Abstract
Background
Despite initial indications that the transcription factor Twist could be used as a breast cancer prognostic marker, there still exists some controversy about its reliability. Thus, the aim of the present study was to assess the relationship between Twist expression and prognosis in breast carcinoma.
Materials and methods
We identified eligible studies that reported an association between Twist expression and breast cancer prognosis by searching the literature in PubMed, Embase, the Cochrane Library, and Web of Science databases, through June 5, 2017. Studies investigating Twist protein or mRNA expression as well as reporting survival data in breast cancer were included. The pooled hazard ratio (HR) and odds radio (OR) with a 95% confidence interval (95% CI) were used to estimate associations.
Results
A total of 2,671 patients from seven included studies were assessed, and the data indicated that increased Twist expression significantly correlated with poor overall survival (OS) (HR, 1.15; 95% CI, 1.00–1.33; P = 0.04) in breast cancer. In addition, we also observed a significant correlation of elevated Twist expression with larger tumor size (OR, 1.92; 95% CI, 1.31–2.81; P = 0.0009), lymph node involvement (OR, 3.81; 95% CI, 1.16–12.54; P = 0.03), higher nuclear grade (OR, 1.45; 95% CI, 1.06–2.00; P = 0.02), and positive human epidermal growth factor receptor 2 (HER2) status (OR, 1.49; 95% CI, 1.06–2.09; P = 0.02). However, no correlation between Twist expression and disease-free survival (DFS), age, estrogen receptor (ER) status, and progesterone receptor (PR) status was observed.
Conclusions
Our results demonstrate that Twist over-expression is a statistically significant indicator of OS in breast cancer. In addition, our meta-analysis shows that increased Twist expression is significantly associated with larger tumor size, lymph node involvement, higher nuclear grade, and positive HER2 status.
Introduction
Breast cancer incidence is high not only in Chinese women but also worldwide, and thus is ranked as one of the most common cancers[1, 2]. Breast cancer is categorized into different subtypes based on the expression of various biomarkers, including estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki67. Expression of these markers plays a vital role in deciding the fundamental therapeutic strategy[3]. As a result, the mortality rate associated with breast cancer has reduced due to significant progress in early diagnosis and development of multiple treatment options. However, a significant percentage of the patient population still fails to respond to these already developed therapies, and many patients develop metastasis, relapse, or display therapeutic resistance. Notable among them is triple-negative breast cancer (TNBC) subtype. Thus, there is an essential need to identify additional novel molecular biomarkers that have the potential to predict therapeutic value across multiple subtypes and serve as therapeutic targets.
Twist is a basic helix-loop-helix (bHLH) transcription factor that has been previously implicated in cell lineage determination and differentiation during embryogenesis. In recent years, Twist has also been shown to contribute to carcinogenesis through triggering epithelial to mesenchymal transition (EMT) and downregulating E-cadherin expression, thereby influencing tumor invasion, metastasis, adverse prognosis, and drug resistance in multiple tumors[4, 5]. Previous studies have demonstrated that Twist interfered the ARF-p53 pathway to prevent c-myc-induced apoptosis and the anti-apoptotic character of Twist was considered the reason for metastatic process[6–9]. Twist expression has been significantly associated with invasion and metastasis of various cancers, including breast cancer[10], non-small cell lung cancer[11], prostate cancer[12], gastric cancer[13], melanoma[14], Sezary syndrome[15], osteosarcoma[16], and hepatocarcinoma[17]. Some studies have indicated that increased Twist expression correlated with worse breast cancer prognosis[18, 19], while other studies showed opposite results[20]. Therefore, to further clarify the prognostic value of Twist in breast cancer, we conducted a new meta-analysis to estimate the association between Twist expression and survival outcomes in breast cancer. In addition, we also assessed the correlation of Twist with clinicopathological features of breast carcinoma.
Materials and methods
Search strategy
Eligible studies through June 5, 2017 were identified using PubMed, Embase, the Cochrane Library, and Web of Science databases. The following MeSH terms were used to search relevant articles: “breast neoplasms” and/or “breast cancer”, and/or “Twist”, and/or “prognosis”. Moreover, the reference lists of eligible studies were further searched manually to identify additional relevant studies.
Inclusion and exclusion criteria
Studies were included in our meta-analysis based on the following criteria: (1) all studies provided information about survival outcome in breast cancer; (2) Twist expression was analyzed in all breast cancer patients; and (3) the hazard ratio (HR) with 95% confidence interval (CI) was either available or sufficient information was available to indirectly estimate it. Studies were excluded from the meta-analysis if they were either (1) duplicate studies, (2) animal or cell studies, (3) HR information was not available and could not be extracted from a Kaplan-Meier curve, or (4) if they were reviews, letters, or only case reports.
