Abstract
The present review is a sequel to the previous review on cancer metabolism published in this journal. This review focuses on the selective antiproliferative and cytotoxic effects of mitochondria-targeted therapeutics (MTTs) in cancer cells. Emerging research reveals a key role of mitochondrial respiration on tumor proliferation. Previously, a mitochondria-targeted nitroxide was shown to selectively inhibit colon cancer cell proliferation at submicromolar levels. This review is centered on the therapeutic use of MTTs and their bioenergetic profiling in cancer cells. Triphenylphosphonium cation conjugated to a parent molecule (e.g., vitamin-E or chromanol, ubiquinone, and metformin) via a linker alkyl chain is considered an MTT. MTTs selectively and potently inhibit proliferation of cancer cells and, in some cases, induce cytotoxicity. MTTs inhibit mitochondrial complex I activity and induce mitochondrial stress in cancer cells through generation of reactive oxygen species. MTTs in combination with glycolytic inhibitors synergistically inhibit tumor cell proliferation. This review discusses how signaling molecules traditionally linked to tumor cell proliferation affect tumor metabolism and bioenergetics (glycolysis, TCA cycle, and glutaminolysis).
Keywords: Triphenylphosphonium cation, Pancreatic ductal adenocarcinoma, Extracellular acidification rate, Oxygen consumption rate, Coenzyme Q10
1. Introduction
Cancer cells demonstrate several distinct biologic hallmarks resulting from genetic and epigenetic changes that transform normal cells into tumor cells with unrestricted growth and movement capabilities [1], [2], [3]. One of the key hallmarks recognized more than 60 years ago [4] is cancer cells’ remarkable tendency to reprogram their metabolic capability. Inhibition of oxidative phosphorylation (OXPHOS) leads to elevated glycolytic metabolism [3]. Metabolic reprogramming is a direct result of oncogene activation, notably KRAS, or inhibition of tumor suppressors such as phosphatase and tensin homolog (PTEN) [5], [6], [7], [8]. Also, inhibiting glycolysis using a pyruvate mimic (e.g., dichloroacetate) shifts cellular metabolism to OXPHOS [9]. Thus, a prudent chemotherapeutic strategy is to combine both inhibitors of glycolysis and mitochondrial metabolism.
Recent research implicates a key role for mitochondrial complex I inhibition in the antitumor effects of drugs including metformin [10], [11]. Cancers with mutations in mitochondrial gene encoding for the complex I protein in the electron transport chain (ETC) are more susceptible to mitochondrial inhibition [12]. Cancer patients with defects in oxidative phosphorylation are more likely to respond positively to treatment with drugs that more selectively inhibit oxidative phosphorylation in tumors. Mitochondrial complex I and respiration play a major role in cancer cell proliferation [1], [13]. Mitochondria are now a focal point of many targeted therapies in studies using cell culture and preclinical xenograft models of various cancers [14], [15]. However, discovery of drugs selectively targeting oxidative phosphorylation in cancer cells has remained a challenge, as classical inhibitors of cellular respiration such as cyanide are well known to poison normal cells too [16]. In this context, the recent developments of delocalized and lipophilic compounds containing the triphenylphosphonium (TPP+) group targeting mitochondria in cancer cells are highly significant from a therapeutic point of view.
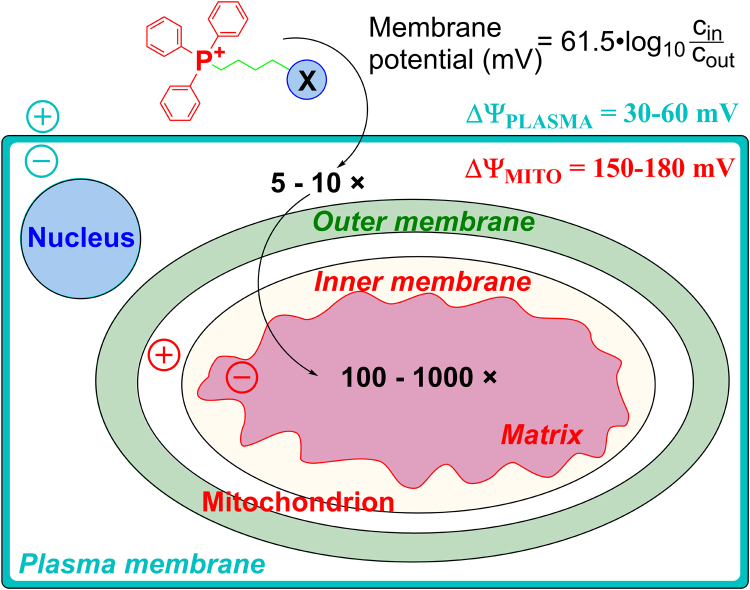
Delocalized lipophilic cationic agents such as rhodamine-123 fluorescent dye were shown to accumulate preferentially in tumor cells (e.g., lung carcinoma cells) and inhibit mitochondrial respiration [17]. Many TPP+-containing compounds were shown to inhibit the proliferation of cancer cells [18]. Key biophysical parameters that account for enhanced cellular uptake of positively charged compounds are increased negative plasma membrane potential and mitochondrial membrane potential [19], [20], [21]. Compared with normal cells, tumor cells exhibit a more negative mitochondrial transmembrane potential [22], which serves as a major driving force for enhanced accumulation of positively charged compounds or drugs (Fig. 1). However, factors contributing to differences in the mitochondrial membrane potential between tumor cells and the corresponding normal cells remain unclear. The positively charged compounds inhibit mitochondrial respiration in cancer cells [1], [14]. One of the strategies to “hypersensitize” tumor cells involves the combined use of mitochondrial inhibitors (oligomycin and antimycin) or delocalized cationic compounds with an antiglycolytic agent, 2-deoxyglucose (2-DG) [23], [24]. Therefore, dual targeting of mitochondrial and glycolytic pathways was proposed as a promising chemotherapeutic strategy [14]. In addition to tumor cells, cardiac muscle cells exhibit the enhanced negative mitochondrial transmembrane potential that contributes to enhanced accumulation and sensitivity to positively charged compounds [20], [25].
Fig. 1.
Cellular uptake of TPP+-linked compounds driven by plasma membrane and mitochondrial membrane potentials (Obtained and Reprinted with permission from Ref. [21]. Copyright 2017 American Chemical Society.).
One of the most important factors is the increased toxicity of this combination to normal cells. For example, mitochondrial inhibitors of OXPHOS, such as oligomycin, in combination with 2-DG, can rapidly eradicate tumor cells [26]. However, because these agents will also affect the mitochondrial and glycolytic metabolism of normal, nontransformed cells, they will lack the tumor cell selectivity that is crucial for their use as chemotherapeutic agents.
2. Selective targeting of mitochondria
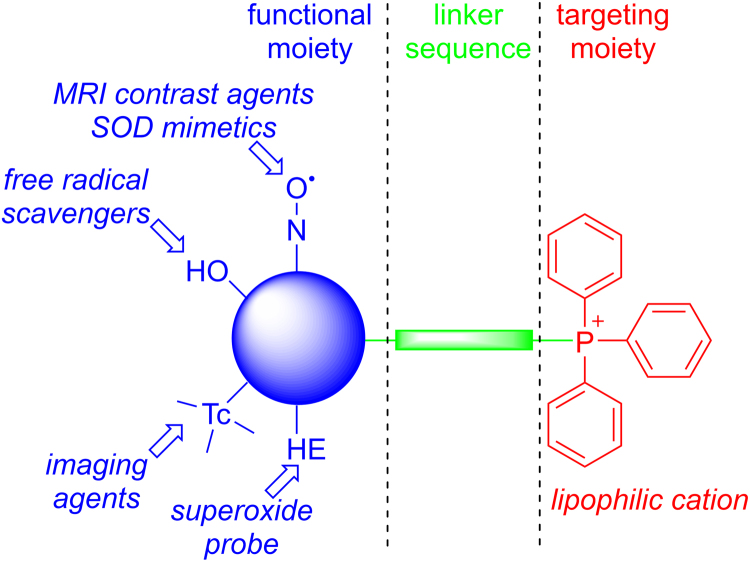
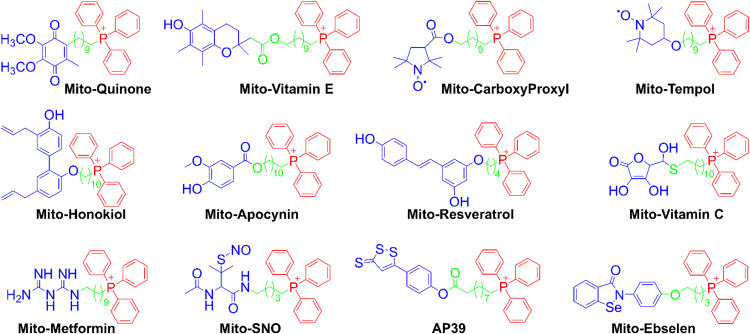
Lipophilic cations were used as carriers to target various biologically active molecules to the mitochondrial inner membrane and matrix driven by mitochondrial membrane potential. An example of a lipophilic-cationic-linked mitochondria-targeted antioxidant is mitoquinone (Mito-Q) that is synthesized by combining the naturally occurring coenzyme Q with a TPP+ moiety through a linker aliphatic chain [27], [28]. Murphy and coworkers developed the methodology to “fine-tune” the chemical structure of Mito-Q by altering the aliphatic chain length such that they are more effectively targeted to the mitochondrial matrix and membranes [29]. We adapted this approach to investigate the mitochondrial mechanism in tumor biology. The anatomy of a typical mito-targeted molecule with a different functional group conjugated to the TPP+ is shown in Fig. 2 [21]. The parent “untargeted” molecule is shown in blue. Typically, the “untargeted” molecule is a nitroxide that exhibits a superoxide dismutase mimetic activity, a phenolic hydroxyl exhibiting a radical scavenging property, a radiolabeled technetium for use in metabolic imaging, or a hydroethidine moiety that forms a diagnostic marker product (e.g., 2-hydroxyethidium) upon reaction with a superoxide radical anion [30], [31]. The functional group is conjugated to a TPP+ cation (shown in red) via an alkyl chain or other linker (shown in green). Depending on the length of the linker alkyl chain (typically n = 2–10 carbons), the mitochondrial uptake and antiproliferative potency in cancer cells may be altered. A few examples of chemical structures of the compounds conjugated to the TPP+ group via an alkyl chain are shown in Fig. 3. This technology also circumvents the poor solubility problems of the “untargeted” molecule such as coenzyme Q10 (CoQ10). Initially, we synthesized mitochondria-targeted therapeutics (MTTs) by conjugating a TPP+ cation to a quinone, nitroxide, or chromanol moiety.
