Abstract
Animal mitochondrial DNA (mtDNA) is commonly described as a small, circular molecule that is conserved in size, gene content, and organization. Data collected in the last decade have challenged this view by revealing considerable diversity in animal mitochondrial genome organization. Much of this diversity has been found in nonbilaterian animals (phyla Cnidaria, Ctenophora, Placozoa, and Porifera), which, from a phylogenetic perspective, form the main branches of the animal tree along with Bilateria. Within these groups, mt-genomes are characterized by varying numbers of both linear and circular chromosomes, extra genes (e.g. atp9, polB, tatC), large variation in the number of encoded mitochondrial transfer RNAs (tRNAs) (0–25), at least seven different genetic codes, presence/absence of introns, tRNA and mRNA editing, fragmented ribosomal RNA genes, translational frameshifting, highly variable substitution rates, and a large range of genome sizes. This newly discovered diversity allows a better understanding of the evolutionary plasticity and conservation of animal mtDNA and provides insights into the molecular and evolutionary mechanisms shaping mitochondrial genomes.
Keywords: mitochondrial DNA, Metazoa, Porifera, Cnidaria, Ctenophora, Placozoa
Introduction
Mitochondria are double membrane-bound organelles found in most eukaryotic cells. Commonly described as cellular “power plants” for their role in the generation of ATP by oxidative phosphorylation, mitochondria perform a variety of additional cellular functions, such as biosynthesis of amino acids and steroids, β-oxidation of fatty acids, FeS metabolism, and initiation of apoptosis (Mcbride et al. 2006; Pagliarini and Rutter 2013; Chandel 2014; Birsoy et al. 2015). However, oxidative phosphorylation appears to be the only function that requires the presence of a mitochondrial genome (mt-genome) (Race et al. 1999), which always codes for some protein subunits of the electron transport chain (Feagin 1992). In addition, nearly all mt-genomes encode ribosomal RNA (rRNA) components of mitochondrial ribosomes, most encode at least some mitochondrial transfer RNA (mt-tRNA), while some encode the ribosomal subunit of RNaseP and proteins involved in mitochondrial transcription, translation, protein import and maturation (Gray et al. 2004).
Extensive biochemical and phylogenetic studies have demonstrated that all mitochondria originated from a single α-proteobacterial ancestor (Gray et al. 1999) from within the order Rickettsiales (Wang and Wu 2015). Given that the modern representatives of Rickettsiales have small circular genomes (Schulz et al. 2015), such a genome was likely present in the bacterial ancestor of mitochondria. During eukaryotic evolution, this ancestral genome lost most of its genes [retaining as few as 4 (Flegontov et al. 2015) or perhaps even 1 (Petersen et al. 2014) and at most 100 (Burger et al. 2013) genes] and gave rise to a remarkable diversity of architectures, including genomes with varying numbers of linear and circular chromosomes ranging in size from only a few kilobases (kb) to several megabases (Mb) (Gray et al. 2004; Gualberto et al. 2014; Smith and Keeling 2015). In a few lineages, mitochondrial DNA (mtDNA) has been lost altogether (Muller et al. 2012), and in one case mitochondria themselves have been completely lost (Karnkowska et al. 2016). A sample of mtDNA diversity can be seen among close unicellular relatives of animals (defined here as protists that are more closely related to animals than fungi), where both linear and multipartite genomes have evolved (Lavrov and Lang 2014). In contrast, previous reviews of animal mtDNA have emphasized its remarkable uniformity (Wolstenholme 1992a; Boore 1999; Saccone et al. 2002) but have been focused primarily on bilaterian animals.
Progress in molecular phylogenetics has brought renewed attention to four major groups of nonbilaterian animals (sponges, placozoans, cnidarians, and ctenophores), which have been traditionally categorized as phyla, but which, along with bilaterians, constitute the main branches of the animal tree from a phylogenetic perspective (Ax 1996) (fig. 1). Each of these lineages encompasses hundreds of millions of years of independent evolution (Dos Reis et al. 2015), and their extant representatives demonstrate distinct possibilities in animal mtDNA evolution. Although some aspects of mtDNA diversity in nonbilaterian animals have been reviewed recently by Breton et al. (2014), Bernt et al. (2013), and Osigus et al. (2013), these studies were published before data from all major nonbilaterian lineages became available. In this review, we put conventional views of animal mtDNA in perspective by providing a comprehensive summary of mt-genome organization in nonbilaterian animals.
