Abstract
Vesicular stomatitis virus (VSV) and rabies and Chandipura viruses belong to the Rhabdovirus family. VSV is a common laboratory virus to study viral evolution and host immune responses to viral infection, and recombinant VSV-based vectors have been widely used for viral oncolysis, vaccination, and gene therapy. Although the tropism of VSV is broad, and its envelope glycoprotein G is often used for pseudotyping other viruses, the host cellular components involved in VSV infection remain unclear. Here, we demonstrate that the host protein leucine-rich repeat-containing G protein–coupled receptor 4 (Lgr4) is essential for VSV and VSV-G pseudotyped lentivirus (VSVG-LV) to infect susceptible cells. Accordingly, Lgr4-deficient mice had dramatically decreased VSV levels in the olfactory bulb. Furthermore, Lgr4 knockdown in RAW 264.7 cells also significantly suppressed VSV infection, and Lgr4 overexpression in RAW 264.7 cells enhanced VSV infection. Interestingly, only VSV infection relied on Lgr4, whereas infections with Newcastle disease virus, influenza A virus (A/WSN/33), and herpes simplex virus were unaffected by Lgr4 status. Of note, assays of virus entry, cell ELISA, immunoprecipitation, and surface plasmon resonance indicated that VSV bound susceptible cells via the Lgr4 extracellular domain. Pretreating cells with an Lgr4 antibody, soluble LGR4 extracellular domain, or R-spondin 1 blocked VSV infection by competitively inhibiting VSV binding to Lgr4. Taken together, the identification of Lgr4 as a VSV-specific host factor provides important insights into understanding VSV entry and its pathogenesis and lays the foundation for VSV-based gene therapy and viral oncolytic therapeutics.
Keywords: cell sorting, G protein-coupled receptor (GPCR), gene knockout, immunohistochemistry, surface plasmon resonance (SPR), virus entry, Lgr4, Pseudotyped virus, VSV
Introduction
The Rhabdovirus family includes vesicular stomatitis virus (VSV)4 as well as significant human pathogens like rabies virus and Chandipura virus (1). As a common laboratory virus used to study the properties of rhabdoviridae viruses, viral evolution, and host immune responses to virus, VSV has been extensively studied and characterized (2, 3). In common with the other members of the Rhabdovirus family, the entry of VSV is facilitated exclusively by the envelope glycoprotein G (VSV-G), which is widely used for pseudotyping other viruses (4–6). These VSV-G-pseudotyped vectors are currently used in effective gene therapy protocols for many human tissues (7, 8). Furthermore, native or engineered forms of VSV can also be used in preclinical models of malignant glioma, melanoma, hepatocellular carcinoma, breast adenocarcinoma, prostate cancer, osteosarcoma, and others (9–11). In addition, VSV-based vaccination has been used for tumor antigens and a range of pathogens such as Ebola and HIV (12, 13).
Glycoprotein G plays a critical role during the initial steps of the infectious cycle through binding with specific receptors and then mediating viral and endosomal membrane fusion (14). The attachment of VSV to the cell surface was initially thought to be facilitated by interacting with the lipid phosphatidylserine (15). Meanwhile, the majority of VSV particles were endocytosed in a clathrin-based, dynamin-2-dependent manner (16, 17). Studies have suggested that the endoplasmic reticulum chaperone gp96 is also essential for VSV infection (18). Subsequently, low density lipoprotein receptor (LDLR) and its family members were proposed to be cell surface receptors for VSV (19, 20), although additional receptors must exist to account for VSV infection of insect cells which lack LDLR.
As the largest cell membrane receptor family, more than 1000 members of the human genome were identified as G-protein coupled receptors (GPCRs), which play an irreplaceable role in signal recognition and transduction (21). Interestingly, more and more GPCRs were found to be involved in facilitating viral propagation and thereby contributing to viral pathogenesis. For example, CXC-chemokine receptor-4 (CXCR4) and CC-chemokine receptor-5 (CCR5) are the most famous cell-fusion cofactors for HIV infection (22, 23). Furthermore, many members of the herpesvirus family hijack GPCRs and associated signaling from their cellular host through encoding viral GPCRs (24). Indeed, some antagonists to histamine receptors, 5-hydroxytryptamine (serotonin) receptors, muscarinic acetylcholine receptor, and adrenergic receptor can effectively block replication of both infectious Ebola virus and Marburg virus, and newly developed CCR5 and CXCR4 antagonists have already shown clinical promise as HIV-entry inhibitors, indicating a broad antiviral activity of the GPCR antagonists (25).
Lgr4, also known as Gpr48 (G-protein-coupled receptor 48), is a member of the glycoprotein hormone receptor subfamily. The extracellular domain (ECD) of LGR4 exhibits a twisted horseshoe-like structure composed of 17 leucine-rich repeats at the N-terminal domain, which has been recognized as the binding site for R-spondins, ligands that enhance Wnt/β-catenin signaling (26–28). Curiously, almost all Toll-like receptors (TLRs) and Nod-like receptors have a domain consisting of multiple leucine-rich repeats, which is believed to be involved in ligand binding. Similar structural patterns suggested a potential role of Lgr4 in innate immune responses. Our previous study has shown that Lgr4 negatively regulates TLR2/4-associated pattern recognition and innate immunity (29). Meanwhile, Lgr4 is also involved in the regulation of tumor growth (30, 31), organ development (32–35), and stem cell functions (36), but Lgr4 function in virus infection has not been previously reported. Here, we demonstrate that Lgr4 serves as a VSV-specific entry factor through binding with viral glycoprotein. Consequently, VSV infection could be restrained by pretreating cells with Lgr4 antibody, soluble LGR4 ECD, and R-spondin 1 suggesting the great potential of Lgr4 as a promising antiviral drug target for VSV infection.
Results
Lgr4 facilities VSV infection both in vivo and in vitro
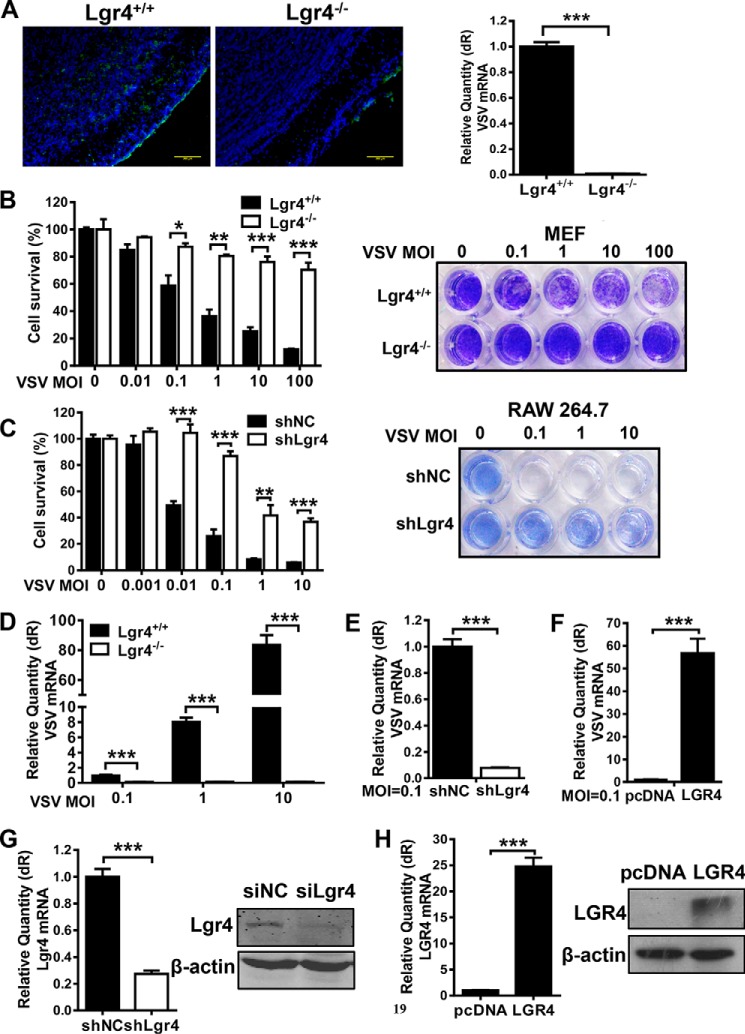
To investigate the significance of Lgr4 in viral infection, we intranasally infected Lgr4+/+ and Lgr4−/− mice with VSV (1 × 106 pfu) for 24 h. As shown in Fig. 1A, the distribution of VSV in Lgr4-deficient olfactory bulbs was reduced significantly compared with wild-type mice through intranasal infection. It suggested that Lgr4 plays a nonredundant role in the VSV infection mouse model. To further determine whether Lgr4 mediated VSV infection of host cells, we challenged mouse embryonic fibroblast (MEF) and RAW 264.7 cells with VSV in a dose-dependent manner to induce cell death and cytopathic effects. VSV-induced cell death (Fig. 1B) was significantly reduced in Lgr4-deficient MEF cells compared with wild-type cells. We observed similar results following Lgr4 knockdown in mouse macrophage-like RAW 264.7 cells (Fig. 1C). To confirm whether Lgr4 enhances VSV infection, we measured cell-associated VSV by quantitative RT-PCR of VSV RNA. Accordingly, the VSV RNA level was also significantly inhibited in Lgr4-deficient (Fig. 1D) and Lgr4-knockdown (Fig. 1E) cells. On the contrary, the VSV RNA level was increased dramatically when we overexpressed LGR4 in RAW 264.7 cells (Fig. 1F). The RNA and protein expression of Lgr4 on both knockdown and overexpression cells are shown in Fig. 1, G and H, respectively. Therefore, Lgr4 plays an important role in rendering mice and cells susceptible to VSV infection.
