | [25] |
| 26 | 11.38 | C39H64O14 | 755.4218 | 755.4218 | 0.00 | 755 [M-H]−, 593 [M-H-C6H10O5]−, 575 [M-H-H2O-C6H10O5]−, 431 [M-H-2C6H10O5]−, 271 [M-H-2C6H10O5-C8H14O2-H2O]−, 253 [M-H-2C6H10O5-C8H14O2-2H2O]−, |
Timosaponin A II or Timosaponin G (C39) or Anemarrhena sapinin III | Spirostanol glycosides | [5, 27] |
| 27 | 12.25 | C16H18O3 | 257.1178 | 257.1190 | 4.67 | 257 [M-H]−, 107 [M-H-C9H10O2]−, 91 [M-H-C9H10O3]−, |
Broussonin A or Broussonin B | Coumarins | [29] |
| 28 | 13.16 | C17H16O2 | 251.1072 | 251.1075 | 1.19 | 251 [M-H]−, 107 [M-H-C10H8O]−, 91 [M-H-C10H8O2]−, |
Hinokiresinol | Coumarins | [29] |
| 29 | 16.17 | C39H64O13 | 739.4269 | 739.4252 | 2.30 | 739 [M-H]−, 577 [M-H-C6H10O5]−, 415 [M-H-2C6H10O5]−, 273 [M-H-2C6H10O5-C8H14O2]−, 255 [M-H-2C6H10O5-C8H14O2-H2O]−, |
Timosaponin AIII | Spirostanol glycosides | [5, 27] |
| 30 | 16.25 | C39H64O13 | 739.4269 | 739.4280 | 1.49 | 739 [M-H]−, 577 [M-H-C6H10O5]−, 415 [M-H-2C6H10O5]−, 273 [M-H-2C6H10O5-C8H14O2]−, 255 [M-H-2C6H10O5-C8H14O2-H2O]−, |
Timosaponin AIV | Spirostanol glycosides | [5, 27] |
| 31 | 16.38 | C39H64O13 | 739.4269 | 739.4286 | 2.30 | 739 [M-H]−, 577 [M-H-C6H10O5]−, 415 [M-H-2C6H10O5]−, 273 [M-H-2C6H10O5-C8H14O2]−, 255 [M-H-2C6H10O5-C8H14O2-H2O]−, |
Smilageninoside | Spirostanol glycosides | [5, 27] |
| 32 | 16.73 | C18H18O2 | 265.1229 | 265.1235 | 2.26 | 265 [M-H]−, 121 [M-H-C10H8O]−, |
Monomethyl-cis-hinokiresinol | Coumarins | [29] |
3.2. Steroidal Saponins
Certain cleavage law exists under a certain MS condition for steroidal saponins. Steroidal saponins can be divided into two types: furostanol glycosides, which bear an open side chain at C-22, and spirostanol glycosides, which contain a closed spiroketal ring at C-22. Based on abundant fragment ions generated by losing sugar chains, side chains and dehydration steroidal saponins were divided into types I to V according to the numbers of hydroxyl group and double bond in the A, B, C, and D rings [23, 24, 27, 30].
First, [M-H-OH]− or [M-H-OCH3]− molecular ion peaks were obtained as the CFs of furostan saponins, which display C22-OH or C22-OCH3 on their structures, because furostan saponins preferentially lose the hydroxyl or methoxy on C-22. In contrast, spirostanol saponins generally produced [M-H]− molecular ion peaks. In the negative ion mode, type I saponins produce the same fragment ions at m/z 273 [C19H29O]−, 255 [C19H27]−, 301 [C21H33O]−, and 283 [C21H31]−; types II to IV produce the same fragment ions at m/z 271 [C19H27O]−, 253 [C19H25]−, 299 [C21H31O]−, and 281 [C21H29]−; and type V saponins produce the same fragment ions at m/z 269 [C19H25O]− and 251 [C19H23]−. Type IV saponins could be determined when fragment ions at m/z 271 [C19H27O]−, 253 [C19H25]−, 299 [C21H31O]−, and 281 [C21H29]− are generated, whereas type II and type III saponins can be identified when fragment ions at m/z 289 [C19H29O2]− and 317 [C21H33O2]− appear in addition to the above-mentioned fragments. Fragment ions at m/z 271 [C19H27O]− and 299 [C21H31O]− resulted from the loss of sugar chains and side chains, followed by further loss of one water molecule for type II and type III saponins and the loss of the sugar chains and side chains only for type IV saponins. All steroidal saponins produce abundant characteristic fragment ions through the loss of the branched glycoside chains and side chains or by dehydration; the ions could be rapidly identified by combining CFs with NLs at m/z 162 (C6H10O5), 132 (C5H8O4), 142 (C8H14O2) and 18 (H2O). We used compounds purpureagitoside, timosaponin H1, and timosaponin AIII to discuss the fragmentation pathways in detail in the successive ESI–MSn experiments [5, 23, 24, 27, 30].
