Abstract
TRIM-NHL proteins are highly conserved regulators of developmental pathways in vertebrates and invertebrates. The TRIM-NHL family member NHL-2 in Caenorhabditis elegans functions as a miRNA cofactor to regulate developmental timing. Similar regulatory roles have been reported in other model systems, with the mammalian ortholog in mice, TRIM32, contributing to muscle and neuronal cell proliferation via miRNA activity. Given the interest associated with TRIM-NHL family proteins, we aimed to further investigate the role of NHL-2 in C. elegans development by using a synthetic RNAi screening approach. Using the ORFeome library, we knocked down 11,942 genes in wild-type animals and nhl-2 null mutants. In total, we identified 42 genes that produced strong reproductive synthetic phenotypes when knocked down in nhl-2 null mutants, with little or no change when knocked down in wild-type animals. These included genes associated with transcriptional processes, chromosomal integrity, and key cofactors of the germline small 22G RNA pathway.
Keywords: NHL-2, TRIM-NHL, RNAi screen, germline, Caenorhabditis elegans, Mutant Screen Report
TRIM-NHL proteins are highly conserved and are required for several biological processes including innate immunity, skeletal muscle plasticity and tumor suppression (Kudryashova et al. 2005; Liu et al. 2014; Nisole et al. 2005; Rajsbaum et al. 2008). In Caenorhabditis elegans, NHL-2 is one of five TRIM-NHL proteins and is orthologous to Brat and Mei-P26 in Drosophila melanogaster and TRIM32 in Mus musculus and Homo sapiens (Hammell et al. 2009; Loedige et al. 2014; Neumuller et al. 2008; Schwamborn et al. 2009). In flies, Mei-P26 is required for germline stem cell maintenance by regulating cellular proliferation and cell cycle exit via the microRNA (miRNA) pathway (Neumuller et al. 2008). In addition to this, Brat is required for tumor suppression in the larval brain and co-immunoprecipitates with AGO1, suggesting a miRNA-mediated role (Marchetti et al. 2014). In mammals, TRIM32 negatively regulates the tumor suppressor gene p53 via E3 ubiquitin ligase activity, suggesting that TRIM32 plays a role in tumorigenesis (Horn et al. 2004; Kano et al. 2008; Liu et al. 2014). Interestingly, TRIM32 positively regulates the miRNA pathway, and together with its E3 ligase activity is critical to regulating neuronal differentiation by regulating the transcription factor c-Myc (Schwamborn et al. 2009). These observations suggest diverse miRNA-associated roles for the TRIM-NHL subgroup of proteins in mammals and invertebrates.
In C. elegans, NHL-2 is required for embryonic polarity, asymmetric cell division, and sex determination (Hyenne et al. 2008; McJunkin and Ambros 2017). Moreover, NHL-2 functions as a miRNA cofactor to regulate developmental timing and cell fate progression (Hammell et al. 2009; Karp and Ambros 2012). In this role, NHL-2 associates with the DEAD-box RNA helicase CGH-1 to maintain miRNA function, and depletion of cgh-1 in an nhl-2 null background results in a let-7 defective phenotype (Hammell et al. 2009). In addition to this, Hammell et al. (2009) reported a temperature-sensitive reproductive defect in nhl-2 null mutants. Therefore, we employed a synthetic RNAi screening approach to identify genes that result in strong reproductive phenotypes when knocked down in nhl-2 mutants. In total, we identified 42 high-confidence candidate genes that produced defective reproductive phenotypes when knocked down in nhl-2 null animals, but had little or no effect on wild-type animals. These candidate genes are associated with core biological functions such as cell cycle regulation, transcriptional processes, and DNA repair. We also identified several core cofactors of the germline-specific 22G RNA pathway. These genes represent potential candidate genes that associate with NHL-2 to maintain optimal reproduction, either through germline-specific small 22G RNAs, or through novel mechanisms.
Materials and Methods
Maintenance of C. elegans strains
C. elegans strains were obtained from the C. elegans Genetics Centre (CGC, Saint Paul, MN) and cultured under standard conditions (Brenner 1974). Strains used in this study were N2 (Bristol) as wild type and nhl-2(ok818), which was backcrossed eight times. Animals were grown while feeding on Escherichia coli OP50, seeded on NGM plates at 20°, unless otherwise stated.
