Abstract
There have been no reports describing the effects of cancer cell–derived extracellular vesicles (EVs) on three-dimensional organoids. In this study, we delineated the proneoplastic effects of esophageal adenocarcinoma (EAC)–derived EVs on gastric organoids (gastroids) and elucidated molecular mechanisms underlying these effects. EVs were identified using PKH-67 staining. Morphologic changes, Ki-67 immunochemistry, cell viability, growth rates, and expression levels of miR-25 and miR-210, as well as of their target mRNAs, were determined in gastroids co-cultured with EAC-derived extracellular vesicles (c-EVs). C-EVs were efficiently taken up by gastroids. Notably, c-EV–treated gastroids were more crowded, compact, and multilayered and contained smaller lumens than did those cultured in organoid medium alone or in EAC-conditioned medium that had been depleted of EVs. Moreover, c-EV–treated gastroids manifested increased proliferation and cellular viability relative to medium-only or EV-depleted controls. Expression levels of miR-25 and miR-210 were significantly higher, and those of PTEN and AIFM3 significantly lower, in c-EV–treated versus medium-only or EV-depleted control groups. Inhibitors of miR-25 and miR-210 reversed the increased cell proliferation induced by c-exosomes in co-cultured gastroids by lowering miR-25 and miR-210 levels. In conclusion, we have constructed a novel model system featuring the co-culture of c-EVs with three-dimensional gastroids. Using this model, we discovered that cancer-derived EVs induce a neoplastic phenotype in gastroids. These changes are due, at least in part, to EV transfer of miR-25 and miR-210.
Introduction
Organoids are currently defined as three-dimensional structures derived from pluripotent stem cells grown from organ-specific tissues that self-organize through self-renewal and tenogenic differentiation [1]. Organoids have thus far been successfully established from murine intestine [2], liver [3], pancreas [4], colon [5], and prostate [6], [7], as well as from human small intestine, colon [5], stomach [8], and prostate [6], [7]. These organoids can be cultured long term and resemble their patient tissue origins based on whole-genome sequencing [7], suggesting that they have stable phenotypes and genetic characteristics. Previously, researchers studied gastrointestinal (GI) tract cancers using cell lines and xenografts; however, these previous models were flawed because cell lines were immortalized, while xenografts had organotypic properties. In contrast, GI tissue–derived organoids possess cell type–specific features which can be used to model organogenesis, infection, and malignancy, as well as in drug efficacy and toxicity studies [9], [10], [11], [12].
Extracellular vesicles (EVs) are secreted from various cells under normal and pathological conditions [13], [14]. One type of EV is the exosome, which originates from the fusion of multivesicular bodies with the plasma membrane [15] and which has a diameter ranging from 30 to 150 nm. Microvesicles, or “ectosomes,” are another type of EV ranging in size from 50 to 2000 nm; these structures bud directly from the cell's plasma membrane [16]. A third type of EV comprises apoptotic bodies, which have a larger size, ranging from 1 to 4 mm. Apoptotic bodies are released when apoptotic membrane blebbing occurs during the late stages of programmed cell death [17]. In the current study, we isolated EVs from conditioned medium derived from esophageal adenocarcinoma (EAC) cells using a 0.22-μm filter; thus, the EVs we extracted consisted principally of exosomes and some small microvesicles in this manuscript.
EVs consist of a complex structure enriched in proteins, lipids, and nucleic acids—including messenger RNAs (mRNAs), microRNAs (miRNAs), and DNA [18]. MiRNAs are small noncoding nucleotides, approximately 18 to 23 nucleotides in length, that are widely present in eukaryotic cells. MiRNAs regulate cell proliferation, differentiation, and apoptosis by degrading or inhibiting translation of target mRNA transcripts, leading to inhibition of target gene expression [19]. MiRNAs are involved in embryonic development, normal metabolism, and many human diseases, including tumorigenesis [20]. In the current study, we focused on EV miRNAs as potential transmitters of EV function.
The membrane structure of EVs can block degradation of protein or nucleic acid molecules in EV cargoes from enzymes present in serum or other bodily fluids: for this reason, they have been considered in biomarker development. In addition, due to their stability, they are important mediators in cell-to-cell communication, impacting target cells and regulating their function via autocrine or paracrine mechanisms [21], a mechanism that is also implicated in many human diseases [22], [23].
Many scientists have performed research on EVs in cell lines [22], [23]. In the current study, in contrast, we sought to determine mechanisms mediating the interaction of EVs with three-dimensional GI organoids in co-culture. To our knowledge, there have been no reports describing this particular type of interaction. We therefore established and investigated a novel model system based on co-culturing of EVs derived from EAC cells with normal human gastric epithelial organoids (gastroids). In addition, we evaluated the effects of these EVs on inducing a neoplastic phenotype in gastroids. Finally, we assessed whether EV-induced neoplastic changes in gastroids were related to aberrant expression of EV microRNAs, specifically miR-25 and miR-210.
Materials and Methods
Cell Culture and Components of the Final Human Organoid Culture Medium
Human OE33 and SKGT4 EAC cell lines, Het-1A normal esophageal epithelial cells, and human L-Wnt3A and 293T-HA-Rspo1-FC cells were obtained from American Type Culture Collection between 2012 and 2016 and authenticated between 2013 and 2017 by small tandem repeat profiling. All cell lines were cultured for less than 2 months after thawing early-passage cells and were tested for mycoplasma prior to use. We prepared human organoid culture medium consisting of Wnt3A-conditioned medium (CM), R-spondin 1-CM, epidermal growth factor (EGF) (Invitrogen), and additional growth factors, as described [2], [5].
