Abstract
Lymphocytes are immune cells that are critical for the maintenance of adaptive immunity. Differentiation of lymphoid progenitors yields B-, T-, and NK-cell subtypes that individually correlate with specific forms of leukemia or lymphoma. Therefore, it is imperative a precise method of cell categorization is utilized to detect differences in distinct disease states present in patients. One viable means of classification involves evaluation of the cell surface proteome of lymphoid malignancies. Specifically, this manuscript details the use of an antibody independent approach known as Cell Surface Capture Technology, to assess N-glycoproteome of four human lymphocyte cell lines. Altogether, 404 cell surface N-glycoproteins as markers for specific cell types involved in lymphocytic malignancies, including 82 N-glycoproteins that had not been previously been described for B- or T-cells within the Cell Surface Protein Atlas. Comparative analysis, hierarchical clustering techniques, and label free quantitation was used to reveal proteins most informative for each cell type. Undoubtedly, the characterization of the cell surface proteome of lymphoid malignancies is a first step towards improving personalized diagnosis and treatment of leukemia and lymphoma.
Keywords: N-glycoproteins, Cell surface proteins, Plasma membrane
Leukemia and non-Hodgkin lymphoma are two of the most deadly cancers in the United States [1]. The classification of these hematopoietic neoplasms requires numerous considerations related to; clinical features, cellular morphology, immunophenotype and genetic markers present in the malignant cells. [2]. Overall, the phenotypic and genetic diversity of these diseases manifests as variations in their aggressiveness and treatability. Improved strategies to classify and sub-classify distinct hematopoietic malignancies has positively impacted patient survival, and in some cases this relies on the measurement of informative cell surface markers (e.g. immunoglobulin mutational status and CD38 expression in chronic lymphocytic leukemia [3]). Cell surface proteins have long been useful in lymphoid cell typing and classification of distinct cell subtypes (e.g. progenitor vs. immature B-cell vs. memory B-cell [4–6]). Further, localization to the cell surface is a common property for many proteins that are established drug targets [7]. Consequently, efforts to more comprehensively define the cell surface proteome of healthy and diseased lymphoid cells will most certainly contribute to a better understanding of disease biomarkers, which can subsequently improve diagnosis and treatment of lymphoid malignancies.
In this study, the cell surface N-glycoproteome of four human malignant lymphocyte cell lines was characterized using an antibody-independent strategy known as Cell Surface Capture Technology (CSC-Technology) [8]. This technique uses affinity enrichment and mass spectrometry to identify cell surface N-glycoproteins while simultaneously determining N-glycosite usage and membrane topology. The characterization of cell surface N-glycoproteins is invaluable as these proteins constitute an estimated 90% of the cell surface proteome [9]. This workflow can be summarized as: 1) extracellular oligosaccharides on viable cells are biotinylated using membrane-impermeable reagents, 2) glycoproteins are digested, 3) biotinylated glycopeptides are captured using immobilized streptavidin, and 4) peptide-N-glycosidase F (PNGase F) is used to cleave the oligosaccharide from the peptide backbone to release the formerly N-glycosylated peptides. During this process, the mass of the asparagine residue at the site of N-glycosylation is modified (from asparagine to aspartic acid), generating a convenient “mass tag” that is used to filter the final dataset for peptides of interest (i.e. peptides must contain deamidated asparagine within the original sequence motif NxS/T/C). We and others have previously applied CSC-Technology to a wide range of mouse and human cell lines and primary cells [10–18] and many of these datasets are part of the Cell Surface Protein Atlas (CSPA [19]). The CSPA is a public resource containing 1492 experimentally determined human cell surface N-glycoproteins and new candidates for cell type specific makers [10, 11]. Here, we offer an additional set of lymphocyte cell surface proteins to complement those already included in the CSPA [20–23]. These data were then used to classify N-glycoprotein surfaceome proteins according to their ability to distinguish among the cell types investigated in this study. Such data are useful for comparative purposes with other CSC-Technology data and can identify candidates as new immunophenotyping and drug targets [10, 11, 24].