Figure 1.
Lgr4 facilities VSV infection both in vivo and in vitro. A, left: immunofluorescence analysis of VSV (green) in the olfactory bulb. 8-Week-old Lgr4+/+ and Lgr4−/− C57BL6 mice were infected intranasally with VSV (1 × 106 pfu) for 24 h. Mice were sacrificed and olfactory bulb tissue sections were stained with VSV-G-mAb (1:200 dilution). DAPI (blue), scale bar, 200 μm. Right graph: Q-PCR analysis of VSV RNA in Lgr4+/+ and Lgr4−/− olfactory bulbs. Representative images of n = 3 mice are shown. B and C, survival ± S.D. of MEF (B) and RAW 264.7 cells (C) was determined by MTS staining (left) and crystal violet-stained plates (right) after treating with VSV at the indicated m.o.i. for 24 h. D–F, quantitative RT-PCR of VSV RNA in MEF (D), Lgr4 knockdown (E), and LGR4 overexpressing (F) RAW 264.7 cells after challenge by VSV at the indicated m.o.i. for 12 h. G, mRNA and protein expression of Lgr4 in Lgr4 knockdown RAW 264.7 cells. H, mRNA and protein expression of LGR4 in LGR4 overexpressing RAW 264.7 cells. Data are representative of at least three independent experiments. Asterisks indicate statistical significance: ***, p < 0.001; **, p < 0.01l; *, p < 0.05.
Lgr4-mediated VSV infection is dispensable for antiviral innate immune responses
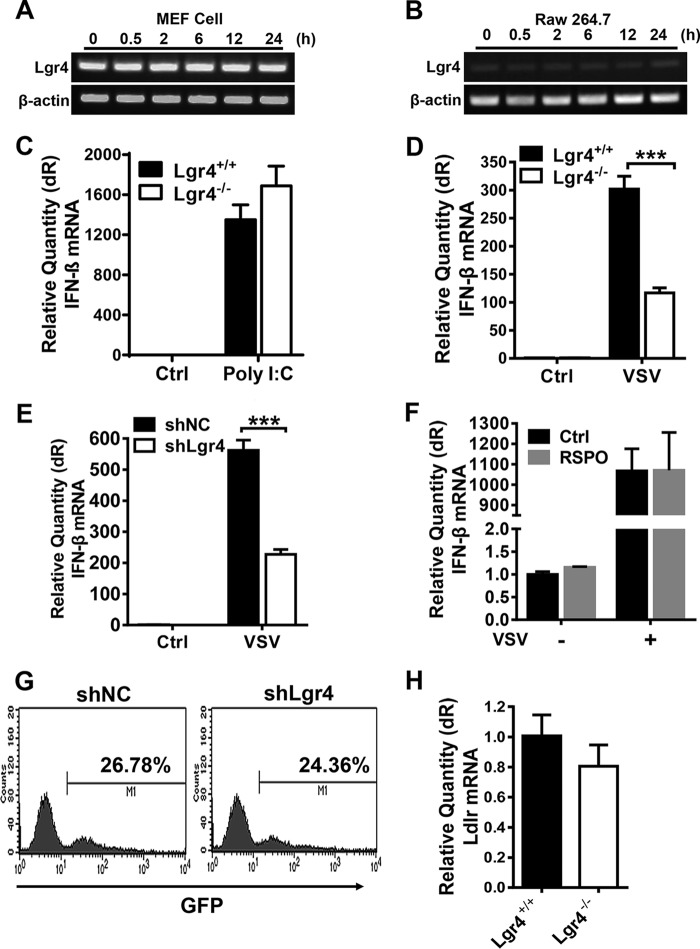
To explore the role of Lgr4 in antiviral immunity, we detected the expression of type I interferon and phagocytosis in Lgr4-deficient and Lgr4-knockdown cells. As shown in Fig. 2, A and B, the expression of Lgr4 is little changed following VSV infection suggesting that Lgr4 is not a virus-induced or IFN-induced gene. Furthermore, the interferon response was intact in Lgr4-deficient peritoneal macrophages exposed to poly(I:C) (Fig. 2C). However, the VSV-induced expression of IFN-β was dramatically reduced in Lgr4-deficient (Fig. 2D) or Lgr4-knockdown (Fig. 2E) cells, which is consistent with decreased intracellular VSV in these cells. To further examine whether Lgr4 signaling might directly potentiate IFN production, we activated peritoneal macrophages with R-spondin 1 (an endogenous ligand to Lgr4). As shown in Fig. 2F, the RNA expression of IFN-β was little changed by R-spondin 1 in both VSV-infected or uninfected cells, suggesting a negligible role of Lgr4 in IFN production. Macrophage-mediated endocytosis of particles was also little changed in Lgr4 knockdown cells (Fig. 2G). Furthermore, we also examined the expression of a known VSV receptor, Ldlr. The expression of Ldlr was little changed in Lgr4−/− MEFs (Fig. 2H). Taken together, these results indicate that Lgr4 facilitated VSV infectivity but is dispensable for innate antiviral immune responses and Ldlr expression.
Figure 2.
Lgr4-mediated VSV infection is dispensable for antiviral innate immune responses. A and B, gene expression of Lgr4 following VSV infection (m.o.i. = 1) of MEF (A) and RAW 264.7 cells (B) at the indicated times after infection. β-Actin was used as a control for RT-PCR. C, Q-PCR analysis of IFN-β mRNA expression in macrophages transfected with poly(I:C) (1 μg/ml) for 4 h. D, Q-PCR analysis of IFN-β mRNA expression from Lgr4+/+ and Lgr4−/− MEF cells infected with VSV at m.o.i. = 1 and harvested 12 h post-infection. E, Q-PCR analysis of IFN-β mRNA expression from control (shNC) and Lgr4 knockdown (shLgr4) RAW 264.7 cells infected with VSV at m.o.i. = 1. F, Q-PCR analysis of IFN-β mRNA expression from peritoneal macrophages stimulated by R-spondin1 (100 ng/ml) with or without VSV (m.o.i. = 1). G, internalization of FITC-dextran in shNC and shLgr4 RAW 264.7 cells was determined by flow cytometry. H, Q-PCR analysis of Ldlr mRNA expression from Lgr4+/+ and Lgr4−/− peritoneal macrophages. Data are representative of at least three independent experiments. Asterisks indicate statistical significance: ***, p < 0.001.
