Abstract
The data presented here are related to the research article entitled “Selective expression of the transcription elongation factor ELL3 in B cells prior to ELL2 drives proliferation and survival” (Alexander et al., 2017) [1]. The cited research article characterizes Eleven-nineteen Lysine-rich Leukemia 3 (ELL3) expression in the B cell compartment and functional dependence in B lymphoma cell lines. This data report describes the mRNA expression pattern in a panel of cell lines representing the B cell compartment, supplementing the protein expression data presented in the associated research report. In addition, a reanalysis is presented of publicly available mRNA expression data from primary murine B cells to reveal dynamic regulation of the ELL family members post LPS stimulation (Barwick et al., 2016) [2]. The effect of ELL3 depletion on cell morphology, latent Epstein Barr Virus (EBV) lytic replication and differentiation markers in a Burkitt's lymphoma (BL) cell line cells are presented.
Abbreviations: ELL, Eleven-nineteen Lysine-rich Leukemia; EBV, Epstein Barr Virus; BL, Burkitt's Lymphoma
Keywords: ELL3, Transcription elongation, B-cell Lymphoma, Cell division, EBV
Specifications Table
| Subject area | Immunology and Molecular Biology |
| More specific subject area | Transcriptional elongation |
| Type of data | Figures and Images |
| How data was acquired |
|
| Data format | Analyzed |
| Experimental factors |
|
| Experimental features |
|
| Data source location | H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA |
| Data accessibility | Data is within this article |
Value of the data
-
•
This data describes an expression pattern of ELL family members that is replicated in both human and murine B cell compartment
-
•
The data shows the role of ELL3 in the morphology of B cells and reveals disruption of cell division
-
•
The data reveals the impact of ELL3 depletion on B cell differentiation markers and latent EBV gene expression.
1. Data
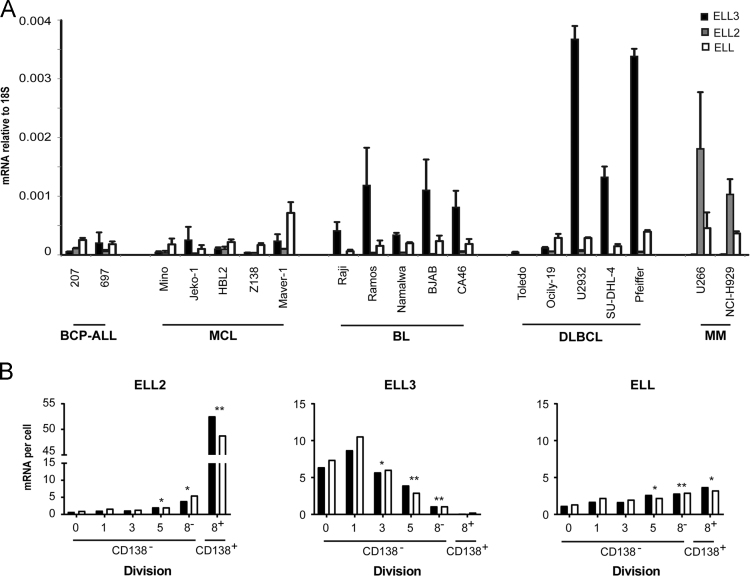
The mRNA levels of ELL, ELL2 and ELL3 in a B cell lymphoma cell line panel is depicted in Fig. 1A. Fig. 1B depicts the average mRNA per murine primary B cell following LPS stimulus and cell sorting based on cell division and plasma cell marker CD138 based on data from GSE70294 [2]. The effect of ELL3 depletion is shown in Fig. 2; including observations of cell morphological changes, PRDM1 mRNA expression, EBV lytic replication factors expression, B cell factors BCL6, PAX5, MYC, and immunoglobulin isoforms.
Fig. 1.
ELL family member transcript levels in murine primary B cells and human B cell lymphoma cell lines. A. Relative mRNA expression profile of ELL family members across human B cell lines. The cell line name and lymphoma subtype are indicated on the x-axis. Data represents the average of 3 independent experiments; errors bars represent SD. B. Expression of ELL family members mRNA in murine B cells during proliferation and differentiation in response to LPS stimulation in vivo. Data was extracted from GSE70294 [2] and presented as mRNA copies per cell. The x-axis represents the number of cell divisions. CD138 positivity is indicated by the (+) and represents the fully differentiated plasma cells. Data represents one experiment with biological duplicates. *p<0.05, **p<0.01 (two-tailed t-test).