Synchronized populations of animals
To synchronize each strain, gravid adults were washed from NGM plates using M9 buffer (86 mM NaCl, 42 mM Na2HPO4, 22 mM KH2PO4, and 1 mM MgSO4), and animals were bleached by the addition of 1 ml of bleaching stock solution to each tube (1 ml of 10 M NaOH, 4 ml household bleach, and 9 ml of ddH2O). Tubes were then vortexed for 4 min and centrifuged at 1000 × g. Bleach was removed by four wash steps with sterile M9 buffer and eggs were allowed to hatch at 20° on a rotator.
Liquid-based RNAi screen
The method used to screen for genes that produce synthetic phenotypes when knocked down in nhl-2(ok818) null mutants (referred to from here on as nhl-2(0)) was essentially as outlined by Lehner et al. (2006), with slight modifications as follows. RNAi was performed by feeding in duplicate 96 well plates for each strain using the ORFeome library (Reboul et al. 2003). Approximately 10 L1 animals of each strain in 10 μl M9 buffer with 0.01% Triton X-100 were added to each well using a multi-channel pipette. Plates were then placed in a shaking incubator at 150 rpm for 4 days at 20° and scored under a dissecting microscope for the following worm phenotypes: sterile (no progeny), embryonic lethality (unhatched embryos), low progeny (<20 progeny), very low progeny (<5 progeny), and egg-laying defective (eggs hatching inside animal). Phenotypes that were observed in the nhl-2(ok818) background, but not in wild-type animals, were considered as genes of interest. Positive hits were then rescreened using RNAi via plate feeding. Bacterial clones were grown in a solution of 2x TY media plus 100 μg/ml ampicillin overnight, then seeded onto NGM plates (3% bacto-agar, 86 mM NaCl, 42 mM Na2HPO4, 22 mM KH2PO4, and 1 mM MgSO4) containing 100 μg/ml ampicillin plus 4 mM IPTG. Approximately 10 synchronized L1 animals of each strain were then pipetted on to each plate in duplicates and grown at 20° for 4 days. Phenotypes were then scored under a dissecting microscope.
Brood size assay
Brood size assays were performed for wild-type and nhl-2(ok818) strains with animals feeding on E. coli OP50. Synchronized populations of each strain were grown at 20° until the fourth larval stage (L4). Individual L4 animals were then placed on preseeded NGM plates, transferred to new plates every 12 hr, and scored for progeny after 48 hr. This process was repeated until each worm failed to lay new progeny. Total progeny included viable progeny and unhatched embryos.
Germline dissection and immunostaining
Germline immunostaining was performed as described by Navarro et al. (2001). Anti-Phospho-Histone H3 antibody was applied at a 1:300 dilution (Abcam, Cambridge, England) and a secondary antibody and DAPI were applied at 1:1000 (Thermo Scientific, MA, USA). Slides were examined using an Olympus IX81microscope attached to an X-Cite series 120Q fluorescent light box.
Bioinformatics
Potential homologs of hits from the RNAi screen were identified through BLASTP searches using H. sapiens, D. melanogaster, and Saccharomyces cerevisiae predicted proteomes available on the NCBI. Gene ontology (GO), was used to assign genes using GO Term Mapper (http://go.princeton.edu/cgi-bin/GOTermmapper) (Harris et al. 2004).
Plasmid preparation and DNA Sequencing
To sequence genes from the secondary screen, bacteria expressing clones from the ORFeome library were grown overnight in 2x TY media plus 100 μg/ml ampicillin; plasmids were extracted by using the PureYield Miniprep System (Promega, Wisconsin, USA). Purified plasmids were then quantified by using a NanoDrop 2000 spectrophotometer (Thermo-Fischer Scientific, Massachusetts, USA) and sent to Macrogen for sequencing (http://dna.macrogen.com/eng/).