Lgr4-mediated virus infection is dependent on VSV-G
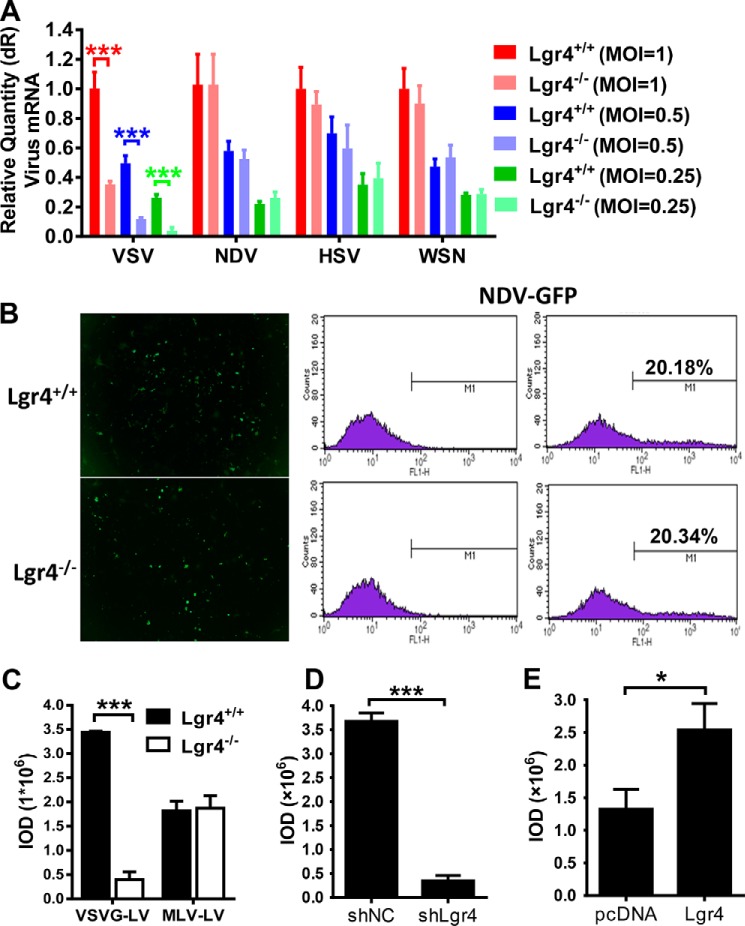
Binding and internalization into host cells are the initial steps in virus infection. To investigate the specificity of the role of Lgr4 in virus infection, we challenged wild-type and Lgr4-deficient peritoneal macrophages with different kinds of virus such as influenza virus (A/WSN/33, ssRNA virus), Newcastle disease virus (NDV, ssRNA virus), and Herpes simplex virus (HSV-1, dsDNA virus). To our surprise, only VSV infection was reduced in Lgr4-deficient cells; infection with WSN, NDV, and HSV-1 were all unaffected by Lgr4 (Fig. 3A). We also used fluorescence microscopy and FACS to detect NDV infection in Lgr4 wild-type and knock-out MEF cells. No significant difference in NDV infection was observed due to loss of Lgr4 (Fig. 3B). As a commonly used envelope protein in pseudotyped lentivirus, VSV-G is widely used in packaging lentivirus to infect a broad array of species and cell types. To examine Lgr4 specificity in viral vector tropism, we packaged lentivirus with either VSV-G (VSVG-LV) or MLV Env-glycoproteins (MLV-LV), which differs from VSVG-LV only in its coat protein, to infect MEFs. Then we transduced VSVG-LV and MLV-LV into Lgr4+/+ and Lgr4−/− MEFs. As shown in Fig. 3C, only VSVG-LV transduced GFP expression was reduced in Lgr4-deficient MEF cells. In contrast, the GFP fluorescence intensity induced by MLV-LV was changed little. VSVG-LV fluorescence intensity was also decreased in Lgr4-knockdown RAW 264.7 cells (Fig. 3D). Furthermore, overexpression of Lgr4 in RAW 264.7 cells significantly increased the GFP fluorescence intensity (Fig. 3E). These data confirmed that Lgr4 specifically increased VSV-G-mediated infection.
Figure 3.
Lgr4-mediated virus infection is dependent on VSV-G. A, Q-PCR analysis of indicated virus in Lgr4+/+ and Lgr4−/− macrophages infected with VSV, NDV, HSV-1, or WSN at an m.o.i. of 1, 0.5, and 0.25 for 8 h. B, quantitative flow cytometry analysis of Lgr4+/+ and Lgr4−/− MEFs infected with NDV-GFP (m.o.i. = 10) for 24 h. C–E, Lgr4+/+ and Lgr4−/− MEFs were infected with VSV-G protein-coated lentivirus or MLV protein-coated lentivirus expressing GFP for 48 h (C). Lgr4 control and knockdown RAW 264.7 cells were infected with VSV-G protein-coated lentivirus expressing GFP for 48 h (D). Control and Lgr4 overexpressing RAW 264.7 cells were infected with VSV-G protein-coated lentivirus expressing GFP for 48 h (E). Digital images were captured using an Olympus IX71 inverted fluorescence microscope with DP2-BSW imaging software. Results are shown as the mean ± S.D. (n = 3). The mean fluorescence density was determined by IOD. Data are representative of at least three independent experiments. Asterisks indicate statistical significance: ***, p < 0.001; *, p < 0.05.
VSV entry is decreased in Lgr4-deficient cells
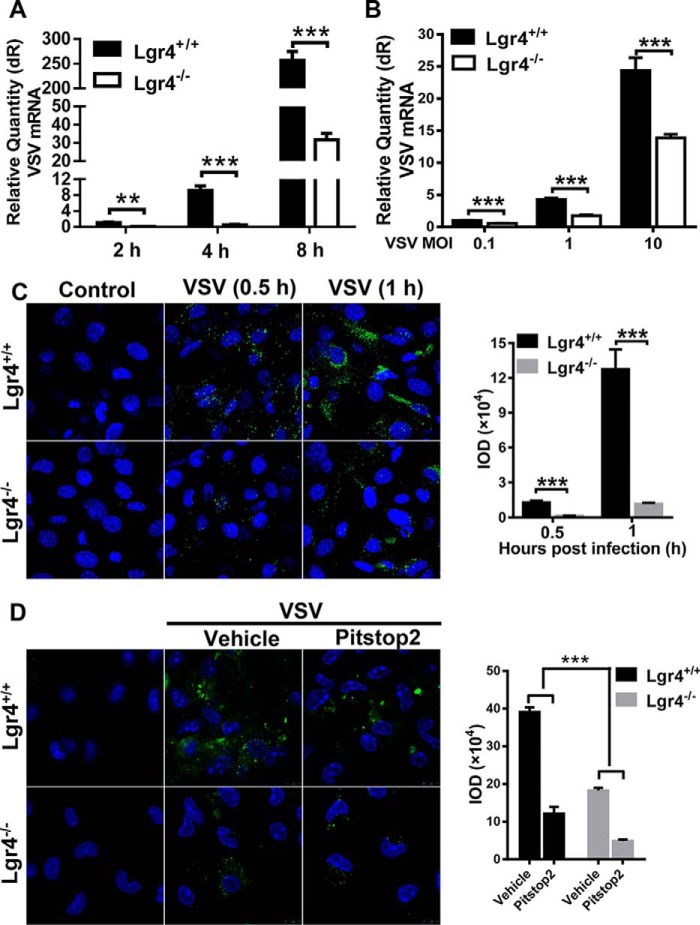
To explore the stage of VSV infection influenced by Lgr4, we infected wild-type and Lgr4-deficient peritoneal macrophages cells with VSV in a time- and dose-dependent manner. As shown in Fig. 4A, VSV RNA was reduced substantially in Lgr4-deficient cells at time points between 2 and 8 h post-infection. Similarly, although VSV RNA in peritoneal macrophages increased as m.o.i. was raised, the viral load in Lgr4-deficient cells was obviously lower than that of wild-type cells at any given dose (Fig. 4B). To further confirm that the entry of VSV was influenced by Lgr4, we challenged wild-type and Lgr4-deficient MEF cells with VSV for a very short time and the attached virus was detected by immunofluorescence. Compared with wild-type MEF cells, Lgr4-deficient MEF cells internalized significantly less VSV at both 0.5 and 1 h (Fig. 4C). Following receptor binding, most incoming VSV were present in pits and vesicles with electron-dense coats, implying a dominant role for clathrin-mediated endocytosis (37). Because Lgr4 loss did not affect NDV/HSV/WSN infection (Fig. 3A), Lgr4 is unlikely to play a role in global endocytosis; however, to examine whether it may function specifically in VSV endocytosis, we tested whether Lgr4 deficiency affects VSV infection in cells with impaired endocytosis. It has been shown that Pitstop2 specifically inhibits the association between the terminal domain of clathrin and amphiphysin (38). So we used Pitstop2 to inhibit clathrin-mediated endocytosis in VSV infection to explore the role of Lgr4 in VSV endocytosis. Consistent with a previous study (39), we found that VSV infection in MEFs was significantly inhibited by Pitstop2 (Fig. 4D). Furthermore, because Lgr4 deficiency significantly reduced VSV infection even in MEFs where endocytosis was blocked by Pitstop2 treatment, it is likely that Lgr4 functions at a different stage of VSV infection than endocytosis. Taken together, these results implied that Lgr4 has a major role in very early stages of VSV infection, such as viral attachment.