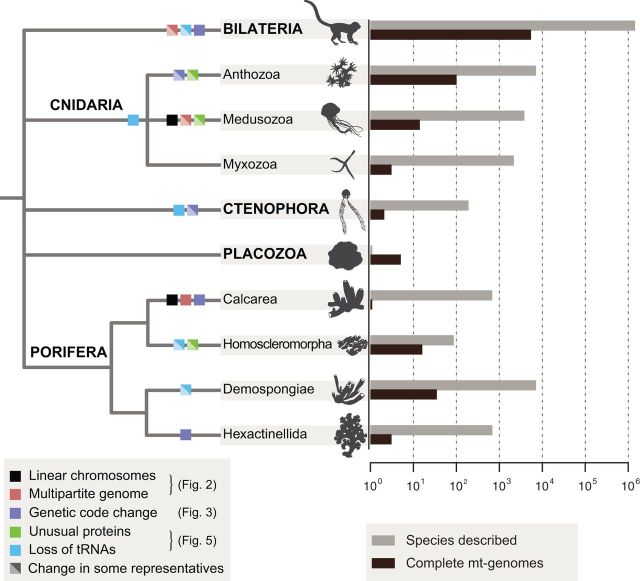
Fig. 1.—
Mitochondrial genomes in Metazoa. Consensus view of animal relationships with mapped changes in mt-genome architecture and gene content. Diagonally split boxes indicate changes that occur only in some representatives of the group. Black bars to the right indicate numbers of complete mtDNA sequences in the NCBI RefSeq database as of April 9, 2016 (for data on nuclear genomes, see [Dunn and Ryan 2015]). Gray bars indicate estimated numbers of described species and were obtained from the Global Invertebrate Genomics Alliance (GIGA Community of Scientists 2014). Only the taxa that are mentioned in the text are included. See associated figures for a more detailed depiction of each of these changes. Branch lengths were chosen for illustration purposes only.
Typical Animal Mitochondrial DNA: The Birth of a Myth
The first determined animal mtDNA sequence came from human (Anderson et al. 1981) and that from Drosophila followed soon after (Clary and Wolstenholme 1984). Though the gene order was different between mammalian and Drosophila mtDNA, resulting in distinct patterns of transcription (Berthier et al. 1986), most of the genetic and genomic features were remarkably similar. Both mt-genomes were small (∼16 kb), circular-mapping molecules, containing the same set of 37 genes (13 protein genes encoding subunits of complexes I, III, IV, and V of the oxidative phosphorylation (OXPHOS) system, 2 rRNA genes, and 22 tRNA genes) with a high degree of synteny. Both genomes encompassed a single large noncoding region, but otherwise contained very few intergenic nucleotides. A modified genetic code was inferred in both species, with AUA and UGA codons specifying methionine and tryptophan, respectively, and AGR codons specifying either serine (in Drosophila) or termination (in human). Other unusual genetic and genomic features of bilaterian mtDNA included unorthodox start codons, incomplete stop codons, diminished genes for tRNAs and rRNAs [including a characteristic trnS(gcu) that codes for a tRNA with a DHU-arm replacement loop], the absence of introns, and a high rate of sequence evolution (Clary and Wolstenholme 1984; Wolstenholme 1992a).
Given the more than one billion years of combined evolution that separates humans and Drosophila (Dos Reis et al. 2015) and their radically different external morphology, the similarity in their mtDNA was astounding. In addition, earlier electron microscopy studies had shown that the sizes of mtDNA in a variety of animal species were similar (Borst and Kroon 1969). Thus, the notion of “typical animal mtDNA” was born, with the assertion that “all metazoa, from Platyhelminthes to mammals possess a mt-genome that consists of a single circular molecule ranging in size from 14.5 to 19.5 kb” and that both gene content and, to some extent, gene order were conserved in all animal mt-genomes (Clary and Wolstenholme 1985). The notion of a conserved, nearly “frozen” (Saccone et al. 2002) genome was reinforced by the observation that most of the mutations in the human mt-genome are associated with disease (Linnane et al. 1989; Wallace 1992).