Statistical analysis and software
Generation of graphs and statistical analysis was performed using the Prism 5 software package (GraphPad Software, California, USA). Images were processed using Adobe Photoshop (Adobe Systems, California, USA). Annotation of photos and generation of cartoon images used Adobe Illustrator (Adobe Systems).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Reproductive capacity of nhl-2(0) mutants
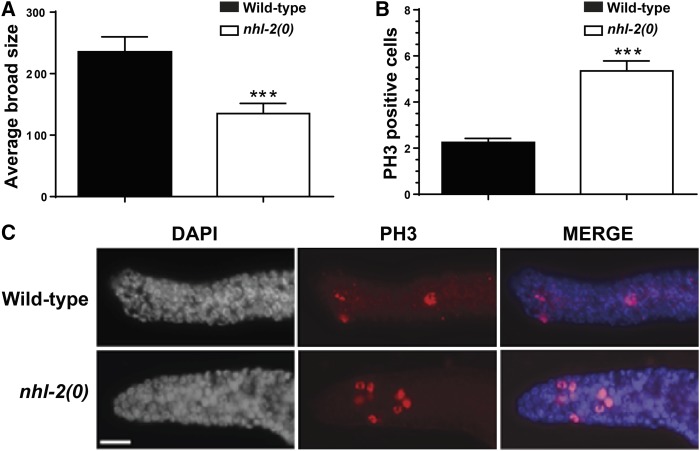
The predicted nhl-2(0) mutant exhibits low penetrant heterochronic deficiencies and temperature-sensitive reproduction defects (Hammell et al. 2009). However, little evidence to date shows that NHL-2 plays a role in the C. elegans germline. To investigate the requirement of NHL-2 for germline function, a brood size assay was conducted in nhl-2(0) mutants and compared to wild-type animals. The nhl-2(0) mutants showed significantly reduced brood size when compared to wild-type animals at 20° (Figure 1A), suggesting that NHL-2 is required for normal reproductive function. To investigate potential nhl-2(0) germline defects, 1-day-old nhl-2(0) worms were dissected and the germlines immunostained with DAPI and an antibody specific for phosphorylation of histone 3 at serine 10 (PH3). This histone modification is required for correct chromosomal condensation during mitosis and is a suitable marker for mitotic proliferation (Hsu et al. 2000; Van Hooser et al. 1998). The nhl-2(0) germline morphology appeared grossly normal in appearance; however, analysis of the mitotic region of the germline revealed significantly higher numbers of cells that were positive for PH3 in nhl-2(0) mutants when compared to wild-type animals (Figure 1, B and C). This combination of reduced brood size and elevated mitotic proliferation prompted the use of an RNAi screen to identify potential cofactors that associate with NHL-2 to maintain normal germline function.
Figure 1.
Brood size and elevated germ cell proliferation in nhl-2(0) mutants. (A) nhl-2(0) mutants display significantly reduced brood size when compared to wild-type animals at 20°. *** P <0.001, error bars represent SEM. n = 10. (B) Quantification of mitotic cells positive for PH3. n = 20. *** P <0.001, error bars represent SEM. (C) Represented images of germlines stained with PH3. Bar, 10 µm. n = 20.
Partial genome-wide RNAi screening in nhl-2(0) mutants
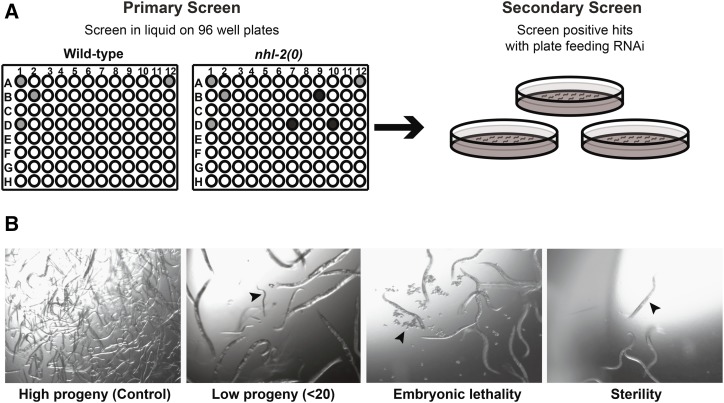
The RNAi screen used the ORFeome library that consists of 11,942 C. elegans open reading frames (∼55% of the C. elegans genome) (Reboul et al. 2003). Liquid feeding is the most efficient delivery method for large-scale RNAi screens (reviewed in Ahringer (2006)). Therefore, we optimized the appropriate volume of bacteria in 96-well plates to ensure that worms had sufficient food throughout the screen. Additionally, the appropriate orbital speed for shaking plates was assessed to guarantee maximum aeration for each well, but also to prevent cross-contamination between wells that contained different RNAi clones. Following this, our primary liquid feeding RNAi screen was performed at 20°, which resulted in 166 primary hits (Supplemental Material, Table S1 in File S1). These hits were defined as genes that gave synthetically defective reproductive phenotypes when knocked down in nhl-2(0) mutants when compared to knockdown of each gene in wild-type animals. These primary hits underwent a secondary screen where RNAi was delivered by plate feeding, resulting in 42 hits that displayed the same phenotypes as previously found in our primary screen (Figure 2). This final value of 42 genes resulted in a hit rate of 0.3% of the total ORFeome library. The most common phenotype identified was reduced progeny (24 hits), followed by embryonic lethality (14 hits) and sterility (four hits). Clones were sequenced for final confirmation prior to any further analysis. While some of the remaining 124 hits displayed abnormal germline phenotypes when knocked down in