Data extraction
Data extraction from the eligible studies was performed independently by two authors (Weiqiang Qiao and Heyang Liu). The extracted information included: author of publication, year, country, number of patients, age, time of follow-up, clinical outcome, survival analysis, Twist expression, detection method, its cut-off values, antibody, proportion of tumors with Twist over-expression, and correlation between Twist mRNA and protein levels. The HR information was directly recorded if present, or extracted from the Kaplan-Meier curves using Engauge Digitizer Version 4.1 (http://markummitchell.github.io/engauge-digitizer/) software. The quality of the studies was assessed using the modified Newcastle Ottawa Scale (NOS)[21], which classified the studies into eight categories and scored them. A score of 9 represented a maximum score; however, a score of 7 or higher indicated high quality.
Statistical analysis
The complete meta-analysis was performed using Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guideline[22] (S1 File). In studies where HR value was not available, HR was calculated from Kaplan-Meier curves, as suggested by Tierney et al.[23]. The Cochran Q test and I2 statistics were applied to detect heterogeneity. A P value of < 0.05 and I2 value of > 50% represented strong heterogeneity[24]. The fixed effects model was applied for meta-analysis if very low or no heterogeneity was observed. In contrast, the random effects model was used when notable heterogeneity existed between different studies[25]. In addition, sensitivity analysis was also conducted to assess the robustness of the data. The Meta-regression analysis was applied to estimate the sources of heterogeneity. Moreover, potential publication bias was tested using Begg’s test[26]. Overall, all analyses were conducted using Review Manager version 5.3 (Cochrane Collaboration, Copenhagen, Denmark), and STATA version 14.0 (Stata Corporation, TX, USA) software. All statistical tests were two-sided, and a P value of < 0.05 represented a statistically significant difference.
Results
Identification of relevant studies
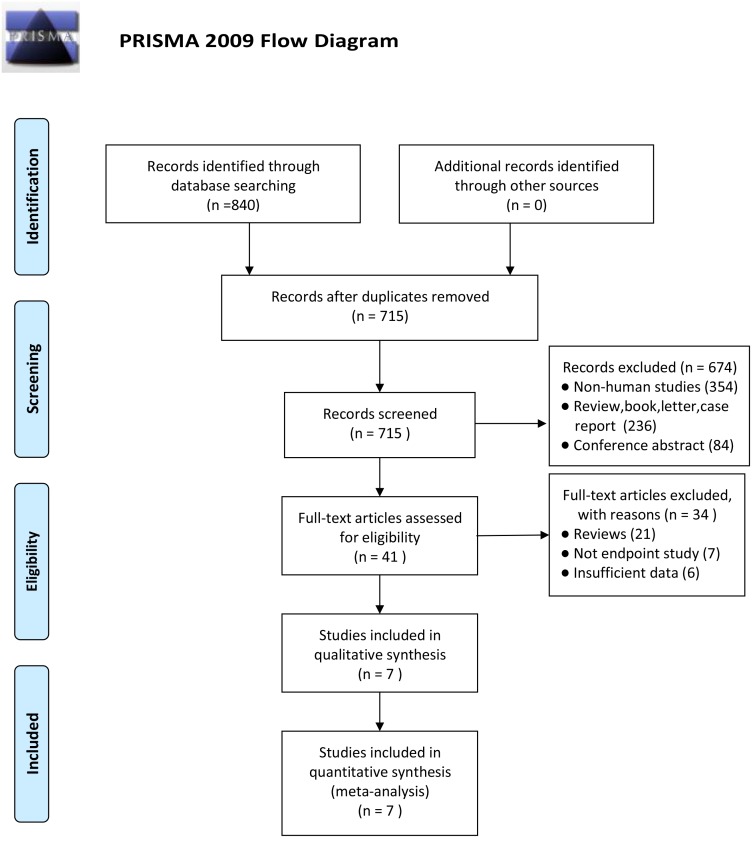
A total of 840 initial studies were identified based on the search strategy (S2 File). Among these, 125 duplicates studies were excluded. After screening the titles and abstracts of the remaining studies, 674 articles were further excluded, thus leaving 41 studies for full review. Based on the exclusion criteria, another 34 studies (21 review articles, 7 with no endpoint, and 6 with insufficient data) were removed. The remaining 7 eligible studies were included in our meta-analysis, which included 2,671 patients[18–20, 27–30]. The flow chart for study selection is outlined in Fig 1.
Fig 1. Flow chart of study selection process.
Characteristics of eligible studies
The primary characteristics of all the included studies are shown in Tables 1 and 2. All of these articles were primarily published between the years 2011 and 2015. The included studies used different techniques to measure Twist expression. Seven studies analyzed Twist expression using immunochemical (IHC) staining, while 3 studies used reverse transcriptase polymerase chain reaction (RT-PCR). There was some inconsistency regarding Twist expression, as a few studies analyzed Twist, while others specifically measured Twsit1 levels. In addition, we also observed variation in the cut-off values for Twist expression among different studies. Overall, all included studies tested the correlation between Twist expression and breast tumor prognosis. Importantly, all the included studies were observational studies, and thus their quality was assessed using the NOS criteria. Our analysis revealed that all studies were of high quality with a score of ≥ 7 score, as shown in S1 Table.