To date, complete mt-genome sequences from over 6,000 animals have been made available, of which about 97% are from bilaterian animals (http://www.ncbi.nlm.nih.gov/genome/organelle/, last accessed April 9, 2016). Although the majority of bilaterian mt-genomes do fit the description presented above, several noticeable exceptions have been found. First, unusual mtDNA organizations have been discovered in several lineages, with multipartite mt-genomes found in the mesozoan genus Dicyema (Watanabe et al. 1999) (>3 circles), the rotifer genus Brachionus (Suga et al. 2008; Hwang et al. 2014) (2 circles), the nematode genus Globodera (Armstrong et al. 2000; Gibson et al. 2007) (6 circles), the booklice genus Liposcelis (2 circles) (Wei et al. 2012), the thrips species complex Scirtothrips dorsalis (Dickey et al. 2015) (2 circles), and several independent cases in parasitic lice (up to 20 minichromosomes) (Cameron et al. 2011; Jiang et al. 2013; Dong et al. 2014) (fig. 2). In addition, a transient linear genome has been reported in the isopod Armadillidium vulgare (Marcadé et al. 2007). Second, the size of bilaterian mtDNA was shown to be more variable than originally thought, ranging from just over 11 kb in chaetognaths (Helfenbein et al. 2004; Papillon et al. 2004; Faure and Casanova 2006; Miyamoto et al. 2010) to over 50 kb in the ark shell Scapharca broughtonii (Liu et al. 2013). Third, several changes in gene content have been reported, including losses of atp8 in most nematodes (Okimoto et al. 1992; Sultana et al. 2013; but see Lavrov and Brown 2001) and flatworms (Le et al. 2000; Von Nickisch-Rosenegk et al. 2001; Sola et al. 2015), the loss of atp6, atp8 and most tRNA genes in Chaetognatha (Helfenbein et al. 2004; Papillon et al. 2004; but see Barthelemy and Seligmann 2016), and the gain of a putative novel gene in bivalve molluscs with doubly uniparental inheritance (Breton et al. 2011; Zouros 2013). Furthermore, it has been proposed that additional proteins may be encoded in most if not all animal mt-genomes as (alternative) open-reading frames (ORFs) within standard mitochondrial genes (Guo et al. 2003; Faure et al. 2011; Capt et al. 2015). Fourth, additional genetic codes have been inferred in several groups as shown in figure 3 (Watanabe and Yokobori 2011; Abascal et al. 2012). Fifth, gene order has been found to be much more fluid in some groups (e.g. most tunicates) (Singh et al. 2009; Gissi et al. 2010; Rubinstein et al. 2013) than others. Finally, a highly unusual system was found in bivalve molluscs, which inherit not one but two mt-genomes: one transmitted maternally to all offspring and the second exclusively from father to sons (Zouros et al. 1992; reviewed by Doucet-Beaupré et al. 2010).
