Abstract
Since sperm size and form do not necessarily provide information on internal sperm structures, novel sperm markers need to be found in order to conduct assisted reproductive therapies (ART) successfully. Currently, the priority of andrologists is not only to select those sperm able to fertilize the oocyte, but also a high quality of sperm that will guarantee a healthy embryo. Evidence of this shows us the importance of studying sperm intensively on genetic and epigenetic levels, because these could probably be the cause of a percentage of infertility diagnosed as idiopathic. Thus, more attention is being paid to posttranslational modifications as the key for better understanding of the fertilization process and its impact on embryo and offspring. Advances in the discovery of new sperm markers should go hand in hand with finding appropriate techniques for selecting the healthiest sperm, guaranteeing its non-invasiveness. To date, most sperm selection techniques can be harmful to sperm due to centrifugation or staining procedures. Some methods, such as microfluidic techniques, sperm nanopurifications, and Raman spectroscopy, have the potential to make selection gentle to sperm, tracking small abnormalities undetected by methods currently used. The fact that live cells could be analyzed without harmful effects creates the expectation of using them routinely in ART. In this review, we focus on the combination of sperm epigenetic status (modifications) as quality markers, with non-invasive sperm selection methods as novel approaches to improve ART outcomes.
MeSH Keywords: Andrology; Epigenomics; Microfluidics; Nanoparticles; Reproductive Techniques, Assisted; Spectrum Analysis, Raman
Background
Widespread Assisted Reproductive Therapies (ART), such as in vitro fertilization (IVF) and mainly intracytoplasmic sperm injection (ICSI), have opened up a new perspective on infertility treatment. The successful outcomes of ART are based on selecting the most suitable gametes with which to conduct the process. And, despite the selection of oocytes in the female being limited due to the small number of oocytes with each individual feature, in the case of males, it is possible to perform strict selection to choose the finest spermatozoa with which to improve the outcome of ART.
Sperm heterogeneity in ejaculate is widely recognized. This diversity can be found at different levels and characteristics such as shape, motility, DNA content, or membrane composition [1,2]. Recently it has been shown that sperm are also epigenetically heterogeneous, although few reports have addressed this approach [3–5]. Epigenetic alterations of the genome and associated posttranslational modifications of DNA-binding histones equally affect gamete development, maturation, and embryogenesis. Therefore, posttranslational modification (methylation, acetylation, phosphorylation, ubiquitination, and SUMOylation) and noncoding RNAs have been revealing such key markers of fertilization and embryo development abilities in sperm [6–9]. Their evaluation is paramount to provide relevant information for ART. Furthermore, the study of sperm epigenetic markers is also crucial because it has been suggested that they are involved in the etiology of male idiopathic infertility [10].
On the other hand, it might be advisable to follow up sperm changes derived from the use of ART and its consequences. The application of ART includes stages outside the male and female reproductive tracts, during which spermatozoa are subjected to procedures aimed at maximizing reproductive success [11]. This in vitro environment can affect sperm characteristics and function. It is currently also known that epigenetic changes are more frequent than DNA mutations, and their contributions during the performance of human ART is not well defined. Although many researchers have reported a higher incidence of imprinting disorders in ART offspring [12–14], a clear relationship has not been demonstrated. Recently, it has been reported that epigenetic alterations could rather arise from the process of gametogenesis rather than ART itself [15,16]. Thus, it can therefore be detected in the semen of the father. However, we should not overlook the effect of our manipulation with gametes on the stability of the epigenetic process. Based on these facts, sperm handling should be sensitive and invasiveness should be the last resort.
As mentioned above, the selection of viable sperm is an essential procedure to carry out ART. Thus far, centrifugation over discontinuous density gradient (DG) has remained the most popular method, together with swim-up [17–21]. Both methods select morphologically normal and motile sperm, and, although these spermatozoa are expected to be epigenetically stable, few studies have been carried out in this direction [22]. Furthermore, there is evidence to support the fact that sperm morphology does not always reflect DNA status [23,24]. It is therefore crucial to find alternative sperm selection methods which allow us to isolate sperm based on novel parameters to better monitor sperm function.
For detection of distinct epigenetic markers, staining of the cell is usually required, together with flow cytometry or immunocytochemistry [25]. These approaches are valid for identifying mistakes in posttranslational modifications, but the sperm cannot subsequently be used in ART. Estimation of markers based on the epigenome for evaluation of sperm quality, in combination with harmless procedures that are able to detect them, could be one of the solutions to help infertile couples. In this context, non-invasive and label-free methods, such as microfluidics, nanopurification, and Raman spectroscopy, are promising.
Microfluidics
The female reproductive tract has many functions, including the facilitation of sperm migration to the site of fertilization, as well as the selection of healthy sperm [26]. The mechanisms by which the female genital tract selects spermatozoa still remain unknown, but it selects against spermatozoa containing damaged DNA, acting as a biological sensor to screen spermatozoa. This fact demonstrates the existence of unrecognized processes for detection and interpretation of markers on the sperm surface that link phenotypic and genotypic qualities of each individual sperm [27]. It has also been reported that the oviduct responds to sperm presence by modifying the oviductal environment [28].