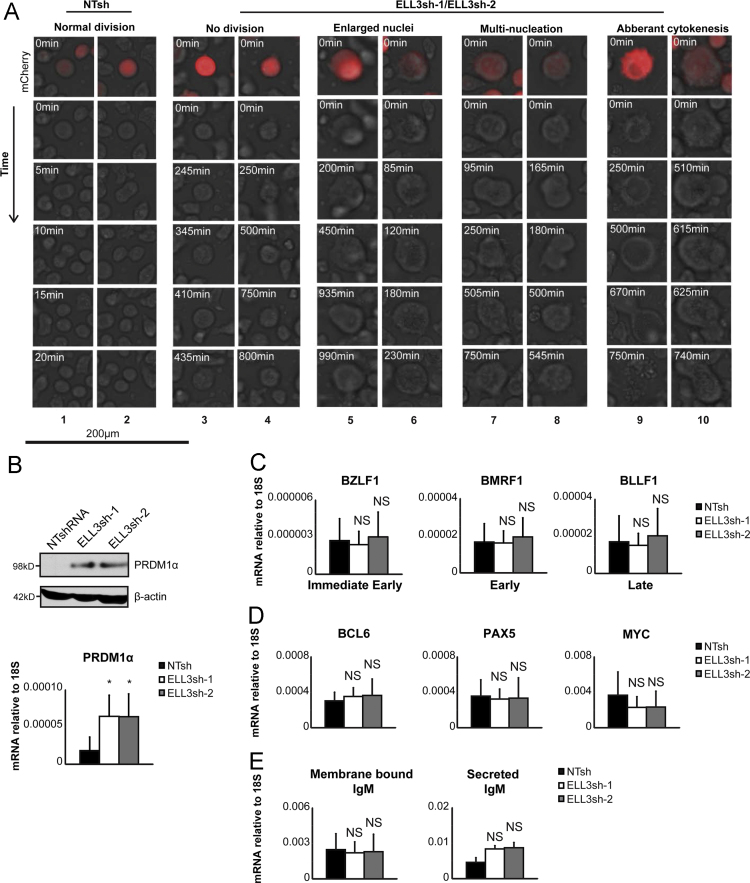
Fig. 2.
Effects of ELL3 depletion on EBV lytic replication, B cell differentiation and morphology. ELL3 expression was depleted in Namalwa cells by transduction with either NTsh, ELL3sh-1, or ELL3sh-2 shRNA expression constructs co-expressing mCherry. A. At day 6 post transduction, cells were subjected to time lapse imaging. Images were taken every 5 min. over 24 h. Data depicts representative images of the control and ELL3-depleted cells. The mCherry fluorescence signal (red) was used to identify shRNA transduced cells. Subsequent images are from the same cell but only imaged with phase to facilitate observation of morphological changes. Time of acquisition is indicated in each image. B. Protein and relative PRDM1α mRNA levels detected by RT-qPCR and immunoblot at 5 days post transduction. C. The relative mRNA quantitation of the EBV genes; BZLF1, BMRF1 and BLLF1. D. The relative mRNA quantitation of B cell factors; BCL6, PAX5 and MYC. E. The relative mRNA quantitation of membrane bound and secreted IgM. Data in panels B through E is presented as the average of 5 independent experiments; errors bars represent SD. *p<0.05; NS is not significant (two-tailed t-test).
2. Experimental design, materials and methods
2.1. Cell culture
Cell lines and cell culture details are as described previously [1].
2.2. Lentiviral shRNA knockdown
The shRNA vectors MISSION® TRC2 pLKO.5-puro ELL3shRNA (ELL3sh-1, TRCN0000289149; and ELL3sh-2, TRCN0000296220), and MISSSION® TRC2 pLKO.5-puro Non-Mammalian control shRNA (NTsh; SHC202; Sigma Aldrich, St. Louis, MO) were purchased. The existing puromycin resistance gene in these vectors was replaced with the mCherry tag from the pLVmCherry vector (Addgene, Cambridge, MA). We produced lentiviral particles with the jetPRIME transfection reagent (Polyplus transfection, Illkirch, France) in HEK-293T using 3rd generation lentiviral packaging mixture (Applied Biological Materials Inc., Richmond, Canada). Cells were transduced at 5×107 cells/ml for 2 h at 1500g, room temperature (RT) in the presence of 0.6 µg/ml polybrene (Merck Millipore, Billerica, MA). Functional assessments were done five days after transduction.