Fig. 2.—
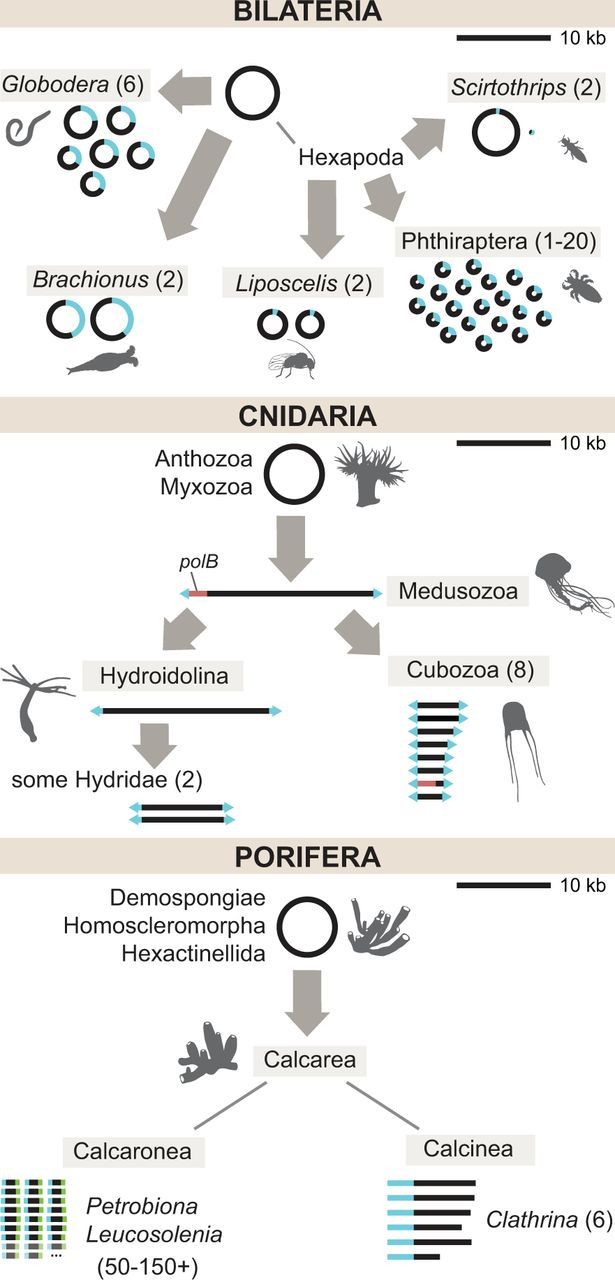
Evolution of linear and multipartite mitochondrial DNA in animals. Gray arrows indicate independent reorganization events in the common ancestor of the indicated group. Blue segments indicate regions of homology shared across chromosomes. Numbers in parentheses indicate the range of chromosome numbers (greater than 1) found in the group. Top: Multipartite mtDNA in Bilateria. Not shown: An unknown number of mini-circle chromosomes in Dicyema (Watanabe et al. 1999). Middle: Linear and multipartite mtDNA in Cnidaria. The red segment denotes the presence of a polB coding sequence in some Medusozoa. Arrowheads at the ends of linear chromosomes signify inverted terminal repeat sequences. Bottom: Multipartite linear mtDNA in Porifera. Faded chromosomes and ellipses indicate the inferred presence of an uncertain number of additional chromosomes. Blue and green segments in Leucosolenia and Petrobiona show distinct end sequences shared across chromosomes, but which are not homologous between the two genera. Genomes are drawn to scale with respect to the 10 kb scale bar in the upper right part of each panel.
Fig. 3.—
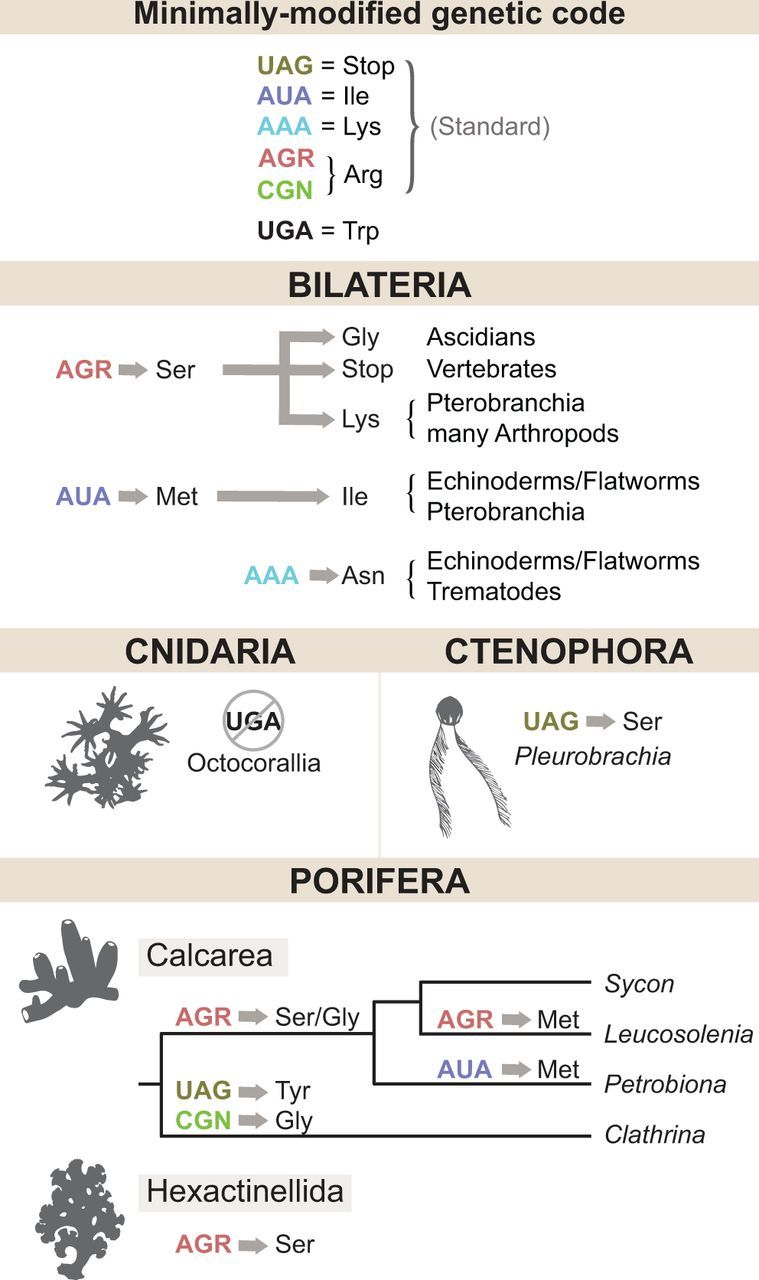
Changes in the animal mitochondrial genetic code. Inferred changes in the identities of six codons (listed in the top panel) are shown by arrows. Consecutive changes are shown by a sequence of arrows. Amino-acid identities are designated by the IUPAC three-letter code. A crossed out circle indicates a codon that is no longer used.
With the notable exceptions of those in Chaetognatha and Tunicata, most of the unusual examples of bilaterian mt-genomes have limited taxonomic distribution and have been commonly interpreted as somewhat curious exceptions to the otherwise valid description of “typical animal mtDNA” that has been propagated widely throughout the literature. However, the absence in Cnidaria of almost all of the genetic novelties usually associated with animal mtDNA had already been described by the early 1990s (Wolstenholme 1992b; Beagley et al. 1995). Data from other nonbilaterian phyla (Porifera, Ctenophora, and Placozoa) reinforce the notion that bilaterian mtDNA is not representative of all animals.
