Abstract
Over the past few years, noncoding RNAs (ncRNAs) have been extensively studied because of the significant biological roles that they play in regulation of cellular mechanisms. ncRNAs are associated to higher eukaryotes complexity; accordingly, their dysfunction results in pathological phenotypes, including cancer. To date, most research efforts have been mainly focused on how ncRNAs could modulate the expression of protein-coding genes in pathological phenotypes. However, recent evidence has shown the existence of an unexpected interplay among ncRNAs that strongly influences cancer development and progression. ncRNAs can interact with and regulate each other through various molecular mechanisms generating a complex network including different species of RNAs (e.g., mRNAs, miRNAs, lncRNAs, and circRNAs). Such a hidden network of RNA-RNA competitive interactions pervades and modulates the physiological functioning of canonical protein-coding pathways involved in proliferation, differentiation, and metastasis in cancer. Moreover, the pivotal role of ncRNAs as keystones of network structural integrity makes them very attractive and promising targets for innovative RNA-based therapeutics. In this review we will discuss: (1) the current knowledge on complex crosstalk among ncRNAs, with a special focus on cancer; and (2) the main issues and criticisms concerning ncRNAs targeting in therapeutics.
1. Introduction
When the Human Genome Project (HGP) began in the late 1990s, researchers hypothesized that our genome comprised about 100,000 protein-coding genes [1]. Over the years, this estimate has been continuously downsized. In 2001, the International Human Genome Sequencing Consortium (IHGSC) published the initial sequence of the human genome and proposed that the number of protein-coding genes was about 30,000 [2]. At the same time, Celera Genomics (a competitor group of IHGSC) estimated this number at 26,000 [3]. In 2004, when the final draft of the human genome was published, this number was further reduced to 24,500 [4], but in 2007 an additional analysis established that it was around 20,500 [5]. More recently, new studies updated the number of human protein-coding genes to 19,000 [6]. This estimate is particularly surprising, because it would suggest that less than 2% of the whole human genome encodes for proteins; accordingly, the keystone of Homo sapiens complexity could lie in the 98% of our DNA (the genome dark matter), which does not encode proteins but would be endowed with critical regulatory functions. In the last decade, two important scientific initiatives supported by the US National Institutes of Health (i.e., the projects ENCODE and Roadmap Epigenomics) reported seminal data on hundreds of thousands of functional regions in the human genome, whose function is to supervise gene expression [7, 8]. These data suggested that much more space in our genome is committed to regulatory than to structural functions. Moreover, these studies proposed that about 80% of the human genome is dynamically and pervasively transcribed, mostly as non-protein-coding RNAs (ncRNAs). The biological relevance of the noncoding transcriptome has become increasingly undeniable over the last few years. Studies of comparative genomics showed that the relative proportion of genome space, occupied by the proteome-encoding genome as opposed to the regulatory (non-protein-encoding) genome is very variable among evolutionarily distant species; for instance, the protein-coding genome represents almost the entire genome of the unicellular yeast Saccharomyces cerevisiae, whereas it constitutes only 2% of mammalian genomes [9]. Moreover and intriguingly, the noncoding transcriptome is frequently altered in major diseases, including cancer [10–12]. These observations strongly suggest that ncRNAs are closely related to the complexity of higher eukaryotes and that their dysfunction may result in pathological phenotypes. RNA is a structurally versatile molecule, able to perform several molecular functions. By simple base pairing with other nucleic acids, RNA can recognize and bind both DNA and RNA targets in a very specific manner and regulate their transcription, processing, editing, translation, or degradation. An intriguing field for future explorations is the tridimensional folding of RNA molecules, which confers them allosteric properties: this increases the range of potential molecular interactors (including proteins); additionally, dynamic conformational changes can be triggered by ligand binding. Moreover and different from proteins, RNA can be rapidly transcribed and degraded making it a very dynamic molecule that can be quite rapidly synthesized without additional time and energetic costs of translation [13]. For all these reasons, over the past few years, ncRNAs have been extensively studied because of the significant biological roles that they play in regulation of cellular mechanisms. Noncoding RNA genes can generally be divided into two major categories by their transcript sizes: (1) long noncoding RNAs (lncRNAs) are longer than 200 nucleotides; and (2) small noncoding RNAs have a length equal to or lower than 200 nucleotides [i.e., microRNAs (miRNAs), small interfering RNAs (siRNAs), small nuclear RNAs (snRNAs or U-RNAs), small nucleolar RNAs (snoRNAs), PIWI-interacting RNAs (piRNAs), and tRNAs] [14]. To date, most research efforts have been focused on how ncRNAs (in particular, miRNAs) modulate the expression of protein-coding genes and their roles in human pathophysiology. However, recent evidence has shown the existence of unexpected interplay among ncRNAs, which influences cell physiology and diseases. In addition to the canonical multilayered control of expression of protein-coding genes (briefly described below), ncRNAs can interact with and regulate each other through various molecular mechanisms generating a complex network including different species of RNAs. In such a regulatory network, ncRNAs also compete among each other for binding to mRNAs, thus acting as competing endogenous RNAs (ceRNAs). In this review, we will summarize the current knowledge on the complex crosstalk among ncRNAs (including miRNAs, lncRNAs, and circRNAs) and how they could reciprocally interact to regulate cancer progression and dissemination.
1.1. miRNAs
miRNAs are 18–25 nucleotides long, evolutionarily conserved, single-stranded RNAs, which negatively modulate the expression of their target mRNAs (more than 60% of protein-coding genes) by binding to the 3′-UTR of specific mRNA targets, leading either to their translational repression, cleavage, or decay [15–17]. This binding occurs through a specific miRNA region (named seed region), which is a contiguous string of at least 6 nucleotides beginning at position two of the 5′ of the molecule [18]. The block of translation is due to the inhibition of mRNA 5′-cap recognition and interference on the interaction between the mRNA and the 60S ribosomal subunit, while mRNA degradation is promoted by mechanisms of decapping and deadenylation [19]. These molecular mechanisms are mediated by an RNA-induced silencing complex (RISC) that includes proteins belonging to the Argonaute (AGO) family; specifically, RISC endonuclease activity depends exclusively on AGO2 protein [20]. A single miRNA can control the expression of several mRNAs, and a single mRNA may be targeted by more than one miRNA, thus creating a complex interplay of cooperative regulation [21]. To date, more than 2500 mature miRNAs have been included in the miRbase database [22].
Extensive studies have shown that miRNAs control pivotal cellular processes, (e.g., cell proliferation, differentiation, migration, cell death, and angiogenesis), thus contributing to the pathogenesis of diseases such as cancer. Indeed, several miRNAs have been identified as potential oncogenes or tumor suppressors in cancer development and progression [23]. In the last two decades, their mutations and altered expression were reported to be causally related to the neoplastic features of the cells, thus providing new perspectives for the understanding of the complex regulatory networks that rule tumor biology [24]. miRNA dysfunctions exert a pleiotropic effect on the expression of their mRNA targets impairing the functioning of biological networks. It has been convincingly demonstrated that different cancer histotypes display specific miRNA expression patterns: this phenomenon would be helpful to improve diagnosis of poorly differentiated tumors and predict prognosis in cancer [25, 26]. Moreover, multiple experimental evidence has shown that miRNAs can be also secreted by cancer cells into bodily fluids, sending oncogenic signals through circulation, which could advantageously mold the extracellular tumor environment [27]. These discoveries gave a new intriguing diagnostic and prognostic role to circulating miRNAs, paving the way for their potential use as noninvasive molecular RNA markers in clinical management of cancer patients [28–30].
1.2. lncRNAs
lncRNAs are the most heterogeneous class of non-protein-coding RNAs with lengths ranging from 200 nt to 100,000 nt. They include transcripts that may be classified as (a) intergenic lncRNAs, (b) intronic lncRNAs, (c) sense or antisense transcripts, (d) pseudogenes, and (e) retrotransposons [14]. Currently, LNCipedia 4.0 records more than 118,000 human lncRNAs, which are usually expressed in a developmental and tissue-specific manner [31]. lncRNAs regulate gene expression at different levels, including chromatin modification, alternative splicing, and protein localization and activity [32]. Such a wide range of mechanisms is due to their ability to bind to DNA, RNAs, and proteins. lncRNAs, thanks to their binding to promoter DNA, can prevent the access of transcription factors to their own promoter binding sites and impede the transcription of specific genes (e.g., DHFR) [33]. Some lncRNAs (e.g., HOTAIR) are associated with chromatin-modifying complexes (e.g., polycomb repressive complex 2) to regulate epigenetic silencing of target genes [34]. Much evidence has also shown that lncRNAs may work as molecular scaffolds to connect two or more proteins in functional complexes or can serve to localize protein complexes to appropriate cellular compartments [35]. Antisense lncRNAs can target, by direct sequence complementarity, their antisense mRNAs and, accordingly, modulate alternative splicing processes or protect 3′-UTR from miRNA binding, increasing the stability of mRNAs (e.g., ZEB2-AS1, BACE1-AS) [36, 37]. Several recent studies have shown that lncRNAs are critically involved in a wide range of biological processes, such as cell cycle regulation, pluripotency, differentiation, and cell death [38–41]. Dysregulation of lncRNA activity has been frequently reported in association to diseases, including several types of cancer. Specifically, upregulated lncRNAs in cancer seem to possess tumor-promoting abilities, whilst downregulated lncRNAs exhibit tumor-suppressive roles [42–47]. Although several lncRNAs have been reported to be dysregulated in neoplastic phenotypes, their mechanistic role in cancer biology has not been satisfactorily explained for most of them. However, scientific evidence strongly suggests a promising role for lncRNAs as cancer-related biomarkers and potential targets for innovative therapeutic approaches.
