Abstract
MicroRNAs (miRNAs) are small noncoding endogenous RNAs, typically 21–23 nucleotides long, that regulate gene expression, usually post-transcriptionally, by binding to the 3′-UTR of target mRNA, thus blocking translation. The expression of several miRNAs is significantly altered during cardiac hypertrophy, myocardial ischemia, fibrosis, heart failure, and other cardiac myopathies. Recent studies have implicated miRNA-9 (miR-9) in myocardial hypertrophy. However, a detailed mechanism remains obscure. In this study, we have addressed the roles of miR-9 in muscle development and function using a genetically tractable model system, the indirect flight muscles (IFMs) of Drosophila melanogaster. Bioinformatics analysis identified 135 potential miR-9a targets, of which 27 genes were associated with Drosophila muscle development. Troponin-T (TnT) was identified as major structural gene target of miR-9a. We show that flies overexpressing miR-9a in the IFMs have abnormal wing position and are flightless. These flies also exhibit a loss of muscle integrity and sarcomeric organization causing an abnormal muscle condition known as “hypercontraction.” Additionally, miR-9a overexpression resulted in the reduction of TnT protein levels while transcript levels were unaffected. Furthermore, muscle abnormalities associated with miR-9a overexpression were completely rescued by overexpression of TnT transgenes which lacked the miR-9a binding site. These findings indicate that miR-9a interacts with the 3′-UTR of the TnT mRNA and downregulates the TnT protein levels by translational repression. The reduction in TnT levels leads to a cooperative downregulation of other thin filament structural proteins. Our findings have implications for understanding the cellular pathophysiology of cardiomyopathies associated with miR-9 overexpression.
Keywords: Drosophila melanogaster, Troponin-T, hypercontraction, miRNA, indirect flight muscles
Muscle contraction is crucially dependent on the proper assembly, maintenance, and function of myofibrils (Beall and Fyrberg 1991; Gordon et al. 2000). Myofibril assembly is a highly complex and coordinated process that requires the maintenance of appropriate stoichiometries of structural proteins and protein complexes, such as the acto-myosin complex and the Tn–Tropomyosin complex (Laing and Nowak 2005; Firdaus et al. 2015). Defects, genetically caused or otherwise, in muscle development, structure, or function, result in a number of disorders and diseases, collectively referred to as myopathies (Charge and Rudnicki 2003; Selcen 2011; Gautam et al. 2015). Congenital myopathies, including genetic heart diseases, comprise a wide variety of muscle disorders that are mostly due to mutations in the contractile proteins (Chawla 2011; Selcen 2011). For example, mutations of cardiac TnT, α-Tm, and myosin cause hypertrophic cardiomyopathy (HCM) (Watkins et al. 1995; Karibe et al. 2001). Stoichiometric imbalances of structural proteins and altered isoform expression leading to myocardial damage are also seen in secondary cardiomyopathies, resulting from infection or other factors (Thierfelder et al. 1994; Watkins et al. 1996; Sisakian 2014). To achieve the appropriate stoichiometric balances, many levels of regulation are required. While transcriptional regulation of these components is extremely important, the significance of the roles of microRNAs (miRNAs) in myopathies in general, and in hypertrophy in particular, are becoming increasingly recognized (Eisenberg and Psaty 2009; Parkes et al. 2015).
miRNAs are small noncoding endogenous RNAs, typically 22–30 nucleotide long, that exert subtle control over gene expression, transcriptionally or post-transcriptionally. This makes them an indispensable part of the regulatory network in almost all complex events in organisms ranging from plants to mammals, and even their viruses (Bartel and Chen 2004; Allen and Howell 2010; Carthew and Sontheimer 2009). In muscles, many miRNAs—including both the muscle-specific “myomiRs” such as miR-1, miR-133, and miR-206 (McCarthy 2008), as well as those more widely expressed, such as miR-24, miR-29, and miR-181 (Erriquez et al. 2013)—are involved in the regulation of both myoblast proliferation and differentiation (Chen et al. 2008). Importantly, the expression of several miRNAs is significantly altered during cardiac hypertrophy, myocardial ischemia, fibrosis, heart failure, and other cardiac myopathies (Latronico and Condorelli 2011; Oliveira-Carvalho et al. 2012). miR-9, a miRNA of recognized neural functions (Gladka et al. 2012; Krichevsky et al. 2003), reportedly plays a regulatory role in myocardial hypertrophy and is antagonistic to myocardin, a positive mediator of cardiac hypertrophy (Wang et al. 2010). In addition, miR-9 was implicated in the regulation of platelet-derived growth factor receptor-β, a regulator of cardiomyocyte angiogenesis (Zhang et al. 2011). Both studies reported downregulation of miR-9 upon activation of the hypertrophic response. Recently, clinical studies revealed that miR-9 expression levels are significantly lower in hypertensive patients as compared to healthy controls, and appear to be correlated with ventricular mass (Kontaraki et al. 2014). Thus, miR-9 appears to have more than one role in cardiac function. It is thus imperative to characterize its roles in the context of muscle development and function. There is high evolutionary conservation of the ultrastructure of striated muscles, their component proteins, and the mechanisms that regulate the assembly of sarcomeres and myofibril formation throughout vertebrates; similarly, there is substantial similarity between the muscles of invertebrates with complex locomotion (Taylor 2006). The Drosophila IFMs provide a good system with which to study muscle development, function, and associated diseases (Sparrow et al. 2008). This is particularly true with respect to the investigation of cardiomyopathic disorders, as the IFMs exhibit properties such as stretch activation and asynchronous contraction that are physiologically similar to those of cardiac muscles (Vigoreaux 2001). Specific mutations of the Drosophila contractile machinery (Kronert et al. 1995; Nongthomba et al. 2003), signaling cascades (Gajewski et al. 2006), and connective tissues (Pronovost et al. 2013) lead to muscle hypercontraction, associated with decreased structural integrity of the sarcomeres, which is similar to that seen in many myopathic conditions of higher organisms, including humans. In particular, the Drosophila miR-9a is an exact copy of the human miR-9 (Yuva-Aydemir et al. 2011).
In this study, we have investigated the regulatory role of miR-9a in Drosophila for IFM development and functioning. We show that Drosophila miR-9a plays a novel role in the regulation of TnT, a major structural protein, during myofibrillogenesis. This finding will lead to a better understanding of how human miR-9 may be involved in the pathogenesis of cardiac hypertrophy.
Materials and Methods
Fly strains and crosses
All flies were maintained on standard cornmeal-agar-yeast medium. Canton-S was used as a control for most of the experiments unless specified. Crosses were performed at 25°, unless otherwise indicated. UH3-Gal4 (X chromosome) expression from 36 hr after puparium formation (APF) onwards becomes IFM-specific (Singh et al. 2014). UAS-miR-9a [third chromosome, Bloomington Drosophila Stock Center (BDSC) #41138] and UAS-miR-SP-9a (third chromosome, kind gift from David Van Vactor, Harvard Medical School) were used for overexpression and knocking-down of miR-9a, respectively. UAS-SlsRNAi (third chromosome), UAS-mbcRNAi (third chromosome), and UAS-NeuralizedRNAi (third chromosome) were procured from the BDSC, and UAS-TnTRNAi (third chromosome) was from VDRC, Vienna (v27853). The green fluorescent protein (GFP) construct [sls-GFP (third chromosome)] has been described in Morin et al. (2001). All chromosomes and gene symbols are as mentioned in FlyBase (http://flybase.org), unless specifically described.