Table 1. Characteristics of eligible studies.
| Publication | Year | Country | Cancer subtype | No. of patients | Median age (years) | Median follow-up (years) | Outcome | Survival analysis | NOS (score) |
|---|---|---|---|---|---|---|---|---|---|
| Markiewicz | 2012 | Poland | II-III BC | 36 | 56.5 | 4.2 | DFS, OS | multivariate | 8 |
| Montserrat | 2011 | Spain | invasive BC | 76 | 67 | 4.5 | DFS, OS | multivariate | 8 |
| Riaz | 2012 | Netherlands | primary BC | 1427 | 55 | 8.7 | DFS, OS | multivariate | 8 |
| Soini | 2011 | Finland | invasive BC | 387 | NR | NR | OS | univariate | 7 |
| Xu | 2014 | China | primary BC | 137 | NR | 5 | DFS, OS | multivariate | 8 |
| Zhang | 2015 | China | invasive BC | 408 | 50 | 1.3 | DFS, OS | univariate | 7 |
| Zhao | 2013 | China | primary BC | 200 | 50 | NR | OS | univariate | 7 |
BC, breast cancer; NR, not reported; DFS, disease-free survival; OS, overall survival; NOS, Newcastle Ottawa Scale
Table 2. Methods of quantitative Twist measurement of eligible studies.
| Publication | Year | Twist phenotype | Detection method | Twist expression | Antibody | Cut-off value (low/high level) | High Twist expression | Correlation between Twist mRNA and protein levels | |
|---|---|---|---|---|---|---|---|---|---|
| Markiewicz | 2012 | Twist1 | RT-PCR, IHC | mRNA, protein | anti-Twist1 (ab50581, Abcam) | NR | 66%(29/44) | kappa coefficient | P = 0.002 |
| Montserrat | 2011 | Twist | RT-PCR, IHC | mRNA, protein | anti-Twist polyclonal antibodies | low(≤10%),high (>10%) | 16%(12/76) | Spearman rank test | P = 0.009 |
| Riaz | 2012 | Twist1 | RT-PCR, IHC | mRNA, protein | envision mouse kit, DAKO | NR | NR | Spearman rank test | P < 0.004 |
| Soini | 2011 | Twist | IHC | protein | mouse monoclonal anti-twist antibody | low(≤5%),high (>5%) | 35%(135/387) | NR | NR |
| Xu | 2014 | Twist1 | IHC | protein | anti-Twist1 (ab50887, Abcam, MA) | high (staining score≥3) | 46.7%(64/137) | NR | NR |
| Zhang | 2015 | Twist | IHC | protein | mouse monoclonal antibody | NR | 53%(220/408) | NR | NR |
| Zhao | 2013 | Twist | IHC | protein | anti-Twist polyclonal antibody | high (staining score≥6) | 75.5%(151/200) | NR | NR |
NR, not reported; RT-PCR, reverse transcriptase polymerase chain reaction; IHC, immunohistochemistry
Correlation analysis of Twist expression with disease free survival & overall survival
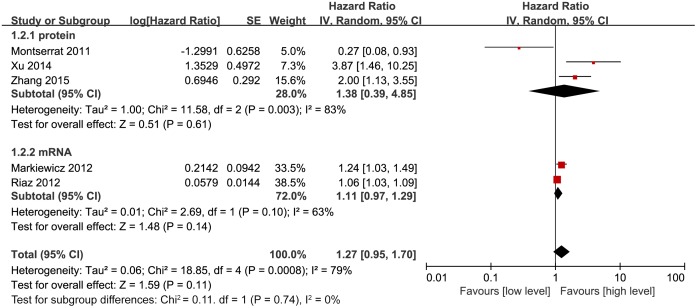
Among the 7 included studies, 5 estimated the correlation between Twist expression and disease-free survival (DFS) in breast cancer patients. Our meta-analysis used the random effect model due to the high heterogeneity (P = 0.0008, I2 = 79%) between studies and revealed that Twist expression did not correlate with DFS in breast cancer patients (HR, 1.27; 95% CI, 0.95–1.70; P = 0.11; Fig 2). Next, we assessed this correlation based on Twist expression (mRNA or protein) stratification, but this analysis also did not show any association between Twist expression and DFS. The HR value based on Twist protein expression was 1.38 (95% CI, 0.39–4.85; P = 0.61; Fig 2), while the HR for mRNA levels was 1.11 (95% CI, 0.97–1.29; P = 0.14; Fig 2).
Fig 2. Forest plot depicting association between Twist expression and DFS in breast cancer.