1.3. circRNAs
Circular RNAs (circRNAs) represent a recently discovered class of noncoding RNAs, composed of single-stranded, covalently closed, exonuclease-resistant circular transcripts [48]. Although the existence of circular RNAs has been known since the 70s [49], for a long time such molecules were considered only by-products of pre-mRNA processing and therefore interpreted as artifacts of aberrant RNA splicing [50]. However, recent advances in RNA sequencing technologies have revealed a ubiquitous, and in some cases abundant, expression of endogenous circRNAs in mammalian genomes [51, 52]. circRNAs are a circularized isoform of linear protein-coding genes generated through backsplicing, a molecular process that is different from the canonical splicing of linear RNAs. Circular RNA biogenesis can occur both from exons (exonic circRNAs or ecircRNAs), through different mechanisms of backsplicing and introns (intronic circRNAs or ciRNAs), when lariat introns escape typical debranching processes [53]. Currently, about 35,000 circRNAs are reported in the circBase database [54], but molecular functions and biological processes, in which they are involved, remain elusive for most of them. Recent emerging evidence convincingly suggests that circRNAs may play an important role in RNA-RNA interactions. In some instances, circRNAs exhibit multiple binding sites for the same miRNA and represent a potential molecular sponge for sequestering the most abundant miRNAs [55]. In other words, circRNAs may negatively regulate the function of miRNAs, and, thus, protect miRNA targets, by acting as competing endogenous RNAs. As some papers would suggest that ceRNA role of circRNAs could not be their main function in cell biology, other molecular functions have been proposed for circRNAs (a) to bind and sequester RNA binding proteins (RBPs) [56–58] and (b) to be translated into proteins when recognized by ribosomes in the presence of internal ribosome entry sites (IRESs) [59, 60]. As circRNAs are potentially able to control different layers of gene expression, it is not surprising that their dysregulation is associated with human pathologies, including cancer [61–63]. Most reports that connect circRNAs and tumors mainly concern comparative gene expression profiling studies between tumor and normal samples. These investigations have shown that circRNAs are frequently downregulated in several types of cancer (e.g., colorectal cancer, ovarian cancer, and gastric cancer) [64–66]. Just few of these studies attempted to functionally explain how abnormal expression of circRNAs could impair physiological cell homeostasis and thus promote cancer phenotypes [67–69].
2. Noncoding RNAs: Different Ways to Interplay among Each Other
Interplay between ncRNAs obviously occurs because of sequence complementarity; for instance, ncRNAs may share miRNA response elements (MREs) with mRNAs and thus be targeted in the same manner [70]. The effects of miRNAs binding to other ncRNAs (i.e., lncRNAs and circRNAs) could be twofold: on the one hand, miRNAs could be sequestered and prevented from acting on the protein-coding mRNAs; on the other hand, miRNA binding to lncRNAs and circRNAs could promote their decay, similarly to mRNAs. In the next paragraphs, we will discuss the different mechanisms of ncRNA interaction and their influence on cancer biology.
2.1. miRNAs Induce Degradation of lncRNAs
Several papers have reported that miRNAs can bind lncRNAs and promote their degradation contributing to cancer processes (Table 1). lncRNAs are structurally similar to mRNAs; indeed, they have 5′-caps and 3′-poly(A) tails [71]; accordingly, the proteins involved in the regulation of decapping, deadenylation, and degradation of mRNAs may also control the turnover of lncRNAs by binding of specific miRNAs.
Table 1.
miRNAs inducing degradation of lncRNAs.
| miRNA | lncRNA/circRNA target | Tumor | miRNA role | PMID |
|---|---|---|---|---|
| let-7b | lincRNA-p21 | Cervical carcinoma | Tumor suppressor | 22841487 |
| let-7b, let-7i | HOTAIR | Cervical carcinoma | Tumor suppressor | 24326307 |
| miR-1 | UCA1 | Bladder cancer | Tumor suppressor | 25015192 |
| miR-9 | MALAT1 | Hodgkin lymphoma, glioblastoma | Tumor suppressor | 23985560 |
| miR-21 | CASC2 | Renal cell carcinoma | Oncogene | 27222255 |
| miR-21 | CASC2 | Glioblastoma | Oncogene | 25446261 |
| miR-21 | GAS5 | Breast cancer | Oncogene | 23933812 |
| miR-34a | HOTAIR | Prostate cancer | Tumor suppressor | 23936419 |
| miR-101 | MALAT1 | Esophageal squamous cell carcinoma | Tumor suppressor | 25538231 |
| miR-125b | HOTTIP | Hepatocellular carcinoma | Tumor suppressor | 25424744 |
| miR-125b | MALAT1 | Bladder cancer | Tumor suppressor | 24396870 |
| miR-141 | H19 | Gastric cancer | Tumor suppressor | 26160158 |
| miR-141 | HOTAIR | Renal carcinoma | Tumor suppressor | 24616104 |
| miR-217 | MALAT1 | Esophageal squamous cell carcinoma | Tumor suppressor | 25538231 |
| miR-671 | CDR1AS | Glioblastoma | Oncogene | 26683098 |
This table reports for each miRNA: (1) its lncRNAs/circRNA target; (2) tumor where such interaction was reported; (3) its function in cancer (oncogene or tumor suppressor); and (4) bibliographic reference reported as Pubmed ID (PMID).
UCA1 (urothelial cancer associated 1), an lncRNA upregulated in several tumors (i.e., bladder cancer, tongue squamous cell carcinoma, breast cancer, and ovarian cancer) [72–75], possesses two predicted binding sites for miR-1, a well-known tumor suppressor miRNA. The binding of miR-1 to UCA1 has been confirmed by luciferase reporter assay in bladder cancer and, accordingly, in vitro upregulation of miR-1 induced UCA1 downregulation and caused a decreased cell growth and migration and also an augmented apoptosis. Such functional effects were reverted after UCA1 overexpression and silencing of AGO2, suggesting that miR-1 was able to downregulate UCA1 expression in an AGO2-mediated manner [76].
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is one of the most studied and abundant lncRNAs: its expression was initially associated with metastasis in non-small-cell lung carcinoma (NSCLC) [77], but then its deregulation has been reported in several other neoplastic diseases [78–80]. The 3′ end of MALAT1 is cleaved by RNase P and RNase Z, producing a tRNA-like ncRNA, called mascRNA (MALAT1-associated small cytoplasmic RNA), which will be exported into the cytoplasm [81], while most of the MALAT1 molecules are localized to nuclear speckles where they regulate alternative splicing of specific pre-mRNAs [82]. Moreover, MALAT1 may bind CBX4 (chromobox 4), a component of polycomb repressive complex 1 (PRC1), and modulate its localization in interchromatin granules, leading to activation or inhibition of gene expression [83]. Through these molecular mechanisms, MALAT1 controls the expression of several genes related to cell cycle and metastatic processes, thus influencing cell proliferation, migration, and invasion. Recent publications reported that MALAT1 is a target of a number of tumor suppressor miRNAs, which could induce its degradation and suppress its oncogenic effects. Leucci et al. reported miRNA-mediated regulation of MALAT1 in the nucleus of Hodgkin lymphoma and glioblastoma cell lines through direct binding of miR-9 to two different MREs in an AGO2-dependent manner [84]. There is evidence of a posttranscriptional regulation of MALAT1 by miR-101 and miR-217 in esophageal squamous cell carcinoma (ESCC) cells [85]. MiR-101 and miR-217 are functionally involved in several cancers as tumor suppressors and exhibited a significant negative correlation with MALAT1 in ESCC tissue samples and adjacent normal tissues. Enforced expression of miR-101 and miR-217 significantly repressed MALAT1 expression, leading to inhibition of cell growth, invasion, and metastasis in ESCC cells [85]. In bladder cancer, MALAT1 is inversely expressed with miR-125b. This miRNA was partially complementary with MALAT1 and bound it in in vitro models. MiR-125b was downregulated in bladder cancer, and its overexpression decreased the expression of MALAT1, causing an inhibition of bladder cancer cell proliferation, motility, and activation of apoptosis [86].
Additionally, miR-125b was also identified as a posttranscriptional regulator of HOTTIP (HOXA distal transcript antisense RNA) in hepatocellular carcinoma (HCC) [87]. HOTTIP is one of the most upregulated lncRNAs in HCC, also in early stages of HCC onset, and maps in antisense position to the distal end of the HOXA gene cluster. HOTTIP promotes tumor growth and metastasis in vitro and in vivo through regulation of the expression of its neighboring HOXA genes (e.g., HOXA10, HOXA11, and HOXA13). MiR-125b has been reported to be frequently downregulated in HCC, and a negative correlation of expression between miR-125b and HOTTIP existed in such cancer. The interaction between miR-125b and HOTTIP was validated by luciferase reporter assay; this was confirmed by ectopic expression of miR-125b that induced downmodulation of HOTTIP [87].
HOTAIR (HOX antisense intergenic RNA) is one of the most intensively studied lncRNAs, as it is frequently associated with different neoplasias. HOTAIR exerts its oncogenic functions by working as a scaffold to assemble polycomb repressive complex 2 (PRC2) on the HOXD gene cluster and inducing the transcriptional silencing of multiple metastasis suppressor genes (e.g., the protocadherin gene family) [34, 88]. HOTAIR is posttranscriptionally destabilized by several tumor suppressor miRNAs in different cancers. Chiyomaru et al. reported a functional binding between miR-34a and HOTAIR in prostate cancer cell lines treated with genistein, an isoflavone with antitumor activity: miR-34a directly bound to two MREs within HOTAIR RNA and lowered its levels [89]. Yoon et al. reported that human antigen R (HuR), let-7b, let-7i, and AGO2 cooperatively bind HOTAIR and promote HOTAIR decay, thus inhibiting the processes of ubiquitination and proteolysis of Ataxin-1 and Snurportin-1, promoted by HOTAIR [90]. Interestingly, HuR and let-7b/AGO2 complex also decreased the stability of lincRNA-p21, an oncogenic lncRNA that reduced translation of beta-catenin and JUNB (JunB proto-oncogene, subunit of transcription factor AP-1) mRNAs in human cervical carcinoma HeLa cells [91]; even if in other experiments HuR was not able to transfer let-7b to AGO2 [92]. In another paper by Chiyomaru et al., it was reported that HOTAIR expression is negatively correlated to that of miR-141 in renal carcinoma cells (RCC) [93]. MiR-141 belongs to the miRNA-200 family, which has been reported to inhibit epithelial-mesenchymal transition (EMT) by ZEB1 (zinc finger E-box-binding homeobox 1) repression and E-cadherin upregulation [94]. MiR-141 was able to target and cleave HOTAIR in an AGO2-dependent manner, and such molecular action downregulated the expression of ZEB2 (zinc finger E-box-binding homeobox 2) induced by HOTAIR [93].