The human lymphocyte cell lines used here included cell types representative of Burkitt’s lymphoma (Ramos [25]), chronic lymphocytic leukemia (HG-3 [26]), B-cell acute lymphoblastic leukemia (RCH-ACV [27]), and T-cell acute lymphoblastic leukemia (Jurkat [28]). Cells were cultured and passaged following manufacturer protocols specific for each cell line. Cells were inspected visually prior to passaging for bacterial contamination, tested quarterly for mycoplasma contamination, and authenticated by short tandem repeat analysis. For this application of CSC-Technology, biological replicates were defined as separate cultures of a cell line that had undergone >5 passages since the split (~2–3 weeks of aseptic, separate cultures). CSC-Technology [8] was performed as previously described [11, 13] and highlighted above. Following PNGase F treatment, the formerly N-glycosylated peptides were analyzed using a Dionex UltiMate 3000 RPLCnano system (Thermo, Waltham, MA) in line with a Velos Orbitrap mass spectrometer (Thermo). MS1 and MS2 data were acquired in the Orbitrap. Data were analyzed using ProteomeDiscoverer 2.1 (Thermo) with all three technical replicates for each biological replicate searched in unison. Data were searched against the human UniProt database using Sequest HT, MS Amanda, Mascot search algorithms. The exported peptide lists were then manually reviewed and proteins lacking at least one peptide with a deamidated asparagine within the N-linked glycosylation consensus sequence were disqualified. Peptide data were exported in tab-delimitated format for further statistical and informatics analyses. Methods for cell culture, CSC-Technology, mass spectrometry, data processing, and bioinformatics are described in detail in the online supporting information.
In total, 404 cell surface N-glycoproteins were identified for the four human lymphocyte cell lines, including 108 cluster of differentiation (CD) antigens (Supporting Information Tables 1 and 2), known lymphocyte cell surface markers (e.g. CD5, CD30), and 82 proteins not previously characterized for the 16 lymphocyte cell types that are included in the CSPA (e.g. CD1A, CD9, HLA-A, ROBO2). As expected for datasets enriched for cell surface proteins, 10% of identified proteins were annotated as transporters and 19% were annotated as receptors. Additionally, gene ontology (GO) analysis revealed that 16% of identified proteins were associated with immune function and antigen processing, cellular defense response, and B cell mediated immunity were overrepresented in the dataset (Supporting Information Fig. 1). Overall, CSC-Technology reliably identified many cell surface N-glycoproteins expected to be present on the surface of these lymphocyte lines.
Previous studies have shown that CSC-Technology data can be informative for the selection of markers that can be utilized in discriminating cell types [10, 11, 15]. Here, we applied bioinformatic clustering approaches to identify cell surface proteins capable of discriminating B- and T-cell lines (see Supporting Information for details). First, two-step clustering was applied using the protein expression data obtained from the 12 samples (three biological replicates for each of the four cell lines). As expected, the optimal number of fixed clusters determined by two-way clustering was four, matching the number of cell lines analyzed, and the biological replicates formed a unique cluster for each cell line (Fig. 1A). Hierarchical clustering analysis corroborated these results; each cell type grouped into a distinct cluster (Fig. 1B). Extending our hierarchical clustering approach CSPA lymphocyte data corroborated the capacity of CSC-Technology data to classify and distinguish among cell types and revealed the consistency of datasets among different laboratories (e.g. Jurkat; Supporting Information Fig. 2).
Figure 1.
(A) Results of the two-step cluster analysis demonstrating a favorable cluster ratio (<3) and silhouette measure (>0.5) at four fixed clusters. (B) Dendrogram of the hierarchical clustering results (BL: Burkitt’s lymphoma, B-ALL: B cell Acute lymphocytic leukemia, CLL: chronic lympocytic leukemia, T-ALL: T cell Acute lymphocytic leukemia. (C) Graphical distribution of the 247 proteins observed in at least two biological replicates of a cell line (generated using Cytoscape [33]). Color indicates whether the protein is a CD antigen and size is proportional to number of drugs developed against it, as cataloged in ChEMBL, PharmGKB, and Therapeutic Targets Database [34–36].
In a complementary approach to clustering, candidate proteins potentially informative of cell identity were prioritized and filtered based on their detection across each cell line. This analysis was filtered to include the 247 N-glycoproteins identified in at least two of three biological replicates for at least one cell line. Visualization of the expression patterns for these proteins across the examined cell lines revealed numerous non-CD antigens that are informative for these cell types (Fig. 1C), which suggests that future analyses will benefit from technologies that extend beyond CD antibody panels. Moreover, the vast majority of identified cell surface proteins are not known targets for available drugs (Fig. 1C). Altogether, these findings reveal that there is a treasure trove of potentially “druggable” cell surface proteins that can be used to improve personalized therapeutics to combat specific lymphoid malignancies.