Generation of UAS-TnT lacking the miR-9 binding site
Two transgenic fly lines [UAS-TnT (10a) and UAS-TnT (10b)] were generated for the overexpression of either the adult isoform (10a) or the pupal isoform (TnT-10b). The TnT transcripts (both 10a and 10b) were amplified using cDNA extracted from wild-type thoraces, using primers designed to target the 5′- and 3′-UTRs but to exclude the miR-9a binding site. The primers were also modified to incorporate EcoRI and KpnI restriction sites for subcloning into the pUAST overexpression vector (TnT FP with EcoRI site: 5′-GAACCGCAGAATTCGCTCCTAC-3′, TnT RP with KpnI site: 5′-GTGAAGGAAAGTGGTACCCGAG-3′). The transcripts were cloned into a TA vector and the clones were screened for the presence of 10a or 10b transcripts using reverse primers specific for the alternatively spliced exon (TnT FP + TnT10a RP: 5′-TTGTGCGCTGAGTGAATC-3′ and TnT FP + TnT10b RP: 5′-CGGTGTATTGCTCCTTCT-3′). The presence of the 10a or 10b transcripts in the respective clones was confirmed by sequencing. The sequenced clones and pUAST plasmid were digested with EcoRI and KpnI, the released inserts and cut pUAST vector were ligated, and the transgenic constructs were cloned and confirmed by sequencing. The construct, either pUAST-TnT-10a or pUAST-TnT-10b, along with л-helper plasmid (encoding for a transposase) was injected into embryos of white-eyed w1118 flies using the Olympus CK-X31- Narishige IM-9B microinjection system. The adult flies (G0) that emerged were then crossed with white-eyed flies to produce the F1 generation. The transgenic flies were identified by their red eye phenotype.
Behavioral test
Flight ability was assayed using 2–3-d-old individual flies as described previously (Drummond et al. 1991) and the flies were categorized as up-flighted, horizontally-flighted, down-flighted, or flightless. Each fly was flight tested three times.
Polarized microscopy
For polarized microscopy, 2–3-d-old flies were bisected and processed using a protocol described previously (Nongthomba and Ramachandra 1999). Images were captured using an Olympus SZX12 microscope fitted with an Olympus C -5060 camera.
Hematoxylin and eosin staining
Hematoxylin and eosin staining of transverse sections of the adult thorax was done as previously described (Pantoja et al. 2013). Sections were mounted using Dibutylphthalate Polystyrene Xylene (DPX) mounting medium (Qualigens, Mumbai) and analyzed by light microscopy. Images were acquired using a Leica DFC300FX camera and processed using inbuilt software.
Confocal microscopy
The bisected fly hemithoraces were processed for immunohistochemical analysis as described previously (Rai et al. 2014). Following blocking, samples were treated differently depending on the type of analysis required.
Scanning electron microscopy (SEM)
SEM analysis of IFMs was used to visualize sarcomeric structure. Three-d-old flies were dehydrated using an alcohol series (50, 70, 80, 90, 95, and 100%). Samples were incubated for 10 min in each dilution with the final dehydration step in 100% alcohol repeated twice. After twice incubating each sample in hexamethyldisilazane for 45 min, they were dried in a desiccator for 24 hr. The head, wings, abdomen, and legs were then removed, transferred to a glass slide, and bisected sagittally. Bisected thoraces were mounted onto an aluminum stub with carbon tape and surface coated by gold sputtering (20 nm thick film) using a Baltec sputter to avoid charging. A Zeiss, Ultra 55, Field Emission SEM with Secondary Electron Detector was used for imaging with an accelerating voltage of 2–5 keV and 8 mm working distance.
RNA and PCR
RNA was isolated from the IFMs of 2–3-d-old flies. IFMs were removed from the bisected thoraces at 4° and immersed in Trizol (Sigma). Next, RNA was extracted with the help of Trizol (Sigma) as per the manufacturer’s protocol. cDNA was made using 1–2 µg of extracted RNA and a cDNA synthesis kit (Fermentas). The following primers were used: RP49 (FP: 5′-TTCTACCAGCTTCAAGATGAC-3′, RP: -5′-GTGTATTCCGACCACGTTACA-3′); upheld (up) (FP: 5′-CTCGGGTGTCTCGGGCTCAC-3′ RP: 5′-CTCGAACGAGAAGATCTGGA-3′); and Opa1-like (FP: 5′-AACGGTGGAGCCAGTTCTCG-3′; RP: 5′-TGATCTCCGTCTGCAGCGTC-3′). Quantitative PCR was carried out using DyNAmoTM HS SYBR green mix (F-410L; Thermo Scientific). Fluorescence intensities were recorded and analyzed in an ABI Prism 7900HT sequence detection system (SDS 2.1; Applied Biosystems). The relative changes in gene expression were estimated after normalization to the expression of a housekeeping gene, RP49. For semiquantitative PCR, reactions were set up using the 2× PCR Mastermix (Fermentas) and PCR amplification was carried out using a Mastercycler Nexus (Eppendorf).
Northern blotting
Detection of miRNA was carried out using the northern blotting protocol of Varallyay et al. (2008). RNA was extracted as described earlier, and quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Equal concentrations of samples were loaded on the gel. RNA bands were visualized under a UV transilluminator (JH BIO Innovations Pvt. Ltd.) and transferred onto a Nitrocellulose membrane (Millipore) by semidry transfer. Locked nucleic acid (LNA) Probe for miR-9a (5′-TCATACAGCTAGATAACCAAAGA-3′) and control probe for U6snoRNA (5′-GTCATCCTTGCGCAGGGGCCATGC-3′) was labeled using 1 µl T4 polynucleotide kinase (PNK), 1 µl [γ-32P] ATP, and 2 µl PNK buffer, and the final volume was made up to 20 µl. Following incubation at 37° for 60 min, the probe was purified using a Sephadex G-50 column. The membrane was washed twice with 0.5× TBE (Tris Borate-EDTA), allowed to cross-link under UV light for 60 sec, and then incubated at 40° for hybridization with LNA probe in 1× Perfect Hyb Plus buffer (Sigma) for 2–3 hr. The probed membrane was then washed three times with 2× SSC, 0.1% SDS buffer at 40° and exposed to photographic film. The exposed film was scanned using a Phosphor image scanner (GE Typhoon 9500).
Western blotting
Dissected adult IFMs were homogenized in sample buffer (312.5 mM Tris-HCl pH 6.8, 10% SDS, 0.5% Bromophenol Blue, 50% glycerol, and 25% β-mercaptoethanol) and denatured for 3 min at 95°. Samples were run on a 12.5% resolving gel and western blotting was carried out as described previously (Nongthomba et al. 2007). The primary antibodies used to detect specific proteins were: Drosophila anti-TnT raised in rat 1:1000 (gift from John Sparrow, UK), Drosophila anti-TnI raised in rabbit 1:1000 (gift from Alberto Ferrus, Spain), Drosophila anti-Flightin raised in rabbit 1:1000 (gift from Jim O. Vigoreaux, Vermont), Drosophila anti-Actin raised in rabbit 1:1000 (gift from John Sparrow, UK), and Drosophila anti-α-Tubulin raised in mouse 1:1000 (DHSB).
Bioinformatics analysis
miR-9a target prediction was done using five different target prediction software suites. These were Miranda (http://www.microrna.org/, Enright et al. 2003), Pic-Tar (http://pictar.bio.nyu.edu, Krek et al. 2005), the method used by Stark et al. (2003) (http://www.russell.embl.de/miRNAs/), EMBL target prediction, and Target scan fly (http://www.targetscan.org/, Lewis et al. 2003). All five different algorithms predict miRNA targets, and hence their results should be nonoverlapping. We shortlisted targets recognized by three or more software suitesas significant matches. These shortlisted genes were cross-referenced to a Drosophila IFM microarray dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70252) to check for their expression levels in adult IFMs. The miR-9a targets thus obtained were then functionally annotated using DAVID (http://david.abcc.ncifcrf.gov/, Huang et al. 2008) and PantherDB software (http://www.pantherdb.org/about.jsp, Mi et al. 2005).
Data availability
We have provided the details of all the web addresses of data resources that we made use of in this study. Other data that support our findings have been included as Supplemental Material and are described in the Results section. Supplemental Figure legends are available in File S1.