Classification and Phylogeny of Nonbilaterian Animals
Nonbilaterian animals display less morphological and species diversity than bilaterian animals and have been traditionally subdivided into four phyla: Porifera, Cnidaria, Ctenophora, and Placozoa, with about 8,500, 11,000, 150, and 1 described species, respectively (Zhang 2011; WoRMS Editorial Board 2015). In addition, Myxozoa, a large (∼1,300 species) group of oligo-cellular parasites of fish and annelids with previously unknown phylogenetic affinity, is now placed inside Cnidaria (Nesnidal et al. 2013; Feng et al. 2014; Foox and Siddall 2015; Takeuchi et al. 2015). The relationships among nonbilaterian phyla and bilaterian animals remain contentious (Pisani et al. 2015; Whelan et al. 2015), but are largely irrelevant to the present study. Thus, for the purpose of this review we depict them as a soft polytomy (fig. 1).
Cnidaria. Cnidarians are subdivided into two major lineages: Anthozoa (cnidarians with only a polypoid body form) and Medusozoa (cnidarians with a medusa stage in their life cycle) (Daly et al. 2007; Zapata et al. 2015), but should now also include Myxozoa. Anthozoa is subdivided into Hexacorallia (e.g. sea anemones, hard corals), Octocorallia (soft corals), and Ceriantharia (tube anemones) (Stampar et al. 2014). Within Medusozoa four major clades are recognized: Scyphozoa (jellyfishes), Staurozoa (stalked jellyfishes), Cubozoa (box jellyfishes), and Hydrozoa (hydroids, fire corals, freshwater jellyfishes, siphonophores, etc.) (Collins 2002). Finally, Myxozoa are separated into two classes: Malacosporea and Myxosporea, with the latter containing the majority of species (Fiala et al. 2015).