It is known that many of these characteristics of the oviductal environment are impossible to reproduce during ART. However, some features, such as unidirectional, laminar or gradient flow, containment in a 3-D physical environment, and changing the chemical composition in the medium, can be obtained in a microfluidic environment [29].
These microfluidic devices use microchannels to sort sperm according to normal morphology, motility, and higher DNA integrity for IVF techniques [30,31]. Important advantages have been shown compared to traditional selection techniques, such as the potential to work with small sperm sample volumes, short processing times, and the ability to manipulate single cells in a non-invasive manner. In addition, the potential to be a versatile tool for selection applications or fundamental studies on sperm has also been shown [24,32]. Furthermore, the yield of the selected sperm by microfluidic technique was estimated at 41%, what is comparable to the recovery rate of currently used conventional methods [33].
Another of the advantages of using a microfluidic device in ART is its one-step sorting protocol without the need to centrifuge. Eliminating the centrifugation step minimizes the exposure of sperm to reactive oxygen species (ROS), preserving the integrity of the chromatin [34]. DNA fragmentation is significantly decreased in treated sperm with the microfluidic sperm sorting system [35,36]. Wang et al. [37] compared the swim-up method with a microfluidic device, resulting in a significantly lower rate of DNA damage (16.4% swim-up vs. 8.4% microfluidic). A recent microfluidic study [30] utilized polycarbonate filter paper with different-sized pores to determine how the different pore sizes affect sperm retrieval, ROS production, and DNA integrity. The device was efficient in selecting motile and morphologically normal sperm. In addition, there was significantly less ROS production and improved sperm retrieval using the microfluidics versus the swim-up method [30]. Another research group created a device that uses a radial array of microchannels to select the most motile sperm, and they were able to obtain 80% improvement in sperm DNA integrity after sorting [38].
Decreasing levels of DNA fragmentation and ROS can prevent epigenetic changes, since it has been observed that, in sperm damaged by oxidative stress, impaired DNA sequence prevents the process of DNA methylation [39]. Other studies have suggested that oxidative stress due to smoking affects protamine protein levels and histone retention rates in mature sperm [40,41]. The fact that the epigenetically aberrant sperm populations were preferentially found in men with severe sperm abnormalities and that non-imprinted genes are also affected, suggests that there might be a link between phenotype and epigenotype [4]. However, sometimes normal morphology is not necessarily associated with DNA integrity [24]. Moreover, epigenetic markers had significant variation between samples from different men, as well as significant variation within the same semen sample. This reinforces that epigenetic patterns may be used as biomarkers for sperm quality [42].
Future studies should investigate creating sperm-sorting devices that can isolate spermatozoa to limit genetic diseases, while also maintaining sperm viability. New sperm sorting techniques have been shown to be able to improve DNA integrity, morphology, and motility, but there are still some conflicts as to whether these are significant improvements over the conventional centrifuged-based techniques [43].
In summary, microfluidic technology could offer a new non-invasive method for selecting sperm with good molecular characteristics to prepare sperm for IVF or ICSI, which would greatly improve the current point-of-care ART. Although newer microfluidic-based sperm sorting methods showed promising results, these devices should be thoroughly analyzed for their clinical utility to continue progress in the fields of microfluidics and andrology.
Sperm Nanopurification
Nanotechnologies and their application have undergone huge expansion in different disciplines, including the medical field. The suggestion exists for the use of nanoparticles in drug delivery, diagnostics, and, in the case of sperm, as a non-invasive sperm selection method [44,45]. While methods such as Raman spectroscopy, described below, can function with any sperm molecules, nanoparticles for sperm selection interact with external molecules of the sperm membrane or acrosome. This could seem to be a disadvantage, but as mentioned before, we are witness to sperm ability reflecting their inner condition on the sperm surface [46,47].
Nanotechnologies function with different nanoparticle materials, but some harmful side effects can be found [48,49]. To date, the most successful method for sperm selection using nanoparticles is magnetic-activated cell sorting (MACS), which separates apoptotic and non-apoptotic spermatozoa [50,51]. However, in recent years, a new method based on magnetic nanoseparation of abnormal sperm has been implemented and was applied in cattle, utilizing ferritin nanoparticles coated with specific antibodies [52]. The sperm nanopurification process is relatively easy to implement. First, a mixture of sperm with ferritin nanoparticles is created and coated with antibodies against specific molecules, especially those which are expressed on the surface of defective sperm [52]. Spermatozoa with these molecules interact with the coated nanoparticles, and a magnetic separator is placed at the bottom of a tube for final nanopurification. Thus, the pellet containing unhealthy sperm attached to nanoparticles is removed. Utilizing this technique for bull semen purification, 25–30% spermatozoa were eliminated and it was possible to obtain double the number of artificial insemination doses per semen collection [47,52]. Success in sperm nanopurification in cattle has been reported, increasing the number of pregnancies after insemination and IVF [52].