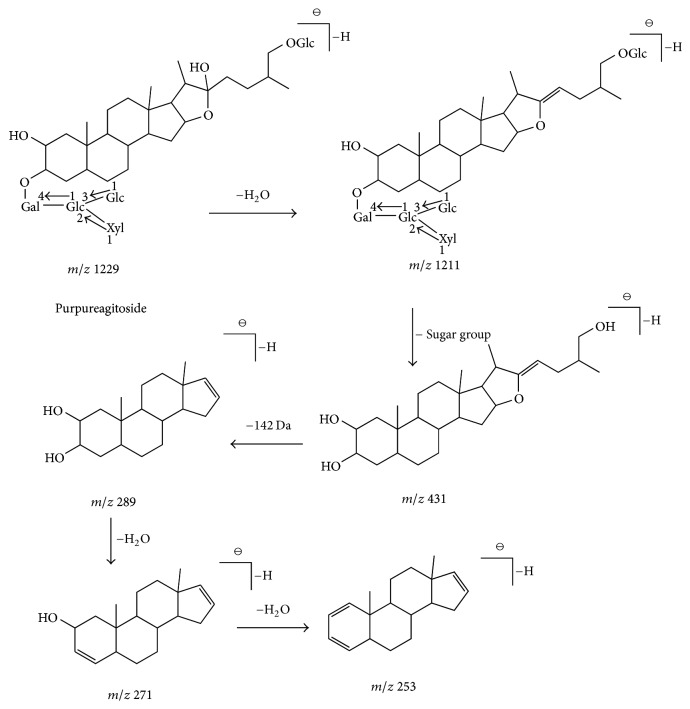
Compound 8 presented a retention time of 5.09 min and formula of C56H94O29. Six fragment ions at m/z 1229 [M-H]−, 1211 [M-H-H2O]−, 289 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]−, 271 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]−, 253 [M-H-3H2O-C5H8O4-4C6H10O5-C8H14O2]−, and 281 [M-H-3H2O-C5H8O4-4C6H10O5-C6H10O2]− were detected in negative ion mode. The compound exhibit a molecular ion peak 1229 [M-H]−. The main fragment ions at m/z 289, 271, 253, and 281 were generated from the losses of sugar chain, along with the cleavage of the E-ring and dehydration. The fragment ion at m/z 289 displayed the sequential losses of 132 Da for a xyl molecule, 4 × 162 Da for four hexose molecules, and 142 Da (C8H14O2) based on the fragment ion at m/z 1211 [M-H-H2O]−. The fragment ion at m/z 271 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]− was derived from the loss of one water molecule on the basis of the losses of sugar chains and side chains. According to NLs rules, Compound 8 can be inferred as furostanol glycosides. The fragment ion at m/z 1211 [M-H-H2O]− indicated the presence of hydroxyl at the position of C-22 due to the fact that furostanol glycosides preferentially lose hydroxyl located in C-22. Compound 8 can be identified as purpureagitoside through comparison with the literature [5, 27]. The fragmentation patterns of purpureagitoside are presented in Figure 2.
Figure 2.
The fragmentation patterns of furostan saponins (type III), taking purpureagitoside as the example: the ion at m/z 1229 was the molecular ion [M-H]−, the fragmentation at m/z 1211 indicated a loss of 18 Da (H2O) on the basis of the parent ion, the fragment ions m/z 431 displayed the loss of sugar group, the fragment ion (m/z 289) was formed by the cleavage of the E-ring on the basis of the fragment ion (m/z 431), and the fragment ions (m/z 271 and 253) were obtained by a loss of 18 Da (H2O) or 2 × 18 Da (H2O) on the basis of the fragment ion (m/z 289).
Compound 13, with a retention time of 8.12 min and a formula of C56H92O28, exhibited fragment ions at m/z 1211 [M-H]−, 1193 [M-H-H2O]−, 271 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]−, and 253 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]− in the MS spectrum. The compound produced molecular ions at m/z 1211 [M-H]−. The main fragment ions at m/z 271 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]− and 253 [M-H-2H2O-C5H8O4-4C6H10O5-C8H14O2]− were both derived from the losses of sugar chain, 142 Da (C8H14O2), and water molecule in the MS2 spectrum, whereas the fragment ion of m/z 289 was not detected according to CFs and NLs rules shown in Table 1. Compound 13 was revealed as a furostanol glycoside (type IV). The fragment ion at m/z 1193 [M-H-H2O]− suggests that one hydroxyl group exists at the C-22 position of the compound because furostanol glycosides preferentially lose hydroxyl in C-22. The fragment ion at m/z 271 [M-H-H2O-C5H8O4-4C6H10O5-C8H14O2]− was produced by the losses of sugar chains and side chains only. Thus, compound 13 was identified as timosaponin H1 [5, 27].
