Abstract
Compared to the evolutionarily related bacterial ribosome, the reduced ribosomal RNA content and 36 additional proteins of mammalian mitochondrial ribosomes (mitoribosomes) generate new complexities for assembly. However, the molecular details of mitoribosomal biogenesis remain elusive. Here, we report the structures of two late-stage assembly intermediates of the human mitoribosomal large subunit (mt-LSU) isolated from a native pool within a human cell line and solved by electron cryomicroscopy to ~3 Å resolution. Comparison of the structures reveals insights into the timing of rRNA folding and protein incorporation during the concluding steps of ribosomal maturation, and the evolutionary adaptations that overcome structural diversification of mitoribosomes to preserve biogenesis. Furthermore, the structures redefine the role of the ribosome silencing factor (RsfS) family as multi-functional biogenesis factors and identify two new assembly factors (L0R8F8 and mt-ACP) not previously implicated in mitoribosomal biogenesis.
Introduction
Mammalian mitochondrial ribosomes (mitoribosomes) synthesize essential subunits of the oxidative phosphorylation machinery. Although they share a common ancestor with bacterial ribosomes, mammalian mitoribosomes have half the length of ribosomal RNA (rRNA) and 36 additional proteins1,2. While mt-rRNA is encoded by the mitochondrial genome, all 82 mitoribosomal proteins are imported from the cytoplasm. The architectural changes and the need to coordinate two genetic systems generate additional complexities for the assembly of mitoribosomes. Work in bacteria has shown that ribosomal proteins are recruited to rRNA as it folds and actively remodel it upon binding3,4. A repertoire of trans-acting assembly factors increases the efficiency of this process. Similar mechanisms are proposed to drive mitoribosomal assembly5. Although many assembly factors found in mitochondria have homologs in bacteria, the greater complexity of mitoribosomal assembly implies the involvement of as-yet-unknown mitochondria-specific auxiliary factors6.
Defects in mitochondrial translation, including the auxiliary factors necessary for mitoribosomal assembly, are associated with degenerative pathologies. Furthermore, unraveling the molecular details of human mitoribosomal assembly will aid the development of new anti-cancer therapies7 and antibiotics that disrupt bacterial ribosomal assembly8 with fewer off-target effects.
Results
Cryo-EM structures of mitoribosomal assembly intermediates
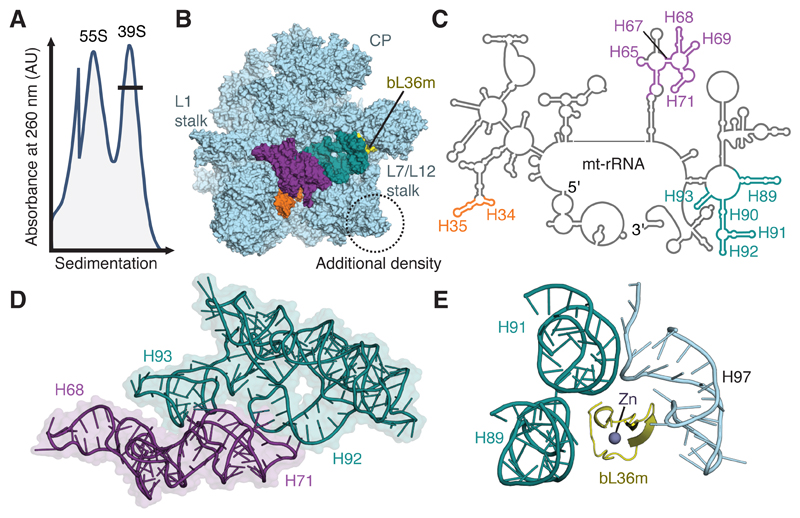
We noted that mitoribosomal material prepared from a HEK293S-derived human cell line9 contained high levels of mitoribosomes and a pool of species of a similar size to the large mitoribosomal subunit (mt-LSU) (Fig. 1A). Mass-spectrometry analysis indicated that this pool contained several mitoribosomal assembly factors (Supplementary Table 1). Initial visualization of this sample by cryo-EM revealed that unlike the mt-LSU of intact mitoribosomes1,10, this population had extra density adjacent to uL14m and unexpectedly poor density at the intersubunit interface. To investigate the molecular details of this complex, we collected a second dataset (Table 1) that was classified initially with an emphasis on isolating particles with additional density (Supplementary Fig. 1). Although the map reached a nominal resolution better than 3.0 Å, the interfacial region still displayed considerably worse local resolution (Supplementary Fig. 2). Further classification revealed two well-defined subclasses (Supplementary Fig. 1) that differ in the presence of density consistent with folded mt-rRNA at the intersubunit interface. Although the more highly populated subclass (at 3.0 Å resolution) seems to lack interfacial mt-rRNA, nebulous density, best seen in 2D slices through the map (Supplementary Fig. 1B), suggests that the missing mt-rRNA is not cleaved but adopts multiple conformations. Relative to the subclass with fully folded mt-rRNA (at 3.1 Å resolution), helices H34-35, H65, H67-71 and H89-93 are absent (Fig. 1B-C). In the mature mitoribosome, these interconnected sections of mt-rRNA (Fig. 1D) comprise over a fifth of the total mt-rRNA and form the peptidyl-transferase center (PTC), which catalyzes peptide-bond formation, and intersubunit bridges with the small subunit (mt-SSU). The remaining rRNA adopts a conformation largely unchanged from that in the intact mitoribosome.