As the most suitable antibodies, those against the ligand of lectins PNA (peanut agglutinin from Arachis hypogaea) and PSA (Pisum sativum agglutinin) that are more abundant on the acrosome membrane of defective sperm [46,53] were selected, as well as ubiquitin, which is a small chaperone molecule known mainly from posttranslational modifications called ubiquitination. While monoubiquitination participates mainly in the regulation of gene transcription, cell signalling, and silencing of X chromosomes [54,55], polyubiquitination plays a role in protein turnover by the ubiquitin proteasome system. Ubiquitin as a quality control marker is secreted by epididymal epithelium to eliminate defective spermatozoa by subsequent phagocytosis [56–58]. Nevertheless, some of the defective spermatozoa tagged by extracellular/cell surface ubiquitination are carried over into the ejaculate. This fact supports the utilization of ubiquitin as an appropriate sperm marker [58,59]. The estimation of ubiquitin for sperm selection was confirmed by sperm ubiquitin-tag immunoassay (SUTI). The intensity of ubiquitination in sperm correlated with the fertilization rate and success of ART, such as ICSI and IVF [59–61]. However, SUTI has been combined with flow cytometry or fluorescence microscope requiring sperm staining, which makes subsequent use of these sperm for fertilization in a harmless way unlikely.
The expectation of the application of sperm nanopurification in human reproductive medicine is based on the current information about its non-invasiveness. To date, no harmful side effects have been noted to be caused by using ferritin nanoparticles or the magnetic field, which is a crucial argument for its use in humans [52]. Furthermore, ubiquitin seems to be the proper marker for sperm selection, not only for its accessibility on the sperm membrane, but mainly for its ability to reflect the DNA status [60]. All this information supports the application of sperm nanopurification in connection with ubiquitin in human reproductive medicine and, due to its non-invasiveness, in the treatment of couples who have problems conceiving.
Raman Spectroscopy
Sperm heterogeneity is widely accepted [4,5], and is based on several sperm features, including slight molecular differences which determine the sperm function. Changes in DNA packaging or epigenetic modifications can be important [62–64]. However, these small but important details are difficult to evaluate by standard methods, without disturbing or even destroying the sperm cell.
Raman spectroscopy (RS) is an optical laser-based technique which provides information on the vibrational energies of the biomolecular constituents of the sample [66]. Any changes in structure are translated into distinct Raman spectra from each molecule or tissue, without use of fixatives or labels [66–68]. However, a laser with suitable wavelengths and intensities to guarantee its invasiveness is required [68]. The combination of Raman spectroscopy with confocal microscopy, called Raman microspectroscopy, allows us to obtain 3D spatial resolution and makes possible investigations into single live cell tracking in situ changes in cell components [68,69].
Although RS has been used in different biomedical fields [70,71], in reproductive medicine and mainly in andrology, it has started to be utilized in recent years [71–73]. Using RS, researchers have characterized different sperm regions such as the head, acrosome, middle piece, and flagellum, as well as inner organelles such as the nucleus and mitochondria [71–73]. This success is a prerequisite for deeper sperm characterization on a molecular level.
Currently, Raman spectroscopy is used mainly in the detection of sperm nuclear damage [67], and also in sex sperm selection [71]. The PO4 backbone of DNA is characterized by peak intensity from 1055 cm−1 to 1095 cm−1 [67,69,73]. Any changes in the intensity are signs of DNA damage, and it is possible to make a map of sites with DNA fragmentation. The results that were obtained by Raman microspectroscopy in the field of DNA evaluation are in correlation with the DNA fragmentation index (DFI), which is associated with infertility and spontaneous abortions [66,69,74]. For sex sperm selection, variations of Raman peaks from DNA content and sex membrane proteins were reported in bovine sperm [71,75]. These results showed, with more than 90% accuracy, that RS may be applied in the near future for sperm sexing in a label-free and non-invasive manner, compared with the sex sorting by flow cytometry used to date [72,76]. Recently, the damage induced by sperm sorting and freezing-thawing procedures have been quantified by laser tweezers Raman spectroscopy [72]. These authors reported a variation of DNA, lipid, carbohydrates, and protein contents in sperm during flow cytometry process. Liu et al. [77] also showed the ability of Raman microspectroscopy to distinguish zona pellucida-bound sperm from unbound sperm, identifying differences in the intensity of Raman spectra on the acrosome region.
Thus, Raman spectroscopy allows us to evaluate DNA and RNA, as well as histones and protamines [78–80]. Poplineau et al. [78] used laser tweezers Raman spectroscopy to isolate a living human cell and monitor epigenetic modifications. These researchers treated Jurkat cells with histone deacetylase inhibitors, which resulted in an increase of histone code acetylation and changes in chromatin. The aforementioned effect was analyzed by different methods (laser tweezers Raman spectroscopy, electrophoresis, and nuclear image cytometry). Laser tweezers Raman spectroscopy showed the best discrimination between treated and control cells.
Noticeably, according to recent studies, if epigenetic modifications of sperm could be evaluated by non-invasive Raman spectroscopy, similarly to Jurkat cells, they would be suitable biomarkers of sperm quality in routine ART, providing relevant information on male infertility.
Conclusion and Perspectives
Correct sperm evaluation and selection of the most representative markers may be the key to resolving male infertility problems. The most feasible option for finding informative biomarkers that will be a guarantee of successful fertilization and proper embryo and offspring development could be found in the epigenome. Many researchers have suggested the possible usefulness to ART of analyzing epigenetic markers in routine sperm analysis [6,11,15,16]. However, further studies are required to confirm the relationship between sperm epigenetic features and the outcome of ART, as well as alterations in offspring generated by ART.