Fig. 2.
Anatomy of TPP+-based mitochondria-targeted agents (Obtained and Reprinted with permission from Ref. [21]. Copyright 2017 American Chemical Society.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
Fig. 3.
Examples of the TPP+-conjugated compounds for their mitochondrial delivery. Color coding represents the three parts of the mitochondria-targeted molecules: functional moiety (blue), linker (green), and targeting moiety (red). (Obtained and Reprinted with permission from Ref. [21]. Copyright 2017 American Chemical Society.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
The mitochondrial inner membrane contains ETC proteins that regulate cell respiration or oxidative phosphorylation and also regulate transport of metabolites between the mitochondrial matrix and the cytosol. Mitochondrial drug targeting includes the ETC, mitochondrial permeability transition, Bcl-2 family proteins, and mitochondrial DNA.
3. Monitoring cancer cell mitochondrial bioenergetics: oxygen consumption rate and extracellular acidification rate
As discussed in previous publications [3], [32], cancer cells change and adapt depending on the metabolic or bioenergetic requirements needed to sustain their unrestricted growth. To meet the needs of rapid proliferation, cancer cells change their substrate preference, including increased glucose, glutamine, and/or lipid metabolism. Thus, the metabolic phenotypes (glycolytic, aerobic, or glutaminolytic) of cancer cells vary, and measuring or monitoring the parameters linked to the hallmarks of cancer (metabolic reprogramming, metabolic phenotype, and substrate preference) will provide increased understanding of tumor cells’ metabolic needs, which will help in the design of metabolic therapies. Two key, readily measurable bioenergetics parameters that link metabolic reprogramming, metabolic phenotype, and substrate preference in cancer cells are glycolytic function or extracellular acidification rate (ECAR) and mitochondrial respiration or oxygen consumption rate (OCR).
The Agilent Seahorse Extracellular Flux (XF) Analyzer is a tool used to measure OCR in culture in real time and has facilitated the study of cellular metabolism in a high throughput fashion. Typically, for determination of mitochondrial function in intact cells, OCR is measured in response to consecutive addition of (i) oligomycin, the inhibitor of adenosine triphosphate (ATP) synthase, (ii) the mitochondrial uncoupler carbonyl cyanide p-triflouromethoxyphenylhydrazone (FCCP), and (iii) complex I and III inhibitors (rotenone and antimycin A, respectively). The changes in both glycolytic and mitochondrial activity in tumor cells in response to treatment can be measured in real time [33], [34].
The Seahorse XF Analyzer is a fully integrated 24- or 96-well instrument that simultaneously measures the cellular OCR due to OXPHOS and ECAR associated with glycolytic metabolism. Each XF assay kit contains a disposable sensor cartridge, embedded with pairs of fluorescence biosensors for oxygen and pH that are coupled to a fiber optic waveguide (532 nm/650 nm for oxygen sensors and 470 nm/530 nm for pH sensors). Each well is equipped with four reagent delivery chambers for injecting substrates and inhibitors into wells during assays. The red and blue dots at the tips of the sensor probes represent the fluorescent sensors for either oxygen or protons. These measurements can be made repeatedly over time in the same cell population [33], [34], [35].
The measurement of mitochondrial respiration has been a reliable assay for assessing mitochondrial function [35]. Using sequential injection of ETC inhibitors (oligomycin, FCCP, and antimycin A), one can measure and quantify multiple mitochondrial parameters such as basal OCR, ATP-linked OCR, proton leak, maximal respiratory capacity, and mitochondrial reserve respiratory capacity. Briefly, the measurement of mitochondrial function in control and treated cells involves three steps. In step 1, after basal OCR is measured, oligomycin (an inhibitor of mitochondrial ATP synthase) is added. The decrease in OCR resulting from this treatment is attributed to the oxygen consumption used to generate ATP. In step 2, the proton ionophore FCCP is added. This uncouples the electron transfer along ETC from the ATP synthesis in complex V and allows for unrestricted electron flux through the ETC, leading to an increase in OCR. The stimulated oxygen consumption is attributed to the maximal mitochondrial respiration rate. In step 3, the complex III inhibitor, antimycin A, is added to determine the oxygen consumption via ETC-independent mechanisms.
Basal respiration is defined as mitochondrial OCR obtained by subtracting the residual OCR after administering ETC inhibitors from the total cellular oxygen consumption in the absence of modulators of mitochondrial function. The residual (ETC-independent) OCR is typically referred to as non-mitochondrial oxygen consumption. Coupled respiration is calculated by subtracting the residual respiration after adding oligomycin from basal respiration. The calculation of the proton leak is based on the difference between OCR measured after oligomycin treatment and non-mitochondrial OCR. Maximal respiration is measured after the addition of FCCP, a potent protonophore that uncouples mitochondrial ATP generation from oxygen consumption.
4. Synergistic effects of mitochondria-targeted drugs and glycolytic inhibitor: cell proliferation and cytotoxicity
Both mitochondrial and antiglycolytic drugs have different molecular targets; one would expect that combining both drugs would elicit synergistic effects. To test the synergy between agents, the effect of their combination on the extent of colony formation can be compared with the dose response to single agents. The ability to form colonies is one of the hallmarks of cancer cells [36]. Breast cancer cells (MCF-7, MDA-MB-231) were treated with 2-DG at several concentrations and colony formation was monitored [14]. No significant decrease in colony formation was observed. In contrast, there was a decrease in colony formation of breast cancer cells treated with mitochondria-targeted carboxy-proxyl (Mito-CP) or Mito-Q. Both Mito-CP and Mito-Q potently inhibited the survival fractions of breast cancer cells as compared with MCF-10A (noncancerous) cells in the presence of 2-DG. Incubation with the Dec-TPP+ and varying levels of 2-DG had only a minor effect on the survival fraction in breast cancer cells.
5. Synergistic effects of mitochondria-targeted compounds and 2-DG: ATP measurements and bioenergetic function
Intracellular ATP levels were measured in MCF-7 and MCF-10A cells treated with 2-DG and Mito-CP, in combination and alone. Results show that, in the presence of both 2-DG and Mito-CP, ATP levels decrease more in MCF-7 cells than in MCF-10A cells [14]. Results also suggest that the extent of ATP depletion in normal and breast cancer cells depends on the MTT and cell type. In nearly all cases, the combined use of both a mitochondria-targeted drug and 2-DG potentiated intracellular ATP depletion. Untargeted compounds (i.e., parent compounds that are not tethered to the TPP+ group) did not elicit the same effect in the presence of 2-DG.
Mito-CP or Mito-Q and 2-DG dramatically decreased intracellular ATP levels in both MCF-7 and in MCF-10A cells, and the cell survival (clonogenic analyses following several days after treatment of cells with drugs) results showed that the extent of inhibition was greater in MCF-7 cells than in MCF-10A cells. This suggested that normal cells such as MCF-10A cells recovered from the combinatorial treatment (mitochondrial and glycolytic inhibition), whereas the breast cancer (MCF-7 or MDA-MB-231) cells failed to recover during the same treatment conditions.
To better relate bioenergetics changes with clonogenic survival, an experimental protocol for bioenergetics function measurements had to be devised that is similar to that used to measure clonogenic survival. Both MCF-7 and MCF-10A cell lines were treated with Mito-CP, Mito-Q, and 2-DG for 6 h followed by washout of treatments and addition of fresh media. Mitochondrial bioenergetics parameters, OCR and ECAR, were obtained in MCF-7 and MCF-10A cells using a Seahorse XF-24 Analyzer. Both Mito-CP and Mito-Q in the presence of 2-DG (after a 36 h treatment) inhibited basal OCR and OCR linked to ATP production to a significantly greater extent in MCF-7 cells, as compared with MCF-10A cells. Note that after a 6 h exposure, the same treatment (Mito-CP or Mito-Q plus 2-DG) inhibited basal OCR in both MCF-7 and MCF-10A cells. Clearly, the normal cells are more adept at recovering from inhibition of mitochondrial function than MCF-7 cells. Thus, it appears that mitochondria-targeted drugs cause an irreversible inhibition of mitochondrial function in cancer cells in contrast to normal cells. The reversible inhibition of mitochondrial function induced by mitochondria-targeted drugs (Mito-CP, Mito-chromanol) in normal cells differs from the irreversible mitochondrial function induced by rotenone (a potent complex I inhibitor) in both cancer and normal cells. Synergistic exacerbation in cytotoxicity occurs in combination with inhibitors of glycolysis and drugs targeting mitochondria [37].
6. Overcoming multidrug resistance using MTTs?
Recently, it was proposed that mitochondria-targeted drugs could counteract ABCA1-dependent resistance of the lung carcinoma cells [38]. Multi-drug resistance-1 (MDR-1) is a multidrug transporter p-glycoprotein that pumps out positively charged chemotherapeutic drugs from cancer cells resulting in chemotherapeutic causing drug resistance of many tumors [19], [39]. Consequently, the chemotherapeutic drug loses its efficacy. However, ATP is required for the pumping mechanism of MDR-1. Mitochondria-targeted drugs (Mito-CP, Mito-chromanol, or metformin analogs) could hinder the pump activity by depleting intracellular ATP levels [14]. Thus, one of the key advantages of using mitochondria-targeted drugs in combination with conventional drugs (cis-platin or doxorubicin) that induce multidrug resistance through elevated expression of MDR-1 is their ability to decrease or deplete intracellular ATP levels. Reports also indicate that ATP regulates chemoresistance in colon cancer cells [40]. However, the uptake of hydrophobic MTTs such as Mito-Q in the brain was shown to be decreased compared with other organs because of the presence of the p-glycoprotein in the blood brain barrier [41].
Solid tumors influence and are influenced by adjacent cells in the oxygen- and nutrient-deprived microenvironment surrounding the transformed cancer cells [42] because of an abnormal vascularization of solid tumors and a lack of adequate blood supply to tumors. Cells residing in such environments are slowly growing with altered phenotypic characteristics. These phenotypic alterations make them resistant to chemotherapeutic agents that rely on DNA replication and cell division as an antitumor mechanism. Tumor cells in a metabolically compromised microenvironment may respond differently to metabolic inhibitor drugs targeting oxidative phosphorylation.