Figure 1. Purification and structural characterization of native mitoribosomal assembly intermediates.
(A) Differential centrifugation separates intact mitoribosomes (55S) from a pool of mt-LSU-like complexes (39S). This pool contains two well-defined assembly intermediates that differ in the presence of folded interfacial rRNA. (B) View of the assembly intermediate with folded interfacial rRNA viewed from the intersubunit interface. Both intermediates feature additional density (circled) relative to the mt-LSU of intact 55S mitoribosomes. However, one class displays unfolded interfacial rRNA with density for H34-35 (orange), H65 and H67-71 (purple), and H89-93 (teal) absent, along with protein bL36m (yellow). Landmark features of the mitoribosome are labeled, including the two stalks and central protuberance (CP). (C) Secondary structure diagram for mt-rRNA with sections of unfolded rRNA colored according to B. (D) Interconnectivity of mt-rRNA helices H67-71 (purple) and H89-93 (teal). (E) In the mature mitoribosome, bL36m coordinates H89 and H91 that are absent in the reconstruction with H97.
Table 1.
Cryo-EM data collection, refinement and validation statistics.
| 39S intermediate with folded rRNA (PDB ID: 5OOL) (EMD-3842) |
39S intermediate with unfolded rRNA (PDB ID: 5OOM) (EMD-3843) |
|
|---|---|---|
| Data collection | ||
| Microscope Camera Magnification Voltage (kV) Electron dose (e–/Ǻ2) Defocus range (μm) Pixel size (Ǻ) Initial particles (no.) Final particles (no.) |
Titan Krios Falcon II 130,841 300 39 -1.5 to -3.5 1.34 600,949 134,685 |
Titan Krios Falcon II 130,841 300 39 -1.5 to -3.5 1.34 600,949 379,869 |
|
Model composition Nonhydrogen atoms Protein residues RNA bases Ligands (Zn2+/Mg2+) Refinement |
99,025 8,230 1,497 3/93 |
90,747 8,135 1,148 3/49 |
| Resolution (Å) FSC (entire box) FSC (around atoms) |
3.06 0.76 0.82 |
3.03 0.77 0.83 |
| Map sharpening B factor (Ǻ2) Average B factor (Ǻ2) |
-85.0 58.9 |
-95.0 59.5 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.016 |
| Bond angles (°) Validation MolProbity score Clashscore Poor rotamers (%) Ramachandran plot Favored (%) Allowed (%) Disallowed (%) |
1.02 1.66 5.73 0.48 95.7 3.82 0.48 |
1.27 1.70 5.94 0.90 94.4 5.54 0.06 |
All mt-LSU proteins are present except bL36m, which is absent only in the class with unfolded interfacial mt-rRNA. In the mature mitoribosome, bL36m stabilizes tertiary interactions between H89, H91, and H97 (Fig. 1E). The recruitment of bL36 and the folding of the PTC occur late in the assembly of bacterial ribosomes11,12, suggesting that the native pool of mt-LSU present in human mitochondria is formed by two late-stage biogenesis intermediates. Furthermore, the recruitment of bL36m and the folding of mt-rRNA may be interdependent as H71 and H89-93 have increased susceptibility to chemical probes in ribosomal subunits purified from bL36-deficient strains of Escherichia coli13, mirroring those that lack density in our structure. Correct folding of this region may rely on auxiliary factors14 or post-transcriptional modifications. Of the rRNA nucleotides missing from the structure, A2617, U3039 and G3040 are methylated15,16 and U3067 is isomerized to a pseudouridine17.