Correct chromatin structure involves the precise realization and timing of posttranslational modifications and protamination, essential for healthy progenitive sperm. Accordingly, deeper insight into DNA structure and epigenome generally seems to be promising in the clarification of processes for sperm fertilization and embryo development [6–8,10,81,82]. Posttranslational processes have an evident impact on embryo imprinting, and defects of imprinting participate in different offspring disorders, such as Angelman, Beckwith-Wiedemann, Prader-Willi, Silver-Russell, Goiter, Kabuki, and Claes-Jensen X-linked mental retardation syndrome [12,13,83,84]. For all these reasons, the development of adequate therapeutic options and sperm selection technologies based on epigenetic quality are crucial for improving ART outcomes [4].
Although the importance of epigenetics is obvious, it is currently not used in sperm selection methods or other ART techniques. The latest knowledge forces us to consider new perspectives for tracking these novel sperm parameters. The above-mentioned techniques (microfluidic device, sperm nanopurification, and Raman spectroscopy) could be options (Figure 1). Furthermore, a combination of microfluidic and Raman spectroscopy is possible and could increase sperm selection efficiency in some infertility cases [24,85].
Figure 1.
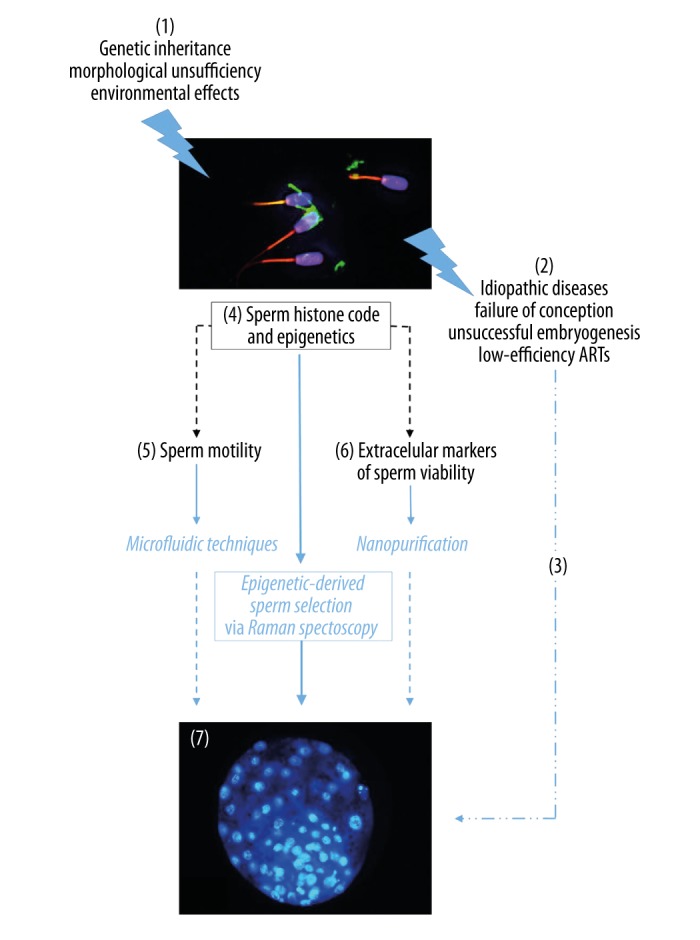
Raman spectroscopy for sperm epigenetics as an innovative tool for sperm selection resulting in successful embryonic development. When various environmental pressures or genetic burden (1), showed as idiopathic infertility followed by conception failure (2), decrease a chance for successful conception (3). On the other hand, sperm population is predestined for strict selection more than oocytes. Therefore, qualified selection of markers of sperm fertilization ability is crucial for advanced approaches to live sperm selection. Based on current knowledge, sperm epigenetics (4) seems to be a general phenomenon regulating clinical obvious features (motility (5) and sperm viability (6)) as well as invisible sperm quality leading to correct embryonic development (7). Raman spectroscopy is a versatile tool for advanced epigenetic-derived sperm selection. Therefore, Raman spectroscopy-selected sperm can improve ART outcomes, even with low-quality ejaculate.
These methods are not only non-invasive, they also have a huge potential for identification of epigenetic markers and use in successful ART. Actual studies of potential techniques show good results that are in correlation with the DNA fragmentation index (%DFI) and success of fertilization, ICSI, and others [32,36,52]. In addition, another use could be to mimic the natural sperm environment and natural sperm selection in the female genital tract, such as microfluidics [86,87].
However, although the sperm separation methods currently in use cannot use epigenetic markers for selecting undamaged sperm, Raman spectroscopy could be a promising method of identifying small differences in the chemical structure which are associated with changes in cell physiological properties (Figure 1). Furthermore, the most recent information on the application of Raman spectroscopy in evaluation of the histone acetylation state of cancer cells, suggests it could be used in sperm and expanding research to other posttranslational modifications, such as phosphorylation and DNA methylation. The level of histone phosphorylation detected by flow cytometry correlates with DNA damage and infertile male patients [88]. In addition, the DNA methylation pattern varies between fertile normospermic males and IVF patients, even if it seems to be a proper indicator of embryo quality after IVF [89]. However, it would be better if Raman spectroscopy could be used for these objectives and this seems to be possible.