Expression of miR-141 was also found to be negatively correlated to that of lncRNA H19 (H19, imprinted maternally expressed transcript) in gastric cancer [95]. H19, an oncofetal lncRNA, is highly expressed during embryogenesis [96] and is upregulated in several cancers, including gastric cancer [97]. H19 acts as the primary miRNA precursor of miR-675, which in turn targets and represses RB1 (RB transcriptional corepressor 1) mRNA [98]. Overexpression of H19 enhances tumor cell growth and induces EMT; additionally, H19 modulates miRNA processing through its interaction with proteins involved in this molecular process (i.e., Drosha, Dicer). MiR-141 was shown to bind H19 in gastric cancer, and suppress H19 expression and its tumor-promoting functions [95].
MiR-21 is the most commonly upregulated miRNA in cancer: its genetic locus is often amplified in solid tumors, and its expression is promoted by a variety of cancer-related stimuli [99]. MiR-21 enhances cell proliferation, migration, and invasion by targeting several tumor suppressor genes, such as CCL20, CDC25A, PDCD4, and PTEN [100–103]. Recent findings showed that some lncRNAs could be added to the repertoire of miR-21 targets. Zhang et al. reported that expression of miR-21 and lncRNA GAS5 (growth arrest-specific 5) is negatively correlated in breast cancer and that miR-21 binds a miR-21-binding site in exon 4 of GAS5, thus inducing AGO2-mediated suppression of GAS5 [104]. GAS5 is an lncRNA with tumor-suppressive properties: its overexpression sensitizes cancer cells to UV or doxorubicin and decreases tumor proliferation and cell invasion. Interestingly, GAS5 also negatively regulated miR-21 at the posttranscriptional level through the RISC complex, suggesting the existence of a reciprocal negative feedback loop between GAS5 and miR-21 [104]. In two different studies on renal cell carcinoma and glioblastoma, it has been shown that miR-21 targeted and suppressed the expression of the tumor suppressor lncRNA CASC2 (cancer susceptibility candidate 2) in an AGO2-dependent manner [105, 106]. Indeed, the overexpression of miR-21 abrogated the inhibition of proliferation, migration, and the induction of apoptosis promoted by CASC2. Notably, when CASC2 was upregulated, miR-21 expression decreased: this suggests reciprocal repression between miR-21 and CASC2 [106].
The first experimental evidence that lncRNAs may be targeted by miRNAs was reported for the antisense transcript of the cerebellar degeneration-related protein 1 (CDR1, also known as CiRS-7 or CDR1AS), which is a circular RNA produced by a backsplice event [107]. MiR-671, a nuclear-enriched miRNA, induced cleavage of CDR1AS in an AGO2-dependent manner. Repression of miR-671 promoted the upregulation of both CDR1AS and CDR1, suggesting that CDR1AS was able to stabilize the sense transcript CDR1. Currently, this represents the only report on circRNA targeted and degraded by a miRNA. The interaction between miR-671 and CDR1AS could affect the biopathological molecular asset of glioblastoma multiforme (GBM), the most prevalent and aggressive cancer originating in the central nervous system, mainly in the brain. Indeed, Barbagallo et al. demonstrated that miR-671-5p is significantly upregulated in GBM. Enforced expression of miR-671-5p increased migration and decreased proliferation rates of GBM cell lines, suggesting its potential role as a novel oncomiRNA in GBM [108]. Expression of miR-671 was inversely correlated to that of CDR1AS and CDR1 in GBM biopsies and the expression of CDR1AS and CDR1 decreased when the miR-671 mimic was used, suggesting that the interaction of these molecules could be functionally altered in a GBM model [108].
2.2. lncRNAs as Decoys of miRNAs
The most explored mechanism of functional interactions between lncRNAs and miRNAs is based on sharing the same miRNA target sequence in both lncRNAs and mRNAs. In this way, lncRNAs are able to sequester miRNAs away from mRNAs, functioning as “miRNA sponges” or “miRNA decoys.” Through such a competitive endogenous mechanism of interaction, lncRNAs decrease the quantity of available miRNAs and increase, accordingly, translations of their mRNA targets. lncRNAs, working as competitive endogenous RNAs, have been extensively described in molecular circuits involved in tumors (Table 2).
Table 2.
lncRNAs acting as decoy of miRNAs.
| lncRNA | miRNA target | Tumor | lncRNA role | PMID |
|---|---|---|---|---|
| CCAT1 | let-7 | Hepatocellular carcinoma | Oncogene | 25884472 |
| EWSAT1 | miR-326/−330-5p cluster | Nasopharyngeal carcinoma | Oncogene | 27816050 |
| FER1L4 | miR-106a-5p | Gastric cancer | Tumor suppressor | 26306906 |
| FTH1P3 | miR-224-5p | Squamous cell carcinoma | Oncogene | 28093311 |
| FTX | miR-374a | Hepatocellular carcinoma | Tumor suppressor | 27065331 |
| GAS5 | miR-135b | Non-small cell lung cancer | Tumor suppressor | 28117028 |
| H19 | let-7a, let-7b | Breast cancer | Oncogene | 28102845 |
| HOST2 | let-7b | Epithelial ovarian cancer | Oncogene | 25292198 |
| HOTAIR | miR-1 | Hepatocellular carcinoma | Oncogene | 27895772 |
| HOTAIR | miR-152 | Gastric cancer | Oncogene | 26187665 |
| HULC | miR-372 | Liver cancer | Oncogene | 20423907 |
| lincRNA-RoR | miR-145 | Breast cancer | Oncogene | 25253741 |
| lincRNA-RoR | miR-145 | Endometrial cancer | Oncogene | 24589415 |
| LOC100129148 | miR-539-5p | Nasopharyngeal carcinoma | Oncogene | 28328537 |
| MALAT1 | miR-1 | Breast cancer | Oncogene | 26676637 |
| MALAT1 | miR-145 | Cervical cancer | Oncogene | 26311052 |
| NEAT1 | miR-449-5p | Glioma | Oncogene | 26242266 |
| PVT1 | miR-152 | Gastric cancer | Oncogene | 28258379 |
| PVT1 | miR-186 | Gastric cancer | Oncogene | 28122299 |
| RMRP | miR-206 | Gastric cancer | Oncogene | 27192121 |
| SPRY4-IT1 | miR-101-3p | Bladder cancer | Oncogene | 27998761 |
| TUG1 | miR-145 | Bladder cancer | Oncogene | 26318860 |
| TUG1 | miR-299 | Glioblastoma | Oncogene | 27345398 |
| TUG1 | miR-300 | Gallbladder carcinoma | Oncogene | 28178615 |
| TUG1 | miR-9-5p | Osteosarcoma | Oncogene | 27658774 |
| TUSC7 | miR-10a | Hepatocellular carcinoma | Tumor suppressor | 27002617 |
| TUSC7 | miR-211 | Colon cancer | Tumor suppressor | 23558749 |
| TUSC7 | miR-23b, miR-320d | Gastric cancer | Tumor suppressor | 25765901 |
| UCA1 | miR-143 | Breast cancer | Oncogene | 26439035 |
| UCA1 | miR-16 | Bladder cancer | Oncogene | 26373319 |
| UCA1 | miR-204-5p | Colorectal cancer | Oncogene | 27046651 |
| UCA1 | miR-216b | Hepatocellular carcinoma | Oncogene | 25760077 |
| UCA1 | miR-485-5p | Epithelial ovarian cancer | Oncogene | 26867765 |
| UCA1 | miR-507 | Melanoma | Oncogene | 27389544 |
| XIST | miR-139-5p | Hepatocellular carcinoma | Oncogene | 28231734 |
| XIST | miR-181a | Hepatocellular carcinoma | Tumor suppressor | 28388883 |
| XIST | miR-34a-5p | Nasopharyngeal carcinoma | Oncogene | 27461945 |
| XIST | miR-92b | Hepatocellular carcinoma | Tumor suppressor | 27100897 |
This table reports for each lncRNA: (1) miRNA sponged; (2) tumor where such interaction was reported; (3) its function in cancer (oncogene or tumor suppressor); and (4) bibliographic reference reported as Pubmed ID (PMID).
EWSAT1 (Ewing sarcoma-associated transcript 1) is an lncRNA with oncogenic functions in Ewing's sarcoma and nasopharyngeal carcinoma (NPC). EWSAT1 has two MREs for the miR-326/330-5p cluster and promoted the development and progression of tumors functioning as a ceRNA for these miRNAs, which in turn induced the expression of Cyclin D1, target of miRNAs from the miR-326/330-5p cluster [109].
Xia et al. showed that both lncRNA FER1L4 (FER-1-like family member 4, pseudogene) and PTEN (phosphatase and tensin homolog) mRNA had binding sites for oncomiR miR-106a-5p and were downregulated in gastric cancer [110]. As FER1L4 behaved as a ceRNA for miR-106a-5p, FER1L4 downregulation released miR-106a-5p that targeted PTEN mRNA, reducing its expression. Dysregulation of FER1L4-miR-106a-5p-PTEN axis increased cell proliferation by promoting the G0/G1 to S phase transition [110].
FTH1P3 (ferritin heavy chain 1 pseudogene 3) has been shown to function as a molecular sponge for miR-224-5p in oral squamous cell carcinoma (OSCC) [111]. Overexpression of FTH1P3 promoted proliferation and colony formation in OSCC cells and the upregulation of FZD5 (frizzled class receptor 5), target of miR-224-5p and an oncogene involved in activation of Wnt/β-catenin signaling.