MALSU1, L0R8F8 and mt-ACP bind both assembly intermediates
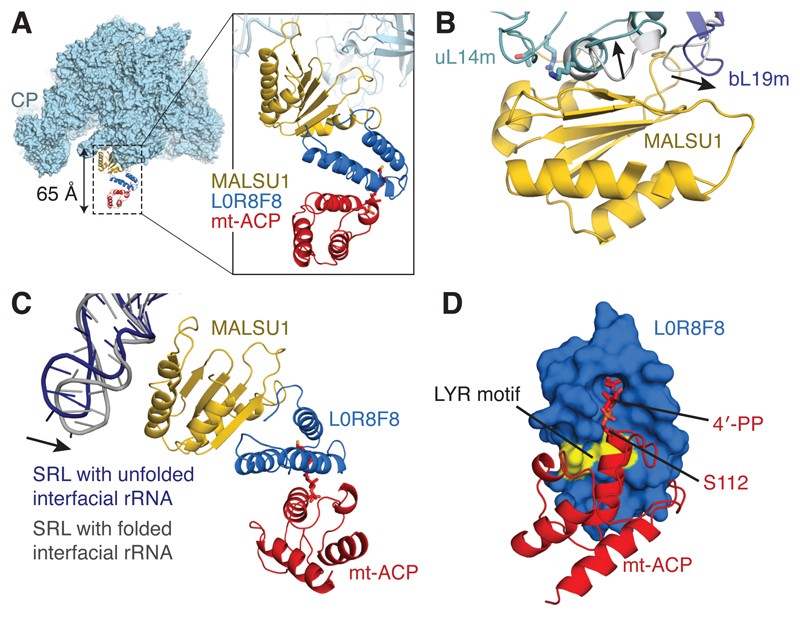
The density adjacent to uL14m, present in both subclasses, was partly assigned to the protein mitochondrial assembly of ribosomal large subunit 1 (MALSU1) (Fig. 2A), which belongs to the ribosome silencing factor (RsfS) family18 (Supplementary Fig. 3). Its bacterial counterpart is known to interact with uL14 through co-localization experiments18,19 and a low-resolution cryo-EM reconstruction of the Mycobacterium tuberculosis RsfS:LSU complex20. The well-defined density for the globular domain of MALSU1 (Supplementary Fig. 3B-C) allowed an atomic model to be built (residues 91-201). The face of the 5-stranded β-sheet of MALSU1 packs perpendicularly against the terminal helix of uL14m, consistent with mutations known to disrupt binding between bacterial RsfS and uL1418 (Fig. 2B). To accommodate MALSU1, both uL14m and the neighboring bL19m display conformational changes from their positions in the mature mitoribosome (Fig. 2B). MALSU1 also forms electrostatic interactions with the sarcin-ricin stem-loop (SRL; H95) of mt-LSU rRNA (Fig. 2C). The SRL forms tertiary interactions with H90-92 in the mature mitoribosome, and interactions with MALSU1 may act to position this helix prior to the folding of the interfacial mt-rRNA. Folding of the mt-rRNA causes a 15° swing of the SRL towards MALSU1 (Fig. 2C).
Figure 2. A module of MALSU1–L0R8F8–mt-ACP binds both mt-LSU assembly intermediates.
(A) Location of the MALSU1–L0R8F8–mt-ACP module, shown here bound to mt-LSU with unfolded interfacial rRNA (viewed from the side). The module extends from the surface of the mt-LSU by ~65 Å. (B) Binding of MALSU1 induces conformational changes in uL14m and bL19m from their positions in the 55S mitoribosome (shown in grey, with the direction of movement indicated with arrows). Residues of uL14 (T97, R98, and K114) that, when mutated to alanine, disrupt binding of RsfS are mapped to the uL14m structure (T117, R118, and K136) and are shown in stick representation. (C) MALSU1 interacts electrostatically with the SRL (H95) of the mt-rRNA. The SRL makes a closer association with MALSU1 when the interfacial rRNA is folded. (D) The interaction between L0R8F8 and mt-ACP involves the LYR motif of L0R8F8 (colored yellow) and the 4´-phosphopantetheine modification of mt-ACP serine 112.
Depletion of MALSU1 from human mitochondria causes aberrantly assembled mitoribosomes to accumulate21, supporting the interpretation of these complexes as mitoribosomal biogenesis intermediates. However, MALSU1 accounts for only half of the extra density. To identify the additional factor(s) we inspected the fit of a library of 14,000 unique protein domains into this map (local resolution 3.5-5.0 Å) using a density-based fold-recognition pipeline that we had developed to interpret the map of the yeast mt-LSU22,23 (Supplementary Fig. 4A). This method singled out the fold of acyl carrier protein (ACP) as being most consistent with the apical end of the density (Supplementary Fig. 4B). Mitochondrial ACP (mt-ACP), identified by mass spectrometry of our sample (Supplementary Table 1), fits the density well (Supplementary Fig. 4C) but incompletely, with three helices left unaccounted for between MALSU1 and mt-ACP (Supplementary Fig. 4D), suggesting the presence of a bridging protein.