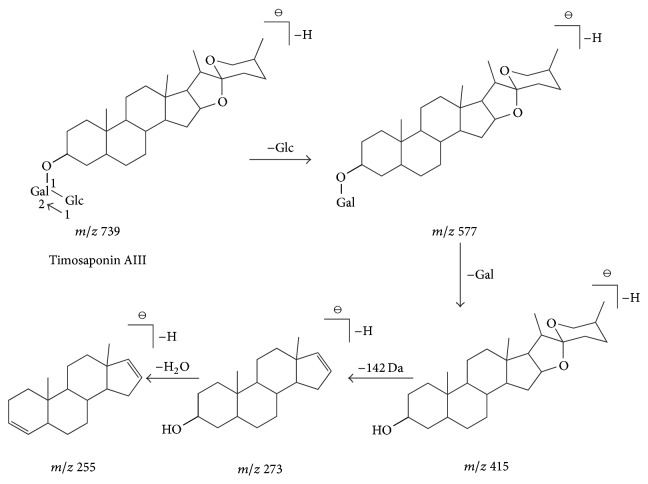
Compound 29 presented a retention time of 16.17 min and a formula of C39H64O13. Several main fragment ions at m/z 739 [M-H]−, 577 [M-H-C6H10O5]−, 415 [M-H-2C6H10O5]−, 273 [M-H-2C6H10O5-C8H14O2]−, and 255 [M-H-2C6H10O5-C8H14O2-H2O]− were obtained in the mass spectrum. The compound was judged to be type I saponins according to the CFs of m/z 273 [M-H-2C6H10O5-C8H14O2]− and 255 [M-H-2C6H10O5-C8H14O2-H2O]−. The ion at m/z 577 [M-H-C6H10O5]− was produced by the loss of a hexose unit (Glc, 162 Da) from the molecular ion at m/z 739 [M-H]−. The fragment ion at m/z 415 [M-H-2C6H10O5]− could be explained by the ejection of a hexose unit (Gal, 162 Da) from the ion at m/z 577. The mass-to-charge ratio difference between the two fragment ions (m/z 415 and 273) was 142 Da, thereby suggesting that the fragment ion at m/z 273 was formed by the cleavage of the E-ring. Fragmentation at m/z 255 was obtained by the fragment ion m/z 273 to lose a molecule H2O. According to CFs and NLs rules shown in Table 1, compound 29 was revealed as spirostanol saponins (type I). These results are consistent with the fragmentation pathway of timosaponin AIII [5, 27]. A proposed mechanistic pathway for fragments formed in MS is shown in Figure 3.
Figure 3.
The fragmentation pathways of spirostanol saponins, taking purpureagitoside as the example: the ion at m/z 739 was the molecular ion [M-H]−, the ion at m/z 577 [M-H-C6H10O5]− was produced by the loss of a hexose unit (Glc, 162 Da), the fragment ion at m/z 415 [M-H-2C6H10O5]− could be explained by ejecting a pentose unit (Gal, 162 Da) from the ion at m/z 577, the fragment ion at m/z 273 was formed by the cleavage of the E-ring, and the fragmentation of m/z 255 was obtained by the fragmentation m/z 273 to lose a water molecule.
3.3. Flavonoids
Flavonoids were divided into mangiferin-type flavonoids, chalcone-type flavonoids, flavanones, homoisoflavonoids, and icariin-type flavonoids according to their chemical structures. Mangiferin-type flavonoids shared the same aglycone structure, the aglycone and sugar of which formed C-glycosides, and C-glycosides tended to lose CH2O units during MS, and 2/3 and 3/4 CH2O units could be eliminated from sugar molecules through 0,3X/0,2X cleavage [5, 25]; therefore, under the negative ion mode, the C-glycosides easily lost the neutral fragments at m/z 90 (C3H6O3) and 120 (C4H8O4), enabling the rapid and efficient identification of mangiferin-type flavonoids based on the two characteristic neutral loss ions. The structures of chalcone-type flavonoids possessed the same B ring and C ring cracking, with fragment ions at m/z 119 [C8H7O]− or 146 [C9H6O2]− produced by A and B ring cleavage during MS (negative ion mode). The compounds can be screened depending on these two characteristic fragment ions. Homoisoflavonoids exhibit the same B ring structure and reverse Diels-–Alder (RDA) of the C ring mainly occurred, producing fragment ion at m/z 133 [C9H9O]−, which could be recognized as the characteristic fragment ion of these compounds. Flavanones formed two main complementary fragments at m/z 119 [C8H7O]− and 165 [C8H5O4]− by C ring for RDA cleavage. Anthocyanin aglycones possess the same aglycone structure and could be rapidly identified according to their aglycone fragment of m/z 367 [C21H19O6]− and neutral loss molecules of m/z 162 (C6H10O5) and 132 (C5H8O4) [5, 25, 29, 31, 32].