Results
Knockdown of miR-9a in the IFMs during myofibrillogenesis does not affect muscle structure and function
To investigate the role of miR-9a during myofibrillogenesis, we performed IFM-specific knockdown of miR-9a from 36 hr APF. Sarcomeres of the IFMs are established by an organized assembly of their structural proteins between 37 and 46 hr APF (Reedy and Beall 1993; Nongthomba et al. 2004). miR-9a is expressed in developing IFMs and the muscle attachment sites (Yatsenko and Shcherbata 2014), and expression is highly reduced in adult IFMs (Supplemental Material, Figure S1A). The knockdown of miR-9a during myofibril assembly had no detrimental effect on flight (Figure S1B). Flies with knockdown of miR-9a (UH3 > miR-SP-9a) exhibited close to normal flight ability, with 74.2% of flies capable of upward flight and 25.8% exhibiting horizontal flight (n = 31), which is similar to wild-type, where 80.6% of flies were up-flighted and 19.4% were horizontally-flighted (n = 31) (Figure S1B). Further, both wild-type (Figure S1C) and miR-9a knocked-down flies (Figure S1D) had six normal dorsal longitudinal muscles (DLM) fibers in each hemithorax with normal sarcomeric structures (Figure S1, C’–D’’).
Overexpression of miR-9a in the IFMs during myofibrillogenesis causes hypercontraction
To determine whether the inherently low expression of miR-9a during myofibrillogenesis (Figure S1A) is important for the critical roles of its targets during this stage of muscle development, we overexpressed miR-9a in the IFMs throughout myofibrillogenesis. While miR-9a expression was barely detectable in wild-type IFMs, UH3-Gal4-mediated (Singh et al. 2014) miR-9a overexpression clearly increased miR-9a levels (Figure S1A) and adversely affected both wing posture and flight performance (Figure 1, A–B’ and E).
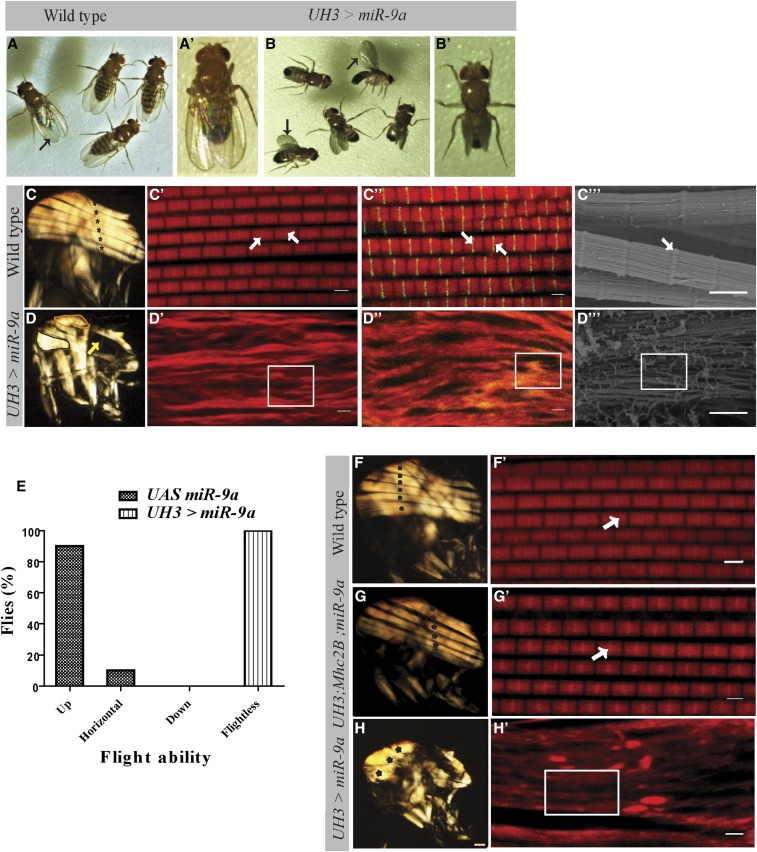
Figure 1.
IFM-specific overexpression of miR-9a causes muscle hypercontraction. (A) Wild-type adult flies with normal wing posture. (A’) Regular wing position along the body axis in a wild-type fly. (B) Adult flies overexpressing miR-9a have upheld wings. (B’) Upright wing phenotype in a fly overexpressing mir-9a. (C) Polarized light micrograph of dissected wild-type adult hemithorax showing six DLMs (asterisks). (C’ and C’’) Adult myofibrils stained with Phalloidin-TRITC (F-actin) and Sls-GFP (arrows indicate Z-disc). (C’’’) SEM micrograph of wild-type myofibril (arrow indicates Z-disc). (D) Polarized light image of hemithorax of adult male with overexpression of miR-9a showing broken muscles (arrow) and clumped muscle (outlined in black and orange). (D’ and D’’) Loss of sarcomeric structural integrity (box) in flies overexpressing miR-9a. (D’’’) SEM micrograph showing absence of proper sarcomeres (box) after miR-9a overexpression (bar, 2 µm). (E) Flight assay of the miR-9a overexpression flies. (F) Images of wild-type hemithorax showing six DLMs (asterisks) under polarized light. (F’) Phalloidin-TRITC-stained myofibril showing normal sarcomeric structure (arrow) in wild-type flies. (G) Adult flies carrying a myosin MhcP401S mutation in the overexpression of miR-9a background show six DLMs (asterisks) in the hemithorax and (G’) normal sarcomeres (arrows) in the myofibrils. (H) Polarized light image of hemithorax from flies with overexpression of miR-9a. (H’) Abrogated sarcomeric structure (box) in the myofibrils of flies overexpressing miR-9a (bar, 2 µm). DLMs, dorsal longitudinal muscles; GFP, green fluorescent protein; IFM, indirect flight muscles; miR-9a, microRna-9a; SEM, scanning electron microscope; TRITC, tetramethylrhodamine.
The hemithoraces of wild-type flies showed presence of six DLMs (asterisks) with conventional sarcomeric structure, with well demarcated Z-discs (arrows) (Figure 1, C–C’’’). Whereas flies overexpressing miR-9a exhibited broken muscle fibers that appeared to be hypercontracted and pulled toward attachment sites (highlighted in Figure 1, D–D’’’), control wild-type adult hemithoraces showed the typical six well-organized DLM fascicles (Figure 1C) and, at higher magnification, individuals showed well-arranged fibers (Figure 1C’). However, overexpression of miR-9a resulted in abnormal muscle and loss of myofibril integrity in the IFMs, with defects in sarcomeric organization and no organized Z-discs (Figure 1, D–D’’’). Unlike those of the wild-type flies (Figure S2, A and A’), flies overexpressing miR-9a had severe muscle disorganization (Figure S2B), with whole fascicles missing (black arrowheads, Figure S2B’). Flies of both sexes, overexpressing miR-9a, also exhibited a complete loss of flight ability [100% flightless (n = 35)] unlike their control counterparts (+; +; UAS miR-9a/+) [90% up-flighted and 10% horizontally-flighted (n = 31)] (Figure 1E).
The loss of muscle integrity and sarcomeric structure caused by miR-9a overexpression (Figure 1, D–D’’) is very similar to the hypercontraction phenotype reported earlier (Nongthomba et al. 2003). This phenotype is usually associated with mutations in genes encoding sarcomeric structural proteins. It is well established in Drosophila IFMs that mutation in some of the structural proteins like TnT, Actin, and TnI can lead a the coordinated reduction in other thin filament proteins. These result in loss of sarcomeric structure and muscle hypercontraction, as a consequence of mis-regulated acto-myosin interaction (Nongthomba et al. 2003, 2004, 2007; Firdaus et al. 2015). The hypercontraction phenotype can be suppressed in flies with the MhcP401S mutation (Figure 1, G and G’) (Nongthomba et al. 2003). This mutation is in the Actin binding head region of the myosin heavy chain, which prevents the interaction of the thin and thick filaments, thus reducing muscle contraction. Flies carrying MhcP401S mutation in the miR-9a overexpression background showed suppression of the muscle structural defects associated with the miR-9a overexpression (Figure 1, F–H). The DLMs and sarcomeric structure in adult flies (Figure 1, G and G’) were comparable to those of wild-type controls (Figure 1, F and F’). Compared to flies with overexpression of miR-9a alone (Figure 1, H and H’), the hypercontraction suppressed flies possessed six DLMs (asterisks) (Figure 1G) with close to normal sarcomeric structures (Figure 1G’, white arrow shows a Z-disc), confirming that the miR-9a muscle phenotype results from unregulated acto-myosin interactions.