It has been demonstrated that lncRNA GAS5 acts as a tumor suppressor in NSCLC by targeting and suppressing miR-135b [112]. GAS5 is downregulated in NSCLC and its expression is inversely correlated to that of miR-135b. After exposure to irradiation, expression of GAS5 and miR-135b was altered, as GAS5 was overexpressed whereas miR-135b was downregulated. Ectopic overexpression of GAS5 led to miR-135b downregulation, repression of cell proliferation, invasion, and improved radiosensitivity [112].
High expression of lncRNA H19 in breast cancer stem cells (BCSCs) is functionally critical for stemness maintenance [113]. In these cells, H19 functions as a molecular sponge for let-7a/b, leading to upregulation of pluripotency factor LIN28, a let-7 target that is highly abundant in BCSCs. Intriguingly, H19 is reciprocally repressed by its targets let-7a/b, but this negative feedback loop can be interfered with by LIN28 because of its ability to inhibit let-7a/b expression [113]. Let-7b expression is also buffered by lncRNA HOST2 (human ovarian cancer-specific transcript 2) in ovarian cancer cells. By binding to let-7b, HOST2 negatively regulates its availability and induces the expression of its oncogenic targets that enhance cell growth and motility in ovarian cancer [114].
Let-7 decoy by lncRNAs was also reported by Deng et al. Upregulation of lncRNA CCAT1 (colon cancer associated transcript 1) in HCC tissues was associated with increased cell proliferation and migration [115]; these oncogenic activities were mediated by its molecular sponge function for let-7: inhibition of let-7 caused upregulated expression of let-7 targets: HMGA2 (high mobility group AT-hook 2) and MYC (MYC proto-oncogene, bHLH transcription factor). Interestingly, other studies reported that MYC, by binding to CCAT1 promoter, induces CCAT1 transcription in colon cancer and gastric carcinoma [116, 117], suggesting the existence of a positive feedback loop between CCAT1 and MYC mediated by let-7 decoy.
Recent works reported the inhibitory effect of HOTAIR on miRNAs functions in different neoplasias. Su et al. found that HOTAIR was highly expressed in HCC tissues and promoted HCC cell proliferation and progression of tumor xenografts [118]. These oncogenic effects were partially due to HOTAIR ability of repressing miR-1 expression. Moreover, also miR-1 was able to negatively regulate HOTAIR expression, thus generating a reciprocal repression feedback loop between these two ncRNAs [118]. Other experimental evidence showed that HOTAIR was capable of binding and downregulating miR-152 in gastric cancer [119]. HOTAIR overexpression in gastric cancer tissues led to decreased expression of miR-152 and to upregulation of its target, HLA-G (human leukocyte antigen G), which in turn facilitated tumor escape mechanisms [119]. Downregulation of miR-152 in gastric cancer could be also caused by PVT1 (plasmacytoma variant translocation 1), an oncogenic lncRNA that acts as a precursor of six miRNAs (i.e., miR-1204, miR-1205, miR-1206, miR-1207-5p, miR-1207-3p, and miR-1208) [120]. Indeed, PVT1 had three MREs for miR-152 and suppressed its expression inducing the upregulation of miR-152 targets (i.e., CD151, FGF2) [121]. Upregulation of PVT1 in gastric cancer was also associated with inhibition of miR-186 function. Indeed, PVT1 bound miR-186 and induced upregulation of HIF-1α (Hypoxia-inducible factor 1-alpha subunit), a target of miR-186 which was related to poor prognosis and invasiveness in gastric cancer [122].
Wang et al. studied in liver cancer the molecular sponge action of lncRNA HULC (highly upregulated in liver cancer). HULC was able to downregulate several miRNAs, including miR-372. Repression of miR-372 enhanced the translation of its target gene, PRKACB (protein kinase cAMP-activated catalytic subunit beta), which in turn promoted phosphorylation of protein CREB1 (cAMP responsive element-binding protein-1) and affected deacetylation and methylation of histones [123]. This process resulted in alterations of chromatin organization and increased expression of HULC, thus showing that HULC was involved in an autoregulatory loop that mantained its abundant expression in liver cancer [123].
Jin et al. reported an association between MALAT1 upregulation and tumor growth and metastasis in triple-negative breast cancer (TNBC) tissues [124]. These tumorigenic properties of MALAT1 were mediated by its ability to decoy miR-1 and, consequently, increase the expression of miR-1 target, SNAI2 (snail family transcriptional repressor 2), also named Slug, an oncogene involved in regulation of cancer cell invasion. Moreover, overexpression of miR-1 was able to reduce MALAT1 expression, demonstrating a reciprocal negative loop between lncRNA and miRNA [124]. The miRNA sponge function of MALAT1 was also reported for cervical cancer [125]. Indeed, MALAT1 levels were found to be more abundant in radio-resistant than in radio-sensitive cancers. Moreover, expression of MALAT1 and of its potential binding partner, miR-145, reverted in response to irradiation. The authors demonstrated that there was a reciprocal repression between MALAT1 and miR-145, which regulated the molecular mechanisms of radio-resistance of cervical cancer [125].
Notably, tumor suppressor miR-145 was frequently reported to be buffered by lncRNAs in cancer models. MiR-145 negatively regulated cell invasion in TNBC, and its downregulation was related to overexpression of lincRNA-RoR (long intergenic ncRNA Regulator of Reprogramming), which acted as competitive endogenous RNA for miR-145 [126]. LincRNA-RoR-mediated downregulation of miR-145 led to upregulation of ARF6 (ADP-ribosylation factor 6), which is strongly involved in metastatic processes; indeed, ARF6 affected E-cadherin localization and impaired cell-cell adhesion, promoting cell invasion in TNBCs [126]. Zhou et al. reported a further effective interaction between lincRNA-RoR and miR-145 in endometrial cancer. Linc-RoR functioned as a miR-145 sponge by repressing the miRNA-mediated degradation of core stem cell transcription factors (i.e., Nanog, Oct4, and Sox2), thereby maintaining the pluripotency of endometrial cancer stem cells [127].
Decoying of miR-145 was performed also by TUG1 (taurine upregulated 1), which is a well-known oncogenic lncRNA, frequently upregulated in cancer and functionally related to several aggressive features of tumors. In bladder cancer, TUG1 decreased the expression of miR-145 and caused upregulation of ZEB2, miR-145 target, promoting EMT, and increasing the metastatic proneness of bladder cancer cells [128]. The ceRNA role of TUG1 was also proved in other tumors. Overexpression of TUG1 was involved in glioblastoma angiogenesis by modulation of endothelial cell proliferation, migration, and tube formation. These cellular processes were mediated by TUG1 interaction with miR-299, which was downregulated in glioblastoma. In fact, knockdown of TUG1-induced upregulation of miR-299 and concomitant decrease of VEGFA (vascular endothelial growth factor A), target of miR-299. These molecular events resulted in a reduced tumor microvessel density in xenograft glioblastoma models [129]. Ma et al. showed that upregulation of TUG1 in gallbladder carcinoma (GBC) was related to GBC cell proliferation and metastasis, and such oncogenic activities were, at least partly, due to the sponge activity of TUG1 that bound miR-300 and negatively regulated its expression [130]. In osteosarcoma, TUG1 acted as a ceRNA by sponging miR-9-5p, inducing the upregulation of transcription factor POU2F1 (POU class 2 homeobox 1) [131]. POU2F1 is frequently upregulated in osteosarcoma and is involved in cell proliferation, differentiation and immune and inflammatory processes. Because POU2F1 is a target of miR-9-5p, silencing of TUG1-inhibited cell proliferation and colony formation, while inducing G0/G1 cell cycle arrest and apoptosis. These cellular processes were mediated by upregulation of miR-9-5p and repression of POU2F1 expression [131].
The tumor suppressor TUSC7 (tumor suppressor candidate 7; also named LOC285194) is an lncRNA transcriptionally induced by TP53 (tumor protein 53); it was initially discovered as depleted in osteosarcoma, inducing abnormal proliferation of osteoblasts, and associated with poor survival of osteosarcoma patients. Competitive endogenous binding between TUSC7 and onco-miRNAs has been frequently reported as associated with cancer-related processes. Wang et al. studied the biopathological meaning of strong downregulation of TUSC7 in HCC [132]. They found that ectopic expression of TUSC7 inhibited cell metastasis, invasion, and EMT, by functioning as a competitive sponge for miR-10a. Moreover, this miRNA was able to promote the EMT process in HCC through directly binding and repressing EPHA4 (EPH tyrosine kinase receptor A4) [132]. Moreover, exon 4 of TUSC7 harbors two binding sites for miR-211 [133]. In colon cancer, miR-211 enhanced cell growth, but this effect was reverted by enforced expression of TUSC7, which buffered the activity of miR-211 [133]. The tumor suppressor role of TUSC7 was also demonstrated in gastric cancer. TUSC7, downregulated in gastric cancer, was an independent prognostic marker of disease-free survival in patients, and its ectopic expression suppressed cancer cell growth both in in vitro and in vivo models, in part by negatively regulating the expression of miR-23 [134].