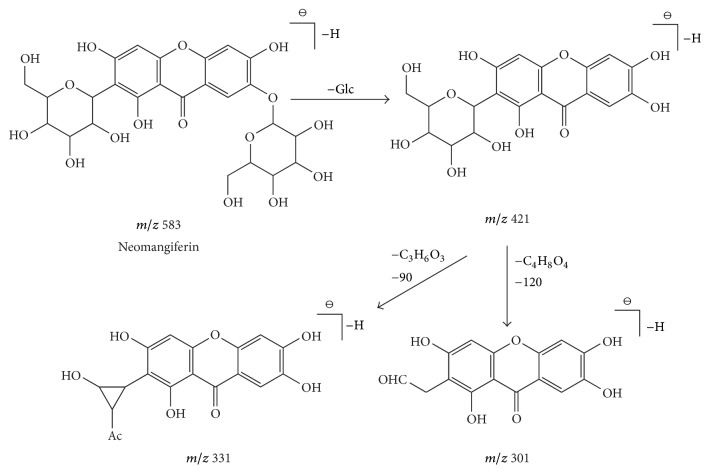
Compound 1 presented a retention time of 3.55 min and a formula of C25H28O16, generating product ions at m/z 583 [M-H]−, 421 [M-H-C6H10O5]−, 331 [M-H-C6H10O5-C3H6O3]−, and 301 [M-H-C6H10O5-C4H8O4]−. The fragment ion at m/z 583 [M-H]− was a molecular ion. In addition, we obtained fragment ions at m/z 421 [M-H-C6H10O5]− that displayed a loss of 162 Da [C6H10O5] based on the parent ion. We also obtained fragment ions at m/z 331 [M-H-C6H10O5-C3H6O3]− and 301 [M-H-C6H10O5-C4H8O4]−, which corroborated with the rules of C-glycoside loss of 90 (C3H6O3) and 120 Da (C4H8O4) from sugar molecules through 0,3X/0,2X cleavage. These results follow the rules of CFs and NLs shown in Table 1. Compound 1 was rapidly and efficiently revealed as mangiferin-type flavonoids. Therefore, compound 1 was determined to be neomangiferin [5, 25]. The specific fragmentation process of neomangiferin is shown in Figure 4.
Figure 4.
The specific fragmentation process of mangiferin flavonoids, take neomangiferin as an example: The fragment ion at m/z 583 [M-H]− was a molecular ion, m/z 421 displayed a loss of 162 Da (C6H10O5) based on the parent ion, the fragment ion at m/z 331 [M-H-C6H10O5-C3H6O3]− conformed to the rule of C-glycosides loss 90 Da (C3H6O3) and m/z 301 [M-H-C6H10O5-C4H8O4]− followed the rules of C-glycosides loss 120 Da (C4H8O4) from sugar molecules through 0,3X/0,2X cleavage.
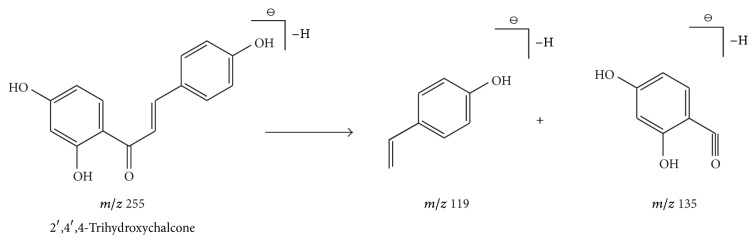
Compound 22 possessed a retention time of 10.69 min and a formula of C15H12O4. In the negative ion mode, several fragment ions at m/z 255 [M-H]−, 135 [M-H-C7H4O2]−, and 119 [M-H-C7H4O3]− were detected; among the fragment ions, those at m/z 135 [M-H-C7H4O2]− and 119 [M-H-C7H4O3]− resulted from the cleavage by separation of A and B rings, and the fragment ion at m/z 119 [M-H-C7H4O3]− corresponded to the typical fragment of chalcone-type flavonoids according to CFs and NLs rules shown in Table 1. Besides, the fragment ion at m/z 255 [M-H]− is a molecular ion. Compound 22 was rapidly revealed as chalcone flavonoids. Finally, compound 22 was identified as 2′,4′,4-trihydroxychalcone [29]. The specific fragmentation process of 2′,4′,4-trihydroxychalcone is shown in Figure 5.