nhl-2(0) mutants, they produced phenotypes that were milder than those reported in our primary screen. This reduction in candidate genes from our primary to secondary screen is common in RNAi screens, and often a larger reduction in candidate genes has been observed in other screens (Maia et al. 2015; da Conceição Pereira et al. 2016; Saur et al. 2013). Nonetheless, the final 42 candidate genes identified through both liquid and plate feeding RNAi approaches produced consistent phenotypes. The robustness of our screen was also supported by the independent identification of mpk-1, which is duplicated in the ORFeome library and consistently resulted in low progeny when knocked down in nhl-2(0) mutants, but not in wild-type animals. We cannot rule out the possibility that some of these hits may not be specific to nhl-2, and represent genes sensitive to germlines with disrupted homeostasis. However, our final screen results represent candidate genes that are worthy of further investigation to elucidate novel roles for NHL-2.
Figure 2.
Genome-wide RNAi screening methodology. (A) Outline of primary liquid RNAi screen. Approximately 10 L1 wild-type or nhl-2(0) worms were deposited into each well of a 96-well plate that contained bacteria expressing dsRNA clones. Black wells represent phenotypes that are present in wild-type animals with no enhancement in nhl-2(0) mutants. Gray wells represent positive hits, where enhanced phenotypes were observed in nhl-2(0) mutants. Positive hits were then validated by plate feeding RNAi (secondary screen). (B) Representative DIC images of phenotypes as observed during the scoring process (arrows).
Characterization of genes
Our primary screen in liquid culture identified several genes associated with pathways that regulate mitotic proliferation. These included the gene encoding the PUF protein, fbf-1, and the Notch signaling components lag-1, nos-3, and sel-12 (Hansen et al. 2004; Henderson et al. 1994; Kadyk and Kimble 1998; Lambie and Kimble 1991; Levitan and Greenwald 1995; Wen et al. 2000; Zhang et al. 1997). However, these genes were excluded from our high-confidence list as they were only mildly enhanced in nhl-2(0) mutants compared to knock down in wild-type animals. In addition to this, we assessed whether the enhanced mitotic proliferation observed in nhl-2(0) mutants was enhanced in the synthetic phenotypes observed in our high-confidence hit list. Unfortunately, no trend was observed when each gene was knocked down in nhl2(0) mutants and immunostained for PH3 (Figure S1 in File S1). This suggests that the genes associated with our final results are not primarily associated with mitotic proliferation.
To determine any trends associated with the 42 high-confidence screen hits, we further classified these based on their homology, their germline enrichment, and their NCBI Clusters of Orthologous Groups (COGs) description (WormBase, WS204). GO terms for molecular function, cellular compartment, and biological process were analyzed by GO Term Mapper (Harris et al. 2004); however, this analysis did not show any clear trend, possibly due to the small number of high-confidence genes (Table S2, Table S3, and Table S4 in File S1). Therefore, genes were grouped based on their reported function, as well as whether each gene was enriched in the germline (WormBase, WS204). This information was then used to group the final secondary screen results based on criteria for each gene (Table 1). However, RNAi by feeding in our laboratory conditions did produce negative results for genes with previously reported lethal phenotypes. An example of this is cgh-1, which results in sterility when knocked down in wild-type animals (Navarro et al. 2001), yet unexpectedly did not register as a positive hit in our screen. This suggests that high-throughput RNAi screening does have limitations. Nonetheless, our final high-confidence gene list showed consistent phenotypes using both liquid and plate feeding RNAi approaches.