To confirm that the muscle defects observed resulted directly from miR-9a overexpression in the IFMs during myofibrillogenesis, we studied the effect of suppressing miR-9a expression in the overexpression background. Knockdown of miR-9a in the overexpression background rescued the flight ability to levels similar to wild-type flies (Figure S1B). The adult flies had a normal arrangement and pattern of DLMs (Figure S2, D–E’). Thus, the hypercontraction was rescued and the IFMs showed ordered sarcomeres (Figure S2, D–E’) in contrast to those with the phenotype from overexpression of miR-9a alone (Figure 1, D and D’).
Overexpression of miR-9a results in downregulation of TnT
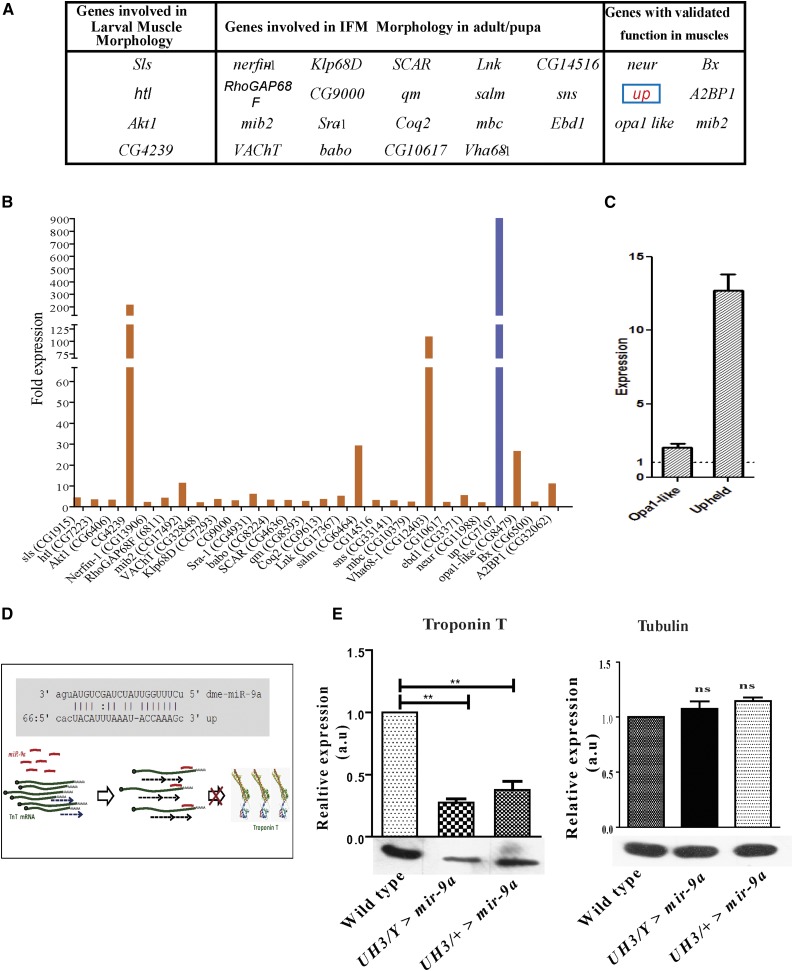
Hypercontraction results from the sarcomeres being unable to withstand the forces produced within them due to changes in the structural proteins of the sarcomere and/or their regulation. Therefore, we investigated if any such proteins are targets of miR-9a. Bioinformatics analysis identified 135 potential miR-9a targets that were then functionally annotated. Five different prediction programs were used (see Materials and Methods) and we chose only those predicted targets that were detected by three or more programs. Functional annotation revealed that these genes are involved in a variety of cellular and developmental functions such as transcriptional regulation, protein degradation, apoptosis, endocytosis, neuronal specification, imaginal disc development, etc., but that 29 genes are involved in muscle development (Figure 2A). All the putative miR-9a targets associated with muscle development proved to be involved in muscle function. Of these miR-9a targets in muscles, 4 genes are reported to be involved in larval muscle development, 19 in the development of pupal and adult muscles (Figure 2A) (Schnorrer et al. 2010), and 6 genes are associated with muscle development (Madan et al. 2017; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70252). These putative targets are expressed in the IFMs as demonstrated using the Drosophila IFM microarray (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70252) (Figure 2B).
Figure 2.
Putative target genes of miR-9a that are involved in muscle development. (A) Lists of genes that are putative targets of miR-9a and their functions in muscles. The upheld gene that encodes TnT is highlighted in red (within blue box) (B) Expression profile of the putative miR-9a targets in the IFMs obtained from the microarray data from IFM of wild-type flies. The upheld gene showed highest expression in IFM (highlighted by the blue bar). (C) Relative expression of the target genes validated by real-time PCR. (D) Schematic representation of miR-9a binding site at 3′-UTR of upheld (TnT) and the mechanism of translation repression of TnT. (E) Quantification of the relative expression of TnT after miR-9a overexpression, using α-Tubulin as loading control (** signifies P value < 0.002). IFM, indirect flight muscles; miR-9a, microRna-9a; PCR, polymerase chain reaction; TnT, Troponin-T; UTR, untranslated region; NS, non-significant.
Among the putative targets of miR-9a genes involved in muscle development and function is the up gene, which codes for all the TnT isoforms in Drosophila melanogaster. up showed a very high level of expression in adult IFMs (blue bar in Figure 2B, expression validated by real-time PCR in Figure 2C). The 3′-UTR of the up gene has the miR-9 binding site sequence (Figure 2D). Given that up mutations have been implicated in muscle hypercontraction (Nongthomba et al. 2003, 2007), we investigated whether miR-9a can cause the downregulation of TnT, possibly through binding to its target sequence at the 3′-UTR of TnT mRNA and suppressing the translational process as represented in Figure 2D. Indeed, there was a significant reduction in TnT protein levels (P value < 0.002) in the DLMs of flies overexpressing miR-9a compared to wild-type (Figure 2E). There was also a concomitant decrease in the levels of other structural proteins (which are not targets of miR-9a) that are part of the thin filament (Actin), including the Tn complex (TnI) (Figure S3A). On the other hand, Flightin, a thick filament component, was not reduced in flies overexpressing miR-9a compared to wild-type flies (Figure S3A). These results were expected since reduction of one thin filament protein is known to lead to a coordinated reduction of other thin filament proteins, but does not affect thick filament components (Nongthomba et al. 2004, 2007).
Repression of TnT by miR-9a is responsible for the hypercontraction phenotype
Since several putative miR-9a targets are important during muscle development, we asked whether the hypercontraction phenotype resulted directly from the downregulation of TnT by miR-9a or was due to the repression of other targets. First, we observed that the knockdown of TnT in IFMs during myofibrillogenesis generated a phenotype very similar to the overexpression of miR-9a. Knockdown of TnT resulted in the disruption of muscle structures (Figure S3, E and E’), which was comparable to the muscle defects seen in overexpression of miR-9a (Figure S3, F and F’). We further tested if the knockdown of some other predicted miR-9a targets can lead to similar defects in muscle structure and function. The downregulation of neuralized, an E3 ubiquitin ligase, failed to show any muscle defects (Figure S3, C and C’). Flies with a knockdown of Sallimus (Sls) showed six DLMs (Figure S3D). However, reduction in Sls, which is a structural component of the Z-disc, did result in some tearing of the sarcomeres (rectangle, Figure S3D’), but this was not comparable to the damage following miR-9a overexpression.