7. Differential susceptibility of pancreatic cancer cells to 2-DG
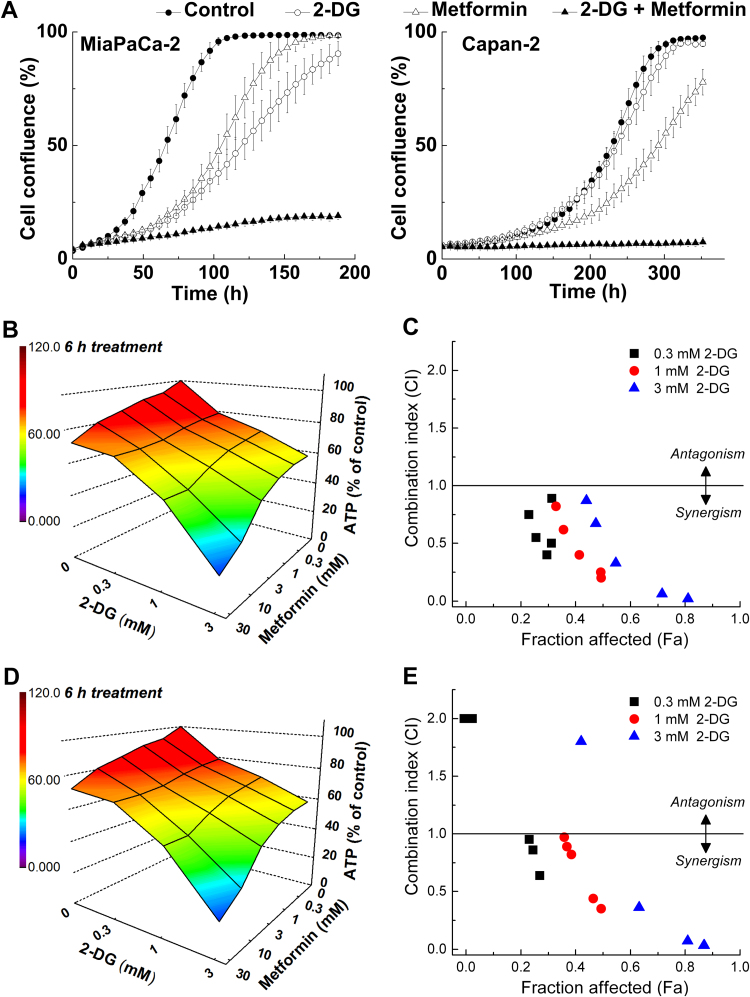
Six different pancreatic cancer cell lines of different genetic backgrounds were treated with 2-DG, and intracellular ATP levels were measured using a luciferase-based assay. These cells were differentially sensitive to 2-DG with respect to ATP depletion. MiaPaCa-2 cells and Capan-2 were the most and least sensitive cell lines, respectively. A three-dimensional heat map representation of intracellular ATP levels as a function of concentration and time of treatment with 2-DG in the most and least sensitive cell lines is shown [43]. Similar heat map studies indicate that different cell lines exhibit different sensitivities to 2-DG [43].
The measurement of the colony-forming ability (clonogenic assay) under the same conditions showed that 2-DG caused a more-extensive decrease in colony formation in MiaPaCa-2 cells than in Capan-2 cells. The cell survival fraction indicates the extreme susceptibility of ASPC-1 and MiaPaCa-2 cells to 2-DG and the resistance of Capan-2 cells to 2-DG.
8. Two-dimensional mapping of bioenergetics profiles: chemotherapeutic predictive values
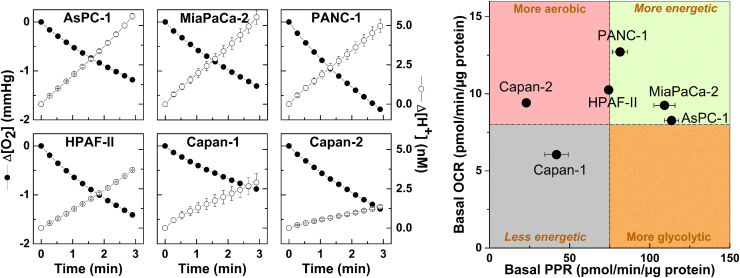
To better understand these results (differential sensitivity of cancer cells to an anti-glycolytic such as 2-DG), we measured the metabolic and energetic requirements. Both oxygen consumption and glycolytic metabolism were measured. The OCR and ECAR values were measured using a Seahorse XF Analyzer and a two-dimensional OCR/proton production rate (PPR) bioenergetics map was constructed (Fig. 4). The PPR parameter is similar to ECAR but is directly related to changes in proton (H+) concentration, while ECAR is related to changes in pH (which is a logarithmic scale). The cell line Capan-2 was the least glycolytic, whereas MiaPaCa-2 cells were the most glycolytic. OCR values for Capan-2 and MiaPaCa-2 cells were nearly the same, but the PPRs, which are a surrogate marker of glycolysis, differ by a factor of five (Fig. 4). These results suggest that monitoring changes in bioenergetic metabolism or bioenergetics profiling in pancreatic ductal adenocarcinoma cells (PDACs) may provide new biomarkers and mechanistic insights into how targeted blockades of glycolytic and mitochondrial metabolism pathways can be used effectively in cancer treatment.
Fig. 4.
Two-dimensional map of bioenergetics in pancreatic cancer cells. (left) Oxygen consumption (ΔO2) and proton production (ΔH+) traces in six pancreatic cancer cell lines as monitored with a Seahorse XF-96 Analyzer. The changes in O2 and H+ concentrations were normalized to 1 µg of protein. (right) Two-dimensional map of OCR and PPR measured in six pancreatic cancer cell lines. (Obtained from Ref. [43], Copyright © 2014, Rights Managed by Nature Publishing Group).
9. Dual targeting of mitochondrial and glycolytic pathways in PDACs: the synergistic effect of metformin and 2-DG
Mitochondria-targeted drugs exhibit either cytostatic or cytotoxic effects in tumor cells. These effects are dependent on their ATP demands. If cellular ATP demands exceed its production/supply in highly proliferating tumor cells (high energy demand), these cells will die. However, in normal cells with nonproliferation and low energy demand, ATP demand is lower and the cells survive. These effects are dependent on the presence of bioenergetic substrates, e.g., on glucose availability.
Metformin is a relatively safe antidiabetic drug, but its antitumor effects are only modest or, in some cases, negligible [44]. The bioavailability of metformin in cancer patients is not robust, and this compromises its antitumor effects. Thus, maximizing metformin's monotherapy will be highly beneficial in cancer treatment. Combining metformin with 2-DG synergistically depleted ATP levels in each of the PDAC cell lines, including poorly glycolytic Capan-1 and Capan-2 cells (Fig. 5) [43]. The heat maps of ATP depletion in response to 2-DG and metformin treatment of MiaPaCa-2 cells show a strong synergy for the combined therapeutic effect (Fig. 5) [43]. These results indicate that metformin treatment sensitizes pancreatic cancer cells to 2-DG. Metformin/2-DG combined therapy is more effective in pancreatic cancer cells that are more resistant to antiglycolytic monotherapy. In the tumor microenvironment, glucose levels are decreased due to glucose's increased consumption from glycolytic metabolism and/or inefficient blood delivery. This may account for metformin's enhanced in vivo antitumor efficacy in some cases. The chemotherapeutic efficacy of several other standard-of-care drugs such as gemcitabine, doxorubicin, and celecoxib was increased in PDACs when used in combination with 2-DG [43].
Fig. 5.
Inhibition of cell proliferation by 2-DG and metformin, and synergistic depletion of ATP by 2-DG and metformin in MiaPaCa-2 cells. (A) Effects of 2-DG and metformin alone and together, on cell proliferation. MiaPaCa-2 and Capan-2 cells were treated with 2-DG (0.5 mM in MiaPaCa-2, 1 mM in Capan-2 cells) or metformin (1 mM) alone and together. Cell proliferation was monitored in real time with the continuous presence of indicated treatments until the end of each experiment. The changes in cell confluence are used as a surrogate marker of cell proliferation. Data shown are the mean ± SD. (n = 6). (B–D) MiaPaCa-2 cells were treated with 2-DG (0.3–3 mM) or metformin (0.3–30 mM) independently and together for 6 h (B) or 24 h (D) and intracellular ATP levels were determined, normalized to total cellular protein amount, and expressed as percentage of untreated cells. A three-dimensional representation showing the concentration-dependent effects of 2-DG or metformin alone and together on intracellular ATP levels in MiaPaCa-2 cells. The combination index-fraction affected (CI-Fa) plots are shown (C,E). Fraction affected parameter is used as a measure of the drug(s) efficiency, with a value of zero indicating the lack of effect on intracellular ATP and the value of 1 indicating total depletion of intracellular ATP. (Obtained from Ref. [43], Copyright © 2014, Rights Managed by Nature Publishing Group).
10. Measurement of IC50 values to inhibit mitochondrial complex I activity in normal and cancer cells
As discussed previously, the mechanism of metformin's antiproliferative effects involved inhibition of mitochondrial complex I. We conjugated metformin to a TPP+ cation to improve its cellular uptake and mitochondrial accumulation [45]. The resulting mitochondria-targeted metformin analog (Mito-Met10) was significantly more potent in inhibiting the proliferation of PDACs, which was later confirmed in an independent study [46]. To directly compare the inhibitory effects of metformin and Mito-Met10 on complex I, pancreatic cancer cells (MiaPaCa-2) were treated with metformin and Mito-Met10 at different concentrations for 24 h, followed by membrane permeabilization and OCR measurements in the presence of complex I substrates using a Seahorse XF Analyzer [45]. Initially, the oxygen consumption traces were obtained for pancreatic cancer cells treated with different concentrations of metformin or Mito-Met10. In order to derive the IC50 values, the mitochondrial complex I-mediated oxygen consumption (last OCR reading prior to injecting succinate) was plotted against metformin or Mito-Met10 concentrations [45]. Mito-Met10 was significantly more potent in complex I inhibition than metformin, with an IC50 value almost three orders of magnitude lower (0.8 mM and 2 µM for metformin and Mito-Met10, respectively). Interestingly, Mito-Met10 seems to exhibit a better selectivity than metformin toward PDAC cells, as much higher concentrations were required to inhibit complex I-mediated oxygen consumption in normal, nontransformed cells, HPNE and IEC-6. Thus, it is evident that mitochondria targeting selectively inhibits complex I activity (as measured by OCR) in cancer cells such as MiaPaCa-2. The close relationship between the IC50 value for complex I-mediated OCR and the IC50 value for the antiproliferative effect (measured from cell survival) in pancreatic cancer cells determined under similar experimental conditions suggests that mitochondrial complex I inhibition may be mechanistically linked to the antiproliferative effects of mitochondria-targeted drugs in cancer biology [45].