Table 1. Genes from secondary screen listed per reported function.
| Gene (Phenotype) | Description (NCBI COGs) | Enriched | Homology | ||
|---|---|---|---|---|---|
| H | D | S | |||
| Transcriptional/nuclear organization hits | |||||
| mdt-8 (Lp) | Mediator of RNA polymerase II transcription subunit 8 | Sp | |||
| mdt-10 (Lp) | Mediator of RNA polymerase II transcription subunit 10 | Sp & Oo | |||
| mdt-27 (Lp) | Mediator of RNA polymerase II transcription subunit 27 | Sp & Oo | |||
| mrg-1 (Ste) | Chromodomain-containing protein | Sp | |||
| ntl-3 (Ste) | CCR4-NOT transcriptional regulation complex | ||||
| C36E8.1 (Ste) | RNA polymerase I transcription factor | Sp & Oo | |||
| spe-44 (Lp) | Nuclear DEAF-1 related transcriptional regulator | Sp & Oo | |||
| pqn-85 (Lp) | Sister chromatid cohesion protein | Sp & Oo | |||
| F58G1.2 (Lp) | Zn-finger protein | Sp | |||
| fem-1 (Lp) | Ankyrin repeat protein | Sp & Oo | |||
| Y54G2A.26 (Lp) | Unknown (transcriptional corepressor in S. pombe). | ||||
| sas-1 (Emb) | Spindle assembly protein | Sp | |||
| Cell cycle regulation | |||||
| cya-1 (Lp) | G2/mitotic-specific cyclin A | Sp & Oo | |||
| orc-5 (Emb) | Origin recognition complex | Sp & Oo | |||
| F35G12.11 (Lp) | Enhancer of rudimentary homolog | Sp | |||
| Motor related proteins | |||||
| arp-11(Emb) | Actin related protein | Sp & Oo | |||
| dli-1 (Emb) | Dynein light intermediate chain | Sp & Oo | |||
| dnc-1a (Lp) | Microtubule-associated protein | Sp & Oo | |||
| mop-25.2a (Lp) | Conserved mouse embryo scaffolding protein | Sp & Oo | |||
| F28D1.2 (Lp) | Myosin 1 homolog | Sp & Oo | |||
| Microtubule-related proteins | |||||
| dnc-1a (Lp) | Microtubule-associated protein | Sp & Oo | |||
| pfd-1 (Lp) | Molecular chaperone Prefoldin subunit 1 | ||||
| pfd-2 (Emb) | Molecular chaperone Prefoldin subunit 2 | Sp & Oo | |||
| Chromosomal integrity and DNA repair | |||||
| smz-2 (Emb) | Sperm meiosis PDZ domain containing protein | Sp | |||
| drh-3 (Emb) | DEAD-box RNA helicase | Sp & Oo | |||
| cde-1 (Emb) | Germline-specific nucleotidyltransferase protein | ||||
| ekl-1 a(Emb) | Tudor domain protein | ||||
| spdl-1 (Lp) | Coiled-coil protein | Sp & Oo | |||
| mop-25.2a (Lp) | Conserved mouse embryo scaffolding protein | Sp & Oo | |||
| rad-51 (Emb) | DNA repair protein | Sp & Oo | |||
| him-14(Emb) | MutS family of DNA mismatch repair protein | Sp & Oo | |||
| dsb-1 (Lp) | Double-strand break factor | Sp & Oo | |||
| Mitochondrial related proteins | |||||
| lpl-1 (Ste) | Lipoate ligase homolog | Sp | |||
| mtch-1 (Lp) | Mitochondrial carrier homolog 1 | Sp&Oo | |||
| Kinase proteins | |||||
| C56A3.8 (Lp) | Phosphatidylinositol 4-kinase type 2-alpha homolog | Sp & Oo | |||
| mpk-1 (Lp) | Mitogen-activated protein kinase | Sp | |||
| Other biological functions | |||||
| C06A8.6 (Lp) | Ortholog of human 1RRC9 | Sp & Oo | |||
| C34D4.4 (Lp) | Ortholog of human TVP23A | Sp & Oo | |||
| R05H5.3 (Emb) | Ortholog of human NXNl2 | Sp & Oo | |||
| unc-11 (Lp) | Clathrin assembly protein | Sp | |||
| T12A2.7 (Lp) | Ortholog of human BCAS2 | Oo | |||
| Proteins with unknown function | |||||
| C46A5.5 (Lp) | Unknown | Sp | |||
Phenotypes in nhl-2(ok818) mutants: Lp, low progeny; Ste, sterile; Emb, embryonic lethal. Gray = BLAST e value <0.1 and >30% query coverage. Black = BLAST e value <0.1 and >20% query coverage. White = BLAST e value >0.1. Sp = sperm enriched, Oo = oocyte enriched. Homology: H = H. sapiens, D = D. melanogaster, S = S. cerevisiae.