Figure 5.
The specific fragmentation process of chalcone flavonoids, take 2′,4′,4-trihydroxychalcone as an example: the fragment ion of m/z 255 [M-H]− is molecular ion and the fragment ions at m/z 135 and 119 resulting from separation of A and B rings.
3.4. Phenylpropanoids
Phenylpropanoid compounds can be classified into lignans and coumarins according to different parent structures. Lignan compounds with the same A ring structure and benzyl groups usually produce the characteristic fragments of m/z 107 [C7H7O]− and 91 [C7H7]− by benzyl cleavage, as well as other fragment ions because of different linking groups to the A ring; then, the compound was determined according to the molecular ion fragments [29, 33].
Compound 27 presented a retention time of 12.25 min and a formula of C16H18O3. In the negative ion mode, several fragment ions of m/z 257 [M-H]−, 107 [M-H-C9H10O2]−, and 91 [M-H-C9H10O3]− were observed; among the fragments, the fragment ion at m/z 107 [M-H-C9H10O2]− was produced as a result of benzyl cleavage, and the fragment ion at m/z 91 [M-H-C9H10O3]− indicated a loss of water molecule from the fragment ion at m/z 107 [M-H-C9H10O2]−. These two fragments show that this compound belongs to coumarin according to CFs rules shown in Table 1. Besides, the fragment ion of m/z 257 [M-H]− as molecular ion was detected; thus, compound 27 was determined as Broussonin A or Broussonin B [29].
Compound 28 possessed a retention time of 13.16 min and a formula of C17H16O2. In the negative ion mode, several fragments were observed at m/z 251 [M-H]−, 107 [M-H-C10H8O]− and 91 [M-H-C10H8O2]− were observed. The fragment ion at m/z 107 [M-H-C9H10O2]− was produced as a result of benzyl cleavage. The fragment ions at m/z 91 [M-H-C9H10O3]− indicated a loss of water molecule based on the fragment ion at m/z 107 [M-H-C9H10O2]−. Besides, the fragment ion at m/z 251 [M-H]− as a molecular ion was detected. According to CFs rules shown in Table 1, compound 28 shared the same fragmentation pathways with compound 27 and belonged to coumarin. Therefore, compound 28 was determined as hinokiresinol [29, 33]. A pathway for fragments formed during MS is shown in Figure 6.
Figure 6.
A fragmentation pathway of lignans formed in MS, take hinokiresinol as an example: the fragment ion of m/z 251 [M-H]− was molecular ion and the fragment ion at m/z 107 was produced as result of benzyl cleavage. The fragment ions m/z indicated a loss of water molecular based on the fragment ion at m/z 107.
3.5. Benzophenones
Benzophenone compounds possess the same A ring structure, which easily loses 94 Da (A ring) and 44 Da (CO2) in the negative ion mode. Therefore, benzophenone compounds in Rhizoma Anemarrhenae could be rapidly screened and identified according to the characteristics neutral loss of 94 Da and 44 Da in combination with molecular ion [26, 34].
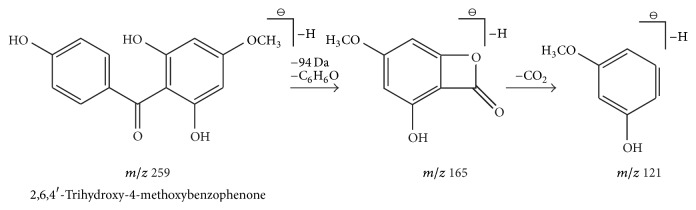
Compound 12 presented a retention time of 7.49 min and a formula of C14H12O5, thereby showing several fragments at m/z 259 [M-H]−, 165 [M-H-C6H6O]−, and 121 [M-H-C6H6O-CO2]. The fragment ion at m/z 165 [M-H-C6H6O]− indicates a loss of 94 Da (C6H6O); the difference between fragment ions (m/z 165 and 121) is exactly 44 Da, indicating a loss of one CO2 molecule. The behavior follows the NLs rules shown in Table 1, and the compound was identified as benzophenone. Besides, m/z 259 [M-H]− is the parent ion; thus, compound 12 was determined as 2,6,4′-trihydroxy-4-methoxybenzophenone [26, 34]. A pathway for fragments formed in MS is shown in Figure 7.