Mt-ACP is a small, abundant and pleiotropic protein that serves as a scaffold for fatty acid synthesis24. Two copies of mt-ACP (known as SDAP-α and SDAP-β) are also found in complex I (NADH:ubiquinone oxidoreductase)25–27, where they are anchored by different leucine–tyrosine–arginine(LYR)-motif proteins; SDAP-α is bound to subunit B14 (NDUFA6) and SDAP-β is bound to subunit B22 (NDUFB9) (Supplementary Fig. 5A). When these modules are superposed onto the mt-ACP present in our structures, the helices of the LYR-motif proteins align with the unassigned density between MALSU1 and mt-ACP (Supplementary Fig. 5B-C). However, since neither NDUFA6 nor NDUFB9 fits perfectly, we analyzed our mass-spectrometry data (Supplementary Table 1) for further LYR-motif-containing proteins. This revealed a single possibility: L0R8F8. L0R8F8 is a eukaryotic-specific protein of just 70 residues, which is synthesized from a bicistronic transcript that also encodes MID51, a transmembrane protein of the outer mitochondrial membrane28. The local resolution of 3.5-4.5 Å in this region allowed a near-complete model for L0R8F8 to be built (Supplementary Fig. 5D-E) and validated (Supplementary Fig. 5G-I). The interaction between L0R8F8 and mt-ACP is mediated by the tyrosine and arginine residues of the LYR motif and the 4-phosphopantetheine (4-PP) modification of mt-ACP (Fig. 2D and Supplementary Fig. 5F) that tethers the growing acyl chain during fatty acid synthesis. The 4-PP modification adopts a “flipped-out” conformation29 and inserts into a hydrophobic pocket of L0R8F8 (Fig. 2D), which resembles the interactions that occur between mt-ACP and the LYR-motif proteins of complex I27. We conclude that L0R8F8 and mt-ACP are assembly factors for the human mitoribosome.
The module may prevent premature subunit association
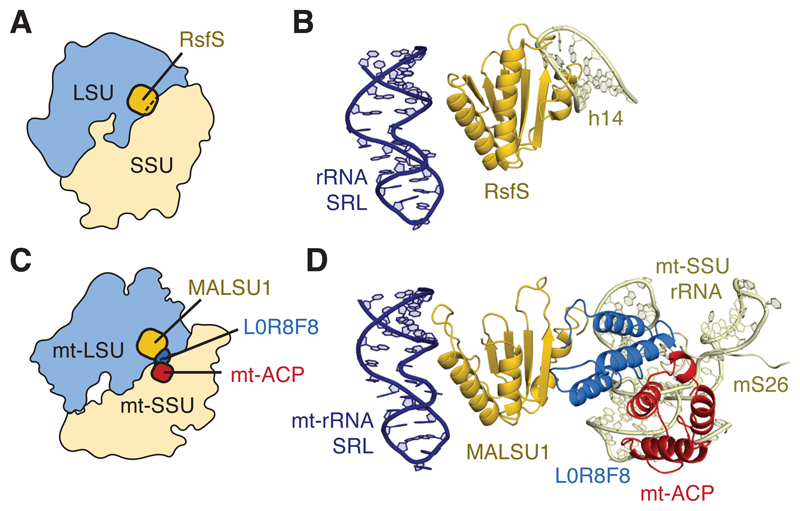
L0R8F8 and mt-ACP may link mitoribosomal biogenesis to fatty acid and iron-sulfur synthesis (in which both mt-ACP30 and LYR-motif proteins31 have been implicated) or have roles in recruiting downstream factors. However, one functional consequence of the MALSU1–L0R8F8–mt-ACP module is that it would sterically obstruct the binding of the mt-SSU. An anti-association function was previously proposed for bacterial RsfS from the observation that by binding to uL14, RsfS would prevent the formation of bridge B8 with helix 14 of 16S rRNA18,20 (Fig. 3A-B). This has parallels in other kingdoms of life, as the structurally and evolutionarily unrelated eIF6 also binds uL14 to sterically hinder the formation of cytosolic ribosomes in eukaryotes and archaea32. However, structural changes in the human mt-SSU, including the loss of h141, means that the globular domain of MALSU1 alone cannot obstruct subunit joining (Fig. 3C). It is possible that the MALSU1–L0R8F8–mt-ACP module co-evolved with architectural changes to the human mitoribosome to maintain this steric block. The module, which spans 65 Å, would clash with h5, h15 and the N-terminus of mS26 of the mt-SSU (Fig. 3D) regardless of the conformation of the mitoribosome (classical, rotated, or rolled)1. Without the acquisition of L0R8F8 loss of h14 may have rendered the anti-association activity of MALSU1 ineffective, potentially causing assembly defects21.
Figure 3. Models of anti-association activity.
(A) Schematic showing the position of RsfS relative to the bacterial ribosome. RsfS overlaps with the position of the ribosomal small subunit (SSU). (B) RsfS would clash with rRNA helix 14 (h14) of the SSU. (C) Schematic showing that MALSU1 alone cannot inhibit mitoribosomal subunit joining by steric hindrance: only together with L0R8F8–mt-ACP does the module bridge the distance between the mt-LSU and mt-SSU in the mature human mitoribosome. (D) L0R8F8–mt-ACP would clash with regions of mt-rRNA around helices h5 and h15 and the N-terminus of mS26 in the mt-SSU, thereby preventing subunit association.