Organoid Culture And Passage
Five normal human gastric tissues were obtained during upper GI endoscopy at the Johns Hopkins Hospital. All patients provided written informed consent under protocols approved by the institutional review board at the Johns Hopkins University School of Medicine. The diagnosis of normal gastric oxyntic mucosa was confirmed based on pathological evaluation. We cultured and passaged gastroids from normal human gastric epithelial tissues as described [5].
For co-culturing, we added 50 μl (concentration 1011/ml) of EVs to 450 μl of organoid culture medium to make a final concentration of 1010 EVs/ml (c-EV group). When gastroids were passaged, we performed co-culture of 20 μl of EVs with 50 μl of crypt-Matrigel suspension and then dispensed this 70-μl mixture into a 24-well plate. Finally, we placed the plate in a 37°C incubator to solidify the Matrigel and then pipetted 470 μl of organoid culture medium (with growth factors) and another 30 μl of EVs into the medium for the c-EV group (total 50 μl exosomes). We also set up control gastroids consisting of 500 μl of organoid culture medium alone (medium-only group), as well as a second EV-depleted [EV(−)] control group consisting of 50 μl of supernatant (from EAC-CM depleted of EVs by ultracentrifugation) + 450 μl of organoid culture medium.
EVs Isolation and Identification
EAC cells were cultured for 48-72 hours to allow them to attain over 85% confluence, at which time CM was harvested and centrifuged at 400 × g for 10 min to remove cells and debris; SKGT4 or OE33 CM was stored at a 1:1 ratio at 4°C. EVs were isolated from the supernatants, which were then ultracentrifuged [24]. The EVs were real-time analyzed using a Nanosight nanoparticle characterization apparatus and Nanoparticle Tracking Analysis software.
PKH-67 Staining of EVs
EVs were labeled using PKH-67 Fluorescent Cell Linker kits (Sigma-Aldrich, MO, USA) according to the manufacturer's instructions to track their movement and location. To determine whether EVs were taken up efficiently by gastroids in vitro, we added PKH-67–labeled EVs or medium only (without EVs) to organoid medium. The green fluorescence signal of PKH-67–labeled EVs in living organoids was excited by laser. We observed the locations of EVs and acquired images under a Zeiss AXIO Observer microscope.
EV Transfection
EV quantity was measured based on BCA measurement. First, we suspended 30 μg of EVs in 250 μl of Opti-MEM after ultracentrifugation. We added 2 μl of miR inhibitor or negative control (scrambled miR inhibitor group, 10 μM and 25 μl Opti-MEM in Tube A), placed 1 μl of RNA iMAX and 25 μl Opti-MEM in Tube B, mixed 250 μl of resuspended exosomes (Opti-MEM) with the contents of Tube A and Tube B, and then maintained both for 6 hours at 37°C in an incubator. Finally, we ultracentrifuged and isolated the EVs again, resuspended the EVs using the same amount of PBS as in Opti-MEM, and added them to organoid medium for co-culture.
Cell Viability Assessment
Gastroids were dissolved and digested into single cells using TrypLE Express (Thermo Fisher); then 3000 cells were counted and applied to each 96-well plate for each group prior to 1 day of gastroid treatment. We set another well with organoid medium only as a blank control; there were five duplicate wells in each group. We seeded single cells from dispersed organoids onto 96-well plates (3000 cells/100 μl/well) using WST-1 cell viability assay kits (Roche, Mannheim, Germany) according to the manufacturer's instructions.
RNA Extraction and Microarray Assays
EV RNA was extracted using a Total EV RNA and Protein Isolation Kit (Thermo Scientific) according to the manufacturer's instructions.
For total RNA from gastroids, pellets were isolated in Trizol reagent (Thermo Scientific) first and then extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer's directions.
Human HT-12 v4 Expression BeadChip arrays (Illumina, CA) were used for microarray hybridizations. Five hundred nanograms of total RNA from each of three groups [500 μl of organoid culture medium alone (medium-only group), 50 μl of supernatant (from EAC-CM depleted of EVs by ultracentrifugation) + 450 μl of organoid culture medium (EV(−) group, c-EV group] was amplified and labeled using the Illumina TotalPrep RNA Amplification Kit (AMIL1791, Ambion, TX) as described by the manufacturer's instructions.
qRT-PCR
Two-step quantitative real-time polymerase chain reaction (qRT-PCR) was carried out using RT primers and SYBR Green Supermix (Bio-Rad, CA, USA). The housekeeping gene GAPDH was used as an internal control. The primers used were as follows: PTEN, F: 5′-GCTACCTGTTAAAGAATCATCTGG-3′ and R: 5′-CATGAACTTGTCTTCCCGT-3′; AIFM3, F: 5′-GCTCATTGTGGGTGCAGGTG-3′ and R: 5′-GGACGGTCGTAGGGAAGGTG-3′; GAPDH, F: 5′-GGTATCGTGGAAGGACTCATGAC-3′ and R: 5′-ATGCCAGTGAGCTTCCCGTTCAG-3′.