Unquestionably, one of the most iconic lncRNA acting as miRNA sponge is UCA1, which was reported to bind and repress several miRNAs in multiple tumors. UCA1 binding to miR-143 was proved in breast cancer, where UCA1 was able to modulate cell growth and apoptosis by downregulating miR-143: this in turn led to upregulation of BCL2 (BCL2, apoptosis regulator) and ERBB3 (erb-b2 receptor tyrosine kinase 3) [135]. The role of UCA1 in bladder cancer was associated with ROS (reactive oxygen species) metabolism [136]. Silencing of UCA1 decreased ROS production and promoted mitochondrial glutaminolysis in bladder cancer cells. In these cells, UCA1 acted as a ceRNA by sponging and downregulating miR-16. This induced the upregulation of GLS2 (Glutaminase 2), one of the miR-16 targets, which enhanced glutamine uptake and the rate of glutaminolysis, which is known to increase in cancer cells. UCA1-induced GLS2 maintained the redox balance and protected cancer cells by reducing excessive ROS production [136]. Oncogenic activity of UCA1 in CRC was the result of its decoy function for miR-204-5p, a critical tumor-suppressive miRNA [137]. UCA1, upregulated in CRC, inhibited miR-204-5p activity, thus promoting the upregulation of miRNA targets CREB1, BCL2, and RAB22A (RAB22A, member RAS oncogene) and regulating cell proliferation and apoptosis [137]. UCA1 upregulation in HCC was associated to cell growth and metastasis; these processes were induced by UCA1 binding to miR-216b and resulted in miR-216b downregulation [138]. Decreased levels of miR-216b led to the derepression of its target FGFR1 (fibroblast growth factor receptor 1) and the activation of ERK pathway [138]. Association between UCA1 and metastatic process was also reported for epithelial ovarian cancer [139]. In fact, UCA1 promoted the expression of MMP14 (matrix metallopeptidase14), a key protein involved in cell invasion, by working as a molecular sponge of miR-485-5p, a miRNA targeting MMP14 [139]. FOXM1 (forkhead box protein M1) is a transcription factor critical for G2/M-phase transition and DNA damage response, and it is also a target of miR-507. UCA1-mediated regulation of FOXM1 was discovered, in melanoma cells, to be based on the ceRNA function of UCA1 for miR-507, resulting in an increased malignant ability of these cells [140].
Finally, a ceRNA role in cancer was also reported for XIST (X-inactivate specific transcript). XIST was the first lncRNA to be functionally characterized, and it is considered the major effector of the X inactivation process during development in female mammals [141]. Its dysregulation was found in several tumors (e.g., breast cancer, glioblastoma, and hepatocellular carcinoma), suggesting that XIST could have a potential diagnostic power in cancer [142–144]. In vitro downregulation or upregulation of XIST was associated with altered cell proliferation, metastasis, and apoptosis in several cancer models. Song et al. discovered that XIST overexpression was related to metastasis and poor prognosis of NPC patients [145]. XIST induced the upregulation of E2F3 (E2F transcription factor 3), which is a critical protein for tumor cell proliferation. The authors demonstrated that XIST-promoted activation of E2F3 was caused by the competitive sponge role of XIST for miR-34a-5p (a well-known tumor suppressor miRNA), which targets E2F3 [145]. On the other hand, Chang et al. showed that XIST acts as tumor suppressor and inhibits metastatization and progression in HCC by binding miR-181a and reducing its availability; XIST induces PTEN upregulation, thus decreasing cell proliferation, invasion, and migration [146].
2.3. circRNAs as miRNA Sponges
circRNAs are considered new potential players among ceRNAs: they may harbor shared MREs and compete for miRNA binding with mRNAs [69]. Indeed, circRNAs competitively suppress the activity of miRNAs by adsorbing and sequestering them. As miRNAs are strongly involved in nearly all aspects of cellular physiology and perform pivotal roles in initiation and progression of cancer, circRNAs could reasonably be considered as a new class of RNA molecules closely associated with regulation of proliferation, differentiation, and metastatic processes (Table 3).
Table 3.
circRNAs acting as miRNA sponges.
| circRNA | miRNA target | tumor | circRNA role | PMID |
|---|---|---|---|---|
| circRNA_0005075 | miR-23b-5p, miR-93-3p, miR-581, miR-23a-5p | Hepatocellular carcinoma | Oncogene | 27258521 |
| circRNA_001569 | miR-145 | Colorectal cancer | Oncogene | 27058418 |
| circRNA_100290 | miR-29 family | Oral cancer | Oncogene | 28368401 |
| Cdr1as | miR-7 | Hepatocellular carcinoma | Oncogene | 27391479 |
| cir-ITCH | miR-7, miR-20a | Colorectal cancer | Tumor suppressor | 26110611 |
| cir-ITCH | miR-7, miR-17, miR-214 | Esophageal squamous cell carcinoma | Tumor suppressor | 25749389 |
| ciR-SRY | miR-138 | Cholangiocarcinoma | Oncogene | 27671698, 23446431 |
| cir-TTBK2 | miR-217 | Glioma | Oncogene | 28219405 |
This table reports for each circRNA: (1) miRNAs sponged; (2) tumor where such interaction was reported; (3) its function in cancer (oncogene or tumor suppressor); and (4) bibliographic reference reported as Pubmed ID (PMID).
Zheng et al. reported that circ-TTBK2 (tau tubulin Kinase 2) is significantly upregulated in glioma tissues and cell lines, differently from its linear counterpart [147]. Overexpression of circ-TTBK2 is associated with increased cell proliferation rate, invasion, and decreased apoptosis. Circ-TTBK2 harbors MREs for miR-217, which has a tumor-suppressive role in glioma cells. In fact, circ-TTBK2 and miR-217 interact with each other in an AGO2-dependent manner and upregulation of circ-TTBK2 induced the malignant behavior of glioma cells via downregulation of miR-217. Thus, HNF1β (HNF1 homeobox B), a direct target of miR-217, was derepressed and bound to the promoter of Derlin-1 increasing its expression. Finally, Derlin-1 was able to promote cell proliferation, migration, and invasion and inhibit apoptosis of glioma cells by activating PI3K/AKT and ERK pathways. Moreover, restoration of miR-217 expression reversed the circ-TTBK2-induced promotion of cancer progression, suggesting a reciprocal negative feedback between circ-TTBK2 and miR-217 [147].
MiR-145 is a well-known tumor suppressor miRNA in CRC targeting the oncogenes ERK5 (mitogen-activated protein kinase 7) and IRS1 (insulin receptor substrate 1); furthermore, its ability to predict survival of CRC patients was also shown. In a study by Xie et al., it was demonstrated that downregulation of miR-145 in CRC was mechanistically explained by the role of circ_001569 acting as a miRNA sponge to directly inhibit miR-145 action [148]. Circ_001569 was found to be upregulated in CRC tissues and correlated with progression and aggressiveness of the disease. Notably, circ_001569 did not directly affect miR-145 expression, but through a sponge mechanism it inhibited its posttranscriptional activity; accordingly, it upregulated its targets E2F5 (E2F transcription factor 5), BAG4 (BCL2-associated athanogene 4), and FMNL2 (formin-like 2), which were responsible for cell proliferation and invasion promotion by circ_001569 [148].
Further work on CRC, investigating the role of cir-ITCH on the biopathology of this cancer, found a potential interaction between cir-ITCH and either miR-7 or miR-20a [149]. Cir-ITCH was downregulated in CRC tissues and its ectopic expression led to decreased cell proliferation. This cellular effect was due to cir-ITCH sponge activity for miR-7 and miR-20a; both can bind the 3′-UTR of ITCH (Itchy E3 Ubiquitin Protein Ligase), which is the linear isoform of cir-ITCH. Cir-ITCH-induced upregulation of ITCH promoted the ubiquitination and degradation of phosphorylated DVL2 (dishevelled segment polarity protein 2) and, accordingly, inhibited the Wnt/β-catenin pathway, by repressing the expression of MYC and CCND1 (cyclin D1) [149]. Interestingly, other authors found very similar findings in ESCC: cir-ITCH worked as a miRNA sponge for miR-7, miR-17, and miR-214, increased ITCH expression, and promoted ubiquitin-mediated DVL2 degradation, thus inhibiting canonical Wnt signaling [150].
Besides the cir-ITCH-induced decoy function for miR-7 described above, sponging of miR-7 by CDR1AS was one of the earliest and the most studied ceRNA mechanisms in ncRNA biology, which is also related to cancer. Expression of CDR1AS was found to be elevated in HCC tissues and inversely correlated to miR-7 expression, which was poorly expressed in the same samples [151]. Despite the oncogenic role of miR-7 (previously reported for CRC and ESCC), this miRNA exhibited tumor-suppressive properties in HCC. CDR1AS has sixty-three MREs for miR-7 and strongly suppresses its activity. Knockdown of CDR1AS promoted the expression of miR-7 and suppressed its targets, CCNE1 (cyclin E1) and PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta): this molecular cascade resulted in a reduction of cell proliferation and invasion in HCC [151].
By expression profiling in OSCC, Chen et al. identified the upregulation of a circRNA named circRNA_100290, which was functionally related to abnormal control of cell cycle and cellular proliferation in OSCC cells [152]. circRNA_100290 worked as a miRNA sponge for several members of the miR-29 family, decreasing the quantity of available miR-29s and, accordingly, promoting translation of one of their targets, CDK6 (cyclin-dependent kinase 6), which in turn could induce transition from G1 to S phase in cancer [152].
The first circular transcript identified was Sry circRNA: its encoding gene maps to the sex-determining region of human Y chromosome and was discovered as highly expressed in adult mouse testis [153]. Initially, Sry circRNA was considered an artifact of aberrant RNA splicing and no specific function was attributed to it. The role of Sry circRNA has recently begun to be investigated. Sry circRNA harbors sixteen putative target sites for miR-138 and its function as a miR-138 sponge was demonstrated by Hansen et al. [55]. Currently, no experimental evidence of Sry circRNA-miR-138 axis dysregulation has been reported in cancer; however, as reviewed by Zhao and Shen, miR-138 could target different cancer-related transcripts [154]. For instance, downregulation of miR-138 promoted the malignant progression in cholangiocarcinoma by its target RhoC (ras homolog gene family, member C) [155]. These observations could suggest that the role of competitive endogenous binding between Sry circRNA and miR-138 would be worthy of in-depth analysis in cancer phenotypes.