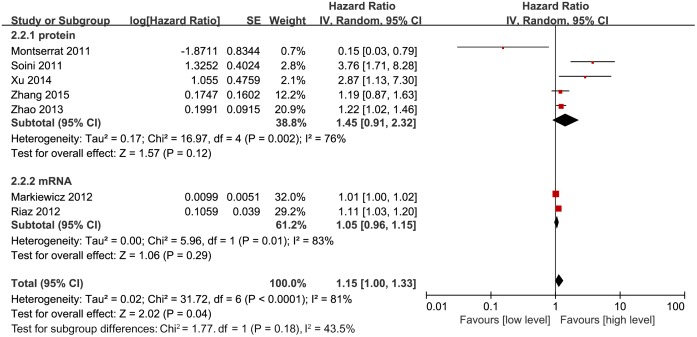
Similarly, we also assessed the correlation of Twist expression with overall survival (OS). All 7 studies had data about pooled HRs for OS. Interestingly, increased Twist expression was significantly associated with worse OS (HR, 1.15; 95% CI, 1.00–1.33; P = 0.04; Fig 3). This analysis was performed using random effect model due to significant heterogeneity (P < 0.0001, I2 = 81%) between the studies. To understand the reasons of high heterogeneity, we performed further subgroup analyses. Surprisingly, there was no significant correlation for either Twist protein (HR, 1.45; 95% CI, 0.91–2.32; P = 0.12; Fig 3) or mRNA (HR, 1.05; 95% CI, 0.96–1.15; P = 0.29; Fig 3) levels with OS.
Fig 3. Forest plot depicting association between Twist expression and OS in breast cancer.
Correlation between Twist expression and other breast cancer clinical parameters
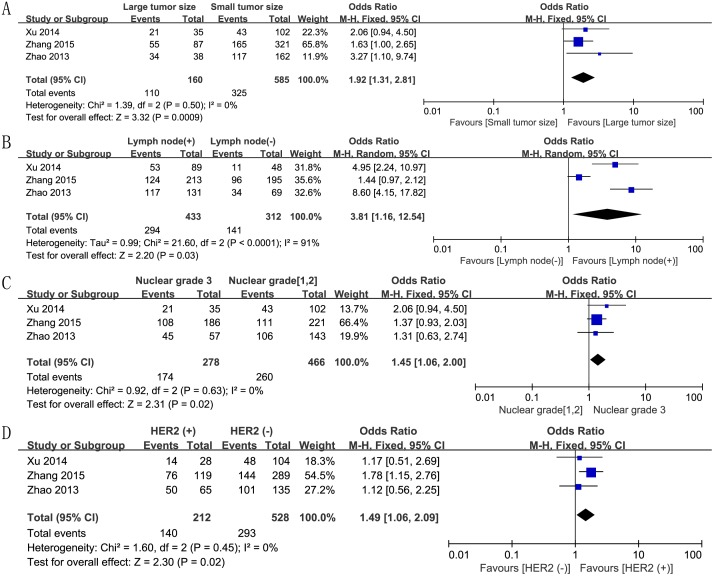
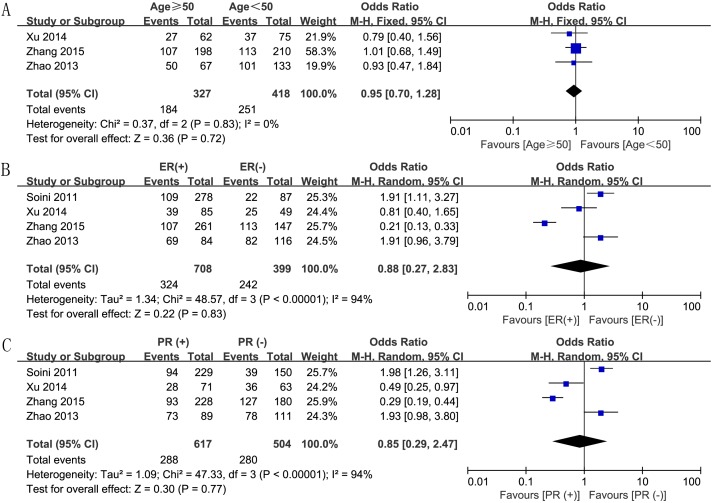
We also assessed the correlation between increased Twist expression and various clinical parameters of breast cancer. First, we analyzed the correlation between Twist expression and breast tumor size, based on data from 3 studies. We analyzed the data using the fixed effect model because there was no heterogeneity (P = 0.50, I2 = 0), and found an OR value of 1.92 (95% CI, 1.31–2.81; P = 0.0009; Fig 4A), thereby establishing a positive association of Twist expression and tumor size. The random effect model based analysis of 3 studies with high heterogeneity (P < 0.0001, I2 = 91%) showed significant association between Twist expression and lymph node involvement (OR, 3.81; 95% CI, 1.16–12.54; P = 0.03; Fig 4B). In addition, the fixed effect model analysis also confirmed significant association between Twist expression and increased nuclear grade (OR, 1.45; 95% CI, 1.06–2.00; P = 0.02; Fig 4C) and positive HER2 status (OR, 1.49; 95% CI, 1.06–2.09; P = 0.02; Fig 4D). However, other clinicopathological characteristics like age (OR, 0.95; 95% CI, 0.70–1.28; P = 0.72; Fig 5A), ER status (OR, 0.88; 95% CI, 0.27–2.83; P = 0.83; Fig 5B), and PR status (OR, 0.85; 95% CI, 0.29–2.47; P = 0.77; Fig 5C) did not show any association with Twist expression.