11. Metabolic reprogramming in cancer cells
11.1. Glutaminolysis
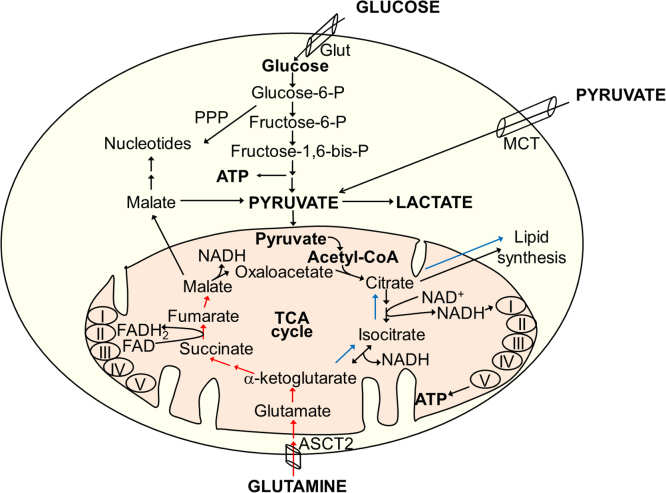
Glutamine is the most abundant amino acid [47]. Glutamine catabolism is another pathway by which cancer cells can derive nutrients and energy. One of the compensatory responses in cancer cells treated with mitochondrial inhibitors is enhanced uptake of glutamine. In cancer cells, this pathway called glutaminolysis is presumably 10-fold higher than that of any other amino acid. Survival of some cancer cells (e.g., melanoma) is linked to glutamine utilization (glutamine addiction) [48]. Glutaminolysis provides precursors for nucleotide, protein, and amino acid (nitrogen source) biosynthesis and substrates for the tricarboxylic acid (TCA) cycle in mitochondria [49]. Glutamine is transported across the plasma membrane and the inner mitochondrial membrane by glutamine transporters and converted to α-ketoglutarate (α-KG) via glutamate. The α-KG, a substrate in the TCA cycle, can undergo oxidative metabolism via succinate (Fig. 6, red arrows) or reductive metabolism via isocitrate (Fig. 6, blue arrows). One of the key aspects of metabolic reprogramming in tumor cells involves glutaminolysis. Recent developments in metabolomics studies have made it possible to investigate the metabolic reprogramming that occurs in tumor cells in response to drug treatments.
Fig. 6.
Metabolic pathways of glucose and glutamine in cells. Red and blue arrows indicate oxidative and reductive pathways of α-KG metabolism. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article)
11.2. Stable isotope tracer studies
The Seahorse technology measures OCR and ECAR that give some information on metabolic reprogramming related to glycolysis and mitochondrial respiration. However, liquid chromatography–mass spectrometry-based stable isotope tracer studies provide more definitive and quantitative information on changes in glycolytic and TCA intermediates in response to metabolic reprogramming. The basic principles of stable isotope studies are described below (Fig. 7) [50], [51], [52].
Fig. 7.
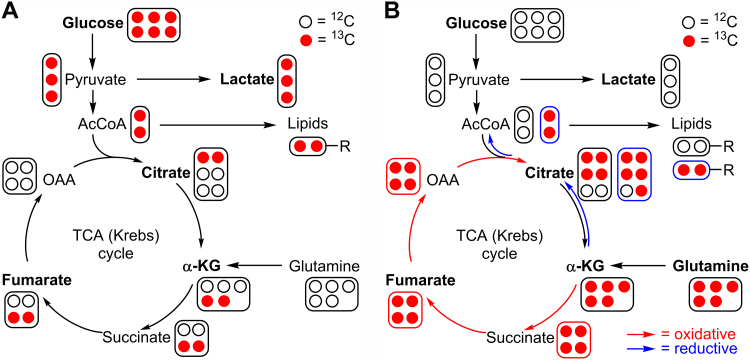
Measuring glucose and glutamine metabolism by LC-MS-based stable isotope tracing. The labeling patterns for Krebs cycle intermediates are shown for one cycle only. (A) Pathways of 13C enrichment from 13C6-glucose. (B) Pathways of 13C enrichment from 13C5-glutamine in oxidative and reductive pathways. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article)
Normal glucose contains six carbon-12 atoms (atomic weight, 12; NMR silent, I = 0). Fully labeled 13C6-glucose (m+6) (Fig. 7, red circles) is used as a tracer (atomic weight, 13; NMR active, I = 1/2), and glycolytic metabolism is assessed by measuring 13C3-lactate (three 13C atoms [m+3]). The labeling patterns for Krebs cycle intermediates (citrate, fumarate) from 13C6-glucose after a single-cycle oxidative pathway are [m+2]citrate and [m+2]fumarate, respectively (Fig. 7A). Inhibitors of mitochondrial respiration often induce a compensatory increase in aerobic glycolysis. Thus, 13C6-glucose will be oxidized to [m+3]pyruvate accompanied by an increase in [m+3]lactate in cancer cells under conditions of decreased respiration.
Glutaminolysis is assessed using a fully labeled 13C5-glutamine (five red circles, Fig. 7B). The extent of 13C-enrichment of α-KG [m+5] is used to monitor total glutaminolysis activity. The labeling pattern of citrate provides insight into the relative extent of oxidative and reductive α-KG metabolism (red and blue arrows, Fig. 7B). The oxidative pathway yields [m+4]citrate and the reductive pathway yields [m+5]citrate.
In summary, the labeling patterns of citrate, α-KG, succinate, and fumarate using 13C6-glucose reveal the impact of inhibitors on glucose-driven mitochondrial TCA cycle and OXPHOS, and the labeling patterns of α-KG, succinate, fumarate, and citrate from 13C5-glutamine can be used to assess the extent of glutaminolysis.
11.3. Metabolic flux profiling
Using the stable isotope tracer technique, it was shown that metabolic reprogramming occurs in melanocytes such that the TCA cycle was fully functional in melanoma cells, even under hypoxic conditions [51], [52]. In addition, measurement of 13C-labeled TCA cycle metabolites in melanoma cells using 13C glucose revealed relatively little 13C-labeling [53]. This suggests that cancer cell metabolism under hypoxia is not restricted to glycolysis. Because glutamine is a frequently used carbon source for cancer cells, it was hypothesized that glutamine is a likely nutrient contributing to TCA cycle function in melanocytes.
This hypothesis was tested using 13C-glutamine. Melanoma cells under hypoxia and normoxia were treated with 13C-glutamine. Formation of TCA cycle metabolites that were labeled with 13C atoms suggests that glutamine is used as a substrate to drive the TCA cycle. Hypoxia markedly increased the contribution from glutamine as indicated by enhanced 13C-labeling of TCA cycle metabolites.
Fig. 7B shows the labeling of metabolites from 13C-glutamine. Typically, the TCA cycle pathways were considered to be oxidative and unidirectional. However, 13C-labeling studies have provided new insights into the reverse flux or the reductive carboxylation of glutamine-derived α-KG. As mentioned previously, the blue arrows in Fig. 7B show the reverse flux from glutamine, and the red arrows denote the oxidative pathway. 13C-labeling of fatty acid proves a direct reverse metabolic route between 13C-glutamine, citrate, and fatty acid synthesis. It was also determined that 13C-glutamine did not affect citrate formed from the oxidative pathway, because there was no 13C-labeling of pyruvate or acetyl-CoA in those studies. The percentage of 13C-labeled fatty acid was increased in melanoma cells treated with 13C-glutamine under hypoxia compared with normoxia. In the presence of 13C-glutamine, the citrate mass unit was increased by five. This was because the reverse flux and hypoxia increased the citrate (m+5) by 38% as compared with normoxia (12%) [51], [52], [53].
12. Intermediacy of an oncometabolite, 2-hydroxyglutarate, formed in the TCA cycle: potential biomarker in precision oncology?
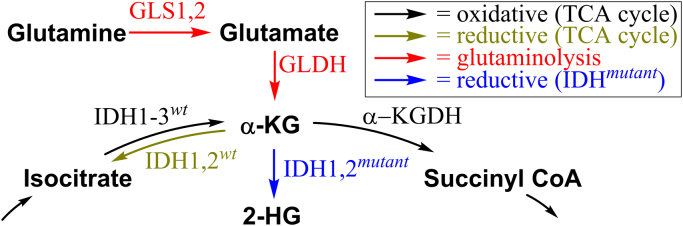
Isocitrate dehydrogenase (IDH) enzymes play a critical role in tumorigenesis [54]. There are several isoforms of IDH, namely IDH1, IDH2, and IDH3. IDH1 catalyzes the reversible conversion of isocitrate to α-KG in the cytosol and peroxisomes, whereas IDH2 performs a similar reaction in mitochondria [55]. IDH3 is responsible for the same reaction within the TCA cycle. In certain types of cancers, for example glioblastoma multiforme, mutations in IDH1 and IDH2 were identified. The mutated enzymes catalyze the reduction of α-KG to D-2-hydroxyglutarate, which is dependent on Mg2+ and NADPH (Fig. 8). Mutated IDH1 and IDH2 are proposed to activate oncoproteins, e.g., via epigenetic mechanisms. Under hypoxic conditions, the other stereoisomer of 2-hydroxyglutarate, L-2HG, is formed and both isomers are considered to act as oncometabolites [56], [57]. 2-Hydroxyglutarate competitively inhibits α-KG-dependent enzymes, including the TET family of enzymes (e.g., 5-methylcytosine hydroxylases and histone lysine demethylases) [58]. 2-hydroxyglutarate-mediated modulation of histone and DNA methylation leads to hypermethylated phenotypes in some cancers [59]. 2-hydroxyglutarate is not detectable in a normal brain, and elevated levels of this oncometabolite were found in tumors with mutant IDH1 compared with wild-type IDH1.
Fig. 8.
Metabolism of α-KG in the presence of wild-type and mutated IDH enzymes.