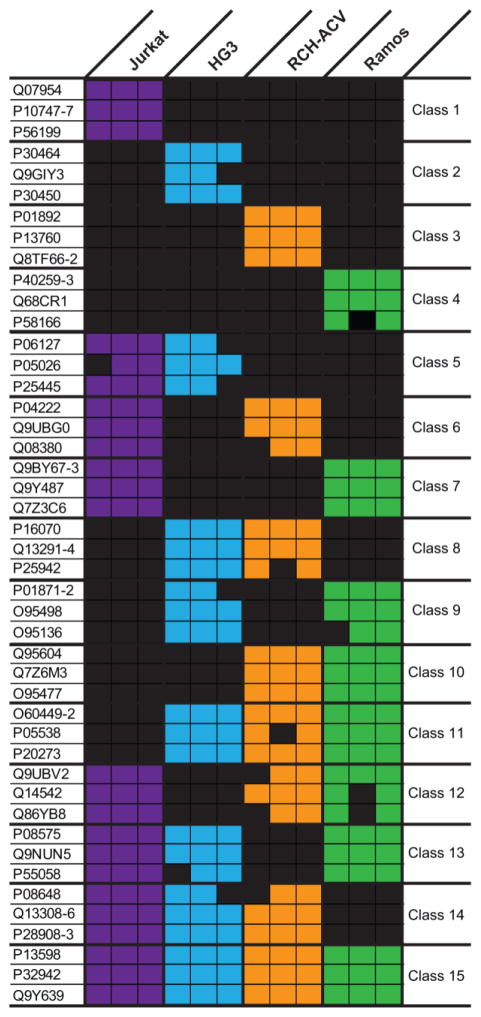
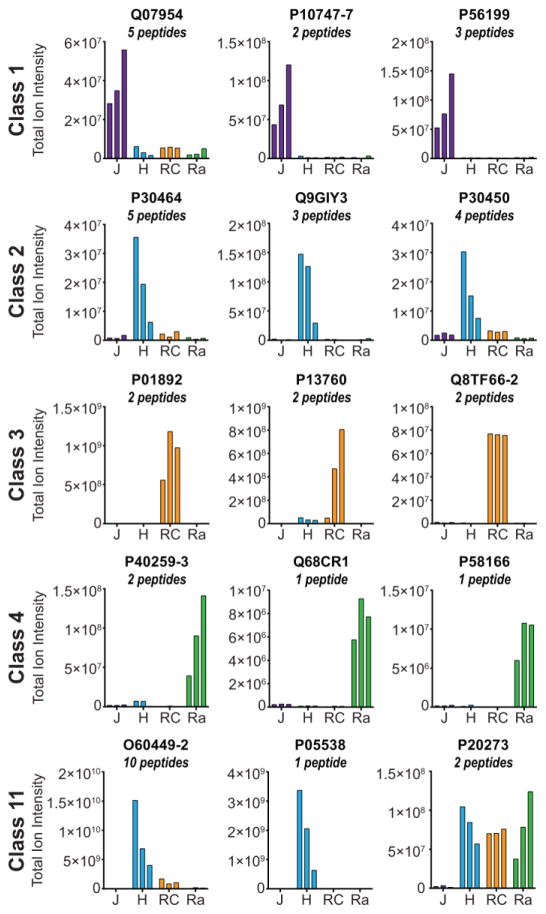
Next, we aimed to determine the most promising candidates that could be used to discriminate each of the lymphocyte cell lines investigated here. For this analysis, the cell surface proteins were binned into 15 distinct classes based on the cell types in which they were identified and the three proteins identified by the highest number of peptide spectrum matches were selected as candidates for cell-type specific markers (Fig. 2 and Supporting Information Fig. 3). These proteins averaged >14 spectral matches per biological replicate To discriminate between missing data and the true absence of a cell surface protein, MS1 peak areas for peptides representing a subset of these proteins of interest were determined using Skyline [29]. Briefly, missing data could represent a lack of peptide identification despite the presence of the peptide precursor; for example, a low abundance precursor may not be selected for fragmentation because the precursor ion did not meet the intensity threshold needed for data-dependent MS2 acquisition as dictated by the top-10 MS/MS method. Using Skyline, it is possible to manually verify the presence of a precursor ion at the MS1 level and thus distinguish between missing data and a true absence. Despite some discrepancies for proteins identified by a single peptide (e.g. P05538), protein abundance was consistent with the binary data (observed vs. not observed) when the abundance values for multiple peptides mapping to the same cell surface protein were combined (Fig. 3).