Progress in Raman spectroscopy suggests that in the near future we will be able to better observe the epigenome of gametes without disturbing or destroying them. The results obtained by these methods show that sperm selected in these ways, in comparison to standard methods, exhibit better quality. Nevertheless, although the results and application in ART seem to be promising, further observations are necessary to confirm their safety in the epigenetic context.
Acknowledgments
Miriam Štiavnická was supported by a Predoctoral Fellowship from Charles University in Prague. Olga Garcia-Alvarez was supported by a Postdoctoral Fellowship from Charles University in Prague. We would like to thank Dr. Young-Joo Yi for kind help and Ms. Veronika Rotova for progress in introduction of Raman spectroscopy.
Footnotes
Source of support: The research discussed in this review was supported by Charles University (PROGRES Q-39) and the National Sustainability Program I (NPU I) No. LO1503, provided by the Ministry of Education, Youth and Sports of the Czech Republic
Competing interests
The authors declared no competing interest.
References
- 1.Jenkins TG, Aston KI, Trost C, et al. Intra-sample heterogeneity of sperm DNA methylation. Mol Hum Reprod. 2015;21:313–19. doi: 10.1093/molehr/gau115. [DOI] [PubMed] [Google Scholar]
- 2.Holt WV, Van Look KJ. Concepts in sperm heterogeneity, sperm selection and sperm competition as biological foundations for laboratory tests of semen quality. Reproduction. 2004;127:527–35. doi: 10.1530/rep.1.00134. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins TG, Aston KI, Pflueger C, et al. Age-associated sperm DNA methylation alterations, possible implications in offspring disease susceptibility. PLoS Genet. 2014;10:e1004458. doi: 10.1371/journal.pgen.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurentino S, Borgmann J, Gromoll J. On the origin of sperm epigenetic heterogeneity. Reproduction. 2016;151:R71–78. doi: 10.1530/REP-15-0436. [DOI] [PubMed] [Google Scholar]
- 5.Schagdarsurengin U, Steger K. Epigenetics in male reproduction, effect of paternal diet on sperm quality and offspring health. Nat Rev Urol. 2016;13:584–95. doi: 10.1038/nrurol.2016.157. [DOI] [PubMed] [Google Scholar]
- 6.Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci USA. 2015;112:13699–704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–96. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siklenka K, Erkek S, Godmann M, et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350:aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 9.Tang D, Huang Y, Liu W, Zhang X. Up-regulation of microRNA-210 is associated with spermatogenesis by targeting IGF2 in male infertility. Med Sci Monit. 2016;18:2905–10. doi: 10.12659/MSM.897340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012;97:267–74. doi: 10.1016/j.fertnstert.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Yu B, Zhou H, Liu M, et al. Epigenetic alterations in density selected human spermatozoa for assisted reproduction. PLoS One. 2015;10:e0145585. doi: 10.1371/journal.pone.0145585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han JY, Park J, Jang W, et al. A twin sibling with Prader-Willi syndrome caused by type 2 microdeletion following assisted reproductive technology. A case report. Biomed Rep. 2016;5:18–22. doi: 10.3892/br.2016.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiura H, Okae H, Chiba H, et al. Imprinting methylation errors in ART. Reprod Med Biol. 2014;13:193–202. doi: 10.1007/s12522-014-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurinczuk JJ, Bhattacharya S. Rare chromosomal, genetic, and epigenetic-related risks associated with infertility treatment. Semin Fetal Neonatal Med. 2014;19:250–53. doi: 10.1016/j.siny.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Pinborg A, Loft A, Romundstad LB, et al. Epigenetics and assisted reproductive technologies. Acta Obstet Gynecol Scand. 2016;95:10–15. doi: 10.1111/aogs.12799. [DOI] [PubMed] [Google Scholar]
- 16.Stuppia L, Franzago M, Ballerini P, et al. Epigenetics and male reproduction: The consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clin Epigenetics. 2015;7:120. doi: 10.1186/s13148-015-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boomsma CM, Heineman MJ, Cohlen BJ, Farquhar C. Semen preparation techniques for intrauterine insemination. Cochrane Database Syst Rev. 2007;17:CD004507. doi: 10.1002/14651858.CD004507.pub3. [DOI] [PubMed] [Google Scholar]
- 18.Henkel RR, Schill WB. Sperm preparation for ART. Reprod Biol Endocrinol. 2003;1:108. doi: 10.1186/1477-7827-1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimer JJ. Sperm preparation methods. J Androl. 2000;21:357–66. [PubMed] [Google Scholar]