The presence of 2-hydroxyglutarate has been used as an intraoperative biomarker of IDH1 mutant brain tumors [60]. Using a sophisticated mass spectrometry technique, investigators identified 2-hydroxyglutarate in IDH1 mutant tumors, enabling clinical decision making with regard to surgical resection of IDH1 tumors because reports exist that aggressive surgical resection of IDH1 mutant tumors result in an improved clinical outcome [61]. Emerging literature indicates that development of targeted therapy against oncogenic IDH1 and IDH2 mutations is a potentially promising avenue of research [62].
13. Regulation and manipulation of signaling pathways and metabolism as a potential anticancer therapy
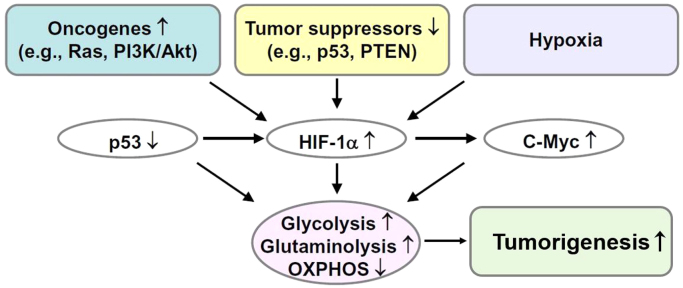
Although tumor metabolism was discovered more than 100 years ago by Otto Warburg, tumor metabolism research did not take off for many decades. This was largely due to the discovery of oncogenes (genes that have the potential to cause cancer) and tumor suppressor genes (genes that encode proteins that prevent cancer) [63]. Emerging research reveals a dramatic resurgence of interest in cancer metabolism and metabolic reprogramming in cancer cells, providing new molecular targets for cancer treatment [64]. Signaling molecules that are traditionally linked to cell proliferation, such as KRAS and PI3K, are now considered to be important regulators of metabolic pathways. There is newfound appreciation to understand how oncogenes and tumor suppressors control tumor cell metabolism and reprogramming [65], [66]. Recent studies indicate that a gain of function in oncogenes and/or inactivation of tumor suppressor genes regulates the high glycolytic metabolism in tumor cells [67].
14. Aerobic glycolysis in cancer cells: PI3K-Akt-mTOR pathway
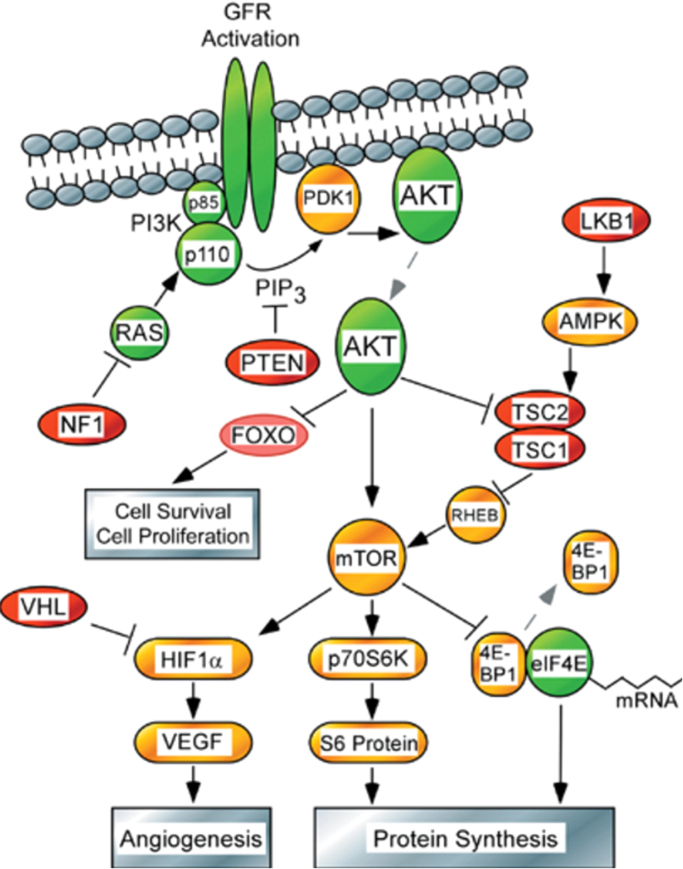
It has been well known for more than 100 years that cancer cells exhibit enhanced rates of aerobic glycolysis when compared with control nontransformed cells [63]. However, the molecular basis for the metabolic switch from oxidative phosphorylation to aerobic glycolysis (the Warburg effect) in cancer cells was only revealed recently [63], [66]. Upregulation of glycolytic enzymes and glucose transporters through activation of Myc, Ras, and Akt and inactivation of p53 is thought to be responsible for enhanced glycolysis in cancer cells (Fig. 9) [68]. The serine/threonine kinase Akt oncogene that is frequently elevated in cancer cells was reported to exert a direct stimulatory effect on glucose metabolism in cancer cells [69]. Akt signaling is upregulated by activation of growth factor receptor tyrosine kinase proteins. The Akt signal transduction pathway includes activation of oncoproteins (shown in green) and inactivation of tumor suppressors (shown in red) (Fig. 10). This is a unique finding in that an oncogene could alter the metabolism of cancer cells.
Fig. 9.
Glycolytic metabolism and activation of signaling pathways.
Fig. 10.
Akt signaling and glycolysis. (Obtained from and Reprinted by permission from Macmillan Publishers Ltd: Ref. [100], copyright 2005). (For interpretation of the references to color in this figure, the reader is referred to the web version of this article)
A major pathway regulating nutrient uptake into cells is the mammalian target of the rapamycin (mTOR) pathway downstream from PI3K/Akt signaling. mTOR is a serine/threonine protein kinase that, as part of mTOR complex I (mTORC1), acts as an important molecular connection between nutrient signals and the metabolic processes indispensable for cell growth. Through a network of downstream signaling pathways, mTORC1 integrates signals from intracellular nutrients and energy with upstream PI3K/Akt growth factor receptor mediated signaling to regulate a battery of anabolic and catabolic processes. Through its control of cellular metabolism, mTORC1 promotes the production of amino acids, fatty acids, and nucleic acids that cumulatively support growth and proliferation of tumor cells. A key response to mTOR signaling is an elevation in the machinery-controlling protein synthesis and cell cycle progression through phosphorylation of translational regulators 4E-binding protein and S6 kinase [3]. mTOR signaling also promotes mitochondrial biogenesis, in part through promotion of PGC-1α [70]. Typically, mTOR, through regulation by AMP-kinase (AMPK) and Akt, is minimally activated, but in cancer, PI3K/Akt overactivation, in conjunction with decreased regulation by AMPK, leads to overactivation of mTOR. Through this pathway, Akt activates glucose uptake and enhanced glycolysis and lactate formation. Akt also increases the mitochondria-bound hexokinase (HK-II) activity and links glucose metabolism to oxidative phosphorylation via stimulation of hexokinase-VDAC interaction at the outer mitochondrial membrane.
AMPK is composed of α, β, and γ subunits that function as master regulators of cellular energy homeostasis [71]. AMPK is typically activated by elevation in cellular AMP or other energy stressors that induce a conformational change in the α-subunit, which in turn exposes threonine-172 (Thr-172) on the γ-subunit for phosphorylation by calcium/calmodulin-dependent protein kinase-kinase 2, or liver kinase B1 [72]. Alternatively, AMPK threonine-172 may be phosphorylated independent from the allosteric changes induced by AMP binding to the γ-subunit [71]. As a sensor of cellular energy, AMPK has an array of effectors that influence cell proliferation and motility. In particular, AMPK activation acts to inhibit mTOR signaling and activate p53, the effects of which restore ATP production through fatty acid oxidation mediated by acetyl-CoA carboxylase, lipogenesis, gluconeogenesis, and in turn cell growth. The antitumor effects of metformin have been linked to activation of AMPK, perhaps through altering levels of AMP, and in turn inhibition of mTOR signaling [73], [74]. Cancer cells also use glucose-dependent metabolism to synthesize lipid precursors that are essential to support membrane biosynthesis. The acetyl-CoA produced from pyruvate is used in the biosynthesis of membrane phospholipids. ATP-citrate lyase (ACL), one of the key Akt-regulated enzymes, is involved in lipid biosynthesis. It was also shown that ACL activation is critical to Akt-mediated tumorigenesis [75]. Thus, Akt regulates several metabolic pathways related to glucose metabolism that are implicated in tumorigenesis [76], [77].
The activation of the Akt signaling pathway is a frequently occurring molecular alteration in malignant tumors. Therefore, targeting the Akt signaling pathway is a rational approach in cancer therapy. However, because of the widespread involvement of Akt signaling in the normal physiological processes (glucose metabolism, insulin signaling, etc.) and the potential toxic side effects of Akt inhibition, tumor cells should be sensitized more selectively than control cells in order to mitigate normal cell toxicity.
15. HIF-1α and glycolysis
Although it is a less efficient form of metabolism, most cancer cells are hypoxic and metabolize pyruvate to lactate for ATP generation [78]. Activation of hypoxia-inducible factor 1 (HIF-1α), a heterodimeric DNA-binding complex composed of two basic helix-loop-helix proteins of the PAS (Per-ARNT-Sim) family, is frequently observed in tumors [79]. HIF-1α is continuously synthesized and degraded, and in normoxic conditions has a relatively short (6 min) half-life. Under hypoxia, or a decreasing oxygen concentration, the rate of HIF-1α degradation is decreased, with prolyl hydroxylation suppressed and HIF-1α proteosomal degradation prevented. Thus, HIF-1α accumulates in hypoxia and translocates to the nucleus, resulting in the activation of target genes responsible for increased glucose uptake and lactate production, and decreased mitochondrial respiration in tumor cells.
HIF-1α regulates multiple enzymes responsible for anaerobic glycolysis in tumor cells such as the upregulation of glucose transporters (Glut-1 and Glut-3) and other glycolytic enzymes. HIF-1α also induces pyruvate dehydrogenase kinase-1 (PDK-1) that phosphorylates the pyruvate dehydrogenase enzyme, leading to its deactivation. Deactivation of pyruvate dehydrogenase prevents pyruvate entry into the TCA cycle. Consequently, the mitochondrial respiration is downregulated.
HIF-1α plays a critical role in mediating physiological and cellular response mechanisms that are necessary for adaptation to hypoxia [80]. Thus, any pharmacological manipulation of the HIF-1α pathway as a therapeutic intervention in cancer treatment should be viewed with caution.