For microRNAs, reverse transcription kits (Applied Biosystems) and TaqMan miRNA-specific primers (Life Technologies) were used to synthesize cDNA. MiRNA-specific cDNA was then used for corresponding TaqMan miRNA-specific assays (Assay IDs: miR-21: 000397, miR-25:000403, miR-93:000432, miR-192:000491, miR-210:000512, RNU6B: 001093, Life Technologies) and with TaqMan Universal PCR Master Mix. All data were analyzed according to the 2−ΔΔCt method.
Organoid Immunohistochemistry (IHC)
For gastroid IHC staining, we embedded organoid specimens in paraformaldehyde-fixed paraffin and cut into 1- to 4-mm sections. Slides were dewaxed and then dehydrated. The sections were incubated with a primary antibody against Ki67 (Ab16667, 1:500 dilution; Abcam, UK) at 4°C overnight. The specimens were washed with PBS and incubated with goat anti-mouse IgG (1:500 dilution; Pierce Biotechnology, Inc., Rockford, IL, USA) for 60 min as secondary antibodies at room temperature. Finally, we stained the organoids using diaminobenzidine tetrahydrochloride (DAB, Sigma, MO, USA) and counterstained them by hematoxylin, and then dehydrated in ethanol. Hematoxylin and eosin (H&E) staining was conducted according to manufacturer's instructions. The slides were read and marked by two independent pathologists, and images were acquired under a microscope.
Statistical Analyses
All experiments were repeated at least three times independently. Data are presented as mean ± SEM. Statistical comparisons were analyzed using Student's t test or one-way ANOVA, with subsequent post hoc multiple comparisons. A value of P < .05 was considered to indicate statistical significance. All tests and P values were two-sided. Statistical analysis was performed using SPSS20.0 software.
Results
Identification of EVs and Screening of EAC-Derived EV miRNAs
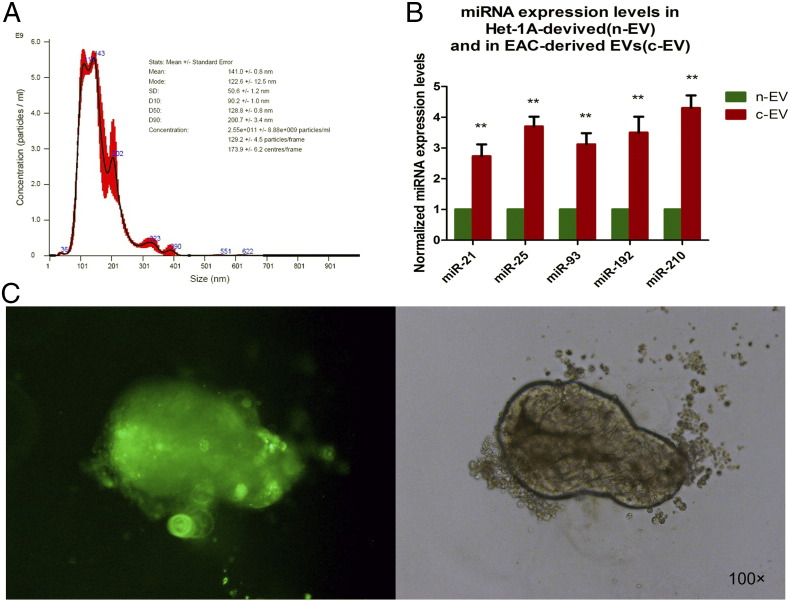
First, we isolated EVs from EAC cells and normal esophageal epithelial cells (Het-1A). As previously described [24], we observed and identified EVs using nanosight tracking analysis (Figure 1A), along with immunoblotting for EV marker proteins (CD63). The average size of these exosomes was 141 ± 0.8 nm; their average concentration was 2.55 × 1011/ml. In the current report, EVs extracted from EAC cells are termed “c-EVs,” while those extracted from Het-1A cells are termed “n-EVs.”
Figure 1.
Characterization, miRNA expression, and PKH-67 staining of EVs. (A) Depiction of isolated EVs by Nanoparticle Tracking Analysis. The curve in the graph was derived by averaging three independent measurements. The average size and average concentration of EAC-derived EVs were 141 ± 0.8 nm and 2.55 × 1011/ml, respectively. (B) Expression of miR-21, miR-25, miR-93, miR-192, and miR-210 in Het-1A–derived (n-EV) and EAC-derived EVs (c-EVs). **P < .01 versus Het-1A group. (C) PKH-67–labeled c-EVs colocalized with EV–co-cultured gastroids, which indicates that the c-EVs were taken up by the gastroids.
An increasing number of studies have established the fact that EV-delivered miRNAs can promote tumor progression and metastasis [25], [26], [27]. MiR-21, miR-25, miR-93, miR-192, and miR-210 in particular are well-known commonly studied oncogenic miRs (oncomiRs), which have also been studied as biomarkers for several types of human cancer [28], [29]. We therefore determined whether EAC-derived EVs contained higher levels of these five onco-miRNAs than did those derived from Het-1A cells by qRT-PCR; results are shown in Figure 1B. Relative to the Het-1A group, there was significantly higher expression of all five miRNAs in the EAC group.