- 20.Ricci G, Perticarari S, Boscolo R, et al. Semen preparation methods and sperm apoptosis, swim-up versus gradient-density centrifugation technique. Fertil Steril. 2009;91:632–38. doi: 10.1016/j.fertnstert.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 21.Xue X, Wang WS, Shi JZ, et al. Efficacy of swim-up versus density gradient centrifugation in improving sperm deformity rate and DNA fragmentation index in semen samples from teratozoospermic patients. J Assist Reprod Genet. 2014;31:1161–66. doi: 10.1007/s10815-014-0287-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henkel R. Sperm preparation, state-of-the-art-physiological aspects and application of advanced sperm preparation methods. Asian J Androl. 2012;14:260–69. doi: 10.1038/aja.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avendaño C, Oehniger S. DNA fragmentation in morphologically normal spermatozoa, how much should we be concerned in the ICSI era? J Androl. 2011;32:356–63. doi: 10.2164/jandrol.110.012005. [DOI] [PubMed] [Google Scholar]
- 24.Samuel R, Badamjav O, Murphy KE, et al. Microfluidics: The future of microdissection TESE? Syst Biol Reprod Med. 2016;62:161–70. doi: 10.3109/19396368.2016.1159748. [DOI] [PubMed] [Google Scholar]
- 25.Krejčí J, Stixová L, Pagáčová E, et al. Post-translational modifications of histones in human sperm. Cell Biochem. 2015;116:2195–209. doi: 10.1002/jcb.25170. [DOI] [PubMed] [Google Scholar]
- 26.Suarez SS. Mammalian sperm interactions with the female reproductive tract. Cell Tissue Res. 2016;363:185–94. doi: 10.1007/s00441-015-2244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt WV, Fazeli A. The oviduct as a complex mediator of mammalian sperm function and selection. Mol Reprod Dev. 2010;77:934–43. doi: 10.1002/mrd.21234. [DOI] [PubMed] [Google Scholar]
- 28.Almiñana C, Caballero I, Heath PR, et al. The battle of the sexes starts in the oviduct, modulation of oviductal transcriptome by X and Y-bearing spermatozoa. BMC Genomics. 2014;15:293. doi: 10.1186/1471-2164-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler MB, Rubessa M. Integration of microfluidics and mammalian IVF. Mol Hum Reprod. 2016 [Epub ahead of print] [Google Scholar]
- 30.Asghar W, Velasco V, Kingsley JL, et al. Selection of functional human sperm with higher DNA integrity and fewer reactive oxygen species. Adv Healthc Mater. 2014;3:1671–79. doi: 10.1002/adhm.201400058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowlton SM, Sadasivam M, Tasoglu S. Microfluidics for sperm research. Trends Biotechnol. 2015;33:221–29. doi: 10.1016/j.tibtech.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 32.de Wagenaar B, Berendsen JT, Bomer JG, et al. Microfluidic single sperm entrapment and analysis. Lab Chip. 2015;15:1294–301. doi: 10.1039/c4lc01425a. [DOI] [PubMed] [Google Scholar]
- 33.Cho BS, Schuster TG, Zhu X, et al. Passively driven integrated microfluidic system for separation of motile sperm. Anal Chem. 2003;75:1671–75. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- 34.Taken K, Alp HH, Eryilmaz R, et al. Oxidative DNA damage to sperm cells and peripheral blood leukocytes in infertile men. Med Sci Monit. 2016;22:4289–96. doi: 10.12659/MSM.898631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuura K, Uozumi T, Furuichi T, et al. Microfluidic device to reduce treatment time of intracytoplasmic sperm injection. Fertil Steril. 2013;99:400–7. doi: 10.1016/j.fertnstert.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Shirota K, Yotsumoto F, Itoh H, et al. Separation efficiency of a microfluidic sperm sorter to minimize sperm DNA damage. Fertil Steril. 2016;105:315–21. doi: 10.1016/j.fertnstert.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Liang GT, Peng YY, et al. Effects of a microfluidic sperm sorter on sperm routine parameters and DNA integrity. Zhonghua Nan Ke Xue. 2011;17:301–4. [PubMed] [Google Scholar]
- 38.Nosrati R, Vollmer M, Eamer L, et al. Rapid selection of sperm with high DNA integrity. Lab Chip. 2014;14:1142–50. doi: 10.1039/c3lc51254a. [DOI] [PubMed] [Google Scholar]
- 39.Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 2009;26:537–44. doi: 10.1007/s10815-009-9346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammadeh ME, Hamad MF, Montenarh M, Fischer-Hammadeh C. Protamine contents and P1/P2 ratio in human spermatozoa from smokers and non-smokers. Hum Reprod. 2010;25:2708–20. doi: 10.1093/humrep/deq226. [DOI] [PubMed] [Google Scholar]
- 41.Yu B, Qi Y, Liu D, et al. Cigarette smoking is associated with abnormal histone-to-protamine transition in human sperm. Fertil Steril. 2014;101:51–57. doi: 10.1016/j.fertnstert.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Houshdaran S, Cortessis VK, Siegmund K, et al. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rappa KL, Rodriguez HF, Hakkarainen GC, et al. Sperm processing for advanced reproductive technologies, Where are we today? Biotechnol Adv. 2016;34:578–87. doi: 10.1016/j.biotechadv.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Doane TL, Burda C. The unique role of nanoparticles in nanomedicine: Imaging, drug delivery and therapy. Chem Soc Rev. 2012;41:2885–911. doi: 10.1039/c2cs15260f. [DOI] [PubMed] [Google Scholar]