Figure 4.
VSV entry is decreased in Lgr4-deficient cells. A, Q-PCR analysis of VSV RNA in Lgr4+/+ and Lgr4−/− peritoneal macrophages at the indicated times (m.o.i. = 1). B, Q-PCR analysis of VSV RNA in Lgr4+/+ and Lgr4−/− peritoneal macrophages infected with VSV for 1 h at 4 °C at the indicated m.o.i. C, immunofluorescence staining of VSV (green) on the surface of Lgr4+/+ and Lgr4−/− MEF cells infected by VSV (m.o.i. = 1000) for 30 min or 1 h at 4 °C. D, immunofluorescence staining of VSV (green) on the surface of Lgr4+/+ and Lgr4−/− MEF cells infected by VSV (m.o.i. = 1000) for 6 h after treatment with pitstop2 (30 μm) 10 min at 37 °C. DAPI, blue. Data are representative of at least three independent experiments. Asterisks indicate statistical significance: ***, p < 0.001; **, p < 0.01.
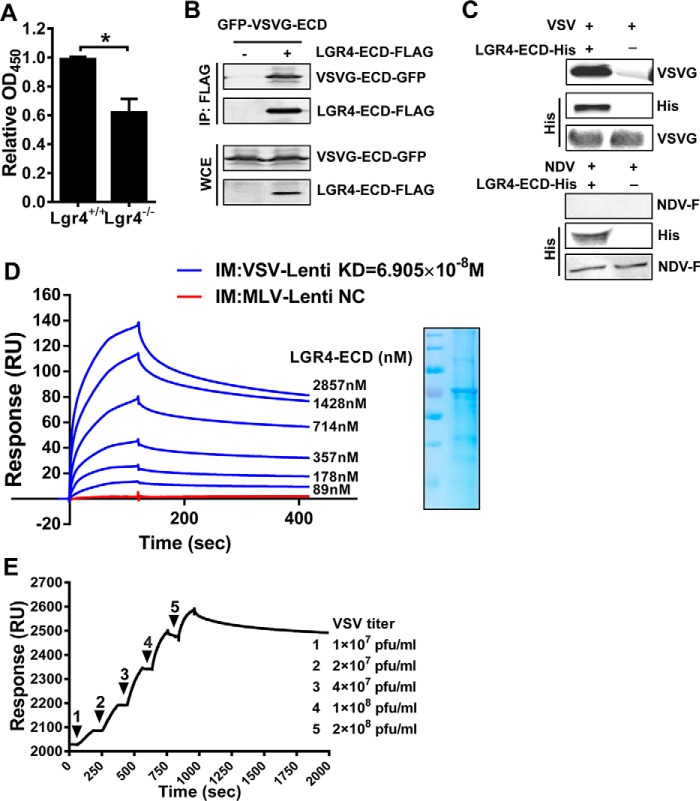
VSV binds host cells through the extracellular domain of Lgr4
To illustrate the potential interactions between Lgr4 and VSV, cell ELISA was performed to detect the VSV attachment to Lgr4 wild-type and deficient cells. As shown in Fig. 5A, the attachment of VSV on the cell membrane was reduced in Lgr4-deficient cells, which is consistent with our previous data. To determine whether Lgr4 and VSV-G are associated in a complex, we expressed FLAG-tagged LGR4 extracellular domain (LGR4-ECD-FLAG) and GFP-tagged VSV-G extracellular domain (VSVG-ECD-GFP) in 293T cells. Immunoprecipitation with anti-FLAG mAb revealed that LGR4 ECD was in a complex with VSV-G (Fig. 5B). Likewise, purified LGR4-ECD-His (purified from Drosophila Schneider2 (S2) cell supernatant) could bind specifically to VSV but not NDV in vitro (Fig. 5C). Furthermore, we immobilized either VSV-G or MLV Env-glycoprotein pseudotyped virus on Biacore sensor chips and analyzed their binding with LGR4-ECD-His, respectively. As shown in Fig. 5D, VSVG-LV bound to LGR4 ECD in a dose-dependent manner (KD = 6.905 × 10−8 m). However, MLV-LV did not bind with LGR4-ECD-His even at 714 nm. When we immobilized LGR4 ECD-His to a Biacore sensor chip and analyzed the binding with VSV, the LGR4 ECD-His bound to VSV in a dose-dependent manner as well (Fig. 5E). Thus, these data further confirmed the interaction between Lgr4 and VSV-G.
Figure 5.
VSV binds host cells through the extracellular domain of Lgr4. A, the binding of VSV on Lgr4+/+ and Lgr4−/− MEF cells was analyzed by cell-based ELISA. B, HEK293T cells were transfected with plasmids encoding LGR4-ECD-FLAG and VSVG-ECD-GFP for 36 h before lysis. Then the complex was immunoprecipitated by anti-FLAG-agarose followed by immunoblot analysis of VSVG-ECD-GFP and LGR4-ECD-FLAG. C, immunoblotting of VSV-G and LGR4-ECD-His after LGR4-ECD-His and associated proteins were pulled down with nickel beads. Reblotting of the membrane with anti-VSV-G mAb was performed as an input control. NDV is a negative control. D, dose-response of LGR4-ECD binding to immobilized VSVG-LV or MLV-LV by surface plasmon resonance in PBS. VSVG-LV or MLV-LV were immobilized on a CM5 sensor chip, and LGR4-ECD was passed at the indicated concentrations. MLV-LV as a negative control (NC) was shown with a concentration of 714 nm LGR4-ECD. SDS-PAGE of LGR4-ECD is shown on the right. IM, immobilize: VSVG-LV (blue), MLV-LV (red). E, dose-response of VSV binding to immobilized LGR4-ECD by surface plasmon resonance in PBS. LGR4-ECD (20 μg/ml) was immobilized on a sensor chip NTA, and VSV was passed at the indicated concentrations. The kinetic constants of binding were obtained using a 1:1 Langmuir binding model via BIAevaluation software. Data are representative of at least three independent experiments. Asterisks indicate statistical significance: *, p < 0.05.