3. Noncoding RNA Network: Future Perspectives for New Therapeutic Approaches
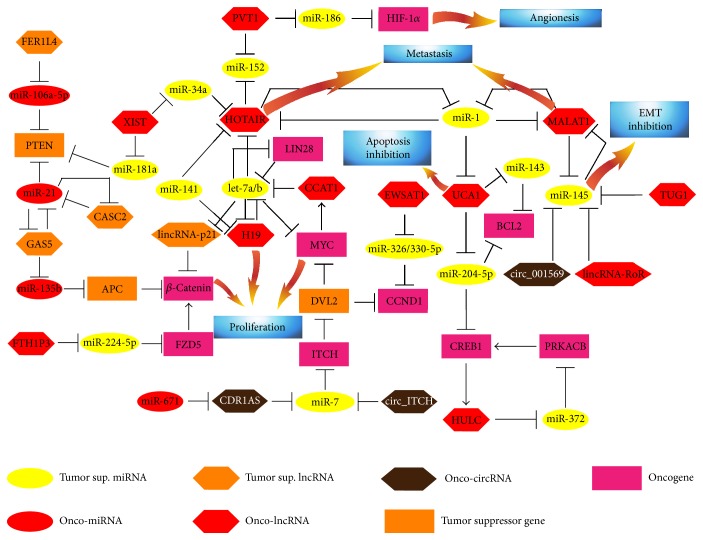
The existence of a complex RNA-based regulatory signaling, which controls cancer-related pathways, is evident from the experimental evidence collected to date. Such a partially hidden network of RNA-RNA interactions pervades and defines the correct functioning of canonical protein-coding pathways, classically involved in proliferation, differentiation, and invasion in cancer (Figure 1()). The complexity of this noncoding landscape is dramatically expanded by the presence of several positive and negative regulatory loops: these make RNA signaling very robust and persistent, though complex and hard to functionally unveil. From a network biology point of view, it is possible to identify some ncRNA hubs that are a crossroad among different RNA-based circuits; accordingly, they represent a keystone of network structural integrity. For instance, the tumor suppressor miR-1 could repress and be sponged by the three most potent oncogenic lncRNAs, HOTAIR, UCA1, and MALAT1, which, in turn, could inhibit dozens of miRNAs with tumor-suppressive properties [76, 118, 124]. The signaling passing through let-7a/b appears extremely complex and pronged. Let-7a/b could be considered a crossroad of multiple interplays among cancer-related ncRNAs: let-7a/b and MYC are reciprocally negatively regulated through lncRNA CCAT1 [115], but MYC expression could be indirectly impaired by miR-7, which, in turn, is buffered by different circRNAs [156]. Moreover, let-7a/b could indirectly suppress the β-catenin pathway, which in a different way could be activated by lncRNA FTHIP3 [111], but also is regulated by molecular axis miR-21-GAS5-miR-135b [112]. This unexpected crosstalking between ncRNA signaling could shed a light on expression relationships among ncRNAs and mRNAs, which have been frequently reported in cancer literature, but to date have not been satisfactorily explained [157–159]. This scenario is made more complex by the tissue-specific expression pattern of all ncRNAs, which could effectively influence the occurrence of specific interactions among ncRNAs. In other words, specific and effective functional interplays among ncRNAs in a particular biological system could occur only if RNA molecules, binding each other, are present at appropriate concentrations. Effectiveness of Ago binding to miRNAs and their targets is dependent on the relative concentration of the miRNA and its target pool [160, 161]. Effective Ago binding occurs when the miRNA : target ratio is close to one but rises dramatically with increasing miRNA : target ratios [162]. Only the most abundant miRNAs show detectable activity, while poorly expressed miRNAs (<100 copies per cell) possess exiguous regulatory properties [163]. However, functional binding between a miRNA and its target can be perturbed by overexpression of other RNAs with multiple shared MREs (e.g., other mRNAs, lncRNAs, and circRNAs) [164]. Such competition among different RNA molecules occurs in a threshold-like manner [165]. Mathematical models predict ceRNA functional effects when miRNA and target levels are near equimolar [166]. However, when the target pool exceeds the threshold set by the buffering miRNA concentration plus the equilibrium dissociation constant (KD) of the miRNA : target interaction, smaller changes in target (i.e., ceRNA) concentration could result in remarkable changes in the concentration of free unrepressed targets [165, 167]. In fact, poorly expressed miRNAs appear to be more susceptible to ceRNA control than more abundant miRNAs. This phenomenon could explain why in in vitro experiments a specific miRNA, when ectopically overexpressed, degraded its lncRNA target, but at the same time the enforced upregulation of lncRNA suppressed miRNA activity (e.g., miR-1/MALAT1, miR-21/GAS5) [104, 124]. Taken together, these considerations strongly suggest that miRNA functionality and the switch to ceRNA-promoted repression of miRNAs would be based on the stoichiometric equilibrium among miRNAs and ceRNAs. Based on these observations, physiological ceRNA expression changes could not affect highly expressed miRNAs; however, the relationship between cellular abundance of RNAs and effectiveness of competitive endogenous interactions remains to be fully unveiled in pathological models, in which strong dysregulation of specific ceRNAs could be present [162, 166, 168].
Figure 1.
Network of noncoding crosstalking in cancer. Molecular interplay among ncRNAs (i.e., miRNAs, lncRNAs, and circRNAs) in cancer. RNA-RNA interactions were retrieved from papers cited in this review. Lines with arrowheads represent expression activation, those with bars represent expression inhibition.
In spite of unclear stoichiometric relationships among ncRNAs in cancer, multiple experimental evidence shows that in vitro and in vivo modulation of ncRNAs strongly impair aggressive properties of cancer cells. The emerging role of ncRNAs as key regulators of cancer-related signaling makes them very attractive and promising targets for novel, potentially groundbreaking therapeutic approaches. RNA-based therapeutics has several advantages compared to other strategies. RNAs are molecules more druggable than proteins, because their targeting is mainly based on nucleic acid complementarity; therefore, an RNA-based drug would be quite easy to design and inexpensive to synthesize (i.e., ASOs, ribozymes, and aptamers) [14]. It is worth stressing that the development of RNA therapeutic strategies has to challenge the redundancy and complexity of the multiple regulatory loops, present in the ncRNA network. It would be quite naive to hypothesize to slow down in vivo tumor progression by targeting a single ncRNA molecule: this would be very hard also for protein-based drugs. This axiom should lead researchers to develop multitargeted RNA therapies to improve their impact on oncogenic signaling. In theory, the β-catenin pathway, frequently hyperactivated in cancers, could be effectively attenuated by simultaneous silencing of miR-21, miR-135b, and FTH1P3 together with restoring physiologic levels of GAS5, CASC2, and miR-224-5p. Furthermore, simultaneous repression of HOTAIR, MALAT1, and UCA1 with reactivation of miR-1 would result in a pleiotropic favorable effect on different cancer-related processes, such as cancer growth, metastatic behavior, and cell death. Such a synergic approach based on simultaneous administration of miRNA mimics and siRNAs against ncRNAs in in vitro and in vivo models has already provided encouraging results. Ideally, such therapeutic approaches would be greatly improved by innovative knockout technologies (such as CRISPR/CAS9), which would avoid potential saturation of RISC complexes, typically occurring by using siRNAs or miRNA mimics [169]. The main issue related to ncRNA therapeutics is to develop efficient delivery systems, which should be able to maintain RNA stability in the circulation and guarantee an effective tissue-specific uptake, as well as minimize off-target side effects. Rapid progress in drug delivery technologies has provided promising chemical and nanotechnological resources well adaptable to RNA therapeutics: chemical modifications of antisense molecules (e.g., steroids and cholesterol) [170], adenoviral vectors [171], cationic liposomes [172], and polymer-based nanoparticles [173]. Recently, an exosomal-based miRNA delivery system has been developed. Such a system appears to be very promising because exosomes are less toxic and better tolerated by the organism and naturally protect their molecular cargo in the blood [174].
4. Conclusions
A better knowledge on the complex interplay among ncRNAs, together with the development of selective methods for RNA delivery to cancer cells, will provide great benefits for cancer treatment. Needless to say, researchers will have to overcome many technical challenges to develop effective RNA-based anticancer strategies realistically applicable to patients. Before ncRNA targeting is pervasively applied in clinical settings, it will be indispensable to organize large collaborative efforts between research institutes and industry to fully realize the clinical potential of this very promising approach.
Acknowledgments
The authors wish to thank the Scientific Bureau of the University of Catania for language support.