PKH-67 Staining of EVs and Their Uptake by Gastroids
We visualized uptake of exosomes by gastroids using PKH-67 dye after co-culturing PKH-67–labeled EVs with gastroids for 48 hours. As shown in Figure 1C, green fluorescence was observed in gastroids that had been cultured with PKH-67–labeled EVs; there was no green fluorescence in the PKH-67–labeled gastroids cultured without any EVs. This finding suggests that EAC-derived EVs (c-EVs) were taken up into gastroids, even penetrating through Matrigel.
Morphologic Features of Gastroids Co-Cultured with EVs
After co-culturing for 6 weeks, gastroids in the medium-only group (Figure 2, A and D) or EV(−) group (Figure 2, B and E) grew as a single thin layer, featuring large lumens and large tubular structures, with budding; in contrast, gastroids in the c-EV group (Figure 2, C and F) were much more crowded, with smaller lumens, more compactness, and multilayered growth. These findings are consistent with neoplastic morphology [30].
Figure 2.
Morphologic features of gastroids co-cultured with EVs. Gastroids in medium-only, EV(−), and c-EV groups (A, B, and C, respectively) were observed under a light microscope (×100). (D, E, and F) Gastroids in corresponding groups were viewed under (×200) magnification observation. Gastroids in medium-only or EV(−) groups (A, B, D, and E) were thin-layer, had large lumens and large tubular structures, and grew with budding; however, gastroids exhibiting a more crowded morphology, with smaller lumens and other neoplastical features, including more compact and multilayered growth, were seen in the c-EV group (C and F).
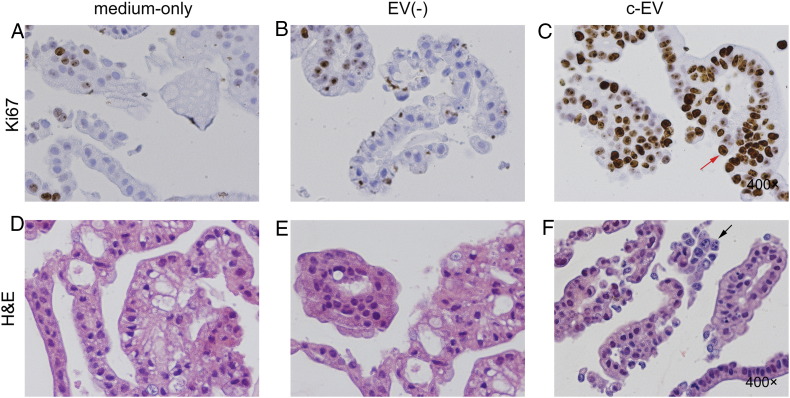
Ki-67 and H&E Staining of Gastroids
After gastroids were co-cultured with c-EVs for 6 weeks, they were mostly Ki-67 positive (Figure 3C), in contrast to the medium-only (Figure 3A) or EV(−) groups (Figure 3B). This finding indicates that c-EVs promoted proliferation of gastroids. Furthermore, H&E staining in the medium-only (Figure 3D) or EV(−) (Figure 3E) groups revealed regular and well-ordered single cell layers in each organoid, whereas irregular, multilayered, and inconsistent structures with deeply staining, large nuclei were observed in the in c-EV group (Figure 3F). This finding indicates that c-EVs induce a neoplastic phenotype in benign gastroids.
Figure 3.
Ki-67 and H&E staining of gastroids. There was low proliferative activity in medium-only (A) and EV(−) (B) groups by Ki-67 staining, in contrast with representative pictures of strongly positive Ki-67 expression in the c-EV group (C, red arrowhead). H&E-staining disclosed a regular and well-ordered single-layer cell structure in the medium-only (D) and EV(−) (E) groups, but gastroids in the c-EV group (F) were lined with an irregular, inconsistent, and multilayered structure and deeply hematoxylin-staining large nuclei (black arrowhead).
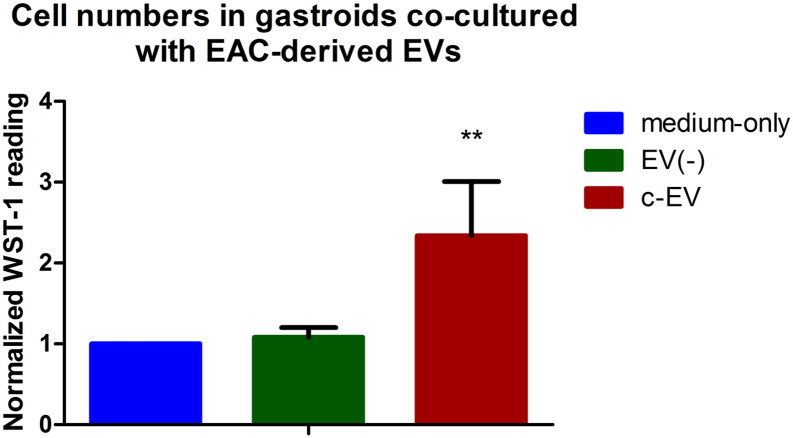
WST-1 Cell Viability Assays in EV–Co-Cultured Gastroids
Based on the above findings, we measured cell growth rate quantitatively using WST-1 cell viability assays. As shown in Figure 4, cell viability was higher in gastroids co-cultured with c-EV than in medium-only or EV(−) groups (P < .05).
Figure 4.