Figure 7.
A fragmentation pathway of benzophenone formed in MS, take 2,6,4′-trihydroxy-4-methoxybenzophenone as an example: m/z 259 [M-H]− is parent ion, the fragment ion at m/z 165 indicated a loss of 94 Da (C6H6O) based on the parent ion, and the fragment ion at m/z 121 indicated a loss of one CO2 molecule based on the fragment at m/z 165.
3.6. Alkaloids
Alkaloids were divided into amide alkaloids with a N-C=O structure and pyridine alkaloids with N atoms in a six-member ring. In the negative ion mode, amide alkaloid with the same part of the structure easily loses a tyramine residue, resulting in neutral loss of molecule 119 (C8H7O). In addition, 15 Da (CH3) can be easily lost based on the parent ion in the high mass-to-charge ratio. Given the same pyridine ring structure of pyridine alkaloids, the characteristic fragment ion at m/z 78 [C5H4N]− was produced by removing the side chain to form the pyridine ring structure, and compounds were further identified through molecular ions [28, 35].
Compound 10 presented a retention time of 6.86 min and a formula of C18H19NO4 and exhibited several fragments at m/z 312 [M-H]−, 297 [M-H-CH3]−, and 178 [M-H-CH3-C8H7O]−. The fragment ion at m/z 297 [M-CH3]− was observed in the high mass-to-charge ratio, displaying a loss of 15 Da (CH3) based on the parent ion. In addition, the fragment ion at m/z 178 [M-H-CH3-C8H7O]− corroborated with the loss of 119 Da (C8H7O) based on the fragment ion at m/z 297. The behavior followed the CFs and NLs rules shown in Table 1, In addition, the fragment ion at m/z 312 [M-H]− was the molecular ion. Thus, compound 10 was identified as N-trans-feruloyltyramine or N-cis-feruloyltyramine [28].
4. Conclusions
Based on the fragmentation patterns of five kinds of compounds in Rhizoma Anemarrhenae (as summarized from the literature), a combination of UPLC-Q-TOF/MS with data postprocessing techniques (CFF and NLF) was developed in this study to rapidly classify and identify 32 compounds in Rhizoma Anemarrhenae extracts. The results provide a basis for the pharmacological and pharmacokinetic studies on Rhizoma Anemarrhenae, overcome the shortage for traditional chemical analysis of TCM to a certain extent, and facilitate the advancement and development of TCM study.
Acknowledgments
This project was supported by The National Natural Science foundation of China (no. 81573825, no. 81273998), Research Programs of Application of Basic and Frontier Technology in Tianjin (15JCYBJC29400), and Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R41).
Contributor Information
Yanyan Xu, Email: xyytjutcm@163.com.
Yubo Li, Email: yaowufenxi001@sina.com.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Yubo Li and Yanyan Xu conceived and designed the study. Lei Yuan, Lanlan Shan, and Yuanyuan Wu carried out the experiments. Lanlan Shan and Yani Zhang analyzed the research data and Lanlan Shan and Lei Yuan wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Committee of National Pharmacopoeia. Pharmacopoeia of the People’s Republic of China, (Chinese edition) Vol. 1. Beijing, China: Chemical Industry Press; 2015. [Google Scholar]
- 2.Zheng J. H. Health pharmacy, (Second Edition) People's health out Edition Agency; 2001. [Google Scholar]
- 3.Wang Y. L., Dan Y., Yang D. W., et al. The genus Anemarrhena Bunge: a review on ethnopharmacology, phytochemistry and pharmacology. Journal of Ethnopharmacology. 2014;153:42–60. doi: 10.1016/j.jep.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Yang B. Y., Zhang J., Liu Y., Kuang H. X. Steroidal saponins from the rhizomes of Anemarrhena asphodeloides. Molecules. 2016;21(8, article 1075) doi: 10.3390/molecules21081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng Y., Zhao L., Lin D., Liu Y., Zhang M., Song S. Determination of the chemical constituents of the different processed products of Anemarrhena asphodeloides Rhizomes by high-performance liquid chromatography quadrupole time-of-flight mass spectrometry. Biomedical Chromatography. 2016;30(4):508–519. doi: 10.1002/bmc.3575. [DOI] [PubMed] [Google Scholar]
- 6.Ma C., Fan M., Tang Y., et al. Identification of major alkaloids and steroidal saponins in rat serum by HPLC-diode array detection-MS/MS following oral administration of Huangbai-Zhimu herb-pair Extract, Biomed. Biomedical Chromatography. 2008;22(8):835–850. doi: 10.1002/bmc.1000. [DOI] [PubMed] [Google Scholar]