During the final stages of maturation, steric hindrance on subunit joining must be alleviated by release of the MALSU1–L0R8F8–mt-ACP module. The presence of this module with both folded and unfolded interfacial rRNA demonstrates that eviction of this module is not a prerequisite step for the folding of interfacial mt-rRNA into a native-like conformation. Rather, it is likely that the module requires active displacement that coincides with the rRNA adopting a folded conformation. This is seen in the eviction of eIF6 from eukaryotic cytosolic ribosomes, where the ribosomal maturation protein SBDS senses the structural integrity of key functional sites by binding the PTC, the SRL and the P stalk before recruiting a GTPase to actively displace eIF633.
Discussion
In summary, we have visualized two late-stage intermediates in the biogenesis pathway of the human mitoribosome. Uniquely among structures of other intermediates of ribosomal biogenesis our structures represent native intermediates and the first intermediates of mitoribosomal assembly. The structures reveal that binding of bL36m and accommodation of interfacial rRNA are among the concluding steps of mitoribosomal biogenesis, and that L0R8F8 and mt-ACP are mitoribosomal assembly factors. This establishes cryo-EM as a tool to investigate native states of macromolecular biogenesis with the potential to detect new proteins and propose their functions.
Online Methods
Cell line
The clonal cell line used is called T501 and constitutively expresses the rat serotonin transporter fused to GFP-His. This cell line was a gift from Chris Tate (MRC Laboratory of Molecular Biology, UK) and is derived from a human embryonic kidney cell line lacking N-acetyl-glucosaminyltransferase (HEK293S TetR GnTI-)9. The cells were not tested for mycoplasma contamination.
Purification of native mitoribosomal complexes
Mitochondria were isolated from healthy (>98% viability) T501 cells. The cells were grown to ~30% of their maximum concentration (<3×106 cells ml-1) in large-scale suspension before mitochondria were isolated following the protocol as described10.
To purify mitoribosomal material, 4 volumes of Lysis buffer (25 mM Hepes-KOH pH 7.45, 100 mM KCl, 25 mM MgOAc, 1.7% Triton X-100, 2 mM DTT) were added to purified mitochondria and incubated for 15 min at 4°C. The membranes were then separated by centrifugation at 30,000 x g for 20 min. The supernatant was loaded on a 1 M sucrose cushion in buffer (20 mM Hepes-KOH pH 7.45, 100 mM KCl, 20 mM MgOAc, 1% Triton X-100, 2 mM DTT). The resuspended pellet was then loaded onto a 10-25% sucrose gradient in the same buffer without Triton X-100 and run for 16 h at 85,000 x g. Fractions corresponding to excess mt-LSU (Fig. 1A, “39S” peak) were collected and sucrose removed by buffer exchange.
Grid Preparation
The purified sample was concentrated to 100 nM for grid preparation. 3 µl of sample was applied on to a freshly glow-discharged holey carbon grid (Quantifoil R2/2 Cu) pre-coated with a home-made continuous carbon film (~30 Å thick) and incubated for 30 seconds at 4°C, 100% humidity in a Vitrobot Mk IV system (FEI). The grids were blotted for 3 seconds prior to plunge cooling in liquid ethane.
Image processing (dataset 1)
The initial dataset was collected at the MRC Laboratory of Molecular Biology on a Titan Krios microscope (FEI) operated at 300 kV and equipped with a Falcon-II direct-electron detector (FEI). Micrographs were obtained from two separate automated data collections (EPU software, FEI) at 104,478 x magnification, yielding a pixel size of 1.34 Å. One-second exposures yielded a total dose of 25 electrons/Å2, with defocus values ranging from -1.5 to -3.5 μm at 0.5 µm intervals. A total of 2,827 micrographs were recorded and kept. Movie frames were aligned and averaged using whole-image movement correction using MOTIONCORR34. Contrast transfer function (CTF) parameters were estimated using Gctf v.0.535. All subsequent image-processing steps were performed in RELION-2.036. From the 2,827 micrographs, 650,329 particles were autopicked in RELION using templates generated from the 2D class averages of a small set of manually picked particles. These particles were subjected to multiple rounds of reference-free 2D classification to discard poorly aligned particles and intact mitoribosomes. The remaining 398,539 particles underwent 3D classification using the map of the human mt-LSU (EMD-2762)10 low-pass filtered to 60 Å as a reference. Well-resolved classes were selected (corresponding to 332,644 particles) and subjected to an initial round of 3D refinement. Movie refinement and “particle polishing”37 was performed to obtain shiny particles that were then re-refined to improve the overall alignment of particles. A single round of focused classification with signal subtraction (FCwSS) without image alignment and a regularization parameter of T=2038 was performed on these particles to improve the local density adjacent to uL14m. The classes containing well-resolved density in this region were combined, which gave a total of 216,218 particles. These particles were subjected to 3D refinement and post-processing, which yielded a map with a global resolution of 3.1 Å based on the FSC=0.143 criterion. This map provided initial insight into the composition of the native pool, but all model building and analysis was performed on the higher-resolution maps generated from dataset 2 (see below).