16. PGC1α, oxidative phosphorylation, and targeted inhibitors
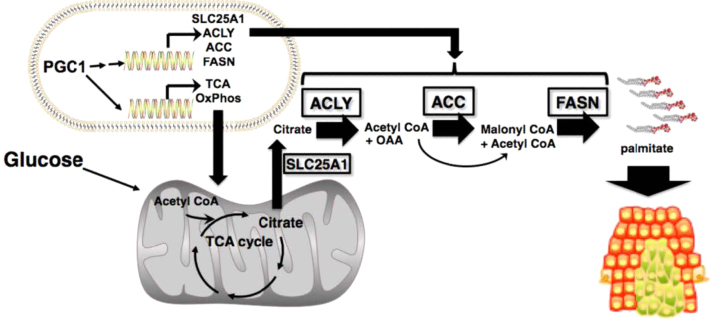
Peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α) is a key regulator of multiple biosynthetic pathways including mitochondrial metabolism and lipogenesis [81]. Recent reports suggest that a subset of human melanomas overexpress PGC1α to support growth and survival through the activation of mitochondrial oxidative metabolism and metabolic compensatory processes through enhanced glutamine utilization [82]. PGC1α induces the expression of mitochondrial OXPHOS and TCA cycle-specific genes, and genes promoting de novo lipogenesis and the pentose phosphate pathway increasing NADPH production for fatty acid synthesis [83]. Although the role of PGC1α in altering the signal transduction pathways for fatty acid synthesis in tumor cells is not yet fully understood, PGC1α remains an attractive therapeutic target for cancer treatment. PGC1α highlights the need to better understand the molecular mechanisms of lipogenesis and its regulation in cancer. Acetyl-CoA generated from glucose is in the mitochondria, whereas the fatty acid synthesis occurs in the cytosol. PGC1α induces the genes responsible for converting the citrate back to oxaloacetate and acetyl-CoA for enhancing fatty acid synthesis in the cytosol (Fig. 11). In some melanoma phenotypes, PGC1α is responsible for increased OXPHOS [84]. These PGC1α-positive cells are more sensitive to inhibitors (e.g., metformin) targeting OXPHOS, as opposed to PGC1α-negative melanoma cells that rely on glycolysis for energy.
Fig. 11.
PGC1α and oxidative phosphorylation. (Obtained from Ref. [83], Copyright © 2012, American Association for Cancer Research).
17. Glutaminolysis, c-Myc, and Rag-mTORC1 signaling
Glutamine, the most abundant amino acid in the blood, is metabolized through glutaminolysis in a two-step process to form α-KG, which is critical for producing oxaloacetate and citrate in the TCA cycle [85]. The enzyme glutaminase, which hydrolyzes glutamine to glutamate and whose activity correlates with tumor growth, is regulated by the oncogene c-Myc. Indeed, Myc-expressing tumor cells undergo apoptosis in the absence of glutamine [86]. Glutamate dehydrogenase binds to leucine and stimulates the conversion of glutamate to α-KG via oxidative deamination. Glutamine also activates the mTOR mediated by leucine and formation of GTP (via the TCA cycle) that is required for activation of mTORC1. Thus, glutaminolysis and mTORC1 play a crucial role in cancer cell metabolism. It was hypothesized that targeted inhibition of both mTORC1 and glutaminolysis may synergistically inhibit tumor cell growth [87].
18. KRAS and tumor cell metabolism
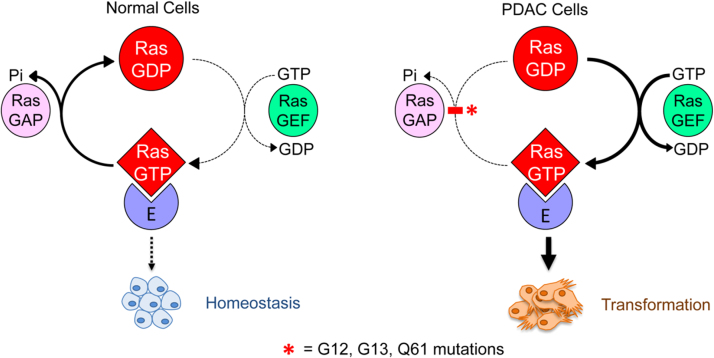
Oncogenic KRAS facilitates reprogramming of tumor cell metabolism [88]. Human Ras proteins are small GTPases that facilitate cycling between an active GTP-bound and an inactive GDP-bound state [89]. In nontransformed cells, active KRAS is rapidly inactivated by guanine-exchange factor proteins into its GDP-bound form, whereas in cancer cells (e.g., PDAC cells), KRAS is predominantly GTP-bound, thereby functioning as a dominant-active protein. KRAS signaling plays roles in cancer cell morphology, proliferation, migration potential, and survival. Metabolically, KRAS mutations decrease mitochondrial OXPHOS while simultaneously increasing glycolysis. Although KRAS mutation occurs in several cancers, it appears with increasing frequency in PDAC, colorectal cancer, and non-small cell lung cancer. As shown (Fig. 12), the direction of signaling in PDAC cells is opposite of that observed normal cells. Oncogenic KRAS promotes glucose uptake through enhanced expression of GLUT1 that accelerates the glycolytic activity [90]. The KRAS oncogene also channels glucose in the pentose phosphate pathway. Signaling by GTP-bound KRAS results in the perpetual signaling of PI3K/Akt/mTOR, which function to exacerbate cancer cell proliferation. Recent work discovered that KRAS signaling in PDAC elevates fluid-phase nutrient uptake through micropinocytosis, providing an alternative source of amino acids needed to sustain the growing tumor [91]. Fig. 12 schematically outlines alterations in signaling by oncogenic KRAS.
Fig. 12.
KRAS in normal and tumor cell metabolism. (Obtained from Ref. [90]; Reprinted from Trends in Biochemical Sciences, 39, Kristen L. Bryant, Joseph D. Mancias, Alec C. Kimmelman, Channing J. Der, KRAS: feeding pancreatic cancer proliferation, Pages No. 91–100, Copyright 2014, with permission from Elsevier).
19. Immunotherapy, mitochondrial bioenergetics, and metabolism
Immunotherapy is an emerging treatment avenue that, in some malignancies, has shown long-lasting remission [92]. Immune cells within the tumor microenvironment are susceptible to the same demand for nutrients, essential metabolites, and oxygen imposed on cancer cells. Moreover, metabolic reprogramming by actively proliferating tumor cells yield by-products such as lactate and kynurenine, which functionally suppress the antitumor immune response [93]. Immune cells share several metabolic features of cancer cells. For example, while memory T cells have quiescent metabolism, the activated T cells switch energy production from OXPHOS to glycolysis [94], [95], [96]. Elevated T cell metabolic activity is necessary at the tumor site to promote tumor killing. Whereas chemotherapy directly kills cancer cells, immunotherapy indirectly kills cancer cells by turning on the immune system. Abnormal glycolytic metabolism normally observed in cancer cells can decrease the cytotoxic T cell activity responsible for destroying cancer cells. Targeting mitochondrial and glycolytic bioenergetics and metabolism in immune cells may be a viable therapeutic strategy in cancer treatment. Both OXPHOS and glycolysis are required for adaptive immune cells (T and B cells), and glycolysis is the predominant form of metabolism in innate immune cells. The goal of immunotherapy is to generate a lasting antitumor effect by inducing antitumor effector T cells that destroy tumor cells. However, the antitumor potency of effector T cells is not maintained for long, so creating long-lasting antitumor memory T cells is an emerging area of research in tumor immune biology. Drugs used in immunotherapy (i.e., checkpoint inhibitors that prevent cancer cells from turning off the immune system) in combination with mitochondria-targeted antiproliferative cationic drugs could have a greater overall antitumor efficacy. Recently, it was shown that mitochondria-targeted antioxidants prevented T cell exhaustion [97].
20. Concluding remarks and future perspectives
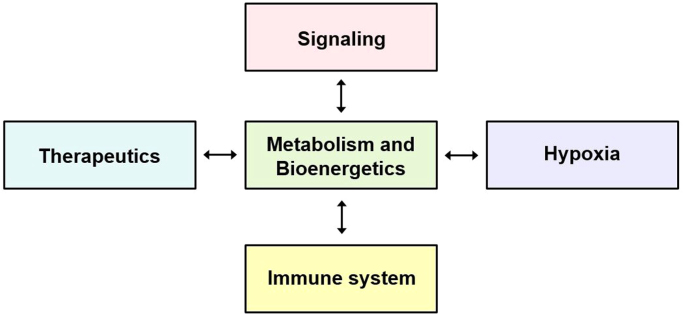
In this review, we discussed the role of MTTs in the selective inhibition of cancer cell proliferation via targeting cancer cell metabolism. These compounds constitute a unique class of drug therapeutics that inhibit mitochondrial respiration, mitochondrial bioenergetics, and redox metabolism in tumor cells at relatively nontoxic concentrations [98]. We also discussed how these compounds may affect oncogenes and tumor suppressor genes that, in turn, regulate metabolic pathways in cancer cells. Bioenergetic mapping of cancer cells may provide new insight into the ways in which blockades of glycolytic, glutaminolytic, and mitochondrial metabolic pathways can enable the selection and use of metabolic inhibitors in combination with conventional standard-of-care drugs in cancer therapy. Based on emerging research, it is clear that all of the aspects discussed here are connected (Fig. 13). Thus, new therapeutics addressing tumor metabolism and bioenergetics, oncogenic signaling, the immune system, and tumor hypoxia [99] are likely to impact future development of anticancer therapy in combination with conventional chemotherapy, radiation therapy, and immunotherapy.
Fig. 13.
The central role of metabolism and bioenergetics.
Acknowledgements
The research reported in this publication was supported by the NIH NCI U01 CA178960 to MD and BK (MPIs) and by NIH NCI R01 CA208648 to Ming You and BK (MPIs). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This review is based on the lectures on cancer biology that BK presented as part of Anna University's Global Initiative Academic Networks (GIAN) program, Chennai, India.