2.3. Immunoblotting
Immunoblotting procedure was as described previously [3]. Primary antibodies include: β-actin (1:12,000 dilution) (AC-15, Sigma Aldrich, St. Louis, MO) and PRDM1 (C14A4). Horse radish peroxidase conjugated secondary antibodies were purchased from GE Healthcare Life Sciences (Pittsburgh, PA).
2.4. Quantitative mRNA analysis
RNA was extracted using the E.Z.N.A. Total RNA Kit I (Omega Bio-Tek, Norcross, GA) and reverse transcribed into cDNA with the qScript cDNA synthesis Kit (Quanta Biosciences Inc., Gaithersburg, MD). 3 µl of one to eleven diluted cDNA was analyzed in duplicate using primers specific to PRDM1α, EBV lytic replication genes (BZLF1, BMRF1 and BLLF1), and B cell and plasma cell factors (BCL6, PAX5, MYC, membrane-bound IgM, secreted IgM) at primer specific annealing temperatures. mRNA expression was analyzed using the ΔΔCt method, with 18 S as a normalization gene [4]. Primer sequences are described in Supplemental Table I and were designed to span exon-exon junctions and to amplify a single PCR product [3, [5], [6], [7], [8], [9]]. The annealing temperatures were experimentally determined using a temperature gradient and high efficiency was validated by PCR of cDNA serial dilutions.
2.5. Microscopy
For time-lapse imaging, cells were plated on a 6-well flat bottom plate at 2×105 cells/ml, placed in Evos Onstage Incubator set at 37 °C and 20%O2 and imaged every 5 min for 24 h on Evos Auto FL Cell Imaging System (Thermo Fisher Scientific Inc., Waltham, MA). All images were taken at 20×magnification using the RFP filter and phase.
Acknowledgements
The authors thank the Analytic Microscopy Cores at the Moffitt Cancer Center & Research Institute for their support and technical assistance. This work was supported by the National Cancer Institute of the National Institutes of Health Grants R01CA164641 to KLW and T32CA115308 to JW and JAR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.09.042.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.09.042.
Transparency document. Supplementary material
Supplementary material
Appendix A. Supplementary material
Supplementary material
References
- 1.Alexander L.E.M.M., Watters J., Reusch J.A., Maurin M., Nepon-Sixt B.S., Vrzalikova K., Alexandrow M.G., Murray P.G., Wright K.L. Selective expression of the transcription elongation factor ELL3 in B cells prior to ELL2 drives proliferation and survival. J. Mol. Immunol. 2017 doi: 10.1016/j.molimm.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barwick B.G., Scharer C.D., Bally A.P., Boss J.M. Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat. Immunol. 2016;17:1216–1225. doi: 10.1038/ni.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith M.A., Maurin M., Cho H.I., Becknell B., Freud A.G., Yu J., Wei S., Djeu J., Celis E., Caligiuri M.A., Wright K.L. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J. Immunol. 2010;185:6058–6067. doi: 10.4049/jimmunol.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 5.Tam W., Gomez M., Chadburn A., Lee J.W., Chan W.C., Knowles D.M. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 6.Ryan J.L., Fan H., Glaser S.L., Schichman S.A., Raab-Traub N., Gulley M.L. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J. Mol. Diagn. 2004;6:378–385. doi: 10.1016/S1525-1578(10)60535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilscher C., Vahrson W., Dittmer D.P. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X., McCarthy P.J., Wang Z., Gorlen D.A., Mertz J.E. Shutoff of BZLF1 gene expression is necessary for immortalization of primary B cells by Epstein-Barr virus. J Virol. 2012;86:8086–8096. doi: 10.1128/JVI.00234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husson H., Carideo E.G., Neuberg D., Schultze J., Munoz O., Marks P.W., Donovan J.W., Chillemi A.C., O'Connell P., Freedman A.S. Gene expression profiling of follicular lymphoma and normal germinal center B cells using cDNA arrays. Blood. 2002;99:282–289. doi: 10.1182/blood.v99.1.282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material