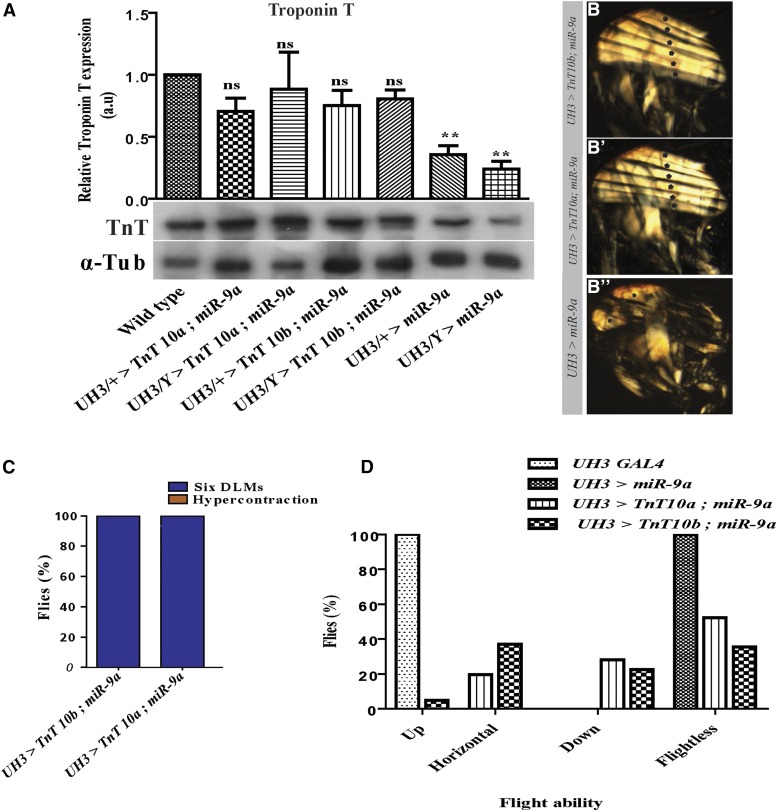
To further confirm that that the muscle hypercontraction resulting from overexpressing miR-9a is a direct result of knockdown of TnT alone, we carried out a rescue of the muscle phenotype by overexpressing TnT devoid of the miR-9a binding site in flies with elevated levels of miR-9a in their IFMs. TnT transgenic lines were driven using UH3-Gal4 and shifted to 29° at 50–52 hr APF. Overexpression of transgenic TnT, either the TnT 10a or 10b isoform, devoid of the miR-9a binding sequence, restored TnT protein levels in these flies compared to flies overexpressing miR-9a alone (Figure 3A). Importantly, the restoration of TnT levels in the background of miR-9a overexpression completely rescued the hypercontraction phenotype. This was evidenced by the presence of six DLMs (Figure 3B’) and the complete absence of any hypercontracted DLMs in the hemithoraces of these flies (Figure 3C). Further, the flight ability of these flies was also partially rescued: control flies [(UH3/+; +; +) Gal4 flies] were 100% up-flighted (n = 31); for TnT 10a, 19.6% of flies were horizontally-flighted, 28.2% down-flighted, and 52.2% flightless (n = 46); and for TnT 10b: 4.8% flies were up-flighted, 37.1% horizontally-flighted, 22.6% down-flighted, and 35.5% were flightless (n = 62); as compared to the flies with overexpression of miR-9a alone, of which were all flightless (n = 60) (Figure 3D). Both of these transgenic lines (TnT 10a and TnT 10b) showed almost complete restoration of muscle integrity and sarcomeric structure (Figure 4, B, B’, D, and D’) comparable to that of wild-type controls (Figure 4, A and A’), in stark contrast to the muscles in the flies overexpressing miR-9a (Figure 4, C and C’).
Figure 3.
Transgenic lines with overexpression of TnT (10a or 10b isoform) restore Troponin-T levels and rescue the muscle hypercontraction phenotype resulting from overexpression of miR-9a. (A) Western blots and the quantification of the relative expression of Troponin-T (loading control α-Tubulin). (B) Polarized light micrograph showing six normal DLMs following the overexpression of TnT-10a or (B’) TnT-10b isoforms in the background of miR-9a overexpression. (B’’) Polarized image showing hypercontracted muscles after miR-9a overexpression. (C) Quantification of the percentage of flies overexpressing miR-9a that present with hypercontraction phenotype after restoration of TnT levels. (D) Flight data for the flies overexpressing TnT 10a or 10b isoforms in the background of overexpression of miR-9a. ** signifies P value < 0.005, DLMs, dorsal longitudinal muscles; miR-9a, microRna-9a; NS, non-significant; PCR, polymerase chain reaction; TnT, Troponin- T.
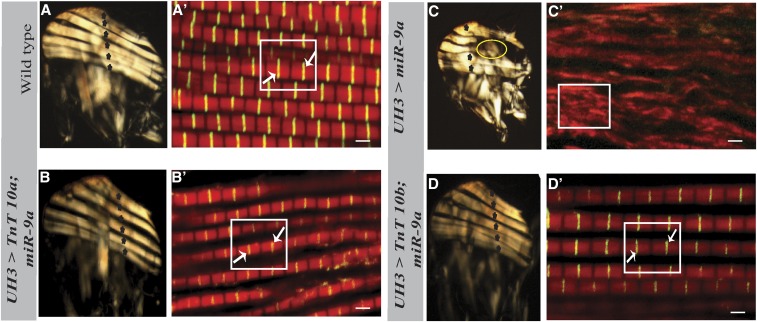
Figure 4.
Rescue of the loss of muscle integrity. (A) Polarized light micrograph of wild-type hemithorax with six DLMs. (A’) Confocal microscopy image of wild-type muscles stained for F-actin along with Sls-GFP. The sarcomeres (box) and Z-discs (white arrows) are highlighted. (B and B’) six DLMs (asterisks) and close to normal sarcomeric structure (box) with Z-discs (white arrows) in muscles of flies expressing TnT-10a in a miR-9a overexpression background. (C and C’) Hypercontracted muscles (circled) and lack of sarcomeres (box) in muscles of flies overexpressing miR-9a. (D and D’) Six DLMs (asterisks) and restored muscle structure (box) with Z-discs (white arrows) in flies overexpressing TnT-10b in a miR-9a overexpression background (Square Box) (Bar, 2 µm). DLMs, dorsal longitudinal muscles; GFP, green fluorescent protein; miR-9a, microRna-9a.
These data argue that TnT is the major target of the miR-9a responsible for the muscle hypercontraction phenotype. We also confirmed that the rescue of the hypercontraction phenotype by expressing TnT isoforms in IFMs is indeed because of the restoration TnT and not due to Gal4 dilution. When UH3-Gal4 was used to drive both UAS-miR-9a and UAS-GFP, the progeny still exhibited hypercontracted muscles in hemithoraces with loss of sarcomeric structure (Figure S4, B and B’), similar to the phenotype that results from driving UAS-miR-9a alone using UH3-Gal4 (Figure 1, D and D’).
Discussion
The present study throws light on a new role played by miR-9a during muscle development and function. Previously, Drosophila miR-9a has only been shown to be involved in neuronal differentiation, wing margin patterning, and myotendinous junction formation (Biryukova et al. 2009; Bejarano et al. 2010; Yatsenko and Shcherbata 2014). We have now shown that miR-9a is involved in the translational regulation of TnT levels during sarcomeric assembly.
TnT is a major target of miR-9a during myofibrillogenesis in the IFMs
In general, miRNAs and their targets have been observed to exhibit mutually exclusive expression (Stark et al. 2005). While, miR-9a is strongly expressed in all developmental stages, its expression is reduced in adult flies, including the adult IFMs where its expression is much reduced compared to earlier developmental stages (Sempere et al. 2003). We have confirmed that miR-9a is barely detectable in adult IFMs (Figure S1A). These data suggest that the expression of some miR-9a targets could be required for IFM development and function.