- 7.Dong H., Yan G. L., Han Y., et al. UPLC-Q-TOF/MS-based metabolomic studies on the toxicity mechanisms of traditional Chinese medicine Chuanwu and the detoxification mechanisms of Gancao, Baishao, and Ganjiang. Chinese Journal of Natural Medicines. 2015;13(9):687–698. doi: 10.1016/S1875-5364(15)30067-4. [DOI] [PubMed] [Google Scholar]
- 8.Wen X. D., Liu E. H., Yang J., et al. Identification of metabolites of Buyang Huanwu decoction in rat urine using liquid chromatography-quadrupole time-of-flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2012;67-68:114–122. doi: 10.1016/j.jpba.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G., Yao S., Zhang X. X., Song H. Rapid screening and structural characterization of antioxidants from the extract of selaginella doederleinii hieron with DPPH-UPLC-Q-TOF/MS method. International Journal of Analytical Chemistry. 2015;2015:9. doi: 10.1155/2015/849769.849769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H., Zhang S., Zhang A., et al. Metabolomic analysis of diet-induced type 2 diabetes using UPLC/MS integrated with pattern recognition approach. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0093384.e93384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., Qian S. H., Qian D. W., Li S. L. Screening of Intestinal Bacterial Metabolites of Platycodin D Using Ultra-Performance Liquid Chromatography/Quadrupole Time-of-Flight Mass Spectrometry. American Journal of Chinese Medicine. 2016;44(4):817–833. doi: 10.1142/S0192415X16500452. [DOI] [PubMed] [Google Scholar]
- 12.Zhang A., Zou D., Yan G., Tan Y., Sun H., Wang X. Identification and characterization of the chemical constituents of Simiao Wan by ultra high performance liquid chromatography with mass spectrometry coupled to an automated multiple data processing method. Journal of Separation Science. 2014;37(14):1742–1747. doi: 10.1002/jssc.201400105. [DOI] [PubMed] [Google Scholar]
- 13.Sandin M., Teleman J., Malmström J., Levander F. Data processing methods and quality control strategies for label-free LC-MS protein quantification. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics. 2014;1844(1):29–41. doi: 10.1016/j.bbapap.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Christin C., Bischoff R., Horvatovich P. Data processing pipelines for comprehensive profiling of proteomics samples by label-free LC-MS for biomarker discovery. Talanta. 2011;83(4):1209–1224. doi: 10.1016/j.talanta.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J. Y., Wang Z. J., Zhang Q., et al. Rapid screening and identification of target constituents using full scan-parent ions list-dynamic exclusion acquisition coupled to diagnostic product ions analysis on a hybrid LTQ-Orbitrap mass spectrometer. Talanta. 2014;124:111–122. doi: 10.1016/j.talanta.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y. Y., Cheng X. L., Wei F., Bai X., Lin R.-C. Application of faecal metabonomics on an experimental model of tubulointerstitial fibrosis by ultra performance liquid chromatography/high- sensitivity mass spectrometry with MSE data collection technique. Biomarkers. 2012;17(8):721–729. doi: 10.3109/1354750X.2012.724450. [DOI] [PubMed] [Google Scholar]
- 17.Yuan L., Zhang Z., Hou Z., et al. Rapid classification and identification of complex chemical compositions in traditional Chinese medicine based on UPLC-Q-TOF/MS coupled with data processing techniques using the KuDieZi injection as an example. Anal. Methods. 2015;7(12):5210–5217. doi: 10.1039/C4AY03103B. [DOI] [Google Scholar]
- 18.Yuan L., Yin J., Tian M., et al. The classification and identification of complex chemical compositions in yanhusuo herb using UPLC-Q-TOF/MS. Anal. Methods. 2016;8(10):2274–2281. doi: 10.1039/C5AY03162A. [DOI] [Google Scholar]
- 19.Qu J., Liang Q., Luo G., Wang Y. Screening and Identification of Glycosides in Biological Samples Using Energy-Gradient Neutral Loss Scan and Liquid Chromatography Tandem Mass Spectrometry. Analytical Chemistry. 2004;76(8):2239–2247. doi: 10.1021/ac030413t. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Zhang Z., Hou Z., et al. A rapid and integrated pyramid screening method to classify and identify complex endogenous substances with UPLC/Q-TOF MS-based metabolomics. RSC Advances. 2015;5(1):202–209. doi: 10.1039/c4ra10719e. [DOI] [Google Scholar]