Ctenophora. Traditionally, ctenophores have been subdivided into two classes based on the presence/absence of their feeding tentacles: Tentaculata (with tentacles) and Nuda (without). Within these classes, 9 orders, 27 families, and about 150 species are currently recognized (Mills 2014). However, the monophyly of both classes and most ctenophore orders is not supported by molecular data and many intraorder relationships remain unresolved (Podar et al. 2001; Moroz et al. 2014; Simion et al. 2015).
Placozoa. Only one placozoan species has been technically described (Schulze, 1883). Nevertheless, molecular studies indicate substantial hidden diversity within this phylum (Eitel et al. 2013). In particular, five substantially different mt-genomes have been published from this phylum (Signorovitch et al. 2007; Miyazawa et al. 2012).
Porifera. Sponges are divided into four major groups (classes): Calcarea (calcareous sponges), Demospongiae (demosponges), Hexactinellida (glass sponges), and Homoscleromorpha (homoscleromorphs) (Gazave et al. 2011). The consensus view on their relationships places demosponges and glass sponges in one group, and homoscleromorphs and calcareous sponges in another (reviewed in Wörheide et al. 2012). Three main lineages are recognized in demosponges: Verongimorpha, Keratosa, and Heteroscleromorpha (Morrow and Cardenas 2015); while calcareous sponges are subdivided into Calcinea and Calcaronea (Manuel 2006; Voigt et al. 2012); glass sponges into Hexasterophora and Amphidiscophora (Dohrmann et al. 2012); and homoscleromorphs into Plakinidae and Oscarellidae (Gazave et al. 2010).
Mitochondrial Genome Architecture in Nonbilaterian Animals
The mt-genomes of most sponges (Wang and Lavrov 2008; Gazave et al. 2010; Haen et al. 2014), anthozoan cnidarians (Brockman and Mcfadden 2012; Figueroa and Baco 2015), ctenophores (Pett et al. 2011; Kohn et al. 2012), and placozoans (Signorovitch et al. 2007) are mapped as monomeric and circular molecules. This organization is also found in choanoflagellates, the closest outgroup to animals, as well as most bilaterian animals and is therefore inferred to be ancestral to Metazoa. However, multipartite and/or linear genomes have evolved in several nonbilaterian lineages, which we will review in more detail below.
Linear Mitochondrial Genomes
Linear mtDNA is found in two major lineages of nonbilaterian animals: Medusozoan cnidarians and calcareous sponges (fig. 2). The occurrence of linear mtDNA in Medusozoa was first inferred based on gel electrophoresis studies (Warrior and Gall 1985; Bridge et al. 1992; Ender and Schierwater 2003). Subsequently, mt-genome sequences were determined for a variety of medusozoan species revealing two specific features associated with linear mtDNA organization in this group: The presence of identical sequences in inverted orientation at the ends of the chromosome(s) (terminal inverted repeats or TIR) and the existence of two additional ORFs (Shao et al. 2006; Kayal and Lavrov 2008; Voigt et al. 2008; Kayal et al. 2012, 2015; Smith et al. 2012) (fig. 2). One of the ORFs shows significant sequence similarity to B-type DNA polymerase (polB), the second is inferred to code for a DNA-binding protein (Shao et al. 2006; Kayal et al. 2012). TIRs are present in all medusozoan mt-genomes and usually include a partial copy of the cytochrome oxidase I gene (cox1). The two ORFs are found in cubozoan, schyphozoan, staurozoan, and some hydrozoan mt-genomes but have been lost in hydroidolinan hydrozoans, which instead have longer TIRs that include duplicated cox1 at each end of their mitochondrial chromosome(s) (Kayal et al. 2012, 2015).
The presence of TIRs and a polB homologue in medusozoan mtDNA suggests that their unusual architecture originated through incorporation of a linear plasmid into the ancestral circular genome, while the finding that all medusozoan polB sequences form a clade indicates a single origin of linear architecture in this group (Kayal et al. 2012). Linear organellar plasmids with identical but inverted sequences at their termini (known as invertrons: Sakaguchi 1990) are common in organelles of plants and fungi (Hausner 2012) and have been implicated in the evolution of a linear genome organization in some fungi (e.g. Fricova et al. 2010). Interestingly, replication of these plasmids is often initiated from a protein bound to the 5′ end of DNA (Klassen and Meinhardt 2007). It has been suggested that the same mode of initiation of replication is retained in mt-genomes that have incorporated such plasmids (Fricova et al. 2010).