References
- 1.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 5.Beckner M.E., Gobbel G.T., Abounader R., Burovic F., Agostino N.R., Laterra J., Pollack I.F. Glycolytic glioma cells with active glycogen synthase are sensitive to PTEN and inhibitors of PI3K and gluconeogenesis. Lab. Invest. 2005;85(12):1457–1470. doi: 10.1038/labinvest.3700355. [DOI] [PubMed] [Google Scholar]
- 6.Chiaradonna F., Sacco E., Manzoni R., Giorgio M., Vanoni M., Alberghina L. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25(39):5391–5404. doi: 10.1038/sj.onc.1209528. [DOI] [PubMed] [Google Scholar]
- 7.Pelicano H., Xu R.H., Du M., Feng L., Sasaki R., Carew J.S., Hu Y., Ramdas L., Hu L., Keating M.J., Zhang W., Plunkett W., Huang P. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J. Cell Biol. 2006;175(6):913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vizan P., Boros L.G., Figueras A., Capella G., Mangues R., Bassilian S., Lim S., Lee W.N., Cascante M. K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res. 2005;65(13):5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- 9.Michelakis E.D., Sutendra G., Dromparis P., Webster L., Haromy A., Niven E., Maguire C., Gammer T.L., Mackey J.R., Fulton D., Abdulkarim B., McMurtry M.S., Petruk K.C. Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2010;2(31):31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- 10.Andrzejewski S., Gravel S.P., Pollak M., St-Pierre J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2014;2:12. doi: 10.1186/2049-3002-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheaton W.W., Weinberg S.E., Hamanaka R.B., Soberanes S., Sullivan L.B., Anso E., Glasauer A., Dufour E., Mutlu G.M., Budigner G.S., Chandel N.S. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birsoy K., Possemato R., Lorbeer F.K., Bayraktar E.C., Thiru P., Yucel B., Wang T., Chen W.W., Clish C.B., Sabatini D.M. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508(7494):108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urra F.A., Munoz F., Lovy A., Cardenas C. The mitochondrial complex(I)ty of cancer. Front Oncol. 2017;7:118. doi: 10.3389/fonc.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng G., Zielonka J., Dranka B.P., McAllister D., Mackinnon A.C., Jr., Joseph J., Kalyanaraman B. Mitochondria-targeted drugs synergize with 2-deoxyglucose to trigger breast cancer cell death. Cancer Res. 2012;72(10):2634–2644. doi: 10.1158/0008-5472.CAN-11-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulda S., Galluzzi L., Kroemer G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 16.Pollak M. Targeting oxidative phosphorylation: why, when, and how. Cancer Cell. 2013;23(3):263–264. doi: 10.1016/j.ccr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Pathania D., Millard M., Neamati N. Opportunities in discovery and delivery of anticancer drugs targeting mitochondria and cancer cell metabolism. Adv. Drug Deliv. Rev. 2009;61(14):1250–1275. doi: 10.1016/j.addr.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Millard M., Pathania D., Shabaik Y., Taheri L., Deng J., Neamati N. Preclinical evaluation of novel triphenylphosphonium salts with broad-spectrum activity. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtoglu M., Lampidis T.J. From delocalized lipophilic cations to hypoxia: blocking tumor cell mitochondrial function leads to therapeutic gain with glycolytic inhibitors. Mol. Nutr. Food Res. 2009;53(1):68–75. doi: 10.1002/mnfr.200700457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summerhayes I.C., Lampidis T.J., Bernal S.D., Nadakavukaren J.J., Nadakavukaren K.K., Shepherd E.L., Chen L.B. Unusual retention of rhodamine 123 by mitochondria in muscle and carcinoma cells. Proc. Natl. Acad. Sci. USA. 1982;79(17):5292–5296. doi: 10.1073/pnas.79.17.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zielonka J., Joseph J., Sikora A., Hardy M., Ouari O., Vasquez-Vivar J., Cheng G., Lopez M., Kalyanaraman B. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 2017;117(15):10043–10120. doi: 10.1021/acs.chemrev.7b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fantin V.R., Berardi M.J., Scorrano L., Korsmeyer S.J., Leder P. A novel mitochondriotoxic small molecule that selectively inhibits tumor cell growth. Cancer Cell. 2002;2(1):29–42. doi: 10.1016/s1535-6108(02)00082-x. [DOI] [PubMed] [Google Scholar]
- 23.Cheng G., Zielonka J., McAllister D.M., Mackinnon A.C., Jr., Joseph J., Dwinell M.B., Kalyanaraman B. Mitochondria-targeted vitamin E analogs inhibit breast cancer cell energy metabolism and promote cell death. BMC Cancer. 2013;13:285. doi: 10.1186/1471-2407-13-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Hu Y.P., Savaraj N., Priebe W., Lampidis T.J. Hypersensitization of tumor cells to glycolytic inhibitors. Biochemistry. 2001;40(18):5542–5547. doi: 10.1021/bi002426w. [DOI] [PubMed] [Google Scholar]
- 25.Lampidis T.J., Savaraj N., Valet G.K., Trevorrow K. Relationship of chemical charge of anticancer agents to increased accumulation and cytotoxicity in cardiac and tumor cells: relevance to multidrug resistance. J. Cell. Pharm. 1989;89:16–22. [Google Scholar]
- 26.Suganuma K., Miwa H., Imai N., Shikami M., Gotou M., Goto M., Mizuno S., Takahashi M., Yamamoto H., Hiramatsu A., Wakabayashi M., Watarai M., Hanamura I., Imamura A., Mihara H., Nitta M. Energy metabolism of leukemia cells: glycolysis versus oxidative phosphorylation. Leuk. Lymphoma. 2010;51(11):2112–2119. doi: 10.3109/10428194.2010.512966. [DOI] [PubMed] [Google Scholar]
- 27.Murphy M.P., Smith R.A. Drug delivery to mitochondria: the key to mitochondrial medicine. Adv. Drug Deliv. Rev. 2000;41(2):235–250. doi: 10.1016/s0169-409x(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 28.Murphy M.P., Smith R.A. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 2007;47:629–656. doi: 10.1146/annurev.pharmtox.47.120505.105110. [DOI] [PubMed] [Google Scholar]
- 29.Asin-Cayuela J., Manas A.R., James A.M., Smith R.A., Murphy M.P. Fine-tuning the hydrophobicity of a mitochondria-targeted antioxidant. FEBS Lett. 2004;571(1–3):9–16. doi: 10.1016/j.febslet.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 30.Debowska K., Debski D., Hardy M., Jakubowska M., Kalyanaraman B., Marcinek A., Michalski R., Michalowski B., Ouari O., Sikora A., Smulik R., Zielonka J. Toward selective detection of reactive oxygen and nitrogen species with the use of fluorogenic probes--Limitations, progress, and perspectives. Pharmacol. Rep. 2015;67(4):756–764. doi: 10.1016/j.pharep.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zielonka J., Joseph J., Sikora A., Kalyanaraman B. Real-time monitoring of reactive oxygen and nitrogen species in a multiwell plate using the diagnostic marker products of specific probes. Methods Enzymol. 2013;526:145–157. doi: 10.1016/B978-0-12-405883-5.00009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeBerardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C.B. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dranka B.P., Hill B.G., Darley-Usmar V.M. Mitochondrial reserve capacity in endothelial cells: the impact of nitric oxide and reactive oxygen species. Free Radic. Biol. Med. 2010;48(7):905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls D.G., Darley-Usmar V.M., Wu M., Jensen P.B., Rogers G.W., Ferrick D.A. Bioenergetic profile experiment using C2C12 myoblast cells. J. Vis. Exp. 2010;46:2511. doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dranka B.P., Zielonka J., Kanthasamy A.G., Kalyanaraman B. Alterations in bioenergetic function induced by Parkinson's disease mimetic compounds: lack of correlation with superoxide generation. J. Neurochem. 2012;122(5):941–951. doi: 10.1111/j.1471-4159.2012.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Enriquez S., Marin-Hernandez A., Gallardo-Perez J.C., Carreno-Fuentes L., Moreno-Sanchez R. Targeting of cancer energy metabolism. Mol. Nutr. Food Res. 2009;53(1):29–48. doi: 10.1002/mnfr.200700470. [DOI] [PubMed] [Google Scholar]
- 38.Sauna Z.E., Smith M.M., Muller M., Kerr K.M., Ambudkar S.V. The mechanism of action of multidrug-resistance-linked P-glycoprotein. J. Bioenerg. Biomembr. 2001;33(6):481–491. doi: 10.1023/a:1012875105006. [DOI] [PubMed] [Google Scholar]
- 39.Dellinger M., Pressman B.C., Calderon-Higginson C., Savaraj N., Tapiero H., Kolonias D., Lampidis T.J. Structural requirements of simple organic cations for recognition by multidrug-resistant cells. Cancer Res. 1992;52(22):6385–6389. [PubMed] [Google Scholar]
- 40.Zhou Y., Tozzi F., Chen J., Fan F., Xia L., Wang J., Gao G., Zhang A., Xia X., Brasher H., Widger W., Ellis L.M., Weihua Z. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012;72(1):304–314. doi: 10.1158/0008-5472.CAN-11-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porteous C.M., Menon D.K., Aigbirhio F.I., Smith R.A., Murphy M.P. P-glycoprotein (Mdr1a/1b) and breast cancer resistance protein (Bcrp) decrease the uptake of hydrophobic alkyl triphenylphosphonium cations by the brain. Biochim. Biophys. Acta. 2013;1830(6):3458–3465. doi: 10.1016/j.bbagen.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaupel P., Kallinowski F., Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49(23):6449–6465. [PubMed] [Google Scholar]
- 43.Cheng G., Zielonka J., McAllister D., Tsai S., Dwinell M.B., Kalyanaraman B. Profiling and targeting of cellular bioenergetics: inhibition of pancreatic cancer cell proliferation. Br. J. Cancer. 2014;111(1):85–93. doi: 10.1038/bjc.2014.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kordes S., Pollak M.N., Zwinderman A.H., Mathot R.A., Weterman M.J., Beeker A., Punt C.J., Richel D.J., Wilmink J.W. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015;16(7):839–847. doi: 10.1016/S1470-2045(15)00027-3. [DOI] [PubMed] [Google Scholar]
- 45.Cheng G., Zielonka J., Ouari O., Lopez M., McAllister D., Boyle K., Barrios C.S., Weber J.J., Johnson B.D., Hardy M., Dwinell M.B., Kalyanaraman B. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Res. 2016;76(13):3904–3915. doi: 10.1158/0008-5472.CAN-15-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boukalova S., Stursa J., Werner L., Ezrova Z., Cerny J., Bezawork-Geleta A., Pecinova A., Dong L., Drahota Z., Neuzil J. Mitochondrial targeting of metformin enhances its activity against pancreatic cancer. Mol. Cancer Ther. 2016;15(12):2875–2886. doi: 10.1158/1535-7163.MCT-15-1021. [DOI] [PubMed] [Google Scholar]
- 47.Curi R., Lagranha C.J., Doi S.Q., Sellitti D.F., Procopio J., Pithon-Curi T.C., Corless M., Newsholme P. Molecular mechanisms of glutamine action. J. Cell. Physiol. 2005;204(2):392–401. doi: 10.1002/jcp.20339. [DOI] [PubMed] [Google Scholar]