Conflicts of Interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Nowak R. Mining treasures from ‘junk DNA’. Science. 1994;263(5147):608–610. doi: 10.1126/science.7508142. [DOI] [PubMed] [Google Scholar]
- 2.Lander E. S., Linton L. M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Venter J. C., Adams M. D., Myers E. W., et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 5.Clamp M., Fry B., Kamal M., et al. Distinguishing protein-coding and noncoding genes in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(49):19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezkurdia I., Juan D., Rodriguez J. M., et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Human Molecular Genetics. 2014;23(22):5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ENCODE Project Consortium, Birney E., Stamatoyannopoulos J. A., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein B. E., Stamatoyannopoulos J. A., Costello J. F., et al. The NIH roadmap epigenomics mapping consortium. Nature Biotechnology. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sana J., Faltejskova P., Svoboda M., Slaby O. Novel classes of non-coding RNAs and cancer. Journal of Translational Medicine. 2012;10:p. 103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T., Alvarez A., Hu B., Cheng S. Y. Noncoding RNAs in cancer and cancer stem cells. Chinese Journal of Cancer. 2013;32(11):582–593. doi: 10.5732/cjc.013.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatima R., Akhade V. S., Pal D., Rao S. M. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Molecular and Cellular Therapies. 2015;3(1):p. 5. doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prensner J. R., Chinnaiyan A. M. The emergence of lncRNAs in cancer biology. Cancer Discovery. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature Reviews Molecular Cell Biology. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragusa M., Barbagallo C., Statello L., et al. Non-coding landscapes of colorectal cancer. World Journal of Gastroenterology. 2015;21(41):11709–11739. doi: 10.3748/wjg.v21.i41.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilczynska A., Bushell M. The complexity of miRNA-mediated repression. Cell Death and Differentiation. 2015;22(1):22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee R. C., Feinbaum R. L., Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 18.Ameres S. L., Zamore P. D. Diversifying microRNA sequence and function. Nature Reviews Molecular Cell Biology. 2013;14(8):475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 19.Valinezhad Orang A., Safaralizadeh R., Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. International Journal of Genomics. 2014;2014:15. doi: 10.1155/2014/970607.970607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carthew R. W., Sontheimer E. J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macfarlane L. A., Murphy P. R. MicroRNA: biogenesis, function and role in cancer. Current Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croce C. M. Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volinia S., Calin G. A., Liu C. G., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jay C., Nemunaitis J., Chen P., Fulgham P., Tong A. W. miRNA profiling for diagnosis and prognosis of human cancer. DNA and Cell Biology. 2007;26(5):293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 27.Ragusa M., Statello L., Maugeri M., et al. Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following cetuximab treatment: biomolecular, genetic and translational implications. Oncoscience. 2014;1(2):132–157. doi: 10.18632/oncoscience.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma R., Jiang T., Kang X. Circulating microRNAs in cancer: origin, function and application. Journal of Experimental & Clinical Cancer Research. 2012;31(1):p. 38. doi: 10.1186/1756-9966-31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Advanced Drug Delivery Reviews. 2015;81:75–93. doi: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Ragusa M., Barbagallo C., Statello L., et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications. Cancer Biology & Therapy. 2015;16(9):1387–1396. doi: 10.1080/15384047.2015.1046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volders P. J., Helsens K., Wang X., et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Research. 2013;41(Database issue):D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie L., Wu H. J., Hsu J. M., et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. American Journal of Translational Research. 2012;4(2):127–150. [PMC free article] [PubMed] [Google Scholar]
- 33.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 34.Li L., Liu B., Wapinski O. L., et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Reports. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engreitz J. M., Ollikainen N., Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nature Reviews Molecular Cell Biology. 2016;17(12):756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 36.Beltran M., Puig I., Pena C., et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes & Development. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T., Huang Y., Chen J., et al. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1AS expression. Molecular Medicine Reports. 2014;10(3):1275–1281. doi: 10.3892/mmr.2014.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosa A., Ballarino M. Long noncoding RNA regulation of pluripotency. Stem Cells International. 2016;2016:9. doi: 10.1155/2016/1797692.1797692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews. Genetics. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 40.Kitagawa M., Kitagawa K., Kotake Y., Niida H., Ohhata T. Cell cycle regulation by long non-coding RNAs. Cellular and Molecular Life Sciences. 2013;70(24):4785–4794. doi: 10.1007/s00018-013-1423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su Y., Wu H., Pavlosky A., et al. Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death & Disease. 2016;7(8, article e2333) doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P., Cao P., Zhu X., et al. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget. 2017;8(20):33137–33143. doi: 10.18632/oncotarget.16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deguchi S., Katsushima K., Hatanaka A., et al. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene. 2017;36(32):4629–4640. doi: 10.1038/onc.2017.88. [DOI] [PubMed] [Google Scholar]
- 44.Yun-Bo F., Xiao-Po L., Xiao-Li L., Guo-Long C., Pei Z., Fa-Ming T. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Medicine. 2016;11(1):163–167. doi: 10.1515/med-2016-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian J., Hu X., Gao W., et al. Identification of the long noncoding RNA LET as a novel tumor suppressor in gastric cancer. Molecular Medicine Reports. 2017;15(4):2229–2234. doi: 10.3892/mmr.2017.6263. [DOI] [PubMed] [Google Scholar]
- 46.Luo G., Liu D., Huang C., et al. LncRNA GAS5 inhibits cellular proliferation by targeting P27Kip1. Molecular Cancer Research. 2017;15(7):789–799. doi: 10.1158/1541-7786.MCR-16-0331. [DOI] [PubMed] [Google Scholar]
- 47.Li Z., Jin C., Chen S., et al. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Molecular and Cellular Biochemistry. 2017;433(1-2):51–60. doi: 10.1007/s11010-017-3015-z. [DOI] [PubMed] [Google Scholar]
- 48.Liu J., Liu T., Wang X., He A. Circles reshaping the RNA world: from waste to treasure. Molecular Cancer. 2017;16(1):p. 58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. Mis-splicing yields circular RNA molecules. The FASEB Journal. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 51.Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2, article e30733) doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeck W. R., Sorrentino J. A., Wang K., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen I., Chen C. Y., Chuang T. J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdisciplinary Reviews RNA. 2015;6(5):563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen T. B., Jensen T. I., Clausen B. H., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 56.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo J. U., Agarwal V., Guo H., Bartel D. P. Expanded identification and characterization of mammalian circular RNAs. Genome Biology. 2014;15(7):p. 409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enuka Y., Lauriola M., Feldman M. E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Research. 2016;44(3):1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen C. Y., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 60.Pamudurti N. R., Bartok O., Jens M., et al. Translation of CircRNAs. Molecular Cell. 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang M., Zhong Z., Lv M., Shu J., Tian Q., Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7(30):47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du W. W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H., Hao X., Wang H., et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cellular Physiology and Biochemistry. 2016;40(6):1334–1344. doi: 10.1159/000453186. [DOI] [PubMed] [Google Scholar]
- 64.Bachmayr-Heyda A., Reiner A. T., Auer K., et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific Reports. 2015;5(1):8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dou Y., Cha D. J., Franklin J. L., et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Scientific Reports. 2016;6(1, article 37982) doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sui W., Shi Z., Xue W., et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncology Reports. 2017;37(3):1804–1814. doi: 10.3892/or.2017.5415. [DOI] [PubMed] [Google Scholar]
- 67.Wan L., Zhang L., Fan K., Cheng Z. X., Sun Q. C., Wang J. J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. BioMed Research International. 2016;2016:11. doi: 10.1155/2016/1579490.1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Scientific Reports. 2016;6, article 30919 doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Memczak S., Jens M., Elefsinioti A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 70.Su X., Xing J., Wang Z., Chen L., Cui M., Jiang B. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chinese Journal of Cancer Research. 2013;25(2):235–239. doi: 10.3978/j.issn.1000-9604.2013.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinn J. J., Chang H. Y. Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 72.Xue M., Li X., Wu W., et al. Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein alpha contributes to bladder cancer cell growth and reduced apoptosis. Oncology Reports. 2014;31(5):1993–2000. doi: 10.3892/or.2014.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang Z., Wu L., Wang L., Yang Y., Meng Y., Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology. 2014;117(1):89–95. doi: 10.1016/j.oooo.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 74.Huang J., Zhou N., Watabe K., et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death & Disease. 2014;5, article e1008 doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L., Cao X., Zhang L., Zhang X., Sheng H., Tao K. UCA1 Overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemotherapy and Pharmacology. 2016;77(3):629–634. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 76.Wang T., Yuan J., Feng N., et al. Hsa-miR-1 downregulates long non-coding RNA urothelial cancer associated 1 in bladder cancer. Tumour Biology. 2014;35(10):10075–10084. doi: 10.1007/s13277-014-2321-2. [DOI] [PubMed] [Google Scholar]
- 77.Ji P., Diederichs S., Wang W., et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 78.Huo Y., Li Q., Wang X., et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8(29):46993–47006. doi: 10.18632/oncotarget.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y., Wu Z., Yuan J., et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Letters. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 80.Lei R., Xue M., Zhang L., Lin Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. OncoTargets and Therapy. 2017;10:35–46. doi: 10.2147/OTT.S112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilusz J. E., Freier S. M., Spector D. L. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tripathi V., Ellis J. D., Shen Z., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.West J. A., Davis C. P., Sunwoo H., et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Molecular Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leucci E., Patella F., Waage J., et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Scientific Reports. 2013;3:p. 2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X., Li M., Wang Z., et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. The Journal of Biological Chemistry. 2015;290(7):3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han Y., Liu Y., Zhang H., et al. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long non-coding RNA MALAT1. FEBS Letters. 2013;587(23):3875–3882. doi: 10.1016/j.febslet.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 87.Tsang F. H., Au S. L., Wei L., et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver International. 2015;35(5):1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 88.Gupta R. A., Shah N., Wang K. C., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chiyomaru T., Yamamura S., Fukuhara S., et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One. 2013;8(8, article e70372) doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoon J. H., Abdelmohsen K., Kim J., et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nature Communications. 2013;4:p. 2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoon J. H., Abdelmohsen K., Srikantan S., et al. LincRNA-p21 suppresses target mRNA translation. Molecular Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoon J. H., Jo M. H., White E. J., et al. AUF1 promotes let-7b loading on Argonaute 2. Genes & Development. 2015;29(15):1599–1604. doi: 10.1101/gad.263749.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiyomaru T., Fukuhara S., Saini S., et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. The Journal of Biological Chemistry. 2014;289(18):12550–12565. doi: 10.1074/jbc.M113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park S. M., Gaur A. B., Lengyel E., Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou X., Ye F., Yin C., Zhuang Y., Yue G., Zhang G. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cellular Physiology and Biochemistry. 2015;36(4):1440–1452. doi: 10.1159/000430309. [DOI] [PubMed] [Google Scholar]