- 45.Vasquez ES, Feugang JM, Willard ST, et al. Bioluminescent magnetic nanoparticles as potential imaging agents for mammalian spermatozoa. J Nanobiotechnology. 2016;4:20. doi: 10.1186/s12951-016-0168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odhiambo JF, Sutovsky M, DeJarnette JM, et al. Adaptation of ubiquitin-PNA based sperm quality assay for semen evaluation by a conventional flow cytometer and a dedicated platform for flow cytometric semen analysis. Theriogenology. 2011;76:1168–76. doi: 10.1016/j.theriogenology.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Sutovsky P, Aarabi M, Miranda-Vizuete A, Oko R. Negative biomarker based male fertility evaluation: Sperm phenotypes associated with molecular-level anomalies. Asian J Androl. 2015;17:554–60. doi: 10.4103/1008-682X.153847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lafuente D, Garcia T, Blanco J, et al. Effects of oral exposure to silver nanoparticles on the sperm of rats. Reprod Toxicol. 2016;60:133–39. doi: 10.1016/j.reprotox.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Moretti E, Terzuoli G, Renieri T, et al. In vitro effect of gold and silver nanoparticles on human spermatozoa. Andrologia. 2013;45:392–96. doi: 10.1111/and.12028. [DOI] [PubMed] [Google Scholar]
- 50.Degheidy T, Abdelfattah H, Seif A, et al. Magnetic activated cell sorting: An effective method for reduction of sperm DNA fragmentation in varicocele men prior to assisted reproductive techniques. Andrologia. 2015;47:892–96. doi: 10.1111/and.12343. [DOI] [PubMed] [Google Scholar]
- 51.Romany L, Garrido N, Cobo A, et al. Obstetric and perinatal outcome of babies born from sperm selected by MACS from a randomized controlled trial. J Assist Reprod Genet. 2016;34:201–7. doi: 10.1007/s10815-016-0838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Odhiambo JF, DeJarnette JM, Geary TW, et al. Increased conception rates in beef cattle inseminated with nanopurified bull semen. Biol Reprod. 2014;91:97. doi: 10.1095/biolreprod.114.121897. [DOI] [PubMed] [Google Scholar]
- 53.Cross NL, Watson SK. Assessing acrosomal status of bovine sperm using fluoresceinated lectins. Theriogenology. 1994;42:89–98. doi: 10.1016/0093-691x(94)90665-6. [DOI] [PubMed] [Google Scholar]
- 54.Mulugeta Achame E, Wassenaar E, Hoogerbrugge JW, et al. The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics. 2010;11:367. doi: 10.1186/1471-2164-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–58. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 56.Da Silva N, Barton CR. Macrophages and dendritic cells in the post-testicular environment. Cell Tissue Res. 2016;363:97–104. doi: 10.1007/s00441-015-2270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richburg JH, Myers JL, Bratton SB. The role of E3 ligases in the ubiquitin-dependent regulation of spermatogenesis. Semin Cell Dev Biol. 2014;30:27–35. doi: 10.1016/j.semcdb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutovsky P, Moreno R, Ramalho-Santos J, et al. A putative, ubiquitin-dependent mechanism for the recognition and elimination of defective spermatozoa in the mammalian epididymis. J Cell Sci. 2011;114:1665–75. doi: 10.1242/jcs.114.9.1665. [DOI] [PubMed] [Google Scholar]
- 59.Sutovsky P, Terada Y, Schatten G. Ubiquitin-based sperm assay for the diagnosis of male factor infertility. Hum Reprod. 2001;16:250–58. doi: 10.1093/humrep/16.2.250. [DOI] [PubMed] [Google Scholar]
- 60.Ozanon C, Chouteau J, Sutovsky P. Clinical adaptation of the sperm ubuquitin tag immunoassay (SUTI): relationship of sperm ubiquitylation with sperm quality in gradient-purified semen samples from 93 men from a general infertility clinic population. Hum Reprod. 2005;20:2271–78. doi: 10.1093/humrep/dei013. [DOI] [PubMed] [Google Scholar]
- 61.Eskandari-Shahraki M, Tavalaee M, et al. Proper ubiquitination effect on the fertilization outcome post-ICSI. Andrologia. 2013;45:204–10. doi: 10.1111/j.1439-0272.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- 62.Kitamura A, Miyauchi N, Hamada H, et al. Epigenetic alterations in sperm associated with male infertility. Congenit Anom (Kyoto) 2015;55:133–44. doi: 10.1111/cga.12113. [DOI] [PubMed] [Google Scholar]
- 63.Manochantr S, Chiamchanya C, Sobhon P. Relationship between chromatin condensation, DNA integrity and quality of ejaculated spermatozoa from infertile men. Andrologia. 2012;44:187–99. doi: 10.1111/j.1439-0272.2010.01128.x. [DOI] [PubMed] [Google Scholar]
- 64.Setti AS, Braga DP, Vingris L, et al. Sperm morphological abnormalities visualised at high magnification predict embryonic development, from fertilization to the blastocyst stage, in couples undergoing ICSI. J Assist Reprod Genet. 2014;31:1533–39. doi: 10.1007/s10815-014-0326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Hong H, Cai W. Imaging with Raman spectroscopy. Curr Pharm Biotechnol. 2010;11:654–61. doi: 10.2174/138920110792246483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang Z, Chen G, Chen X, et al. Rapid and label-free identification of normal spermatozoa based on image analysis and micro-Raman spectroscopy. J Biophotonics. 2014;7:671–75. doi: 10.1002/jbio.201300003. [DOI] [PubMed] [Google Scholar]