- 48.Scott D.A., Richardson A.D., Filipp F.V., Knutzen C.A., Chiang G.G., Ronai Z.A., Osterman A.L., Smith J.W. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J. Biol. Chem. 2011;286(49):42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.-Y., Pfeiffer H.K., Nissim I., Daikhin E., Yudkoff M., McMahon S.B., Thompson C.B. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiller K., Metallo C.M. Profiling metabolic networks to study cancer metabolism. Curr. Opin. Biotechnol. 2013;24(1):60–68. doi: 10.1016/j.copbio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Zamboni N., Fendt S.-M., Ruhl M., Sauer U. 13C-based metabolic flux analysis. Nat. Protoc. 2009;4(6):878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J., Ahn W.S., Gameiro P.A., Keibler M.A., Zhang Z., Stephanopoulos G. 13C isotope-assisted methods for quantifying glutamine metabolism in cancer cells. Methods Enzymol. 2014;542:369–389. doi: 10.1016/B978-0-12-416618-9.00019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filipp F.V., Scott D.A., Ronai Z.A., Osterman A.L., Smith J.W., Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 2012;25(3):375–383. doi: 10.1111/j.1755-148X.2012.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang D., Wang H.L., Sun Z.N., Chung N.W., Shen J.G. A highly selective fluorescent probe for the detection and imaging of peroxynitrite in living cells. J. Am. Chem. Soc. 2006;128(18):6004–6005. doi: 10.1021/ja0603756. [DOI] [PubMed] [Google Scholar]
- 55.Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C., Marks K.M., Prins R.M., Ward P.S., Yen K.E., Liau L.M., Rabinowitz J.D., Cantley L.C., Thompson C.B., Vander Heiden M.G., Su S.M. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Intlekofer A.M., Dematteo R.G., Venneti S., Finley L.W., Lu C., Judkins A.R., Rustenburg A.S., Grinaway P.B., Chodera J.D., Cross J.R., Thompson C.B. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 2015;22(2):304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.H., Ito S., Yang C., Wang P., Xiao M.T., Liu L.X., Jiang W.Q., Liu J., Zhang J.Y., Wang B., Frye S., Zhang Y., Xu Y.H., Lei Q.Y., Guan K.L., Zhao S.M., Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thienpont B., Steinbacher J., Zhao H., D’Anna F., Kuchnio A., Ploumakis A., Ghesquière B., Van Dyck L., Boeckx B., Schoonjans L., Hermans E., Amant F., Kristensen V.N., Koh K.P., Mazzone M., Coleman M.L., Carell T., Carmeliet P., Lambrechts D. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537(7618):63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C.R., Khanin R., Figueroa M.E., Melnick A., Wellen K.E., O'Rourke D.M., Berger S.L., Chan T.A., Levine R.L., Mellinghoff I.K., Thompson C.B. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pope W.B., Prins R.M., Albert Thomas M., Nagarajan R., Yen K.E., Bittinger M.A., Salamon N., Chou A.P., Yong W.H., Soto H., Wilson N., Driggers E., Jang H.G., Su S.M., Schenkein D.P., Lai A., Cloughesy T.F., Kornblum H.I., Wu H., Fantin V.R., Liau L.M. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J. Neurooncol. 2012;107(1):197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santagata S., Eberlin L.S., Norton I., Calligaris D., Feldman D.R., Ide J.L., Liu X., Wiley J.S., Vestal M.L., Ramkissoon S.H., Orringer D.A., Gill K.K., Dunn I.F., Dias-Santagata D., Ligon K.L., Jolesz F.A., Golby A.J., Cooks R.G., Agar N.Y. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc. Natl. Acad. Sci. USA. 2014;111(30):11121–11126. doi: 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohle D., Popovici-Muller J., Palaskas N., Turcan S., Grommes C., Campos C., Tsoi J., Clark O., Oldrini B., Komisopoulou E., Kunii K., Pedraza A., Schalm S., Silverman L., Miller A., Wang F., Yang H., Chen Y., Kernytsky A., Rosenblum M.K., Liu W., Biller S.A., Su S.M., Brennan C.W., Chan T.A., Graeber T.G., Yen K.E., Mellinghoff I.K. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 64.Sever R., Brugge J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015;5(4):a006098. doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones N.P., Schulze A. Targeting cancer metabolism--aiming at a tumour's sweet-spot. Drug Discov. Today. 2012;17(5–6):232–241. doi: 10.1016/j.drudis.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 66.Thompson C.B. Metabolic enzymes as oncogenes or tumor suppressors. N. Engl. J. Med. 2009;360(8):813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sciacovelli M., Gaude E., Hilvo M., Frezza C. The metabolic alterations of cancer cells. Methods Enzymol. 2014;542:1–23. doi: 10.1016/B978-0-12-416618-9.00001-7. [DOI] [PubMed] [Google Scholar]
- 68.Phan L.M., Yeung S.C., Lee M.H. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014;11(1):1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng J.Q., Ruggeri B., Klein W.M., Sonoda G., Altomare D.A., Watson D.K., Testa J.R. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. USA. 1996;93(8):3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 71.Hardie D.G., Ashford M.L. AMPK: regulating energy balance at the cellular and whole body levels. Physiology. 2014;29(2):99–107. doi: 10.1152/physiol.00050.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inoki K., Zhu T., Guan K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 73.Dowling R.J., Zakikhani M., Fantus I.G., Pollak M., Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67(22):10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- 74.Sinnett-Smith J., Kisfalvi K., Kui R., Rozengurt E. Metformin inhibition of mTORC1 activation, DNA synthesis and proliferation in pancreatic cancer cells: dependence on glucose concentration and role of AMPK. Biochem. Biophys. Res. Commun. 2013;430(1):352–357. doi: 10.1016/j.bbrc.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bauer D.E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24(41):6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 76.Elstrom R.L., Bauer D.E., Buzzai M., Karnauskas R., Harris M.H., Plas D.R., Zhuang H., Cinalli R.M., Alavi A., Rudin C.M., Thompson C.B. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64(11):3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 77.Rathmell J.C., Fox C.J., Plas D.R., Hammerman P.S., Cinalli R.M., Thompson C.B. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol. Cell. Biol. 2003;23(20):7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weidemann A., Johnson R.S. Biology of HIF-1alpha. Cell Death Differ. 2008;15(4):621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 80.Papandreou I., Cairns R.A., Fontana L., Lim A.L., Denko N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 81.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vazquez F., Lim J.H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C.B., Granter S.R., Widlund H.R., Spiegelman B.M., Puigserver P. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhalla K., Hwang B.J., Dewi R.E., Ou L., Twaddel W., Fang H.B., Vafai S.B., Vazquez F., Puigserver P., Boros L., Girnun G.D. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011;71(21):6888–6898. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gopal Y.N.V., Rizos H., Chen G., Deng W., Frederick D.T., Cooper Z.A., Scolyer R.A., Pupo G., Komurov K., Sehgal V., Zhang J., Patel L., Pereira C.G., Broom B.M., Mills G.B., Ram P., Smith P.D., Wargo J.A., Long G.V., Davies M.A. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma. Cancer Res. 2014;74(23):7037–7047. doi: 10.1158/0008-5472.CAN-14-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang L., Venneti S., Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 86.Yuneva M., Zamboni N., Oefner P., Sachidanandam R., Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007;178(1):93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duran R.V., Oppliger W., Robitaille A.M., Heiserich L., Skendaj R., Gottlieb E., Hall M.N. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 2012;47(3):349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 88.Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J.H., Lim C., Guimaraes A.R., Martin E.S., Chang J., Hezel A.F., Perry S.R., Hu J., Gan B., Xiao Y., Asara J.M., Weissleder R., Wang Y.A., Chin L., Cantley L.C., DePinho R.A. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.ChemDiv, KRAS Targeted Library, 2014. 〈http://www.chemdiv.com/portfolio/kras-targeted-library/〉.
- 90.Bryant K.L., Mancias J.D., Kimmelman A.C., Der C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014;39(2):91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., Rabinowitz J.D., Metallo C.M., Vander Heiden M.G., Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ribas A. Releasing the brakes on cancer immunotherapy. N. Engl. J. Med. 2015;373(16):1490–1492. doi: 10.1056/NEJMp1510079. [DOI] [PubMed] [Google Scholar]
- 93.Sukumar M., Kishton R.J., Restifo N.P. Metabolic reprograming of anti-tumor immunity. Curr. Opin. Immunol. 2017;46:14–22. doi: 10.1016/j.coi.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buck M.D., O'Sullivan D., Pearce E.L. T cell metabolism drives immunity. J. Exp. Med. 2015;212(9):1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang C.H., Pearce E.L. Emerging concepts in immunotherapy – T cell metabolism as a therapeutic target. Nat. Immunol. 2016;17(4):364–368. doi: 10.1038/ni.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pearce E.L., Poffenberger M.C., Chang C.H., Jones R.G. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fisicaro P., Barili V., Montanini B., Acerbi G., Ferracin M., Guerrieri F., Salerno D., Boni C., Massari M., Cavallo M.C., Grossi G., Giuberti T., Lampertico P., Missale G., Levrero M., Ottonello S., Ferrari C. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med. 2017;23(3):327–336. doi: 10.1038/nm.4275. [DOI] [PubMed] [Google Scholar]
- 98.Vayalil P.K., Oh J.Y., Zhou F., Diers A.R., Smith M.R., Golzarian H., Oliver P.G., Smith R.A., Murphy M.P., Velu S.E., Landar A. A novel class of mitochondria-targeted soft electrophiles modifies mitochondrial proteins and inhibits mitochondrial metabolism in breast cancer cells through redox mechanisms. PLoS One. 2015;10(3):e0120460. doi: 10.1371/journal.pone.0120460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chandel N.S. Cold Springs Harbor Laboratory Press; Cold Spring Harbor, New York: 2015. Navigating Metabolism. [Google Scholar]
- 100.Altomare D.A., Testa J.R. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24(50):7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]