WST-1 cell viability assays in gastroids. Proliferation changes were determined in the medium-only, EV(−), and c-EV groups using WST-1 assays. There was a significant increase in proliferative activity in the c-EV group compared with the medium-only or EV(−) groups (P < .05).
Global Gene Expression Assays in EV–Co-Cultured Gastroids
To obtain clues to potential mechanisms underlying the neoplastic phenotype induced by c-EVs in gastroids, we performed microarray assays of mRNA and miRNA expression. Representative results of these experiments are displayed in Table 1. Notably, several pre-miRNAs were aberrantly upregulated in the c-EV versus the medium-only or EV(−) groups. In particular, miR-25 and miR-210 were the most markedly upregulated (greater than two-fold) in the c-EV group relative to the medium-only and EV(−) groups. This finding is consistent with results of our previous study, wherein we confirmed that miR-25 and miR-210 originated from EVs isolated from EAC cells [24]. This finding suggests that c-EVs delivered miR-25 and miR-210 to gastroids and that these c-EV miRNAs contribute to the observed morphologic changes in gastroids. We therefore focused on these miRNAs for further experiments.
Table 1.
Results of Microarray Analysis in Gastroids Co-Cultured with EVs
| Fold Change (c-EV/Medium-Only) |
Fold Change [c-EV/EV(−)] |
|
|---|---|---|
| hsa-pre-miR-25 | 4.53 | 4.48 |
| hsa -pre-miR-210 | 3.93 | 4.01 |
| hsa-pre-miR-649 | 3.76 | 3.82 |
| hsa -pre-miR-302c | 3.15 | 3.21 |
| hsa-pre-miR-658 | 2.97 | 2.94 |
| hsa-pre-miR-192 | 2.15 | 2.17 |
Profiling of miRNA expression levels in gastroids treated with organoid medium alone (medium-only group), EAC-CM depleted of EVs (EV(−) group), and EAC-derived EVs (c-EV group). Six miRNAs were upregulated more than two-fold in the c-EV group relative to both the medium-only and EV(−) groups.
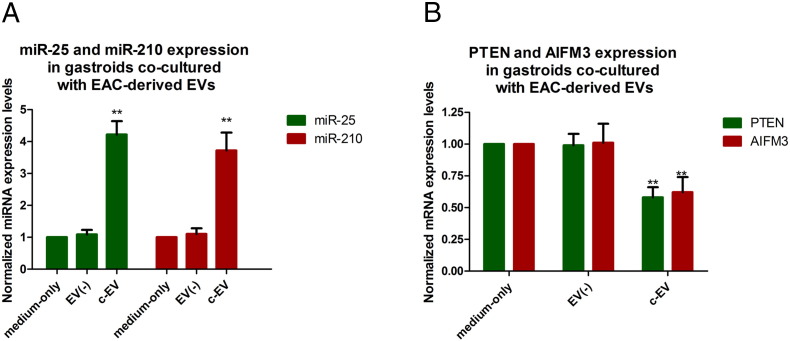
Expression Levels of miR-25, miR-210, and Their Target mRNAs in EV–Co-Cultured Gastroids
To prove that exosomally derived miRNAs exert effects downstream via their target mRNAs, we measured expression of the genes PTEN and AIFM3, which were selected based on Targetscan, miRWalk, and MicroRNA.org as targets of miR-25 and miR-210, respectively. As shown in Figure 5A, miR-25 and miR-210 levels were significantly increased in c-EV–treated gastroids (miR-25 mean fold change normalized to medium-only group, 4.22 ± 0.42, P < .01; miR-210 mean fold change normalized to medium-only group, 3.72 ± 0.56, P < .01). In turn, Figure 5B shows that mRNA levels of PTEN and AIFM3 were significantly decreased in c-EV–treated gastroids (mean fold change of PTEN normalized to medium-only group, 0.58 ± 0.08, P < .01; mean fold change of AIFM3 normalized to medium-only group, 0.62 ± 0.12, P < .01). PTEN and AIFM3 are well-established tumor suppressor genes whose suppression has been shown to contribute to carcinogenesis [31], [32]. Thus, this finding suggests a potential molecular mechanism underlying the observed changes in gastroid phenotype upon treatment with cancer-derived EVs.
Figure 5.
Expression of miR-25, miR-210, and their target mRNAs in EV–co-cultured gastroids. The expression of miR-25, miR-210, and their targets PTEN and AIFM3 was examined by qRT-PCR. (A) miR-25 and miR-210 levels were significantly increased in c-EV–co-cultured gastroids group compared to medium-only or EV(−) groups. (B) mRNA levels of PTEN and AIFM3 were substantially decreased in the c-EV group versus the medium-only or EV(−) groups. Bars represent mean ± SEM. **P < .01.