Fig 4. Forest plots depicting correlations between Twist expression and (A) tumor size (large vs. small), (B) lymph node involvement (positive vs. negative), (C) nuclear grade (3 vs. 1 and 2), and (D) HER2 status (positive vs. negative).
Fig 5. Forest plots depicting correlations between Twist expression and (A) age (≥ 50 vs. < 50), (B) ER status (positive vs. negative), and (C) PR status (positive vs. negative).
Meta-regression analysis to identify confounding variables
We also performed meta-regression analysis to identify variable factors influencing the association of Twist expression with DFS and OS in breast cancer. However, we did not find any evidence of covariates significantly affecting DFS (S2 Table), nor did we identify any significant confounding factors as potential sources of heterogeneity in OS (S3 Table).
Publication bias and sensitivity analysis
Our analysis of publication bias using Begg’s rank correlation test revealed no bias for DFS (P = 0.462) or OS (P = 1.000). Moreover, sensitivity analysis established that the results were stable for both DFS (S1A Fig) and OS (S1B Fig), after excluding one study at a time.
Discussion
In our current meta-analysis, we have tried to exclusively evaluate the actual prognostic value of elevated Twist expression in breast cancer. Two earlier meta-analysis studies also tried to clarify the association of Twist expression in parallel for multiple cancers[31, 32], including breast cancer, where the results were based on data from only 2 studies. One study reported a positive association of Twist expression with worse OS in breast cancer (HR, 2.34; 95% CI, 1.72–3.20; P < 0.001)[31], while the other reported no association (HR, 1.65; 95% CI, 0.19–14.03; P = 0.66)[32]. Thus, these conflicting reports led us to undertake a comprehensive analysis. Our meta-analysis included these two studies as well as an additional five breast cancer studies to determine if there was a significant correlation between Twist expression and breast cancer. We also included studies that specifically examined Twist mRNA expression. In addition, we also examined the association between Twist expression and various breast cancer clinicopathological factors.
Interestingly, our results indicated that higher Twist expression was significantly associated with worse OS in breast cancer, but showed no correlation with DFS. This result was consistent with a previously published study by Wushou et al.[31], which indicated that inhibitors of Twist can be beneficial for improving clinical outcomes in breast cancer treatment. Grzegrzolka et al.[33] demonstrated that higher nuclear Twist expression was associated with worse event-free survival and poor OS in breast cancer patients. Lim et al.[34] revealed that stromal nuclear Twist over-expression was correlated with worse prognosis in terms of disease recurrence and OS in patients with phyllodes tumors of the breast. Xu et al.[35] indicated that increased Twist expression was related to worse distant metastasis-free survival (DMFS) in patients with breast cancer. These findings indicate that future research should focus on testing the efficiency and safety of these inhibitors. In this context, a study by Ranganathan et al.[36] indicated that quercetin downregulated Twist expression through inhibiting the p38MAPK pathway, resulting in breast cancer cell apoptosis. The INK4a/ARF locus was central to apoptosis through p53 pathway to inhibit proliferation[37, 38]. The earlier reports found a novel function of Twist through interfering p14ARF-mediated p53 pathway, leading to developed anti-apoptotic activity[39, 40]. Moreover, Inoue et al.[41] suggested that Dmp1 was a regulator of the ARF-p53 pathway. Another study by Kwilas et al.[42] demonstrated that a poxviral-based cancer vaccine targeting Twist suppressed breast cancer cell growth and metastasis and improved survival outcome in prostate carcinoma. Thus, these studies provided the initial evidence that there is potential benefit in targeting Twist in a therapeutic regimen for treating cancers. However, randomized controlled clinical trials are required to verify if a Twist inhibitor can really serve as a valid therapeutic strategy for cancer. Earlier literature has also reported an association between Twist over-expression and drug resistance against chemotherapeutic drugs, including Taxol in a nasopharyngeal carcinoma cell line[43], and cisplatin and doxorubicin in bladder cancer cells[44]. These observations indicate that inhibiting Twist expression could overcome chemoresistance in human tumors.