We report here that overexpression of miR-9a causes a hypercontraction phenotype in the IFMs. We identified TnT, a structural component of the thin filament of the sarcomere, as a major target for miR-9a in muscles, and have shown that miR-9a overexpression leads to repression of TnT and that this is sufficient to explain the IFM hypercontraction phenotype. In Drosophila, TnT is encoded by the up gene, is a key component in coordinating the Tn–Tm complex (Fyrberg et al. 1994; Nongthomba et al. 2007), and serves as an anchor for the other components of the complex which comprise TnI, TnC, and Tm (Gordon et al. 2000). Whereas the up1 mutation is characterized by the absence of the adult IFM-specific TnT isoform, TnT-10a, resulting in hypercontraction (Nongthomba et al. 2007), the up101 mutation leads to increased calcium sensitivity and irregular acto-myosin interactions, which also cause hypercontraction, producing damaged muscles (Beall and Fyrberg 1991; Nongthomba et al. 2003, 2007). However, previously there has been no report that epigenetic regulation of TnT can also contribute to the maintenance of stoichiometric balance. To the best of our knowledge, this is the first report on the miRNA-mediated post-translational regulation of TnT.
Clearly, downregulation of TnT by miR-9a phenocopies the mutation in up via the same mechanism of stoichiometric imbalance that drives the mis-regulation of the acto-myosin interaction. Importantly, this demonstrates that not only defects in transcriptional control, but also the derailing of other regulatory processes such as miRNA-mediated control, can result in the same defects. Thus, our finding that miR-9a can alter levels of thin filament components via translational control of TnT demonstrates that miRNAs are not just “regulators of regulators,” but can act as direct regulators in coordinating a complex process such as myofibrillar assembly. We show here that a major structural protein, TnT, can in fact be regulated by miRNA.
miR-9 is required for maintaining protein stoichiometry and may have implications in the etiology of myopathies
Studies on IFM mutants indicate that structural integrity of IFMs is highly dependent on interactions between thin and thick filaments, as well as the ratio of individual myofibrillar contractile components. Any change in gene dosage and corresponding protein stoichiometry in the thin filaments translates into defects in normal myofibrillar assembly leading to hypercontraction (Tansey et al. 1991; Nongthomba et al. 2003, 2007). This explains the myofibrillar defects that result from overexpression of TnT (Marco-Ferreres et al. 2005), Mhc (Cripps et al. 1994), and in most of the heterozygotes carrying mutations in genes encoding structural proteins (Prado et al. 1995; Gajewski and Saul 2010).
The phenomena of both hypercontraction and hypertrophy, although observably different, are both responses to the muscle contraction and the imbalance of structural proteins. In hypercontraction, the muscles show properly arranged sarcomeres during early development, but the subsequent uncontrolled acto-myosin interactions lead to stress and muscle tearing (Nongthomba et al. 2003). Hypertrophy, on the other hand, is characterized by an increase in muscle volume. For instance, myocardial hypertrophy is associated with cardiac remodeling where there is an increase of muscle wall thickness, but not through any increase in myocyte number (hyperplasia). However, cardiac hypertrophy is also a physiological response to stress induced by ischemia, mitochondrial defects, and mutations in sarcomeric components, etc. Importantly, mutations in the same gene orthologs that cause hypercontraction in Drosophila are the ones mutated in cardiac hypertrophy patients as well. For example, mutations in the TnT gene are one of the predominant causes of hypertrophy (Seidman and Seidman 2001; Di Pasquale et al. 2012). Most of these TnT mutations exhibit increased calcium sensitivity and activation of muscle contractility (Harada and Potter 2004; Parvatiyar and Pinto 2015; Gilda et al. 2016), a similar pathogenesis to the hypercontraction produced by TnT mutants in Drosophila (Nongthomba et al. 2003, 2007). Viswanathan et al. (2014) have shown that the up101 mutation generates a muscle abnormality similar to human cardiomyopathy through sensitive calcium regulation in the Drosophila heart.
Vertebrate TnT (TNNT) has a vital role in the anchoring of Tn–Tm to Actin and is also essential for Ca+2-mediated activation and inhibition of acto-myosin activity during muscle contraction (Potter et al. 1995; Schiaffino and Reggiani 1996; Domingo et al. 1998; Perry 1998; Oliveira et al. 2000). Mutations in TNNT in Caenorhabditis elegans result in defects in embryonic body wall muscle contractions and sarcomere organization (Myers et al. 1996). Mutations in the cardiac isoform of TnT (TNNT2) are associated with familial HCM, dilated cardiomyopathy, or arthrogryposis (Thierfelder et al. 1994; Kamisago et al. 2000; Sehnert et al. 2002; Sung et al. 2003). TNNT2 was also found to be upregulated in cardiac hypertrophic or myocardial infarction conditions (Salic and De Windt 2012). However, there are still unanswered questions pertaining to the mechanisms by which cardiac TNNT2 upregulation is brought about during hypertrophy. Hence, the upstream players that regulate the level of cardiac TNNT2 during muscle development and function are very important.
Drosophila miR-9a is identical to human miR-9 and the human TnT (TNNT) has significant homology to Drosophila TnT (Lagos-Quintana et al. 2001). Since miR-9a is capable of regulating TnT levels in Drosophila, it is possible that the human miR-9 may also play a role in regulating TNNT levels. Interestingly, sequence analysis of the human skeletal and cardiac isoforms of TNNT reveals that only the cardiac TNNT possesses the miR-9 binding site (Figure S4C) while the skeletal isoforms lack it. Incidentally, bioinformatic analysis failed to find a miR-9 target site in the mRNA sequence of mouse TNNT. It is important to note that the initial report of miR-9’s role in muscle hypertrophy were from studies on mice (Wang et al. 2010), so miR-9a could be playing varied roles in different organisms. Our study suggests that miR-9a might be involved in specifically regulating the levels of cardiac TnT in humans. It would be interesting to know if the increase in cardiac TNNT that occurs in response to hypertrophic stimulus is mediated by miR-9. Many mutations of the human cardiac TnT give rise to the hypertrophic condition (Di Pasquale et al. 2012). However, cardiac hypertrophy is a genetically and clinically heterogeneous disorder and its etiology in many instances has not been determined (Gilda et al. 2016). The present study provides a plausible candidate in the form of miR-9 to explore in the etiology of idiopathic cardiomyopathies. It would be interesting to determine what represses miR-9 during myofibril assembly. Its continuous expression would be deleterious to myofibril assembly through its repression of the expression of very important structural proteins such as TnT. Taurine-upregulated gene-1 (TUG1) negatively regulates miR-9 in a human cancer cell line (Zhao and Ren 2016). Whether TUG1 or similar protein(s) are involved in muscle hypertrophy/hypercontraction and myofibril assembly requires further investigation.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300232/-/DC1.
Acknowledgments
Our sincere thanks to John Sparrow (University of York), Jim O. Vigoreaux (University of Vermont), and Alberto Ferrus (Cajal Institute, Spain) for providing antibodies. The Bloomington Drosophila Stock Center and the National Centre for Biological Sciences stock center, Bangalore, India kindly provided various fly lines. We extend our gratitude to David Van Vactor (Department of Cell Biology, Harvard Medical School) for the UAS-miR-Sp fly lines; Madhangi and John Sparrow for their critical comments and inputs; and anonymous reviewers for their comments and suggestions, which have led to substantial improvements in the manuscript. We also thank Kranthi at the Centre fro Nano Science and Engineering, Bangalore for help with scanning electron microscopy and Divya, Sima, Meenakshi Sen, and Deepti Bapat at the Indian Institute of Science (IISc) Confocal Facility for their patience and interest. We acknowledge the IISc, the Department of Science and Technology (DST FIST, 2008–2013 reference number SR/FST/LSII-018/2007), the University Grant Commission [UGC-SAP to the Department of Molecular Reproduction, Development and Genetics: reference number F.3-47/2009 (SAP-II)], and the Department of Biotechnology (DBT), Government of India (DBT-IISC Partnership Program for Advanced Research in Biological Sciences and Bioengineering Sanction Order number. DBT/BF/PRIns/2011-12/IISc/28.9.2012) for financial assistance.