Inhibition of miR-25 and miR-210 Reverses the Proliferative Effect of c-EV in Co-Cultured Gastroids
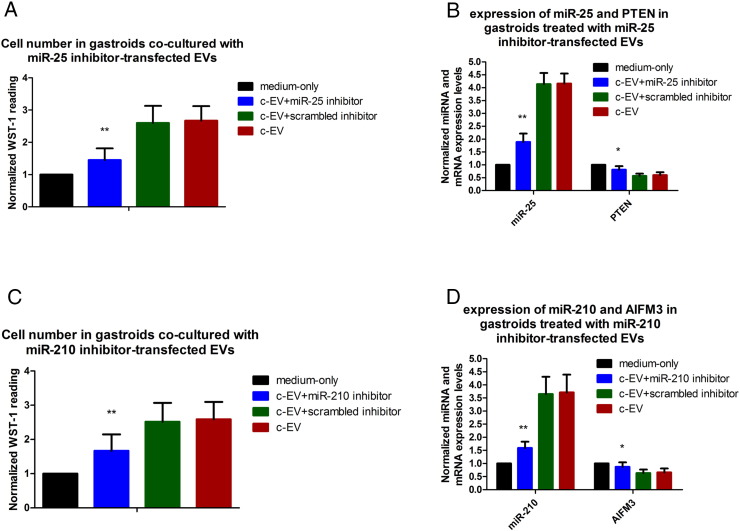
To further investigate whether EV-induced cell proliferation in gastroids was related to EV miR-25 and -210, we transfected miR-25 or miR-210 inhibitors into EVs, respectively, and then co-cultured them with gastroids. There were four groups in this experiment: organoid medium alone group, c-EV group, c-EV + miR25/miR210 inhibitor group, and c-EV + miR25/miR210 scrambled inhibitor group. The cells were then subjected to WST-1 assays and qRT-PCR. Results illustrated that both miR-25 and miR-210 inhibitors reversed the proliferative effects of c-EVs on gastroids (Figure 6, A and C, respectively). Moreover, qRT-PCR showed a significant decrease in miR-25 expression and an increase in PTEN expression in miR-25 inhibitor–transfected c-EV–treated gastroids relative to gastroids treated with c-EVs that had not been transfected with a miR-25 inhibitor or with scrambled inhibitor–transfected c-EVs (Figure 6B); similarly, significant downregulation of miR-210 and upregulation of AIFM3 occurred in gastroids co-cultured with miR-210 inhibitor–transfected EVs versus gastroids treated with untransfected or scrambled inhibitor–transfected c-EVs (Figure 6D). These results imply that c-EVs promote gastroid proliferation, at least in part, by upregulation of c-EV miR-25 and miR-210 and downregulation of their target mRNAs.
Figure 6.
Inhibition of miR-25 and miR-210 reverses the proliferative effect of c-EVs in co-cultured gastroids. (A) Proliferation changes in the medium-only, c-EV + miR-25 inhibitor, c-EV + scrambled inhibitor, and c-EV groups were measured by WST-1 assay after miR-25 inhibitor–transfected EVs were co-cultured with gastroids. (B) Expression of miR-25 and PTEN in these 4 groups was measured using qRT-PCR after miR-25 inhibitor-transfected EVs were co-cultured with gastroids. Proliferative activity (C) and expression of miR-210 and AIFM3 (D) were determined by WST-1 and qRT-PCR, respectively, after miR-210 inhibitor–transfected EVs were co-cultured with gastroids. Bars represent means ± SEM. Versus c-EV + scrambled inhibitor or c-EV groups, **P < .01.
Discussion
Although the precise physiologic functions of EVs are still being elucidated, these structures are widely known to function as communicative vectors in the maintenance of homeostasis. Evidence is rapidly accumulating that cancer cell–derived EVs play significant roles in tumor growth and progression via mechanisms involving immunosuppression, microenvironmental perturbation, neoangiogenesis, metastasis, and others [25], [26], [27]. For example, Chowdhury et al. found that prostate cancer–derived EVs alter the differentiation of bone marrow mesenchymal stem cells into a cancer-associated fibroblast-like state by activating the TGF-beta signaling pathway and promoting tumor proliferation and aggressiveness in a 3D co-culture model [33]. Similarly, in a study by Le et al., highly metastatic breast cancer cells delivered EVs carrying miR-200 family miRNAs to nonmetastatic breast cancer cells, thereby enhancing lung metastasis [34]. This EV transmitted miR-200 induced mesenchymal-to-epithelial transition by inhibiting miR-200 target genes, including zeb2 and sec23, in nonmetastatic breast cancer cell lines and in murine xenografts models [34]. Based on these and other findings, an emerging consensus holds that EVs derived from cancer cells can confer a tumorigenic and/or metastatic phenotype on cells or tissues not previously possessing those characteristics. In accordance with the above studies, we found that co-cultured EVs with normal gastroids promoted their proliferation and induced neoplastic morphology in them (Figure 2, Figure 3).
As intercellular vectors, EVs appear to play a crucial role in communication between tumors and their microenvironment: cancer cells can influence and alter the function of both local and distant nonmalignant recipient cells by delivering cargoes containing oncogenic factors, thereby facilitating tumorigenesis [27]. Accumulating evidence has established that EV-transferred miRNAs are key mediators of cancer cell communication, with complex and powerful components permitting cancer cells to shape their environment [35]. Studies have confirmed horizontal delivery of miRNAs by EVs derived from brain tumors, metastatic breast cancer, melanoma [36], and lung cancer [37]. In the present study, we focused on EV miRNAs as a prominent molecular mechanism underlying neoplastic changes induced in gastroids by co-culture with EVs. According to our results, the miRNAs exhibiting the greatest fold-change among EV miRNAs were miR-25 and miR-210; therefore, these were chosen for further study.