- 21.Zhang J. Y., Zhang Q., Li N., Wang Z. J., Lu J.-Q., Qiao Y.-J. Diagnostic fragment-ion-based and extension strategy coupled to DFIs intensity analysis for identification of chlorogenic acids isomers in Flos Lonicerae Japonicae by HPLC-ESI-MSn. Talanta. 2013;104:1–9. doi: 10.1016/j.talanta.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L. F., Liu Y. Y., Xue F. Q. Bying mass spectrometry in neutral loss scans nitazoxanide in phase II metabolites in dog plasma. Journal of mass spectrometry. 2008;02:101–104. [Google Scholar]
- 23.Zhang T., Liu H., Liu X. T., Xu D. R., Chen X. Q., Wang Q. Qualitative and quantitative analysis of steroidal saponins in crude extracts from Paris polyphylla var. yunnanensis and P. polyphylla var. chinensis by high performance liquid chromatography coupled with mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2010;51(1):114–124. doi: 10.1016/j.jpba.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Liang J., Liu J., et al. Quality control and identification of steroid saponins from Dioscorea zingiberensis C. H. Wright by fingerprint with HPLC-ELSD and HPLC-ESI-Quadrupole/Time-of-fight tandem mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2014;91:46–59. doi: 10.1016/j.jpba.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao X., Li R., Song W., et al. A targeted strategy to analyze untargeted mass spectral data: Rapid chemical profiling of Scutellaria baicalensis using ultra-high performance liquid chromatography coupled with hybrid quadrupole orbitrap mass spectrometry and key ion filtering. Journal of Chromatography A. 2016;1441:83–95. doi: 10.1016/j.chroma.2016.02.079. [DOI] [PubMed] [Google Scholar]
- 26.Berardini N., Carle R., Schieber A. Characterization of gallotannins and benzophenone derivatives from mango (Mangifera indica L. cv. ‘Tommy Atkins’) peels, pulp and kernels by high-performance liquid chromatography/electrospray ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 2004;18(19):2208–2216. doi: 10.1002/rcm.1611. [DOI] [PubMed] [Google Scholar]
- 27.Liu X. W., Zhang F., Gao S. H., Jiang B., Chen W. S. Metabolite profiling of Zi-Shen pill in rat biological specimens by UPLC-Q-TOF/MS. Chinese Journal of Natural Medicines. 2015;13(2):145–160. doi: 10.1016/S1875-5364(15)60019-X. [DOI] [PubMed] [Google Scholar]
- 28.Mhlongo M. I., Piater L. A., Madala N. E., Steenkamp P. A., Dubery I. A. Phenylpropanoid defences in Nicotiana tabacum cells: Overlapping metabolomes indicate common aspects to priming responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS ONE. 2016;11(3) doi: 10.1371/journal.pone.0151350.e0151350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Z. Cong. China Medical Science and Technology Publishing House. 2002. Natural organic mass spectrometry; pp. 670–671. [DOI] [Google Scholar]
- 30.Kang L. P., Zhao Y., Pang X., et al. Characterization and identification of steroidal saponins from the seeds of Trigonella foenum-graecum by ultra high-performance liquid chromatography and hybrid time-of-flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2013;74:257–267. doi: 10.1016/j.jpba.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Yang W. Z., Wu W. Y., Yang M., Guo D. A. Elucidation of the fragmentation pathways of a complex 3,7-O-glycosyl flavonol by CID, HCD, and PQD on an LTQ-Orbitrap Velos Pro hybrid mass spectrometer. Chinese Journal of Natural Medicines. 2015;13(11):867–872. doi: 10.1016/S1875-5364(15)30091-1. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y., Deng J. Internal standard mass spectrum fingerprint: A novel strategy for rapid assessing the quality of Shuang-Huang-Lian oral liquid using wooden-tip electrospray ionization mass spectrometry. Analytica Chimica Acta. 2014;837:83–92. doi: 10.1016/j.aca.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Duffield A. M. Mass spectrometric fragmentation of some lignans. Journal of Heterocyclic Chemistry. 1967;4(1):16–22. doi: 10.1002/jhet.5570040104. [DOI] [Google Scholar]
- 34.Ui J. Y., Ye S. L., Hana J., et al. Identification of antiadipogenic constituents of the rhizomes of Anemarrhena asphodeloides. Journal of Natural Products. 2009;72(10):1895–1898. doi: 10.1021/np900397f. [DOI] [PubMed] [Google Scholar]
- 35.Horai H., Arita M., Kanaya S., et al. MassBank: a public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]