Image processing (dataset 2)
Prior to collecting a second dataset, the sample was optimized by increasing the concentration to 240 nM and adding 2 mM Synercid (Santa Cruz Biotechnology, Inc), which was found to reduce preferential orientation. The second dataset was collected at the Swedish National Facility on a Titan Krios microscope operated at 300 kV and equipped with a Falcon-II detector (FEI). Compared to the first dataset, micrographs were collected at the higher magnification of 130,841 x, yielding a pixel size of 1.06 Å. Defocus values of -0.5 to -3.5 µm at 0.2 µm intervals were specified during automated data collection using EPU software (FEI). For each micrograph, a total of 25 frames were collected over a 1.5 s exposure with a dose rate of 1.56 electrons/Å2/frame. Movies were processed using Motioncor239 for patch-based motion correction and dose weighting. CTF parameters were estimated using Gctf v.0.535. RELION-2.036 was used for all other image processing steps. Templates for reference-based particle picking were obtained from 2D class averages that were calculated from a manually picked subset of the micrographs, and 837,248 particles were autopicked from 3,696 micrographs. Reference-free 2D class averaging was used to discard poorly aligned particles, and the remaining 600,949 particles were subjected to auto-refinement to assign angles in one consensus class, yielding a map with a global resolution of 2.98 Å based on the FSC=0.143 criterion. FCwSS38 without alignment was employed to isolate particles with additional density adjacent to uL14m. This step discarded 90,142 particles and resulted in a map with a global resolution of 2.96 Å. However, density at the intersubunit interface was worse than expected for a map at this resolution. We therefore performed another round of FCwSS without alignment focused on the interface. This isolated two subclasses: with and without folded interfacial rRNA (Supplementary Fig. 1). The density adjacent to uL14m was present in both classes. The particles from both subclasses were subjected to a final 3D refinement and post-processed. The map with folded interfacial rRNA reached a global resolution of 3.1 Å (FSC=0.143 criterion) from 134,685 particles, and the map with unfolded interfacial rRNA reached 3.0 Å from 379,869 particles. During post-processing each density map was corrected for the modulation transfer function (MTF) of the Falcon-II detector, and sharpened by applying a B-factor (given in Table 1) calculated using automated procedures40.
To improve the density adjacent to uL14m to aid model building, we recombined both subclasses and performed FCwSS with a small mask around the appendage to MALSU1. This isolated a class of 224,267 particles, which after refinement and masking yielded a reconstruction with a local resolution of 3.5-5.0 Å, as estimated by ResMap41 (Supplementary Fig. 2).
Model building
Initially, the model of the human mt-LSU (PDB ID: 3J9M)1 was placed into the density map of the assembly intermediate with folded interfacial mt-rRNA using the “fit in map” feature of Chimera42. Differences in the local positions of the mitoribosomal proteins and mt-rRNA helices were corrected using real-space refinement in Coot v0.8.823. Previously unobserved mitoribosomal features were modeled de novo, for example the N-terminus of mL45 (residues 50-91) and the C-terminus of uL23m (residues 126-153). During model building and refinement in Coot, torsion, planar-peptide, trans-peptide and Ramachandran restraints were applied. Trans-peptide restraints were turned off to model cis-prolines.
The model was then fit to the map of the subclass with unfolded interfacial mt-rRNA. Sections of model without density were deleted in Coot. This included substantial regions of mt-rRNA (nucleotides 1931-1971, 2474-2506, 2539-2649, and 2935-3099) as well as shorter sections (nucleotides 2228-2232, 2720-2722, and 3169-3173). Protein bL36m was entirely absent and deleted from the model together with residues 273-288 of uL2m, the N-terminus of mL63, and residues 157-164 of uL22m. These protein sections interact with interfacial rRNA in the mature mitoribosome and likely fold together.
A comparative model for MALSU1 (UniProt ID: Q96EH3) was generated using I-TASSER43. This model was generated using the crystal structure of M. tuberculosis RsfS (PDB ID: 4WCW)20, the crystal structure of protein CV0518 from Chromobacterium violaceum (PDB ID: 2ID1), and the crystal structure of iojap-like protein from Zymomonas mobilis (PDB ID: 3UPS) as templates. The model was placed into the map with Coot and the real-space refined to better fit the density. The N- and C-termini were trimmed, as no density is apparent for the first 90 and the last 33 residues. The absence of fragments in the mass spectrometry analysis for the N-terminus of MALSU1 together with computational predictions suggests that the N-terminus likely forms a cleavable mitochondria-targeting peptide. The C-terminal 33 residues are likely to be flexible and averaged out of the reconstruction.