MiR-25 is a well-characterized oncomiR that is extensively involved in tumor growth and metastasis [38], [39]. Several studies showed that plasma miR-25 levels can serve as a biomarker of poor prognosis in patients with gastric cancer [40], [41]. One well-known target gene of miR-25, phosphatase and tensin homologue (PTEN), represses tumorigenicity by downregulating the highly oncogenic PI3K/Akt signaling pathway [42]. Feng et al. reported that knockdown of miR-25 enhances the sensitivity of liver cancer stem cells, resulting in TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling [31]. Furthermore, Zhang et al. proved that EV-transferred miRNAs derived from astrocyte cells promoted metastasis of brain tumor cells by inhibiting expression of PTEN [43]. In accordance with the report by Zhang [43], we found greater proliferative capacity, upregulation of miR-25, and downregulation of PTEN in c-EV–treated gastroids, which suggest that EVs may promote gastroid proliferation, at least in part, by transmitting EV miR-25, which then targets PTEN (Figure 5, Figure 6B).
MiR-210 is another well-characterized onco-miRNA which correlates with epithelial-to-mesenchymal transition in human gastric, colorectal, and ovarian cancer [44], [45], [46]. Bigagli et al. found that exosomal miR-210 derived from human HCT-8 colon cancer cells promoted the adhesion of neighboring metastatic cells. AIFM3 (apoptosis inducing factor mitochondria associated 3) is a gene with homology to apoptosis-inducing factor (AIF) [29]. Two studies showed that AIF induces apoptosis [47]. Yang et al. demonstrated that downregulation of miR-210 induces apoptosis and improves radiosensitivity in human hepatocellular carcinoma cells by increasing the expression of AIFM3 [48]. Similarly, we discovered that c-EV–treated gastroids exhibit increased proliferative activity, increased miR-210 levels, and diminished AIFM3 levels relative to medium-only or EV(−) groups (Figure 5). Furthermore, there were downregulation of miR-210 and upregulation of AIFM3 in gastroids co-cultured with miR-210 inhibitor–transfected c-EVs in contrast to gastroids treated with untransfected or scrambled inhibitor–transfected c-EVs (Figure 6D). Tumor development and progression can be seen as an imbalance between cellular proliferation and apoptosis [49]. Thus, we speculate that EVs may promote neoplastic change in gastroids via apoptosis suppression, which is in turn mediated by inhibition of AIFM3 by EV transmitted miR-210.
Because of limitations in techniques of EV isolation, it is often difficult to distinguish among different EV types due to lack of specific EV physical and chemical biomarkers; thus, researchers still have difficulty discriminating and identifying functional components in cancer cell–conditioned medium or detailed EV cargo elements (such as proteins, lipids, mRNAs, and miRNAs) [50]. It is still of interest to assess whether EV mRNAs, EV proteins, or other EV oncomiRNAs exert effects on human GI organoids or cause morphologic changes.
In summary, in the current study, we sought to construct a new co-culture model system consisting of EAC-derived EVs and gastroids, as well as to observe the effects and mechanisms of action of EVs on gastroids. We showed that we could establish a model for interaction between EVs and gastroids; furthermore, we discovered that c-EVs promoted proliferation and induced neoplastic phenotypic changes in gastroids. Finally, we found that these effects were mediated, at least in part, by EV miR-25 and miR-210. These results, in the context of the published literature, also suggest that c-EVs may be promising GI cancer biomarkers and potential therapeutic tools, and that miR-25 and miR-210 in particular constitute potential molecular targets in gastric cancer diagnosis and treatment.
Conflict of Interest
The authors declare no conflict of interest.
Footnotes
Acknowledgements: This work was supported by grants from the National Cancer Institute (CA190040, CA211457) and from the program of China Scholarship Council (No. 201406280108). Dr. Meltzer is the Myerberg/Hendrix Professor of Gastroenterology at the Johns Hopkins University School of Medicine and an American Cancer Society Clinical Research Professor.
Contributor Information
Shuixiang He, Email: hesx123@126.com.
Stephen J. Meltzer, Email: smeltzer@jhmi.edu.