Footnotes
Communicating editor: H. Salz
Literature Cited
- Allen E., Howell M. D., 2010. miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21: 798–804. [DOI] [PubMed] [Google Scholar]
- Bartel D. P., Chen C. Z., 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5: 396–400. [DOI] [PubMed] [Google Scholar]
- Beall C. J., Fyrberg E., 1991. Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J. Cell Biol. 114: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F., Smibert P., Lai E. C., 2010. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 338: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukova I., Asmar J., Abdesselem H., Heitzler P., 2009. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev. Biol. 327: 487–496. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J., 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charge S., Rudnicki M. A., 2003. Fusion with the fused: a new role for interleukin-4 in the building of muscle. Cell 113: 422–423. [DOI] [PubMed] [Google Scholar]
- Chawla J., 2011. Stepwise approach to myopathy in systemic disease. Front. Neurol. 2: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lee N. K., Zajac J. D., MacLean H. E., 2008. Generation and analysis of an androgen-responsive myoblast cell line indicates that androgens regulate myotube protein accretion. J. Endocrinol. Invest. 31: 910–918. [DOI] [PubMed] [Google Scholar]
- Cripps R. M., Becker K. D., Mardahl M., Kronert W. A., Hodges D., et al. , 1994. Transformation of Drosophila melanogaster with the wild-type myosin heavy-chain gene: rescue of mutant phenotypes and analysis of defects caused by overexpression. J. Cell Biol. 126: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pasquale G., Urbinati S., Perugini E., Gambetti S., 2012. Interactions between cardiovascular and cerebrovascular disease. Curr. Treat. Options Neurol. 14: 557–593. [DOI] [PubMed] [Google Scholar]
- Domingo A., Gonzalez-Jurado J., Maroto M., Diaz C., Vinos J., et al. , 1998. Troponin-T is a calcium-binding protein in insect muscle: in vivo phosphorylation, muscle-specific isoforms and developmental profile in Drosophila melanogaster. J. Muscle Res. Cell Motil. 19: 393–403. [DOI] [PubMed] [Google Scholar]
- Drummond D. R., Hennessey E. S., Sparrow J. C., 1991. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol. Gen. Genet. 226: 70–80. [DOI] [PubMed] [Google Scholar]
- Eisenberg M. S., Psaty B. M., 2009. Defining and improving survival rates from cardiac arrest in US communities. JAMA 301: 860–862. [DOI] [PubMed] [Google Scholar]
- Enright A. J., John B., Gaul U., Tuschl T., Sander C., et al. , 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erriquez D., Perini G., Ferlini A., 2013. Non-coding RNAs in muscle dystrophies. Int. J. Mol. Sci. 14: 19681–19704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firdaus H., Mohan J., Naz S., Arathi P., Ramesh S. R., et al. , 2015. A cis-regulatory mutation in troponin-I of Drosophila reveals the importance of proper stoichiometry of structural proteins during muscle assembly. Genetics 200: 149–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg C., Parker H., Hutchison B., Fyrberg E., 1994. Drosophila melanogaster genes encoding three troponin-C isoforms and a calmodulin-related protein. Biochem. Genet. 32: 119–135. [DOI] [PubMed] [Google Scholar]
- Gajewski K. K., Saul J. P., 2010. Sudden cardiac death in children and adolescents (excluding sudden infant death syndrome). Ann. Pediatr. Cardiol. 3: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski K. M., Wang J., Schulz R. A., 2006. Calcineurin function is required for myofilament formation and troponin I isoform transition in Drosophila indirect flight muscle. Dev. Biol. 289: 17–29. [DOI] [PubMed] [Google Scholar]
- Gautam R., Vanga S., Madan A., Gayathri N., Nongthomba U., et al. , 2015. Raman spectroscopic studies on screening of myopathies. Anal. Chem. 87: 2187–2194. [DOI] [PubMed] [Google Scholar]
- Gilda J. E., Lai X., Witzmann F. A., Gomes A. V., 2016. Delineation of molecular pathways involved in cardiomyopathies caused by Troponin-T mutations. Mol. Cell. Proteomics 15: 1962–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladka M. M., da Costa Martins P. A., De Windt L. J., 2012. Small changes can make a big difference - microRNA regulation of cardiac hypertrophy. J. Mol. Cell. Cardiol. 52: 74–82. [DOI] [PubMed] [Google Scholar]
- Gordon A. M., Homsher E., Regnier M., 2000. Regulation of contraction in striated muscle. Physiol. Rev. 80: 853–924. [DOI] [PubMed] [Google Scholar]
- Harada K., Potter J. D., 2004. Familial hypertrophic cardiomyopathy mutations from different functional regions of troponin T result in different effects on the pH and Ca2+ sensitivity of cardiac muscle contraction. J. Biol. Chem. 279: 14488–14495. [DOI] [PubMed] [Google Scholar]
- Huang J. C., Frey B. J., Morris Q. D., 2008. Comparing sequence and expression for predicting microRNA targets using GenMiR3. Pac. Symp. Biocomput. 2008: 52–63. [DOI] [PubMed] [Google Scholar]
- Kamisago M., Sharma S. D., DePalma S. R., Solomon S., Sharma P., et al. , 2000. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 343: 1688–1696. [DOI] [PubMed] [Google Scholar]
- Karibe A., Tobacman L. S., Strand J., Butters C., Back N., et al. , 2001. Hypertrophic cardiomyopathy caused by a novel alpha-tropomyosin mutation (V95A) is associated with mild cardiac phenotype, abnormal calcium binding to troponin, abnormal myosin cycling, and poor prognosis. Circulation 103: 65–71. [DOI] [PubMed] [Google Scholar]
- Kontaraki J. E., Marketou M. E., Zacharis E. A., Parthenakis F. I., Vardas P. E., 2014. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: potential markers of target-organ damage. J. Am. Soc. Hypertens. 8: 368–375. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., et al. , 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500. [DOI] [PubMed] [Google Scholar]
- Krichevsky A. M., King K. S., Donahue C. P., Khrapko K., Kosik K. S., 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronert W. A., O’Donnell P. T., Fieck A., Lawn A., Vigoreaux J. O., et al. , 1995. Defects in the Drosophila myosin rod permit sarcomere assembly but cause flight muscle degeneration. J. Mol. Biol. 249: 111–125. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Laing N. G., Nowak K. J., 2005. When contractile proteins go bad: the sarcomere and skeletal muscle disease. Bioessays 27: 809–822. [DOI] [PubMed] [Google Scholar]
- Latronico M. V., Condorelli G., 2011. MicroRNAs in hypertrophy and heart failure. Exp. Biol. Med. (Maywood) 236: 125–131. [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B., 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Madan A., Thimmaiya D., Franco-Cea A., Aiyaz M., Kumar P., et al. , 2017. Transcriptome analysis of IFM-specific actin and myosin nulls in Drosophila melanogaster unravels lesion-specific expression blueprints across muscle mutations. Gene 631: 16–28. [DOI] [PubMed] [Google Scholar]
- Marco-Ferreres R., Arredondo J. J., Fraile B., Cervera M., 2005. Overexpression of troponin T in Drosophila muscles causes a decrease in the levels of thin-filament proteins. Biochem. J. 386: 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. J., 2008. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta 1779: 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., et al. , 2005. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33: D284–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W., 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. D., Goh P. Y., Allen T. S., Bucher E. A., Bogaert T., 1996. Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J. Cell Biol. 132: 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Ramachandra N. B., 1999. A direct screen identifies new flight muscle mutants on the Drosophila second chromosome. Genetics 153: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Cummins M., Clark S., Vigoreaux J. O., Sparrow J. C., 2003. Suppression of muscle hypercontraction by mutations in the myosin heavy chain gene of Drosophila melanogaster. Genetics 164: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Clark S., Cummins M., Ansari M., Stark M., et al. , 2004. Troponin I is required for myofibrillogenesis and sarcomere formation in Drosophila flight muscle. J. Cell Sci. 117: 1795–1805. [DOI] [PubMed] [Google Scholar]