- 96.Lustig O., Ariel I., Ilan J., Lev-Lehman E., De-Groot N., Hochberg A. Expression of the imprinted gene H19 in the human fetus. Molecular Reproduction and Development. 1994;38(3):239–246. doi: 10.1002/mrd.1080380302. [DOI] [PubMed] [Google Scholar]
- 97.Li H., Yu B., Li J., et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsang W. P., Ng E. K., Ng S. S., et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 99.Ribas J., Ni X., Castanares M., et al. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Research. 2012;40(14):6821–6833. doi: 10.1093/nar/gks308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vicinus B., Rubie C., Faust S. K., et al. miR-21 functionally interacts with the 3'UTR of chemokine CCL20 and down-regulates CCL20 expression in miR-21 transfected colorectal cancer cells. Cancer Letters. 2012;316(1):105–112. doi: 10.1016/j.canlet.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 101.Wang P., Zou F., Zhang X., et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Research. 2009;69(20):8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Asangani I. A., Rasheed S. A., Nikolova D. A., et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 103.Roy S., Yu Y., Padhye S. B., Sarkar F. H., Majumdar A. P. Difluorinated-curcumin (CDF) restores PTEN expression in colon cancer cells by down-regulating miR-21. PLoS One. 2013;8(7, article e68543) doi: 10.1371/journal.pone.0068543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Z., Zhu Z., Watabe K., et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death and Differentiation. 2013;20(11):1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cao Y., Xu R., Xu X., Zhou Y., Cui L., He X. Downregulation of lncRNA CASC2 by microRNA-21 increases the proliferation and migration of renal cell carcinoma cells. Molecular Medicine Reports. 2016;14(1):1019–1025. doi: 10.3892/mmr.2016.5337. [DOI] [PubMed] [Google Scholar]
- 106.Wang P., Liu Y. H., Yao Y. L., et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cellular Signalling. 2015;27(2):275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 107.Hansen T. B., Wiklund E. D., Bramsen J. B., et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. The EMBO Journal. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barbagallo D., Condorelli A., Ragusa M., et al. Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7(4):4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song P., Yin S. C. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging. 2016;8(11):2948–2960. doi: 10.18632/aging.101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xia T., Chen S., Jiang Z., et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Scientific Reports. 2015;5, article 13445 doi: 10.1038/srep13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang C. Z. Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene. 2017;607:47–55. doi: 10.1016/j.gene.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 112.Xue Y., Ni T., Jiang Y., Li Y. LncRNA GAS5 inhibits tumorigenesis and enhances radiosensitivity by suppressing miR-135b expression in non-small cell lung cancer. Oncology Research. 2017;25(6):1027–1037. doi: 10.3727/096504017X14850182723737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peng F., Li T. T., Wang K. L., et al. H19/Let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death & Disease. 2017;8(1, article e2569) doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gao Y., Meng H., Liu S., et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Human Molecular Genetics. 2015;24(3):841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 115.Deng L., Yang S. B., Xu F. F., Zhang J. H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. Journal of Experimental & Clinical Cancer Research. 2015;34:p. 18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He X., Tan X., Wang X., et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biology. 2014;35(12):12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 117.Yang F., Xue X., Bi J., et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. Journal of Cancer Research and Clinical Oncology. 2013;139(3):437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 118.Su D. N., Wu S. P., Chen H. T., He J. H. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncology Letters. 2016;12(5):4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song B., Guan Z., Liu F., Sun D., Wang K., Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochemical and Biophysical Research Communications. 2015;464(3):807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 120.Beck-Engeser G. B., Lum A. M., Huppi K., Caplen N. J., Wang B. B., Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5(1):4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li T., Meng X. L., Yang W. Q. Long noncoding RNA PVT1 acts as a “sponge” to inhibit microRNA-152 in gastric cancer cells. Digestive Diseases and Sciences. 2017 doi: 10.1007/s10620-017-4508-z. [DOI] [PubMed] [Google Scholar]
- 122.Huang T., Liu H. W., Chen J. Q., et al. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomedicine & Pharmacotherapy. 2017;88:302–308. doi: 10.1016/j.biopha.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 123.Wang J., Liu X., Wu H., et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Research. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jin C., Yan B., Lu Q., Lin Y., Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biology. 2016;37(6):7383–7394. doi: 10.1007/s13277-015-4605-6. [DOI] [PubMed] [Google Scholar]
- 125.Lu H., He Y., Lin L., et al. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biology. 2016;37(2):1683–1691. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 126.Eades G., Wolfson B., Zhang Y., Li Q., Yao Y., Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Molecular Cancer Research. 2015;13(2):330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou X., Gao Q., Wang J., Zhang X., Liu K., Duan Z. Linc-RNA-RoR acts as a “sponge” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecologic Oncology. 2014;133(2):333–339. doi: 10.1016/j.ygyno.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 128.Tan J., Qiu K., Li M., Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Letters. 2015;589(20, Part B):3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 129.Cai H., Liu X., Zheng J., et al. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36(3):318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 130.Ma F., Wang S. H., Cai Q., et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomedicine & Pharmacotherapy. 2017;88:863–869. doi: 10.1016/j.biopha.2017.01.150. [DOI] [PubMed] [Google Scholar]
- 131.Xie C. H., Cao Y. M., Huang Y., et al. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumour Biology. 2016;37(11):15031–15041. doi: 10.1007/s13277-016-5391-5. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y., Liu Z., Yao B., et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biology. 2016;37(8):11429–11441. doi: 10.1007/s13277-016-4892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu Q., Huang J., Zhou N., et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Research. 2013;41(9):4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Qi P., Xu M. D., Shen X. H., et al. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. International Journal of Cancer. 2015;137(6):1269–1278. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 135.Tuo Y. L., Li X. M., Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. European Review for Medical and Pharmacological Sciences. 2015;19(18):3403–3411. [PubMed] [Google Scholar]
- 136.Li H. J., Li X., Pang H., Pan J. J., Xie X. J., Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Japanese Journal of Clinical Oncology. 2015;45(11):1055–1063. doi: 10.1093/jjco/hyv132. [DOI] [PubMed] [Google Scholar]
- 137.Bian Z., Jin L., Zhang J., et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Scientific Reports. 2016;6(1, article 23892) doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang F., Ying H. Q., He B. S., et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang Y., Jiang Y., Wan Y., et al. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biology. 2016;37(8):10633–10641. doi: 10.1007/s13277-016-4917-1. [DOI] [PubMed] [Google Scholar]
- 140.Wei Y., Sun Q., Zhao L., et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Medical Oncology. 2016;33(8):p. 88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 141.Brown C. J., Hendrich B. D., Rupert J. L., et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 142.Schouten P. C., Vollebergh M. A., Opdam M., et al. High XIST and low 53BP1 expression predict poor outcome after high-dose alkylating chemotherapy in patients with a BRCA1-like breast cancer. Molecular Cancer Therapeutics. 2016;15(1):190–198. doi: 10.1158/1535-7163.MCT-15-0470. [DOI] [PubMed] [Google Scholar]
- 143.Yao Y., Ma J., Xue Y., et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Letters. 2015;359(1):75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 144.Zhuang L. K., Yang Y. T., Ma X., et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death & Disease. 2016;7, article e2203 doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Song P., Ye L. F., Zhang C., Peng T., Zhou X. H. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592(1):8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 146.Chang S., Chen B., Wang X., Wu K., Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17(1):p. 248. doi: 10.1186/s12885-017-3216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zheng J., Liu X., Xue Y., et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. Journal of Hematology & Oncology. 2017;10(1):52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie H., Ren X., Xin S., et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. Cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10(6, article e0131225) doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li F., Zhang L., Li W., et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7, article e0158347) doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 152.Chen L., Zhang S., Wu J., et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 153.Capel B., Swain A., Nicolis S., et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 154.Zhao Z. J., Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biology. 2017;14(5):514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Q., Tang H., Yin S., Dong C. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncology Reports. 2013;29(5):2046–2052. doi: 10.3892/or.2013.2304. [DOI] [PubMed] [Google Scholar]
- 156.Hansen T. B., Kjems J., Damgaard C. K. Circular RNA and miR-7 in cancer. Cancer Research. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 157.Zhou X., Xu X., Wang J., Lin J., Chen W. Identifying miRNA/mRNA negative regulation pairs in colorectal cancer. Scientific Reports. 2015;5, article 12995 doi: 10.1038/srep12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zadran S., Remacle F., Levine R. D. miRNA and mRNA cancer signatures determined by analysis of expression levels in large cohorts of patients. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(47):19160–19165. doi: 10.1073/pnas.1316991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Seo J., Jin D., Choi C. H., Lee H. Integration of microRNA, mRNA, and protein expression data for the identification of cancer-related microRNAs. PLoS One. 2017;12(1, article e0168412) doi: 10.1371/journal.pone.0168412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mullokandov G., Baccarini A., Ruzo A., et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nature Methods. 2012;9(8):840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Arvey A., Larsson E., Sander C., Leslie C. S., Marks D. S. Target mRNA abundance dilutes microRNA and siRNA activity. Molecular Systems Biology. 2010;6:p. 363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bosson A. D., Zamudio J. R., Sharp P. A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Molecular Cell. 2014;56(3):347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Brown B. D., Gentner B., Cantore A., et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nature Biotechnology. 2007;25(12):1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 164.Ebert M. S., Neilson J. R., Sharp P. A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mukherji S., Ebert M. S., Zheng G. X., Tsang J. S., Sharp P. A., van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nature Genetics. 2011;43(9):854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ala U., Karreth F. A., Bosia C., et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(18):7154–7159. doi: 10.1073/pnas.1222509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bosia C., Pagnani A., Zecchina R. Modelling competing endogenous RNA networks. PLoS One. 2013;8(6, article e66609) doi: 10.1371/journal.pone.0066609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jens M., Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nature Reviews Genetics. 2015;16(2):113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 169.Jackson A. L., Linsley P. S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature Reviews Drug Discovery. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 170.Lorenz C., Hadwiger P., John M., Vornlocher H. P., Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorganic & Medicinal Chemistry Letters. 2004;14(19):4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 171.Mowa M. B., Crowther C., Arbuthnot P. Therapeutic potential of adenoviral vectors for delivery of expressed RNAi activators. Expert Opinion on Drug Delivery. 2010;7(12):1373–1385. doi: 10.1517/17425247.2010.533655. [DOI] [PubMed] [Google Scholar]
- 172.Mallick S., Choi J. S. Liposomes: versatile and biocompatible nanovesicles for efficient biomolecules delivery. Journal of Nanoscience and Nanotechnology. 2014;14(1):755–765. doi: 10.1166/jnn.2014.9080. [DOI] [PubMed] [Google Scholar]
- 173.Miele E., Spinelli G. P., Miele E., et al. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. International Journal of Nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ha D., Yang N., Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharmaceutica Sinica B. 2016;6(4):287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]