Human mt-ACP (Uniprot ID: O14561, residues 74-152) was built by placing the model of ovine mt-ACP from complex I (PDB ID: 5LNK, chain X)27 into the density and mutating the residues to match the human sequence. Mt-ACP was identified using a density-based fold-recognition pipeline22,23. In brief, 14,000 unique domains derived from the BALBES database44 were fit to the density using MOLREP45 and ranked based on contrast score, which is the ratio of the top score to the mean score. The top hit with a contrast score of 3.95 belonged to acyl carrier protein (ACP) from Escherichia coli (PDB ID: 3EJB, chain A)46. The fit of this, and the next 9 top-ranked hits, to the density were inspected manually, confirming ACP as the most likely solution.
The model for L0R8F8 (Uniprot ID: L0R8F8) was built de novo in Coot starting from the initial placement of 3 idealized poly(alanine) helices into the density.
Model Refinement
Restrained refinement was performed using PHENIX 1.11.1: phenix.real_space_refine47. Each round of global real-space refinement featured 5 macro-cycles with secondary structure, rotamer, Ramachandran, and Cβ-torsion restraints applied. Secondary structure restraints were determined directly from the model using phenix.secondary_structure_restraints and recalculated for each round of refinement. For the RNA present in the molecule (mt-rRNA and mt-tRNAVal), hydrogen-bonding and base-pair and stacking parallelity restraints were applied. Additional restraints were applied for the 4′-phosphopantetheine modification of mt-ACP (chemical component three-letter code: PNS). Two B-factors were refined per residue in reciprocal space: one for the main chain and one for the side chain. The high-resolution limit was set during refinement to 3.1 Å for both structures (with and without interfacial rRNA).
Model Validation
The final models were validated using MolProbity v.4.3.148 and EMRinger49, with final statistics given in Table 1.
Over-fitting was monitored using cross-validation22 (Supplementary Fig. 2). In brief, for each structure, the coordinates of the final model were perturbed by random displacement up to 0.5 Å from their starting positions using PDBSET and refined against just one of the half maps (half map 1) using real-space refinement in Phenix47. In this refinement, the same parameters and restraints were used as in the final round of refinement of the deposited model. Fourier-shell-correlation curves were then calculated between the model refined against half map 1 and half map 1 (self-validation) and between the same model and half map 2 (cross-validation). The curves are nearly identical (Supplementary Fig. 2A-B) indicating the absence of over-fitting.
As L0R8F8 was built de novo into a region of the map with an estimated local resolution of ~4Å, we performed additional checks to confirm that the model was consistent with prior knowledge. These checks included: (1) consistency of the 3D model with secondary structure predictions (Supplementary Fig. 5G) and (2) consistency of the 3D model with evolutionary couplings (Supplementary Fig. 5G-I). It is expected that residues that are in close spatial proximity in the model would have co-evolved across the L0R8F8 family. To perform these checks, we used the EVcouplings server50. First, an alignment of 3,650 sequences of L0R8F8 homologs was generated before applying a maximum entropy model to identify evolutionarily coupled pairs of columns in the alignments. The evolutionary coupling scores were then ranked using penalized maximum likelihood with a pseudo-likelihood approximation (pseudo-likelihood maximization; PLM) 51. The seven highest-scoring pairs were mapped onto the model of L0R8F8 (Supplementary Fig. 5H-I). Six of the top seven scoring pairs are spatially close, validating the build of the overall fold.
Figures
All figures were generated using PyMOL52 or Chimera42.
A Life Sciences Reporting Summary for this paper is available.
Data Availability
Maps have been deposited with Electron Microscopy Data Bank under accession codes EMD-3842 (39S assembly intermediate with folded interfacial rRNA) and EMD-3843 (39S assembly intermediate with unfolded interfacial rRNA). Models have been deposited with the Protein Data Bank under accession codes 5OOL (39S assembly intermediate with folded interfacial rRNA) and 5OOM (39S assembly intermediate with unfolded interfacial rRNA). All other data that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgements
This work was funded by the Swedish Research Council (NT_2015-04107), the Swedish Foundation for Strategic Research (Future Leaders Grant FFL15-0325), the Ragnar Söderberg Foundation (Fellowship in Medicine M44/16) (A.A.), the UK Medical Research Council (MC_U105184332), the Wellcome Trust (Senior Investigator Award WT096570), the Agouron Institute and the Louis-Jeantet Foundation (V.R.). S.A. was supported by a FEBS Long-Term Fellowship. J.R. and A.A. were supported by Marie Sklodowska Curie Actions (International Career Grant 2015-00579). Funding to M. Minczuk (MC_U105697135) supported the research activities of S.R. and J.R. at the MRC Mitochondrial Biology Unit. Cryo-EM data were collected at the MRC Laboratory of Molecular Biology and the Swedish National Facility. We thank S. Chen, J. Conrad, M. Carroni and C. Savva for help with data collection, S. Peak-Chew, G. Degliesposti, M. Skehel and F. Stengel for mass spectrometry analysis, J. Grimmett, T. Darling and S. Fleischmann for computing support, D. Marks for help with evolutionary couplings, and M. Minczuk for discussions and unpublished data.