- Nongthomba U., Ansari M., Thimmaiya D., Stark M., Sparrow J., 2007. Aberrant splicing of an alternative exon in the Drosophila troponin-T gene affects flight muscle development. Genetics 177: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. M., Nakaie C. R., Sousa A. D., Farah C. S., Reinach F. C., 2000. Mapping the domain of troponin T responsible for the activation of actomyosin ATPase activity. Identification of residues involved in binding to actin. J. Biol. Chem. 275: 27513–27519. [DOI] [PubMed] [Google Scholar]
- Oliveira-Carvalho V., Carvalho V. O., Silva M. M., Guimaraes G. V., Bocchi E. A., 2012. MicroRNAs: a new paradigm in the treatment and diagnosis of heart failure? Arq. Bras. Cardiol. 98: 362–369. [DOI] [PubMed] [Google Scholar]
- Pantoja M., Fischer K. A., Ieronimakis N., Reyes M., Ruohola-Baker H., 2013. Genetic elevation of sphingosine 1-phosphate suppresses dystrophic muscle phenotypes in Drosophila. Development 140: 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes J. E., Day P. J., Chinoy H., Lamb J. A., 2015. The role of microRNAs in the idiopathic inflammatory myopathies. Curr. Opin. Rheumatol. 27: 608–615. [DOI] [PubMed] [Google Scholar]
- Parvatiyar M. S., Pinto J. R., 2015. Pathogenesis associated with a restrictive cardiomyopathy mutant in cardiac troponin T is due to reduced protein stability and greatly increased myofilament Ca2+ sensitivity. Biochim. Biophys. Acta 1850: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. V., 1998. Troponin T: genetics, properties and function. J. Muscle Res. Cell Motil. 19: 575–602. [DOI] [PubMed] [Google Scholar]
- Potter J. D., Sheng Z., Pan B. S., Zhao J., 1995. A direct regulatory role for troponin T and a dual role for troponin C in the Ca2+ regulation of muscle contraction. J. Biol. Chem. 270: 2557–2562. [DOI] [PubMed] [Google Scholar]
- Prado A., Canal I., Barbas J. A., Molloy J., Ferrus A., 1995. Functional recovery of troponin I in a Drosophila heldup mutant after a second site mutation. Mol. Biol. Cell 6: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronovost S. M., Beckerle M. C., Kadrmas J. L., 2013. Elevated expression of the integrin-associated protein PINCH suppresses the defects of Drosophila melanogaster muscle hypercontraction mutants. PLoS Genet. 9: e1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M., Katti P., Nongthomba U., 2014. Drosophila Erect wing (Ewg) controls mitochondrial fusion during muscle growth and maintenance by regulation of the Opa1-like gene. J. Cell Sci. 127: 191–203. [DOI] [PubMed] [Google Scholar]
- Reedy M. C., Beall C., 1993. Ultrastructure of developing flight muscle in Drosophila. I. Assembly of myofibrils. Dev. Biol. 160: 443–465. [DOI] [PubMed] [Google Scholar]
- Salic K., De Windt L. J., 2012. MicroRNAs as biomarkers for myocardial infarction. Curr. Atheroscler. Rep. 14: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisakian H., 2014. Cardiomyopathies: evolution of pathogenesis concepts and potential for new therapies. World J. Cardiol. 6: 478–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C., 1996. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76: 371–423. [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Schonbauer C., Langer C. C., Dietzl G., Novatchkova M., et al. , 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464: 287–291. [DOI] [PubMed] [Google Scholar]
- Sehnert A. J., Huq A., Weinstein B. M., Walker C., Fishman M., et al. , 2002. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 31: 106–110. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Seidman C., 2001. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104: 557–567. [DOI] [PubMed] [Google Scholar]
- Selcen D., 2011. Myofibrillar myopathies. Neuromuscul. Disord. 21: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Sokol N. S., Dubrovsky E. B., Berger E. M., Ambros V., 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-complex gene activity. Dev. Biol. 259: 9–18. [DOI] [PubMed] [Google Scholar]
- Singh S. H., Kumar P., Ramachandra N. B., Nongthomba U., 2014. Roles of the troponin isoforms during indirect flight muscle development in Drosophila. J. Genet. 93: 379–388. [DOI] [PubMed] [Google Scholar]
- Sparrow J., Hughes S. M., Segalat L., 2008. Other model organisms for sarcomeric muscle diseases. Adv. Exp. Med. Biol. 642: 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R. B., Cohen S. M., 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R. B., Cohen S. M., 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Sung S. S., Brassington A. M., Krakowiak P. A., Carey J. C., Jorde L. B., et al. , 2003. Mutations in TNNT3 cause multiple congenital contractures: a second locus for distal arthrogryposis type 2B. Am. J. Hum. Genet. 73: 212–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey T., Schultz J. R., Miller R. C., Storti R. V., 1991. Small differences in Drosophila tropomyosin expression have significant effects on muscle function. Mol. Cell. Biol. 11: 6337–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. V., 2006. Comparison of muscle development in Drosophila and vertebrates, pp. 169–203 in Muscle Development in Drosophila, edited by Sink H. Landes Bioscience, Georgetown, TX. [Google Scholar]
- Thierfelder L., Watkins H., MacRae C., Lamas R., McKenna W., et al. , 1994. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 77: 701–712. [DOI] [PubMed] [Google Scholar]
- Varallyay E., Burgyan J., Havelda Z., 2008. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 3: 190–196. [DOI] [PubMed] [Google Scholar]
- Vigoreaux J. O., 2001. Genetics of the Drosophila flight muscle myofibril: a window into the biology of complex systems. Bioessays 23: 1047–1063. [DOI] [PubMed] [Google Scholar]
- Viswanathan M. C., Kaushik G., Engler A. J., Lehman W., Cammarato A., 2014. A Drosophila melanogaster model of diastolic dysfunction and cardiomyopathy based on impaired troponin-T function. Circ. Res. 114: e6–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Long B., Zhou J., Li P. F., 2010. miR-9 and NFATc3 regulate myocardin in cardiac hypertrophy. J. Biol. Chem. 285: 11903–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins H., Seidman J. G., Seidman C. E., 1995. Familial hypertrophic cardiomyopathy: a genetic model of cardiac hypertrophy. Hum. Mol. Genet. 4: 1721–1727. [DOI] [PubMed] [Google Scholar]
- Watkins H., Seidman C. E., Seidman J. G., Feng H. S., Sweeney H. L., 1996. Expression and functional assessment of a truncated cardiac troponin T that causes hypertrophic cardiomyopathy. Evidence for a dominant negative action. J. Clin. Invest. 98: 2456–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko A. S., Shcherbata H. R., 2014. Drosophila miR-9a targets the ECM receptor Dystroglycan to canalize myotendinous junction formation. Dev. Cell 28: 335–348. [DOI] [PubMed] [Google Scholar]
- Yuva-Aydemir Y., Simkin A., Gascon E., Gao F. B., 2011. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 8: 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chintalgattu V., Shih T., Ai D., Xia Y., et al. , 2011. MicroRNA-9 is an activation-induced regulator of PDGFR-beta expression in cardiomyocytes. J. Mol. Cell. Cardiol. 51: 337–346. [DOI] [PubMed] [Google Scholar]
- Zhao X., Ren G., 2016. LncRNA taurine-upregulated gene 1 promotes cell proliferation by inhibiting microRNA-9 in MCF-7 cells. J. Breast Cancer 19: 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have provided the details of all the web addresses of data resources that we made use of in this study. Other data that support our findings have been included as Supplemental Material and are described in the Results section. Supplemental Figure legends are available in File S1.