- 67.Huser T, Orme CA, Hollars CW, et al. Raman spectroscopy of DNA packaging in individual human sperm cells distinguishes normal from abnormal cells. J Biophotonics. 2009;2:322–32. doi: 10.1002/jbio.200910012. [DOI] [PubMed] [Google Scholar]
- 68.Swain RJ, Stevens MM. Raman microspectroscopy for non-invasive biochemical analysis of single cells. Biochem Soc Trans. 2007;35:544–49. doi: 10.1042/BST0350544. [DOI] [PubMed] [Google Scholar]
- 69.Mallidis C, Sanchez V, Wistuba J, et al. Raman microspectroscopy: Shining a new light on reproductive medicine. Hum Reprod Update. 2014;20:403–14. doi: 10.1093/humupd/dmt055. [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Zhu Y, Li Z. Application of Raman spectroscopy in Andrology: Non-invasive analysis of tissue and single cell. Transl Androl Urol. 2014;3:125–33. doi: 10.3978/j.issn.2223-4683.2014.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Luca AC, Managó S, Ferrara MA, et al. Non-invasive sex assessment in bovine semen by Raman spectroscopy. Laser Physics Letters. 2014:11. [Google Scholar]
- 72.Li XX, Wang M, Chen HH, et al. Flow cytometric and near-infrared Raman spectroscopic investigation of quality in stained, sorted, and frozen-thawed buffalo sperm. Anim Reprod Sci. 2016;170:90–99. doi: 10.1016/j.anireprosci.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Meister K, Schmidt DA, Bründermann E, Havenith M. Confocal Raman microspectroscopy as an analytical tool to assess the mitochondrial status in human spermatozoa. Analyst. 2010;135:1370–74. doi: 10.1039/b927012d. [DOI] [PubMed] [Google Scholar]
- 74.Sánchez V, Redmann K, Wistuba J, et al. Oxidative DNA damage in human sperm can be detected by Raman microspectroscopy. Fertil Steril. 2012;98:1124–29. doi: 10.1016/j.fertnstert.2012.07.1059. [DOI] [PubMed] [Google Scholar]
- 75.Ferrara MA, Di Caprio G, Managò S, et al. Label-free imaging and biochemical characterization of bovine sperm cells. Biosensors (Basel) 2015;5:141–57. doi: 10.3390/bios5020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anel-López L, García-Álvarez O, Parrilla I, et al. Effect of sex-sorting and cryopreservation on the post-thaw sperm quality of Iberian red deer spermatozoa. Theriogenology. 2017;89:206–13. doi: 10.1016/j.theriogenology.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 77.Liu F, Zhu Y, Liu Y, et al. Real-time Raman microspectroscopy scanning of the single live sperm bound to human zona pellucida. Fertil Steril. 2013;99:684–89. doi: 10.1016/j.fertnstert.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 78.Poplineau M, Trussardi-Régnier A, Happillon T, et al. Raman microspectroscopy detects epigenetic modifications in living Jurkat leukemic cells. Epigenomics. 2011;3:785–94. doi: 10.2217/epi.11.102. [DOI] [PubMed] [Google Scholar]
- 79.Schulze HG, Konorov SO, Caron NJ, et al. Assesing differentiation status of human embryonic stem cells noninvasively using Raman microspectroscopy. Anal Chem. 2010;82:5020–27. doi: 10.1021/ac902697q. [DOI] [PubMed] [Google Scholar]
- 80.Sundararajan N, Mao D, Chan S, et al. Ultrasensitive detection and characterization of posttranslational modifications using surface-enhanced Raman spectroscopy. Anal Chem. 2006;78:3543–50. doi: 10.1021/ac051525i. [DOI] [PubMed] [Google Scholar]
- 81.van der Heijden GW, Ramos L, Baart EB, et al. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan S, Schuster A, Tang C, et al. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development. 2016;143:635–47. doi: 10.1242/dev.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berdasco M, Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum Genet. 2013;132:359–83. doi: 10.1007/s00439-013-1271-x. [DOI] [PubMed] [Google Scholar]
- 84.Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- 85.Chrimes AF, Khoshmanesh K, Stoddart PR, et al. Microfluidics and Raman microscopy: Current applications and future challenges. Chem Soc Rev. 2013;42:5880–906. doi: 10.1039/c3cs35515b. [DOI] [PubMed] [Google Scholar]
- 86.Eamer L, Vollmer M, Nosrati R, et al. Turning the corner in fertility: High DNA integrity of boundary-following sperm. Lab Chip. 2016;16:2418–22. doi: 10.1039/c6lc00490c. [DOI] [PubMed] [Google Scholar]
- 87.Lopez-Garcia MD, Monson RL, Haubert K, et al. Sperm motion in a microfluidic fertilization device. Biomed Microdevices. 2008;10:709–18. doi: 10.1007/s10544-008-9182-7. [DOI] [PubMed] [Google Scholar]
- 88.Zhong HZ, Lv FT, Deng XL, et al. Evaluating γH2AX in spermatozoa from male infertility patients. Fertil Steril. 2015;104:574–81. doi: 10.1016/j.fertnstert.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 89.Aston KI, Uren PJ, Jenkins TG, et al. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril. 2015;104:1388–97. doi: 10.1016/j.fertnstert.2015.08.019. [DOI] [PubMed] [Google Scholar]