Furthermore, we also comprehensively investigated the association between elevated Twist expression and clinicopathological parameters of breast cancer. We observed that elevated Twist expression significantly correlated with larger tumor size, lymph node involvement, higher nuclear grade, and positive HER2 status. In contrast, we did not observe a significant relationship of Twist expression with age, ER status, and PR status. Previous studies have demonstrated that larger tumor size, lymph node metastasis, higher nuclear grade, and positive HER2 status are typically poor prognostic indicators of breast cancer[45–48]. Since these clinicopathological parameters showed association with higher Twist expression in our study, we conclude that our meta-analysis further validates that Twist expression is indeed associated with adverse prognosis in breast cancer. Another independent study by Vesuna et al.[49] reported that Twist over-expression was associated with negative ER breast cancer subtype, which is an aggressive prognostic subtype. Besides, Twist expression was notablely higher in TNBC (87.3%, 55/63), followed by the positive HER2 status (71.8%, 51/71), Luminal B (52.1%, 25/48) and Luminal A types (39.4%, 89/226) in terms of molecular subtypes in breast cancer[29] Therefore, collective observations have demonstrated that Twist over-expression could be an appropriate biomarker in breast cancer prognosis.
We should note that our study also had some limitations. First, some HRs were not offered in the original articles, and therefore HRs were extracted from the Kaplan-Meier curves for these studies. This could have impacted the robustness of outcomes. Second, each study varied with regards to Twist detection methods, as well as different variants and cut-off levels. These differences could potentially contribute to strong heterogeneity. Meta-regression analysis was performed to explore the potential sources of heterogeneity, but no significant confounding factors were observed. Finally, the sample size was also relatively small in the included studies, which could have influenced the pooled results.
In conclusion, there was evidence of a just statistically significant difference between high Twist expression and worse OS in breast cancer, however, it may be debated whether it is really clinically relevant, additional well-designed cohort studies are needed to confirm the association. Also, our study found a significant association of Twist expression with breast cancer clinicopathological characteristics, including larger tumor size, lymph node metastasis, higher nuclear grade, and positive HER2 status.
Supporting information
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Program for Science &Technology Innovation teams in Universities of Henan Province (#181RTSTHN026), the innovation team of Henan University of Science and Technology (#2015XTD003) and Henan province’s Key Project of tackle key problems of science and technology (#172102310693). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67(1):7–30. Epub 2017/01/06. doi: 10.3322/caac.21387 . [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66(2):115–32. Epub 2016/01/26. doi: 10.3322/caac.21338 . [DOI] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Annals of oncology: official journal of the European Society for Medical Oncology. 2013;24(9):2206–23. Epub 2013/08/07. doi: 10.1093/annonc/mdt303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–39. Epub 2004/06/24. doi: 10.1016/j.cell.2004.06.006 . [DOI] [PubMed] [Google Scholar]
- 5.Vesuna F, Bergman Y, Raman V. Genomic pathways modulated by Twist in breast cancer. BMC cancer. 2017;17(1):52 Epub 2017/01/15. doi: 10.1186/s12885-016-3033-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, et al. Twist is a potential oncogene that inhibits apoptosis. Genes & development. 1999;13(17):2207–17. Epub 1999/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas AC, Combaret V, et al. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer cell. 2004;6(6):625–30. Epub 2004/12/21. doi: 10.1016/j.ccr.2004.09.033 . [DOI] [PubMed] [Google Scholar]
- 8.Puisieux A, Valsesia-Wittmann S, Ansieau S. A twist for survival and cancer progression. British journal of cancer. 2006;94(1):13–7. Epub 2005/11/25. doi: 10.1038/sj.bjc.6602876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selmi A, de Saint-Jean M, Jallas AC, Garin E, Hogarty MD, Benard J, et al. TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer letters. 2015;357(1):412–8. Epub 2014/12/06. doi: 10.1016/j.canlet.2014.11.056 . [DOI] [PubMed] [Google Scholar]
- 10.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Annals of surgical oncology. 2005;12(6):488–96. Epub 2005/05/03. doi: 10.1245/ASO.2005.04.010 . [DOI] [PubMed] [Google Scholar]