Genes with >1 reported function.
Interestingly, 12 hits were associated with transcriptional/nuclear organization processes, as well as nine genes required for chromosomal integrity or DNA repair. We identified hits associated with components of the DNA damage repair pathway, such as rad-51 and dsb-1 (Stamper et al. 2013; Takanami et al. 2003), as well as the DNA mismatch repair gene him-14 (Zalevsky et al. 1999). Another subset of genes with reported chromosomal association were components of the small 22G RNA pathway. These included the DEAD-box RNA helicase drh-3, the tudor domain protein ekl-1, and the nucleotidyltransferase cde-1 (Gu et al. 2009; Nakamura et al. 2007; van Wolfswinkel et al. 2009). Interestingly, many of the genes associated with chromosomal integrity, DNA repair, and the 22G RNA pathway displayed enhanced synthetic embryonic lethality when knocked down in nhl-2(0) mutants. Given the association of the 22G RNA pathway with centromere formation in early embryogenesis (Claycomb et al. 2009), and the presence of DNA damage repair genes in our screen, it is possible that NHL-2 may have a role associated with embryonic chromosome integrity.
As expected, the majority of genes from our screen were strongly enriched within the germline (Baugh et al. 2003; Reinke et al. 2004). This consisted of 26 genes enriched in both oocytes and sperm, and 10 genes enriched in sperm only. This resulted in a total of 38 genes that were germline enriched and produced synthetically enhanced germline-deficient phenotypes when knocked down in nhl-2(0) mutants. The high rate of genes enriched in sperm and oocytes suggests that these synthetic phenotypes are the result of deficiencies associated with central germline processes.
The majority of NHL-2 screen hits are highly conserved. BLAST results grouped genes based on homology against H. sapiens, D. melanogaster, and S. cerevisiae. Our criteria for homology was as follows: high homology, where genes scored e-values <0.1% and query coverage of >30%; and moderate homology, where genes scored e-values <0.1% with query coverage >20%. Based on these criteria, the homology of our final hits was considerably high, with only four genes being nematode-specific genes. This is highly relevant, as it is anticipated that this investigation will provide additional information that is relevant for orthologs of NHL-2 in higher organisms. In all, this synthetic RNAi screen identified genes associated with candidate pathways that may require NHL-2, which are highly enriched in the germline. Given the diverse nature of TRIM-NHL proteins, including those with multiple functions, this screen may lead to the identification of novel roles for NHL-2.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300166/-/DC1.
Acknowledgments
The pCB19 RNAi negative control was a kind gift from Carolyn Behm (Australian National University). We thank David Piedrafita (Federation University) for his constructive feedback regarding this manuscript. Strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). G.M.D. was supported by a Faculty of Medicine, Nursing and Health Sciences, Monash University PhD scholarship. P.R.B. was funded by the National Health and Medical Research Council of Australia (NHMRC) (grant number 0606575).