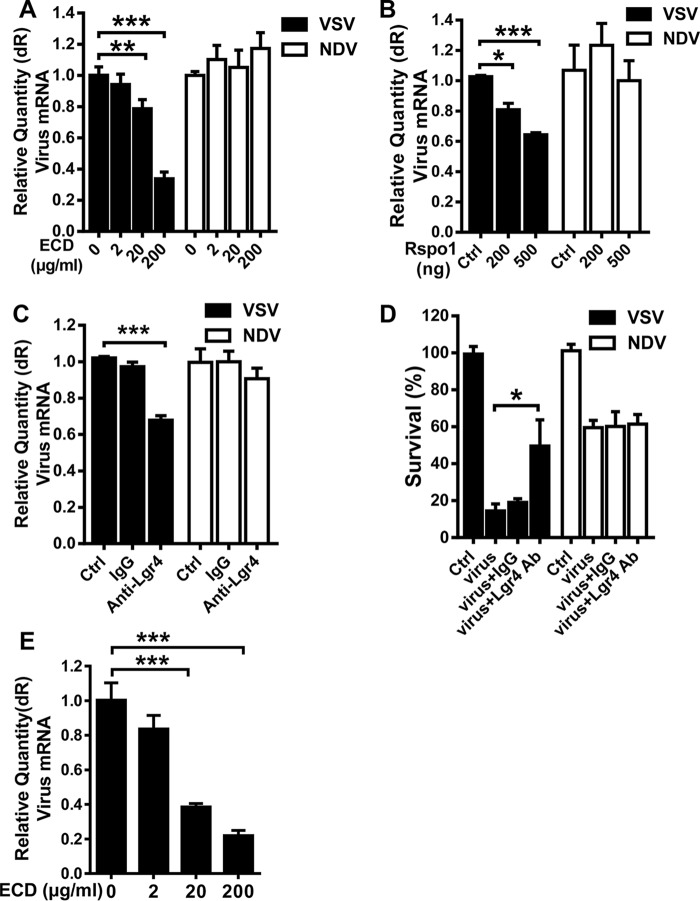
Cells were protected from VSV infection by blocking endogenous Lgr4
To further confirm that VSV infects cells through binding Lgr4, we examined whether the binding of VSV to host cells was blocked by soluble LGR4 ECD, Lgr4 ligand Rspodin1, and an Lgr4-specific antibody. We used NDV as a control virus. Pretreating VSV and NDV with soluble LGR4 ECD decreased VSV infection in a dose-dependent manner, but had no effect on NDV (Fig. 6A). When we pretreated the cells with Rspodin1 to bind with endogenous Lgr4, only VSV infection was reduced in a dose-dependent manner (Fig. 6B). Similar data were also observed in Lgr4 antibody-pretreated cells (Fig. 6C). Accordingly, VSV but not NDV-induced cell death was also diminished by the Lgr4 antibody when compared with control IgG (Fig. 6D). Then we pretreated MDA-MB-231 cells with soluble LGR4 ECD to detect the influence of LGR4 on VSV infection in human cells. As shown in Fig. 6E, VSV infection of MDA-MB-231 cells was decreased by LGR4 ECD in a dose-dependent manner suggesting that LGR4 is also involved in VSV infection of human cells. Therefore, blocking endogenous Lgr4 could be a potential way to prevent VSV infection.
Figure 6.
Cells were protected from VSV infection by blocking endogenous Lgr4. A, VSV or NDV was preincubated with LGR4-ECD (2, 20, and 200 μg/ml) at 4 °C for 4 h then infected MEF cells (m.o.i. = 10) with virus only or preincubated mixture for 1 h at 4 °C. VSV or NDV RNA was quantitated by real-time PCR. B, MEF cells were incubated with R-spondin-1 at different concentrations (200 and 500 ng/ml) at 37 °C for 1 h and then infected by VSV or NDV (m.o.i. = 10) at 4 °C for 1 h. VSV or NDV RNA was quantitated by real-time PCR. C, MEF cells without (Ctrl.) or with control antiserum (IgG) or anti-Lgr4 antiserum (1 h, 37 °C) were subjected to VSV or NDV infection (m.o.i. = 10, at 4 °C for 1 h). Quantitative RT-PCR of VSV or NDV RNA was shown. D, MEF cells treated with control antiserum (IgG) or anti-Lgr4 antiserum (1 h, 37 °C), or kept untreated (Ctrl.) and subjected to VSV or NDV infection (m.o.i. = 1, 24 h). Cell viability (bar plot) was determined by crystal violet staining and quantitated at OD 540 nm, n = 3. E, preincubation of LGR4-ECD (2, 20, and 200 μg/ml) with VSV (m.o.i. = 10) at 4 °C for 4 h was followed by infection of MDA MB 231 cells with the mixture, and VSV RNA was quantitated by real-time PCR. Data are representative of at least three independent experiments. Asterisks indicate statistical significance: ***, p < 0.001; **, p < 0.01; *, p < 0.05.
Discussion
Rhabdoviruses have a characteristic rigid bullet shape with a flat base and a round tip. Similar to other rhabdoviruses, the condensed nucleocapsid of VSV is surrounded by a lipid bilayer containing the viral glycoprotein G that constitutes the spikes that protrude from the viral surface. The envelope of VSV plays a critical role in its infectious cycle through recognizing receptors on the host cell surface and mediating the fusion between viral and cell membranes. Previous studies have suggested that LDLR, gangliosides, and gp96 are all essential for infection with VSV, but the knowledge about the host factors in VSV infection is still far from complete. In this paper, we identified the G-protein-coupled receptor Lgr4 as a VSV-specific entry factor through Lgr4 interaction with viral particles, which extended the understanding of host components involved in VSV infection. Furthermore, the soluble LGR4 ECD, Rspodin1, and LGR4 antibody could inhibit VSV infection through competition with Lgr4, showing great potential for treating vesicular stomatitis as well as influencing the targeting of VSV-based gene therapy and viral oncolytic vectors.
Lgr4 is widely expressed in multiple organs, including lung, skin, brain, bone, and immune cells. Interestingly, the reported VSV receptor LDLR is ubiquitously expressed in both liver and peripheral tissue for maintaining cholesterol homeostasis in mammals, tissues that do not seem relevant during VSV infection (40). Our previous study showed that Lgr4 negatively regulates TLR2/4-associated pattern recognition and innate immunity in bacterial infection. Our study here demonstrates the function of Lgr4 as a co-receptor to promote VSV entry in viral infection, likely through direct binding of VSV-G. However, we still observed limited VSV infection of cells lacking Lgr4, clearly indicating that other viral receptors can partially substitute for the role of Lgr4 in VSV entry. Identifying and characterizing these additional viral receptors remains a key objective for further research.
The identification of key host components involved in virus infection is of significant clinical important in preventing and treating viral infectious diseases. In addition, recombinant VSV and VSV-G-pseudotyped viral vectors have been widely used for viral oncolysis, vaccination, and gene therapy. Our work suggests that the infection efficacy of such vectors can be regulated by modulating the expression of Lgr4. Interestingly, Lgr4 is highly expressed in lung, breast, and colorectal cancer cells (31, 41). Thus, these cells might be preferred targets of VSV-G-based gene therapy as well as VSV-G-based viral oncolysis.
Experimental procedures
Chemicals and reagents
DMEM and other cell culture reagents were purchased from Invitrogen Life Technologies. Poly(I:C) and Alexa Fluor 488-labeled goat antibody to mouse IgG were obtained from Invivogen; DAPI from Sigma. Hyclone fetal bovine serum (FBS) was purchased from GE Healthcare. Monoclonal anti-VSV-G (VSV surface glycoprotein) antibody clone P5D4 (catalogue number V5507), was bought from Sigma. Polyclonal anti-NDV (Newcastle disease virus) antibody was purchased from Bioye (catalogue number HK-10044R). HRP-rabbit polyclonal anti-mouse antibody conjugate (catalogue number ab6728) was purchased from Abcam, and R-spondin-1 (catalogue number 120-38) from PeproTech. Pitstop2 (catalogue number SML1169) was from Sigma.
Cells and virus
RAW 264.7, MDA-MB-231, and HEK-293T cells were obtained from the American Type Culture Collection (ATCC). Primary murine embryonic fibroblasts (MEFs) were prepared from embryos dissected from the same pregnant female at day 13.5 and were cultured in DMEM supplemented with 10% (v/v) FBS. Peritoneal macrophages were harvested from mice 4 days after thioglycollate (BD Biosciences, Sparks, MD) injection. HSV type 1 (KOS strain) was propagated on Vero (African green monkey kidney cells). NDV (which was a gift provided by Professor Jiahuai Han (Xiamen University)) was grown for 2 days at 37 °C in the allantoic cavities of 9-day-old SPF embryonated chicken eggs. VSV (Indiana 1 serotype) was propagated and amplified by infection of a monolayer of BHK-21 cells for 24 h. Filtered supernatants were harvested and subjected to two rounds of centrifugation in 10% sucrose (w/v) on a sterile 1× PBS cushion at 27,000 rpm for 1 h at 4 °C. The pelleted virus was resuspended in sterile 1× PBS and stored at −80 °C. Viral titers were measured by standard plaque assay on Vero cells.