- 11.Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS, Wu KJ. Prognostic significance of hypoxia-inducible factor-1alpha, TWIST1 and Snail expression in resectable non-small cell lung cancer. Thorax. 2009;64(12):1082–9. Epub 2009/09/26. doi: 10.1136/thx.2009.115691 . [DOI] [PubMed] [Google Scholar]
- 12.Alexander NR, Tran NL, Rekapally H, Summers CE, Glackin C, Heimark RL. N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer research. 2006;66(7):3365–9. Epub 2006/04/06. doi: 10.1158/0008-5472.CAN-05-3401 . [DOI] [PubMed] [Google Scholar]
- 13.Luo GQ, Li JH, Wen JF, Zhou YH, Hu YB, Zhou JH. Effect and mechanism of the Twist gene on invasion and metastasis of gastric carcinoma cells. World journal of gastroenterology. 2008;14(16):2487–93. Epub 2008/04/30. doi: 10.3748/wjg.14.2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer research. 2004;64(15):5270–82. Epub 2004/08/04. doi: 10.1158/0008-5472.CAN-04-0731 . [DOI] [PubMed] [Google Scholar]
- 15.van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, et al. Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sezary syndrome identified by gene expression analysis. Cancer research. 2004;64(16):5578–86. Epub 2004/08/18. doi: 10.1158/0008-5472.CAN-04-1253 . [DOI] [PubMed] [Google Scholar]
- 16.Entz-Werle N, Stoetzel C, Berard-Marec P, Kalifa C, Brugiere L, Pacquement H, et al. Frequent genomic abnormalities at TWIST in human pediatric osteosarcomas. International journal of cancer. 2005;117(3):349–55. Epub 2005/05/19. doi: 10.1002/ijc.21068 . [DOI] [PubMed] [Google Scholar]
- 17.Lee TK, Poon RT, Yuen AP, Ling MT, Kwok WK, Wang XH, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12(18):5369–76. Epub 2006/09/27. doi: 10.1158/1078-0432.ccr-05-2722 . [DOI] [PubMed] [Google Scholar]
- 18.Soini Y, Tuhkanen H, Sironen R, Virtanen I, Kataja V, Auvinen P, et al. Transcription factors zeb1, twist and snai1 in breast carcinoma. BMC cancer. 2011;11:73 Epub 2011/02/18. doi: 10.1186/1471-2407-11-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Hu B, Qin L, Zhao L, Wang Q, Wang Q, et al. SRC-1 and Twist1 expression positively correlates with a poor prognosis in human breast cancer. International journal of biological sciences. 2014;10(4):396–403. Epub 2014/04/11. doi: 10.7150/ijbs.8193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montserrat N, Gallardo A, Escuin D, Catasus L, Prat J, Gutierrez-Avigno FJ, et al. Repression of E-cadherin by SNAIL, ZEB1, and TWIST in invasive ductal carcinomas of the breast: a cooperative effort? Human pathology. 2011;42(1):103–10. Epub 2010/10/26. doi: 10.1016/j.humpath.2010.05.019 . [DOI] [PubMed] [Google Scholar]
- 21.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25(9):603–5. Epub 2010/07/24. doi: 10.1007/s10654-010-9491-z . [DOI] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. International journal of surgery (London, England). 2010;8(5):336–41. Epub 2010/02/23. doi: 10.1016/j.ijsu.2010.02.007 . [DOI] [PubMed] [Google Scholar]
- 23.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16 Epub 2007/06/09. doi: 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327(7414):557–60. Epub 2003/09/06. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemporary clinical trials. 2015;45(Pt A):139–45. Epub 2015/09/08. doi: 10.1016/j.cct.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101. Epub 1994/12/01. . [PubMed] [Google Scholar]
- 27.Markiewicz A, Ahrends T, Welnicka-Jaskiewicz M, Seroczynska B, Skokowski J, Jaskiewicz J, et al. Expression of epithelial to mesenchymal transition-related markers in lymph node metastases as a surrogate for primary tumor metastatic potential in breast cancer. Journal of translational medicine. 2012;10:226 Epub 2012/11/20. doi: 10.1186/1479-5876-10-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riaz M, Sieuwerts AM, Look MP, Timmermans MA, Smid M, Foekens JA, et al. High TWIST1 mRNA expression is associated with poor prognosis in lymph node-negative and estrogen receptor-positive human breast cancer and is co-expressed with stromal as well as ECM related genes. Breast cancer research: BCR. 2012;14(5):R123 Epub 2012/09/13. doi: 10.1186/bcr3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang YQ, Wei XL, Liang YK, Chen WL, Zhang F, Bai JW, et al. Over-Expressed Twist Associates with Markers of Epithelial Mesenchymal Transition and Predicts Poor Prognosis in Breast Cancers via ERK and Akt Activation. PloS one. 2015;10(8):e0135851 Epub 2015/08/22. doi: 10.1371/journal.pone.0135851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao M, Hu HG, Huang J, Zou Q, Wang J, Liu MQ, et al. Expression and correlation of Twist and gelatinases in breast cancer. Experimental and therapeutic medicine. 2013;6(1):97–100. Epub 2013/08/13. doi: 10.3892/etm.2013.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wushou A, Hou J, Zhao YJ, Shao ZM. Twist-1 up-regulation in carcinoma correlates to poor survival. International journal of molecular sciences. 2014;15(12):21621–30. Epub 2014/11/28. doi: 10.3390/ijms151221621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, Hu P, Shen H, Yu J, Liu Q, Du J. Prognostic role of Twist or Snail in various carcinomas: a systematic review and meta-analysis. European journal of clinical investigation. 2014;44(11):1072–94. Epub 2014/09/27. doi: 10.1111/eci.12343 . [DOI] [PubMed] [Google Scholar]