Footnotes
Communicating editor: J. Kim
Literature Cited
- Ahringer J., 2006. Reverse genetics (April 6, 2006). WormBook, ed. The C. elegans Research Community WormBook , / 10.1895/wormbook.1.7.1, http://www.wormbook.org. [DOI]
- Baugh L. R., Hill A. A., Slonim D. K., Brown E. L., Hunter C. P., 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Conceição Pereira M., Morais S., Sequeiros J., Alonso I., 2016. Large-scale functional RNAi screen in C. elegans identifies TGF-β and Notch signaling pathways as modifiers of CACNA1A. ASN Neuro 8: 1759091416637025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Jr., Vasale J., Batista P. J., et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V., 2009. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004. Control of the proliferation vs. meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. [DOI] [PubMed] [Google Scholar]
- Harris M. A., Clark J., Ireland A., Lomax J., Ashburner M., et al. , 2004. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 32: D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J., 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924. [DOI] [PubMed] [Google Scholar]
- Horn E. J., Albor A., Liu Y., El-Hizawi S., Vanderbeek G. E., et al. , 2004. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 25: 157–167. [DOI] [PubMed] [Google Scholar]
- Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., et al. , 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291. [DOI] [PubMed] [Google Scholar]
- Hyenne V., Desrosiers M., Labbe J. C., 2008. C. elegans Brat homologs regulate PAR protein-dependent polarity and asymmetric cell division. Dev. Biol. 321: 368–378. [DOI] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kano S., Miyajima N., Fukuda S., Hatakeyama S., 2008. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 68: 5572–5580. [DOI] [PubMed] [Google Scholar]
- Karp X., Ambros V., 2012. Dauer larva quiescence alters the circuitry of microRNA pathways regulating cell fate progression in C. elegans. Development 139: 2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E., Kudryashov D., Kramerova I., Spencer M. J., 2005. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 354: 413–424. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J., 1991. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240. [DOI] [PubMed] [Google Scholar]
- Lehner B., Tischler J., Fraser A. G., 2006. RNAi screens in Caenorhabditis elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions. Nat. Protoc. 1: 1617–1620. [DOI] [PubMed] [Google Scholar]
- Levitan D., Greenwald I., 1995. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature 377: 351–354. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang C., Wang X. L., Ly P., Belyi V., et al. , 2014. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 21: 1792–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Stotz M., Qamar S., Kramer K., Hennig J., et al. , 2014. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 28: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia A. F., Tanenbaum M. E., Galli M., Lelieveld D., Egan D. A., et al. , 2015. Genome-wide RNAi screen for synthetic lethal interactions with the C. elegans kinesin-5 homolog BMK-1. Sci. Data 2: 150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti G., Reichardt I., Knoblich J. A., Besse F., 2014. The TRIM-NHL protein Brat promotes axon maintenance by repressing src64B expression. J. Neurosci. 34: 13855–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2017. A microRNA family exerts maternal control on sex determination in C. elegans. Genes Dev. 31: 422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Ando R., Nakazawa T., Yudazono T., Tsutsumi N., et al. , 2007. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells 12: 997–1010. [DOI] [PubMed] [Google Scholar]
- Navarro R. E., Shim E. Y., Kohara Y., Singson A., Blackwell T. K., 2001. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 128: 3221–3232. [DOI] [PubMed] [Google Scholar]
- Neumuller R. A., Betschinger J., Fischer A., Bushati N., Poernbacher I., et al. , 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S., Stoye J. P., Saib A., 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3: 799–808. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., Stoye J. P., O’Garra A., 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 38: 619–630. [DOI] [PubMed] [Google Scholar]
- Reboul J., Vaglio P., Rual J. F., Lamesch P., Martinez M., et al. , 2003. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35–41. [DOI] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K., 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Saur T., DeMarco S. E., Ortiz A., Sliwoski G. R., Hao L., et al. , 2013. A genome-wide RNAi screen in Caenorhabditis elegans identifies the nicotinic acetylcholine receptor subunit ACR-7 as an antipsychotic drug target. PLoS Genet. 9: e1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn J. C., Berezikov E., Knoblich J. A., 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper E. L., Rodenbusch S. E., Rosu S., Ahringer J., Villeneuve A. M., et al. , 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9: e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanami T., Mori A., Takahashi H., Horiuchi S., Higashitani A., 2003. Caenorhabditis elegans Ce-rdh-1/rad-51 functions after double-strand break formation of meiotic recombination. Chromosome Res. 11: 125–135. [DOI] [PubMed] [Google Scholar]
- Van Hooser A., Goodrich D. W., Allis C. D., Brinkley B. R., Mancini M. A., 1998. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. 111: 3497–3506. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Claycomb J. M., Batista P. J., Mello C. C., Berezikov E., et al. , 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148. [DOI] [PubMed] [Google Scholar]
- Wen C., Levitan D., Li X., Greenwald I., 2000. spr-2, a suppressor of the egg-laying defect caused by loss of sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily. Proc. Natl. Acad. Sci. USA 97: 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen A. J., Duffy J. B., Kemphues K. J., Villeneuve A. M., 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., et al. , 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.