Virus infection
Cells were infected with VSV, lentivirus, A/WSN/33, NDV, or HSV-1 for the indicated times and virus RNA was analyzed by Q-PCR. For GFP-NDV infection, MEF cells were seeded into 12-well plates at a density of 2 × 105 cells/well and allowed to grow overnight; cells were infected by GFP-NDV (1 × 104 pfu/ml) and GFP expression was detected by flow cytometry. Lgr4-deficient mice were constructed by our laboratory (29). All animal experiments were performed with the approval of the Scientific Investigation Board of East China Normal University (M20150401). For in vivo studies of VSV infection, age- and sex-matched groups of littermate mice were intranasally infected with VSV: mice were anesthetized with pelltobarbitalum natricum, a total of 10 μl of PBS buffer containing 1 × 106 pfu VSV was pipetted into both nostrils. Mice were sacrificed 24 h after infection.
Flow cytometry
MEF cells (50,000 cells per 1 ml of DMEM/well) in 12-well plates were incubated with NDV-GFP for 24 h, the cells were then washed three times with PBS and then NDV-GFP was determined using flow cytometry (BD FACSCalibur). Analysis was performed on a window that excluded aggregates, and 10,000 cells were measured.
Measurement of antiviral activity by crystal violet staining and MTS assay
Cells were seeded into 96-well plates at a density of 1 × 104 per 0.1 ml/well overnight and infected with VSV for 24 h, then washed with PBS twice. For crystal violet staining, the cells were stained with crystal violet (5% (w/v) in 66% (v/v) aqueous methanol) for 2 h at room temperature and washed with PBS five times, then photographed. For MTS assays, 20 μl of MTS (Promega) was added to each well and incubated for 2 h at 37 °C. The absorbance was measured at a wavelength of 490 nm using a microplate reader. Cell viability of the untreated group was normalized to 100%.
Viral attachment assays
Cells were grown to ∼90% confluence in 12-well plates. VSV diluted in cold DMEM was added onto cells at the indicated m.o.i. After incubation at 4 °C for 1 h, the wells were washed three times with cold DMEM to remove unbound VSV. For all virus attachment experiments, the RNA in each well was extracted. The viral RNA of the bound virus in each sample was quantified using a quantitative real-time reverse transcription PCR (RT-PCR) protocol (TaKaRa). All experiments were done in triplicate.
Plasmid constructs and transfection
cDNA of human LGR4-ECD (Extracellular domain) and LGR4 were cloned into pcDNA3.1-FLAG eukaryotic expression vectors. cDNA of VSVG-ECD (amino acids 17-467) was cloned into pcDNA3.1-GFP eukaryotic expression vectors. cDNA of human LGR4-ECD (amino acids 25–528) was cloned into the pMT/BiP/V5-His A plasmid for inducible, secreted expression of recombinant proteins in Drosophila Schneider2 (S2) cells. Each construct was confirmed by sequencing. Plasmids were transiently transfected into HEK293T cells and RAW 264.7 cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Protein expression and purification
His-ECD was purified from Drosophila Schneider2 (S2) cell supernatant under native conditions and purified with the nickel-nitrilotriacetic acid system. Briefly, a 1.5 × 15-cm column was packed with 3 ml of nickel-chelating resin and equilibrated with 10 column volumes (40 ml) of 20 mm phosphate buffer, pH 7.8, and 500 mm NaCl. The protein solution was loaded onto the column for 60 min at 4 °C. The column was washed with wash buffer (50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, pH 8.0) three times. The protein was eluted using elution buffer (50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole, pH 8.0). Samples were dialyzed for 24 h at 4 °C with continuous stirring and buffer (PBS) changes every 8 h. The dialyzed protein solution was stored at −80 °C for further analyses.
Preparation of VSV-G and MLV pseudotyped lentivirus
For preparation of the EGFP expressing VSV-G pseudotyped lentivirus VSVG-LV, 293T cells were transfected by a mixture of three expression vectors (pCMV-VSV-G, psPAX2, and pLL3.7), and for MLV-LV (Moloney murine leukemia virus) preparation, a mixture of pHCMV-EcoEnv, psPAX2, and pLL3.7 vectors was used, with Lipofectamine 2000 used as a transfection reagent in both cases. Culture medium containing the resulting viral vectors was collected at 48 h and concentrated 50-fold by ultrafiltration on a 50-kDa cutoff membrane (Millipore). A plasmid encoding the MLV coat protein (pHCMV-EcoEnv) was purchased from Genecopoeia and was used as above instead of the VSV-G plasmid for production of MLV-pseudotyped lentivirus (MLV-LV).
Immunofluorescence assay
For olfactory bulb immunofluorescence, mice were anesthetized with pentobarbital (150 mg/kg) and blood was removed from organs by cardiac perfusion with 10 ml of PBS, followed by perfusion with 10 ml of 4% paraformaldehyde/PBS for fixation. Olfactory bulbs were placed in 4% paraformaldehyde overnight for complete fixation, then submerged in 30% sucrose/PBS overnight for cryoprotection and frozen in O.C.T. compound. 8-μm sagittal sections were cut at −20 °C in a Leica CM1900 cryostat. For MEF cell immunofluorescence, MEF cells were seeded on coverslips in 24-well plates at a density of 5 × 104 cells per well for 12–16 h. Then cells were infected with VSV at an m.o.i. of 1 × 103 for 30 min or 1 h at 4 °C. For endocytosis inhibition assay, the cells were stimulated with 30 μm Pitstop2 for 10 min before the VSV was added at an m.o.i. of 1000 and incubated for 6 h to internalize the attached VSV. Next, the cells were washed with PBS twice and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. For immunofluorescence staining, olfactory bulb biopsy samples and MEF cell samples were permeabilized with 0.1% Triton X-100 in PBS for 5 min. After blocking with 5% BSA in PBS, all samples were stained with an antibody against VSV-G (1:200 dilution) at 4 °C overnight, and washed with PBST three times, followed by incubation with Alexa Fluor 488-labeled goat antibody to mouse IgG (1:2,000 dilution). All objects were mounted with mounting medium (Invitrogen) containing 0.2 mg/ml of DAPI. Olfactory bulbs were examined with a Leica DRM fluorescence microscope. MEF cell confocal images were acquired using a Leica TCS SP5 confocal laser scanning microscope. We used IOD (integrated optical density) to indicate total MEF cell surface fluorescence.
Quantitative RT-PCR
Total RNA from mouse cells or tissues was extracted with TRIzol reagent (Takara) and 500 ng of RNA were used to generate cDNA using a Reverse Transcription Kit (Takara). Gene expression analysis was done using the SYBR Green PCR Master Mix (Takara) according to the manufacturer's instructions and data were normalized by the level of Gapdh or actin expression in each individual sample. The sequences of the primers used in this study for quantitative real-time RT-PCR are listed in Table 1.
Table 1.
Sequences of PCR primers used in this study
| Genes | GenBankTM | Primer sequences (5′-3′) |
|---|---|---|
| Lgr4 | AB209743 | Forward: AAGATAACAGCCCCCAAGAC |
| Reverse: AGGCAGTGATGAACAAGACG | ||
| β-Actin | NM_001101 | Forward: GTACGCCAACACAGTGCTG |
| Reverse: CGTCATACTCCTGCTTGCTG | ||
| VSV | KU296059.1 | Forward: ACGGCGTACTTCCAGATGG |
| Reverse: CTCGGTTCAAGATCCAGGT | ||
| Ldlr | NM_001252659 | Forward: GAGGAACTGGCGGCTGAA |
| Reverse: GTGCTGGATGGGGAGGTCT | ||
| IFNβ | X14455.1 | Forward: CAGCTCCAAGAAAGGACGAAC |
| Reverse: GGCAGTGTAACTCTTCTGCAT | ||
| NDV | Z12110 | Forward: TATACACCTCATCTCAGACAGGGTCAATCA |
| Reverse: GCTCTCTTTAAGTCGGAGGATGTTGGC | ||
| HSV-1 UL30 | NC_001806.2 | Forward: CATCACCGACCCGGAGAGGGAC |
| Reverse: GGGCCAGGCGCTTGTTGGTGTA | ||
| A/WSN/33 | J02177.1 | Forward: GGAACAATTAGGTCAGAAGT |
| Reverse: GTGGCAATAACTAATCGGTCA | ||
| Gapdh | NM_001289746.1 | Forward: ACCCAGAAGACTGTGGATGG |
| Reverse: TTCAGCTCAGGGATGACCTT |
Immunoprecipitation
The transfected cells were lysed in RIPA buffer containing 1 mm phenylmethylsulfonyl fluoride (PMSF) and Roche Complete protease inhibitor mixture (Basel, Switzerland). The lysate was preincubated with FLAG beads after centrifugation at 12,000 × g for 20 min at 4 °C. The complexes captured by FLAG beads were washed three times using RIPA buffer to remove unbound proteins. To detect the binding of LGR4-ECD-His (purified from Drosophila Schneider2 (S2) cell supernatant) and VSV, LGR4-ECD-His and associated proteins were pulled down with nickel beads and incubated with VSV (1 × 107 pfu/ml) for 2 h at 4 °C. All immunoprecipitation samples were then suspended in 2 × SDS buffer and boiled for 10 min. The complex proteins were then analyzed by Western blotting, using specific antibodies (VSV-G, FLAG, GFP, and His 1:1000 dilution).