- 33.Grzegrzolka J, Biala M, Wojtyra P, Kobierzycki C, Olbromski M, Gomulkiewicz A, et al. Expression of EMT Markers SLUG and TWIST in Breast Cancer. Anticancer research. 2015;35(7):3961–8. Epub 2015/07/01. . [PubMed] [Google Scholar]
- 34.Lim JC, Koh VC, Tan JS, Tan WJ, Thike AA, Tan PH. Prognostic significance of epithelial-mesenchymal transition proteins Twist and Foxc2 in phyllodes tumours of the breast. Breast cancer research and treatment. 2015;150(1):19–29. Epub 2015/02/14. doi: 10.1007/s10549-015-3296-4 . [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36(8):1157–66. Epub 2016/08/16. doi: 10.1038/onc.2016.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranganathan S, Halagowder D, Sivasithambaram ND. Quercetin Suppresses Twist to Induce Apoptosis in MCF-7 Breast Cancer Cells. PloS one. 2015;10(10):e0141370 Epub 2015/10/23. doi: 10.1371/journal.pone.0141370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes & development. 2011;25(5):485–98. Epub 2011/02/15. doi: 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cakouros D, Isenmann S, Cooper L, Zannettino A, Anderson P, Glackin C, et al. Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Molecular and cellular biology. 2012;32(8):1433–41. Epub 2012/02/01. doi: 10.1128/MCB.06315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwok WK, Ling MT, Yuen HF, Wong YC, Wang X. Role of p14ARF in TWIST-mediated senescence in prostate epithelial cells. Carcinogenesis. 2007;28(12):2467–75. Epub 2007/08/11. doi: 10.1093/carcin/bgm185 . [DOI] [PubMed] [Google Scholar]
- 40.Shamanin VA, Androphy EJ. Immortalization of human mammary epithelial cells is associated with inactivation of the p14ARF-p53 pathway. Molecular and cellular biology. 2004;24(5):2144–52. Epub 2004/02/18. doi: 10.1128/MCB.24.5.2144-2152.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene. 2007;26(30):4329–35. Epub 2007/01/24. doi: 10.1038/sj.onc.1210226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwilas AR, Ardiani A, Dirmeier U, Wottawah C, Schlom J, Hodge JW. A poxviral-based cancer vaccine the transcription factor twist inhibits primary tumor growth and metastases in a model of metastatic breast cancer and improves survival in a spontaneous prostate cancer model. Oncotarget. 2015;6(29):28194–210. Epub 2015/09/01. doi: 10.18632/oncotarget.4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, Lee DT, et al. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene. 2004;23(2):474–82. Epub 2004/01/16. doi: 10.1038/sj.onc.1207128 . [DOI] [PubMed] [Google Scholar]
- 44.Shiota M, Yokomizo A, Itsumi M, Uchiumi T, Tada Y, Song Y, et al. Twist1 and Y-box-binding protein-1 promote malignant potential in bladder cancer cells. BJU international. 2011;108(2 Pt 2):E142–9. Epub 2010/11/19. doi: 10.1111/j.1464-410X.2010.09810.x . [DOI] [PubMed] [Google Scholar]
- 45.Ono M, Tsuda H, Yoshida M, Shimizu C, Kinoshita T, Tamura K. Prognostic Significance of Progesterone Receptor Expression in Estrogen-Receptor Positive, HER2-Negative, Node-Negative Invasive Breast Cancer With a Low Ki-67 Labeling Index. Clinical breast cancer. 2017;17(1):41–7. Epub 2016/08/02. doi: 10.1016/j.clbc.2016.06.012 . [DOI] [PubMed] [Google Scholar]
- 46.Park S, Lee SK, Paik HJ, Ryu JM, Kim I, Bae SY, et al. Adjuvant endocrine therapy alone in patients with node-positive, luminal A type breast cancer. Medicine. 2017;96(22):e6777 Epub 2017/06/01. doi: 10.1097/MD.0000000000006777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasconcelos I, Hussainzada A, Berger S, Fietze E, Linke J, Siedentopf F, et al. The St. Gallen surrogate classification for breast cancer subtypes successfully predicts tumor presenting features, nodal involvement, recurrence patterns and disease free survival. Breast (Edinburgh, Scotland). 2016;29:181–5. Epub 2016/08/22. doi: 10.1016/j.breast.2016.07.016 . [DOI] [PubMed] [Google Scholar]
- 48.Ehinger A, Malmstrom P, Bendahl PO, Elston CW, Falck AK, Forsare C, et al. Histological grade provides significant prognostic information in addition to breast cancer subtypes defined according to St Gallen 2013. Acta oncologica (Stockholm, Sweden). 2017;56(1):68–74. Epub 2016/10/21. doi: 10.1080/0284186x.2016.1237778 . [DOI] [PubMed] [Google Scholar]
- 49.Vesuna F, Lisok A, Kimble B, Domek J, Kato Y, van der Groep P, et al. Twist contributes to hormone resistance in breast cancer by downregulating estrogen receptor-alpha. Oncogene. 2012;31(27):3223–34. Epub 2011/11/08. doi: 10.1038/onc.2011.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.