Footnotes
Author Contributions
A.B. processed the data, built and refined the model, and wrote the paper. S.R. processed the data and contributed unpublished data. D.K. processed the data. S.A. and X-C.B. collected the data. J.R. contributed unpublished data. A.A. conceived the project and prepared the sample. V.R. initiated the project. All authors contributed to the final version of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greber BJ, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 3.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis JH, et al. Modular Assembly of the Bacterial Large Ribosomal Subunit. Cell. 2016;167:1610–1622.e15. doi: 10.1016/j.cell.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogenhagen DF, Martin DW, Koller A. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 2014;19:618–629. doi: 10.1016/j.cmet.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 6.De Silva D, Tu Y-T, Amunts A, Fontanesi F, Barrientos A. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015;14:2226–2250. doi: 10.1080/15384101.2015.1053672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H-J, Maiti P, Barrientos A. Mitochondrial ribosomes in cancer. Seminars in Cancer Biology. 2017 doi: 10.1016/j.semcancer.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stokes JM, Davis JH, Mangat CS, Williamson JR, Brown ED. Discovery of a small molecule that inhibits bacterial ribosome biogenesis. elife. 2014;3:e03574. doi: 10.7554/eLife.03574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown A, et al. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346:718–722. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, et al. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Research. 2013;41:7073–7083. doi: 10.1093/nar/gkt423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jomaa A, et al. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Research. 2014;42:3419–3435. doi: 10.1093/nar/gkt1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeder C, Draper DE. A small protein unique to bacteria organizes rRNA tertiary structure over an extensive region of the 50 S ribosomal subunit. Journal of Molecular Biology. 2005;354:436–446. doi: 10.1016/j.jmb.2005.09.072. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Research. 2014;42:13430–13439. doi: 10.1093/nar/gku1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baer RJ, Dubin DT. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Research. 1981;9:323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bar-Yaacov D, et al. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 2016;14:e1002557. doi: 10.1371/journal.pbio.1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ofengand J, Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. Journal of Molecular Biology. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- 18.Häuser R, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung S, Nishimura T, Sasarman F, Shoubridge EA. The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol Biol Cell. 2013;24:184–193. doi: 10.1091/mbc.E12-09-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, et al. Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome. Structure. 2015;23:1858–1865. doi: 10.1016/j.str.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rorbach J, Gammage PA, Minczuk M. C7orf30 is necessary for biogenesis of the large subunit of the mitochondrial ribosome. Nucleic Acids Research. 2012;40:4097–4109. doi: 10.1093/nar/gkr1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cronan JE, Fearnley IM, Walker JE. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Letters. 2005;579:4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, et al. Structure of subcomplex Iβ of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc Natl Acad Sci USA. 2015;112:12087–12092. doi: 10.1073/pnas.1510577112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreev DE, et al. Translation of 5' leaders is pervasive in genes resistant to eIF2 repression. elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cronan JE. The chain-flipping mechanism of ACP (acyl carrier protein)-dependent enzymes appears universal. Biochem J. 2014;460:157–163. doi: 10.1042/BJ20140239. [DOI] [PubMed] [Google Scholar]
- 30.Van Vranken JG, et al. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. elife. 2016;5:174. doi: 10.7554/eLife.17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maio N, et al. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 33.Weis F, et al. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K. Gctf: Real-time CTF determination and correction. Journal of Structural Biology. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. elife. 2016;5:19. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai X-C, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. elife. 2015;4:1485. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2013;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 46.Cryle MJ, Schlichting I. Structural insights from a P450 Carrier Protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc Natl Acad Sci USA. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: phenix.real_space_refine. Computational Crystallography Newsletter. 2013;4:43–44. [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks DS, et al. Protein 3D Structure Computed from Evolutionary Sequence Variation. PLoS ONE. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinreb C, et al. 3D RNA and Functional Interactions from Evolutionary Couplings. Cell. 2016;165:963–975. doi: 10.1016/j.cell.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL molecular graphics system. 2002 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Maps have been deposited with Electron Microscopy Data Bank under accession codes EMD-3842 (39S assembly intermediate with folded interfacial rRNA) and EMD-3843 (39S assembly intermediate with unfolded interfacial rRNA). Models have been deposited with the Protein Data Bank under accession codes 5OOL (39S assembly intermediate with folded interfacial rRNA) and 5OOM (39S assembly intermediate with unfolded interfacial rRNA). All other data that support the findings of this study are available from the corresponding authors upon reasonable request.