Surface plasmon resonance
To determine the binding of LGR4-ECD to immobilized VSVG-LV or MLV-LV, we used a BIAcore T200 instrument (GE Healthcare) with a CM5 sensor chip (GE Healthcare). Activation, deactivation, and preparation of the coupled flow cell as well as the ligand-binding assay were performed essentially as described previously (42). Briefly, VSVG-LV or MLV-LV in sodium acetate buffer (3.25 × 107 pfu/ml, pH 4.0) was immobilized on a CM5 sensor chip, and LGR4-ECD-His was passed at the indicated concentrations. Experiments were conducted with PBS, pH 7.4, as the running buffer, and the analyte was injected at a flow rate of 30 μl/min. The association time was 90 s and the dissociation time was 180 s, then the chip was regenerated for 30 s with glycine-HCl (pH 1.5, 10 mm). Equilibration of the chip with the running buffer for another 60 s was performed before the next injection. For surface plasmon resonance analysis of VSV binding to immobilized LGR4-ECD-His, LGR4-ECD-His (20 μg/ml) was immobilized on a sensor chip of NTA (nitrilotriacetic acid) by a His-tag capturing procedure. VSV (molecular mass 2.66 × 108 Da) was suspended at 1 × 107 to 2 × 108 pfu/ml in PBS and passed over the immobilized LGR4-ECD-His in the sensor chip with single injection kinetics method. The kinetic constants of binding were obtained using a 1:1 Langmuir binding model via a BIAevaluation software program.
Competition assays
For the ECD competition assay: MEF cells or MDA-MB-231 cells (1 × 105 cells per 1 ml of DMEM/well) in 12-well plates were cooled to 4 °C. LGR4-ECD (2, 20, and 200 μg/ml) was preincubated with VSV or NDV (m.o.i. = 10) at 4 °C for 4 h, then the mixed incubation liquid was used to infect cells for 1 h at 4 °C. Total RNA was isolated and Q-PCR was performed to quantitate VSV or NDV RNA. For the R-spondin-1 competition assay: MEF cells were incubated with R-spondin-1 (200 ng, 500 ng/ml) at 37 °C for 1 h and then infected by VSV or NDV (m.o.i. = 10) at 4 °C for 1 h. The cells were then washed three times with cold PBS and total RNA was isolated and Q-PCR performed to quantitate VSV or NDV RNA. For the anti-Lgr4 antiserum blocking assay: MEF cells were preincubated with anti-Lgr4 antiserum or control antiserum at 37 °C for 1 h then subjected to VSV or NDV infection at indicated m.o.i. and time.
Cell-based ELISA
Cells were grown to ∼80% confluence in a 96-well plate, then VSV was applied to infect cells. After 1 h at 4 °C the cells were washed three times with cold PBS and then fixed and permeabilized in the wells. Primary antibodies to VSV-G were then added at 1:200 dilution to bind to VSV on the cell surface. After incubation, the unbound primary antibodies were removed by washing with PBST and HRP-labeled secondary antibodies were added. After removing unbound secondary Ab, TMB reaction buffer was added for the colorimetric assays and the signal was read at OD450 nm.
Ethics statement
All animal experiments conformed to the regulations drafted by the Association for Assessment and Accreditation of Laboratory Animal Care in Shanghai and in direct accordance with the Ministry of Science and Technology of the People's Republic of China Animal Care guidelines. The protocol was approved by the East China Normal University Center for Animal Research (AR2013/08002). All surgeries were performed under anesthesia and all efforts were made to minimize suffering.
Statistical analysis
Statistical significance between groups was determined by two-tailed Student's t test and two-way analysis of variance test. Differences were considered to be significant when p < 0.05.
Author contributions
B. D., M. Y. L., and M. Q. conceived and coordinated the study. B. D., N. Z., and H. J. H. designed, performed, and analyzed the experiments and wrote the paper. N. Z. and H. J. H. designed, performed, and analyzed the experiments shown in Figs. 1, 6, and 7. N. Z., B. H. T., Y. L. W., and Q. Q. X. designed, performed, and analyzed the experiments shown in Figs. 2 and 3. H. J. H., Y. Y., and N. N. W. designed, performed, and analyzed the experiments shown in Figs. 4 and 5. L. L. H. designed and constructed vectors for expression of proteins. J. R. X. provided technical assistance for SPR. H. H. H., S. S., and A. C. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by National Natural Science Foundation of China Grants 31000398, 31570896, 81672811, and 31770969, Joint Research Institute for Science and Society (JoRISS) Grant 14JORISS01, and Science and Technology Commission of Shanghai Municipality Grant 15JC1401500. The authors declare that they have no conflicts of interest with the contents of this article.
- VSV
- vesicular stomatitis virus
- LDLR
- low density lipoprotein receptor
- TLR
- Toll-like receptor
- GPCR
- G protein–coupled receptor
- ECD
- extracellular domain
- MEF
- mouse embryonic fibroblast
- m.o.i.
- multiplicity of infection
- Q-PCR
- quantitative PCR
- NDV
- Newcastle disease virus
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- IOD
- integrated optical density.