References
- 1.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 3.Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494(7436):247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greggio C, De Franceschi F, Figueiredo-Larsen M, Gobaa S, Ranga A, Semb H, Lutolf M, Grapin-Botton A. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140(21):4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 6.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155(2):357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med. 2012;18(4):618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 10.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126-136.e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onuma K, Ochiai M, Orihashi K, Takahashi M, Imai T, Nakagama H, Hippo Y. Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc Natl Acad Sci U S A. 2013;110(27):11127–11132. doi: 10.1073/pnas.1221926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luni C, Serena E, Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol. 2014;25:45–50. doi: 10.1016/j.copbio.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Xiao J, Pan Y, Li XH, Yang XY, Feng YL, Tan HH, Jiang L, Feng J, Yu XY. Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis. 2016;7(6):e2277. doi: 10.1038/cddis.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madhavan B, Yue S, Galli U, Rana S, Gross W, Muller M, Giese NA, Kalthoff H, Becker T, Buchler MW. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136(11):2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 15.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Antonyak MA, Zhang J, Cerione RA. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;31(45):4740–4749. doi: 10.1038/onc.2011.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turiak L, Misjak P, Szabo TG, Aradi B, Paloczi K, Ozohanics O, Drahos L, Kittel A, Falus A, Buzas EI. Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice. J Proteomics. 2011;74(10):2025–2033. doi: 10.1016/j.jprot.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao Y, Cheng Y, Yang M, Wang Q, Feng X. MiRNA-194 activates the Wnt/beta-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017;385:117–127. doi: 10.1016/j.canlet.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 21.Meckes DG, Jr., Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107(47):20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Yu J, Zhang H, Wang B, Guo H, Bai J, Wang J, Dong Y, Zhao Y, Wang Y. Exosomes-Derived MiR-302b Suppresses Lung Cancer Cell Proliferation and Migration via TGFbetaRII Inhibition. Cell Physiol Biochem. 2016;38(5):1715–1726. doi: 10.1159/000443111. [DOI] [PubMed] [Google Scholar]
- 23.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Piontek K, Ishida M, Fausther M, Dranoff JA, Fu R, Mezey E, Gould SJ, Fordjour FK, Meltzer SJ. Extracellular vesicles carry microRNA-195 to intrahepatic cholangiocarcinoma and improve survival in a rat model. Hepatology. 2017;65(2):501–514. doi: 10.1002/hep.28735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An T, Qin S, Xu Y, Tang Y, Huang Y, Situ B, Inal JM, Zheng L. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 2015;4:27522. doi: 10.3402/jev.v4.27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vrij J, Maas SL, Kwappenberg KM, Schnoor R, Kleijn A, Dekker L, Luider TM, de Witte LD, Litjens M, van Strien ME. Glioblastoma-derived extracellular vesicles modify the phenotype of monocytic cells. Int J Cancer. 2015;137(7):1630–1642. doi: 10.1002/ijc.29521. [DOI] [PubMed] [Google Scholar]
- 27.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita T, Yip KW, Spence T, Liu FF. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J Human Genet. 2017;62(1):67–74. doi: 10.1038/jhg.2016.87. [DOI] [PubMed] [Google Scholar]
- 29.Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol Therapy. 2016:1–8. doi: 10.1080/15384047.2016.1219815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1-2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int J Oncol. 2016;49(6):2600–2610. doi: 10.3892/ijo.2016.3751. [DOI] [PubMed] [Google Scholar]
- 32.Delettre C, Yuste VJ, Moubarak RS, Bras M, Lesbordes-Brion JC, Petres S, Bellalou J, Susin SA. AIFsh, a novel apoptosis-inducing factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J Biol Chem. 2006;281(10):6413–6427. doi: 10.1074/jbc.M509884200. [DOI] [PubMed] [Google Scholar]
- 33.Chowdhury R, Webber JP, Gurney M, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger mesenchymal stem cell differentiation into pro-angiogenic and pro-invasive myofibroblasts. Oncotarget. 2015;6(2):715–731. doi: 10.18632/oncotarget.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le MT, Hamar P, Guo C, Basar E, Perdigao-Henriques R, Balaj L, Lieberman J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124(12):5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acland P, Dixon M, Peters G, Dickson C. Subcellular fate of the int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343(6259):662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- 37.Munagala R, Aqil F, Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol. 2016;37(8):10703–10714. doi: 10.1007/s13277-016-4939-8. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, Wang W, Zhang L, Zhang X, Tang Q. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335(1):168–174. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 39.Zoni E, van der Horst G, van de Merbel AF, Chen L, Rane JK, Pelger RC, Collins AT, Visakorpi T, Snaar-Jagalska BE, Maitland NJ. miR-25 Modulates Invasiveness and Dissemination of Human Prostate Cancer Cells via Regulation of alphav- and alpha6-Integrin Expression. Cancer Res. 2015;75(11):2326–2336. doi: 10.1158/0008-5472.CAN-14-2155. [DOI] [PubMed] [Google Scholar]
- 40.Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110(9):2291–2299. doi: 10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, Wang W, Li F, Zhang H, Liu J. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med Oncol. 2014;31(10):243. doi: 10.1007/s12032-014-0243-x. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, Li P, Li M, Wang X, Zhang C. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu P, Fan S, Huang L, Yang L, Du Y. MIR210 as a potential molecular target to block invasion and metastasis of gastric cancer. Med Hypotheses. 2015;84(3):209–212. doi: 10.1016/j.mehy.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Wang W, Zhang Y, Chen Y, Hu T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med Oncol. 2014;31(1):799. doi: 10.1007/s12032-013-0799-x. [DOI] [PubMed] [Google Scholar]
- 46.Ding L, Zhao L, Chen W, Liu T, Li Z, Li X. miR-210, a modulator of hypoxia-induced epithelial-mesenchymal transition in ovarian cancer cell. Int J Clin Exp Med. 2015;8(2):2299–2307. [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Q, Lin T, Zhang Y, Zheng J, Bonanno JA. Molecular cloning and characterization of a human AIF-like gene with ability to induce apoptosis. J Biol Chem. 2005;280(20):19673–19681. doi: 10.1074/jbc.M409517200. [DOI] [PubMed] [Google Scholar]
- 48.Yang W, Sun T, Cao J, Liu F, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318(8):944–954. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 49.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 50.Strotman LN, Linder MW. Extracellular Vesicles Move Toward Use in Clinical Laboratories. Clin Lab Med. 2016;36(3):587–602. doi: 10.1016/j.cll.2016.05.004. [DOI] [PubMed] [Google Scholar]