Figure 2.
Classification of 45 cell surface proteins based on their observation among the four cell lines. For each of the 15 protein classes, the three proteins observed in at least two biological replicates with the highest number of peptide spectrum matches are listed. The presence of colored box in a particular biological replicate of a cell line indicates the protein was observed where each column corresponds to a single biological replicate.
Figure 3.
Relative quantitation of three proteins from Classes 1–4 and 11 as defined in Fig. 2. The total MS1 ion intensity for all peptides were summed for each protein and displayed as bars. J: Jurkat, H: HG3, RC: RCH-ACV, and Ra: Ramos.
In summary, these data provide a snapshot of the N-glycoprotein surfaceome of four cell lines derived from human lymphocytic malignancies. While CSC-Technology is valuable for discovering proteins localized to the cell surface, notable limitations include detection of only N-glycoproteins, that detection can be affected by alterations in N-glycosylation status, quantitation can be challenging if relying on a single peptide for a particular protein, and that unique peptides identified for a single protein do not always follow the same quantitative trend. Despite such considerations, these data inform the future development of markers for immunophenotyping and even immunotherapeutics for treating lymphocyte malignancies, especially when combined with other data in the Cell Surface Protein Atlas. These data provide a first step in the iterative strategy for the development of novel cell surface marker “barcodes” [10, 24]. This proposed “barcoding” would first use CSC-Technology to discover cell surface proteins and would be followed by targeted quantitation to rapidly quantify dozens of targets among a broader range of cell types, including primary cells, which are often precluded from the discovery phase of CSC-Technology due to limited supply. Although differences between immortalized cell lines and primary cells are expected, this strategy has the advantage of proceeding without antibody development. As CSC-Technology and similar approaches [30–32] are applied to additional cell types, the growing resource of experimentally identified cell surface proteins is expected to yield new markers. These markers, once validated in human primary cells, will improve our ability to classify and sub-classify distinct cell types, map their stage of differentiation, and aid in the discrimination of related malignancies.
Supplementary Material
Acknowledgments
The authors would like to thank Alexandra J. VanNispen at Northwestern University for her critical review and feedback. This research was supported by the Paul G. Allen Family Foundation (Grant Award 11715) to NLK and RLG, and NIH grants HL126785 and HL134010 to RLG.
Abbreviations
- CSC-Technology
Cell Surface Capture Technology
- CSPA
Cell surface protein atlas
- CD
Cluster of differentiation
Footnotes
Conflict of Interest: The authors have declared no conflict of interest.
References
- 1.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute, Atlanta 2016.
- 2.Arber DA, Orazi A, Hasserjian R, Thiele J, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 3.Damle RN, Wasil T, Fais F, Ghiotto F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 4.Galy A, Travis M, Cen D, Chen B. Human T, B natural killer and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 5.Maurer D, Fischer GF, Fae I, Majdic O, et al. IgM and IgG but not cytokine secretion is restricted to the CD27+ B lymphocyte subset. J Immunol. 1992;148:3700–3705. [PubMed] [Google Scholar]
- 6.van Lochem EG, van der Velden VH, Wind HK, te Marvelde JG, et al. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin Cytom. 2004;60:1–13. doi: 10.1002/cyto.b.20008. [DOI] [PubMed] [Google Scholar]
- 7.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 8.Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nature biotechnology. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 10.Mallanna SK, Cayo MA, Twaroski K, Gundry RL, Duncan SA. Mapping the Cell-Surface N-Glycoproteome of Human Hepatocytes Reveals Markers for Selecting a Homogeneous Population of iPSC-Derived Hepatocytes. Stem Cell Reports. 2016;7:543–556. doi: 10.1016/j.stemcr.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boheler KR, Bhattacharya S, Kropp EM, Chuppa S, et al. A human pluripotent stem cell surface N-glycoproteome resource reveals markers, extracellular epitopes, and drug targets. Stem Cell Reports. 