References
- 1. Rao B. L., Basu A., Wairagkar N. S., Gore M. M., Arankalle V. A., Thakare J. P., Jadi R. S., Rao K. A., and Mishra A. C. (2004) A large outbreak of acute encephalitis with high fatality rate in children in Andhra Pradesh, India, in 2003, associated with Chandipura virus. Lancet 364, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lichty B. D., Power A. T., Stojdl D. F., and Bell J. C. (2004) Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 10, 210–216 [DOI] [PubMed] [Google Scholar]
- 3. Ge P., Tsao J., Schein S., Green T. J., Luo M., and Zhou Z. H. (2010) Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roche S., Albertini A. A., Lepault J., Bressanelli S., and Gaudin Y. (2008) Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell. Mol. Life Sci. 65, 1716–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillies S., and Stollar V. (1980) Generation of defective interfering particles of vesicular stomatitis virus in Aedes albopictus cells. Virology 107, 497–508 [DOI] [PubMed] [Google Scholar]
- 6. Muik A., Kneiske I., Werbizki M., Wilflingseder D., Giroglou T., Ebert O., Kraft A., Dietrich U., Zimmer G., Momma S., and von Laer D. (2011) Pseudotyping vesicular stomatitis virus with lymphocytic choriomeningitis virus glycoproteins enhances infectivity for glioma cells and minimizes neurotropism. J. Virol. 85, 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mátrai J., Chuah M. K., and VandenDriessche T. (2010) Recent advances in lentiviral vector development and applications. Mol. Ther. 18, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yi Y., Noh M. J., and Lee K. H. (2011) Current advances in retroviral gene therapy. Curr. Gene Ther. 11, 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breitbach C. J., De Silva N. S., Falls T. J., Aladl U., Evgin L., Paterson J., Sun Y. Y., Roy D. G., Rintoul J. L., Daneshmand M., Parato K., Stanford M. M., Lichty B. D., Fenster A., Kirn D., Atkins H., and Bell J. C. (2011) Targeting tumor vasculature with an oncolytic virus. Mol. Ther. 19, 886–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Obuchi M., Fernandez M., and Barber G. N. (2003) Development of recombinant vesicular stomatitis viruses that exploit defects in host defense to augment specific oncolytic activity. J. Virol. 77, 8843–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kubo T., Shimose S., Matsuo T., Fujimori J., Sakaguchi T., Yamaki M., Shinozaki K., Woo S. L., and Ochi M. (2011) Oncolytic vesicular stomatitis virus administered by isolated limb perfusion suppresses osteosarcoma growth. J. Orthop. Res. 29, 795–800 [DOI] [PubMed] [Google Scholar]
- 12. Hu B., Tai A., and Wang P. (2011) Immunization delivered by lentiviral vectors for cancer and infectious diseases. Immunol. Rev. 239, 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marzi A., Robertson S. J., Haddock E., Feldmann F., Hanley P. W., Scott D. P., Strong J. E., Kobinger G., Best S. M., and Feldmann H. (2015) EBOLA vaccine: VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349, 739–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Albertini A. A., Baquero E., Ferlin A., and Gaudin Y. (2012) Molecular and cellular aspects of rhabdovirus entry. Viruses 4, 117–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coil D. A., and Miller A. D. (2004) Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J. Virol. 78, 10920–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johannsdottir H. K., Mancini R., Kartenbeck J., Amato L., and Helenius A. (2009) Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83, 440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cureton D. K., Massol R. H., Saffarian S., Kirchhausen T. L., and Whelan S. P. (2009) Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5, e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bloor S., Maelfait J., Krumbach R., Beyaert R., and Randow F. (2010) Endoplasmic reticulum chaperone gp96 is essential for infection with vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 107, 6970–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finkelshtein D., Werman A., Novick D., Barak S., and Rubinstein M. (2013) LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 110, 7306–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amirache F., Lévy C., Costa C., Mangeot P. E., Torbett B. E., Wang C. X., Nègre D., Cosset F. L., and Verhoeyen E. (2014) Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood 123, 1422–1424 [DOI] [PubMed] [Google Scholar]
- 21. Kroeze W. K., Sheffler D. J., and Roth B. L. (2003) G-protein-coupled receptors at a glance. J. Cell Sci. 116, 4867–4869 [DOI] [PubMed] [Google Scholar]
- 22. Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R. E., Hill C. M., Davis C. B., Peiper S. C., Schall T. J., Littman D. R., and Landau N. R. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666 [DOI] [PubMed] [Google Scholar]
- 23. Feng Y., Broder C. C., Kennedy P. E., and Berger E. A. (1996) HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872–877 [DOI] [PubMed] [Google Scholar]
- 24. Sodhi A., Montaner S., and Gutkind J. S. (2004) Viral hijacking of G-protein-coupled-receptor signalling networks. Nat. Rev. Mol. Cell Biol. 5, 998–1012 [DOI] [PubMed] [Google Scholar]
- 25. Cheng H., Lear-Rooney C. M., Johansen L., Varhegyi E., Chen Z. W., Olinger G. G., and Rong L. (2015) Inhibition of Ebola and Marburg virus entry by G protein-coupled receptor antagonists. J. Virol. 89, 9932–9938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu K., Xu Y., Rajashankar K. R., Robev D., and Nikolov D. B. (2013) Crystal structures of Lgr4 and its complex with R-spondin1. Structure 21, 1683–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carmon K. S., Gong X., Lin Q., Thomas A., and Liu Q. (2011) R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 11452–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Lau W., Barker N., Low T. Y., Koo B. K., Li V. S., Teunissen H., Kujala P., Haegebarth A., Peters P. J., van de Wetering M., Stange D. E., van Es J. E., Guardavaccaro D., Schasfoort R. B., Mohri Y., et al. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 [DOI] [PubMed] [Google Scholar]
- 29. Du B., Luo W., Li R., Tan B., Han H., Lu X., Li D., Qian M., Zhang D., Zhao Y., and Liu M. (2013) Lgr4/Gpr48 negatively regulates TLR2/4-associated pattern recognition and innate immunity by targeting CD14 expression. J. Biol. Chem. 288, 15131–15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liang F., Yue J., Wang J., Zhang L., Fan R., Zhang H., and Zhang Q. (2015) GPCR48/LGR4 promotes tumorigenesis of prostate cancer via PI3K/Akt signaling pathway. Med. Oncol. 32, 49. [DOI] [PubMed] [Google Scholar]
- 31. Gong X., Yi J., Carmon K. S., Crumbley C. A., Xiong W., Thomas A., Fan X., Guo S., An Z., Chang J. T., and Liu Q. J. (2015) Aberrant RSPO3-LGR4 signaling in Keap1-deficient lung adenocarcinomas promotes tumor aggressiveness. Oncogene 34, 4692–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kinzel B., Pikiolek M., Orsini V., Sprunger J., Isken A., Zietzling S., Desplanches M., Dubost V., Breustedt D., Valdez R., Liu D., Theil D., Müller M., Dietrich B., Bouwmeester T., et al. (2014) Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice. Dev. Biol. 390, 181–190 [DOI] [PubMed] [Google Scholar]
- 33. Hoshii T., Takeo T., Nakagata N., Takeya M., Araki K., and Yamamura K. (2007) LGR4 regulates the postnatal development and integrity of male reproductive tracts in mice. Biol. Reprod. 76, 303–313 [DOI] [PubMed] [Google Scholar]
- 34. Luo W., Rodriguez M., Valdez J. M., Zhu X., Tan K., Li D., Siwko S., Xin L., and Liu M. (2013) Lgr4 is a key regulator of prostate development and prostate stem cell differentiation. Stem Cells 31, 2492–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo J., Yang Z., Ma Y., Yue Z., Lin H., Qu G., Huang J., Dai W., Li C., Zheng C., Xu L., Chen H., Wang J., Li D., Siwko S., et al. (2016) LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat. Med. 22, 539–546 [DOI] [PubMed] [Google Scholar]
- 36. Wang Y., Dong J., Li D., Lai L., Siwko S., Li Y., and Liu M. (2013) Lgr4 regulates mammary gland development and stem cell activity through the pluripotency transcription factor Sox2. Stem Cells 31, 1921–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Superti F., Seganti L., Ruggeri F. M., Tinari A., Donelli G., and Orsi N. (1987) Entry pathway of vesicular stomatitis virus into different host cells. J. Gen. Virol. 68, 387–399 [DOI] [PubMed] [Google Scholar]
- 38. von Kleist L., Stahlschmidt W., Bulut H., Gromova K., Puchkov D., Robertson M. J., MacGregor K. A., Tomlin N., Pechstein A., Chau N., Chircop M., Sakoff J., von Kries J. P., Saenger W., Kräusslich H. G., et al. (2011) Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 146, 471–484 [DOI] [PubMed] [Google Scholar]
- 39. Sun X., Yau V. K., Briggs B. J., and Whittaker G. R. (2005) Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338, 53–60 [DOI] [PubMed] [Google Scholar]
- 40. Chen Y., Ruan X. Z., Li Q., Huang A., Moorhead J. F., Powis S. H., and Varghese Z. (2007) Inflammatory cytokines disrupt LDL-receptor feedback regulation and cause statin resistance: a comparative study in human hepatic cells and mesangial cells. Am. J. Physiol. Renal Physiol. 293, F680–F687 [DOI] [PubMed] [Google Scholar]
- 41. Yi J., Xiong W., Gong X., Bellister S., Ellis L. M., and Liu Q. (2013) Analysis of LGR4 receptor distribution in human and mouse tissues. PloS One 8, e78144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song Q., Huang M., Yao L., Wang X., Gu X., Chen J., Chen J., Huang J., Hu Q., Kang T., Rong Z., Qi H., Zheng G., Chen H., and Gao X. (2014) Lipoprotein-based nanoparticles rescue the memory loss of mice with Alzheimer's disease by accelerating the clearance of amyloid-beta. ACS Nano 8, 2345–2359 [DOI] [PubMed] [Google Scholar]