2014;3:185–203. doi: 10.1016/j.stemcr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gundry RL, Raginski K, Tarasova Y, Tchernyshyov I, et al. The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Molecular & cellular proteomics : MCP. 2009;8:2555–2569. doi: 10.1074/mcp.M900195-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundry RL, Riordon DR, Tarasova Y, Chuppa S, et al. A cell surfaceome map for immunophenotyping and sorting pluripotent stem cells. Molecular & cellular proteomics : MCP. 2012;11:303–316. doi: 10.1074/mcp.M112.018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kropp EM, Bhattacharya S, Waas M, Chuppa SL, et al. N-glycoprotein surfaceomes of four developmentally distinct mouse cell types. Proteomics Clinical applications. 2014;8:603–609. doi: 10.1002/prca.201400021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallanna SK, Waas M, Duncan SA, Gundry RL. N-glycoprotein surfaceome of human induced pluripotent stem cell derived hepatic endoderm. Proteomics. 2016 doi: 10.1002/pmic.201600397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducret A, Kux van Geijtenbeek S, Roder D, Simon S, et al. Identification of six cell surface proteins for specific liver targeting. Proteomics Clinical applications. 2015;9:651–661. doi: 10.1002/prca.201400194. [DOI] [PubMed] [Google Scholar]
- 17.DeVeale B, Bausch-Fluck D, Seaberg R, Runciman S, et al. Surfaceome profiling reveals regulators of neural stem cell function. Stem Cells. 2014;32:258–268. doi: 10.1002/stem.1550. [DOI] [PubMed] [Google Scholar]
- 18.Danzer C, Eckhardt K, Schmidt A, Fankhauser N, et al. Comprehensive description of the N-glycoproteome of mouse pancreatic beta-cells and human islets. J Proteome Res. 2012;11:1598–1608. doi: 10.1021/pr2007895. [DOI] [PubMed] [Google Scholar]
- 19.Bausch-Fluck D, Hofmann A, Bock T, Frei AP, et al. A mass spectrometric-derived cell surface protein atlas. PloS one. 2015;10:e0121314. doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirkowska P, Hofmann A, Sedek L, Slamova L, et al. Leukemia surfaceome analysis reveals new disease-associated features. Blood. 2013;121:e149–159. doi: 10.1182/blood-2012-11-468702. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann A, Thiesler T, Gerrits B, Behnke S, et al. Surfaceome of classical Hodgkin and non-Hodgkin lymphoma. Proteomics Clinical applications. 2015;9:661–670. doi: 10.1002/prca.201400146. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann A, Gerrits B, Schmidt A, Bock T, et al. Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood. 2010;116:e26–34. doi: 10.1182/blood-2010-02-271270. [DOI] [PubMed] [Google Scholar]
- 23.Bock T, Bausch-Fluck D, Hofmann A, Wollscheid B. CD proteome and beyond - technologies for targeting the immune cell surfaceome. Front Biosci (Landmark Ed) 2012;17:1599–1612. doi: 10.2741/4006. [DOI] [PubMed] [Google Scholar]
- 24.Boheler KR, Gundry RL. Concise Review. Cell Surface N-Linked Glycoproteins as Potential Stem Cell Markers and Drug Targets. Stem Cells Transl Med. 2016 doi: 10.5966/sctm.2016-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein G, Giovanella B, Westman A, Stehlin JS, Mumford D. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5:319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 26.Rosen A, Bergh AC, Gogok P, Evaldsson C, et al. Lymphoblastoid cell line with B1 cell characteristics established from a chronic lymphocytic leukemia clone by in vitro EBV infection. Oncoimmunology. 2012;1:18–27. doi: 10.4161/onci.1.1.18400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack I, Seshadri R, Garson M, Michael P, et al. RCH-ACV: a lymphoblastic leukemia cell line with chromosome translocation 1;19 and trisomy 8. Cancer Genet Cytogenet. 1986;19:261–269. doi: 10.1016/0165-4608(86)90055-5. [DOI] [PubMed] [Google Scholar]
- 28.Schneider U, Schwenk HU, Bornkamm G. Characterization of EBV-genome negative "null" and "T" cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer. 1977;19:621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- 29.Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Molecular & cellular proteomics : MCP. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rugg-Gunn PJ, Cox BJ, Lanner F, Sharma P, et al. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Hoof D, Dormeyer W, Braam SR, Passier R, et al. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res. 2010;9:1610–1618. doi: 10.1021/pr901138a. [DOI] [PubMed] [Google Scholar]
- 32.Parker BL, Palmisano G, Edwards AV, White MY, et al. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Molecular & cellular proteomics : MCP. 2011;10:M110006833. doi: 10.1074/mcp.M110.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P, Markiel A, Ozier O, Baliga NS, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bento AP, Gaulton A, Hersey A, Bellis LJ, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Ji ZL, Chen YZ, TTD Therapeutic Target Database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.