All calcareous sponges sampled to date also have multipartite linear mt-genomes (fig. 2), but appear to utilize a different mechanism for the maintenance and repair of their mtDNA, as they do not contain TIRs and do not appear to encode any DNA polymerase-related proteins. The mtDNA of Clathrina clathrus, a representative of the subclass Calcinea, consists of six linear chromosomes with a peculiar organization, where about a half of each chromosome is noncoding with hairpin-forming repeat sequences at one end, and with a noncoding region at the other end that is similar across chromosomes (Lavrov et al. 2013). In Leucosolenia complicata and Petrobiona massiliana, two representatives of the subclass Calcaronea, most mitochondrial genes are located on individual chromosomes that have similar but not identical sequences at their ends (Lavrov et al. 2016). The mt-genome organization of Sycon ciliatum, another representative of the subclass Calcaronea, appears to be more complex and awaits characterization (Lavrov et al. 2016).
Multipartite Mitochondrial Genomes
As is the case with linear genomes, a multipartite organization has evolved several times in animal mtDNA, with both circular (Armstrong et al. 2000; Gibson et al. 2007; Suga et al. 2008; Wei et al. 2012; Jiang et al. 2013) and linear (Voigt et al. 2008; Kayal et al. 2012; Smith et al. 2012; Lavrov et al. 2013, 2016) chromosome architectures (fig. 2). Among nonbilaterian animals, multipartite genomes have evolved at least three times: Two times in Medusozoa (Voigt et al. 2008; Kayal et al. 2012; Smith et al. 2012) and at least one other time in calcareous sponges (Lavrov et al. 2013, 2016). The first report of a multi-chromosomal mt-genome in animals came from the studies of the hydrozoan cnidarians Hydra attenuata and H. littoralis, in which mtDNA consists of two linear chromosomes, each about 8 kb in size (Warrior and Gall 1985; see also Voigt et al. 2008; Pan et al. 2013). mtDNA from the winged box jellyfish Alatina moseri (Smith et al. 2012) and other representatives of class Cubozoa (Kayal et al. 2012) consists of eight linear chromosomes, 2.9–4.6 kb in size (fig. 2).
A remarkable multipartite organization has evolved in calcaronean sponges (fig. 2). In both L. complicata and P. massiliana, all identified coding sequences are located on individual chromosomes. Furthermore, 147 additional mitochondrial chromosomes were identified in P. massiliana based on sequence conservation in terminal regions. Some of these chromosomes appear to contain tRNA and rRNA genes, but it is not yet clear whether these sequences are functional. Similarly, at least 50 additional mitochondrial chromosomes were identified in L. complicata (Lavrov et al. 2016).
Size Variation and Distribution of Noncoding Nucleotides among Intergenic Regions
The mtDNA of nonbilaterian animals displays at least a 7-fold variation in size (fig. 4). The smallest mt-genomes are found in ctenophores, with those of Mnemiopsis leidyi and Pleurobrachia bachei measuring 10,326 and 11,016 bp, respectively (Pett et al. 2011; Kohn et al. 2012). The small size of these genomes is due to their reduced gene content, highly abbreviated rRNA structures, and extreme scarcity of intergenic nucleotides. On the other end of the spectrum, the mtDNA of Trichoplax adhaerens is 43,079 bp in size (Dellaporta et al. 2006), with several large introns containing intron-associated ORFs. However, the current record in mt-genome size belongs to the multipartite genomes of calcareous sponges, with 6 mitochondrial chromosomes in C. clathrus having a total size of about 51 kb (Lavrov et al. 2013), and 150+ mitochondrial chromosomes in P. massiliana measuring above 77 kb (the exact sizes of these genomes are unknown because of difficulties associated with sequencing the ends of linear chromosomes and/or identifying all chromosomes) (Lavrov et al. 2016).
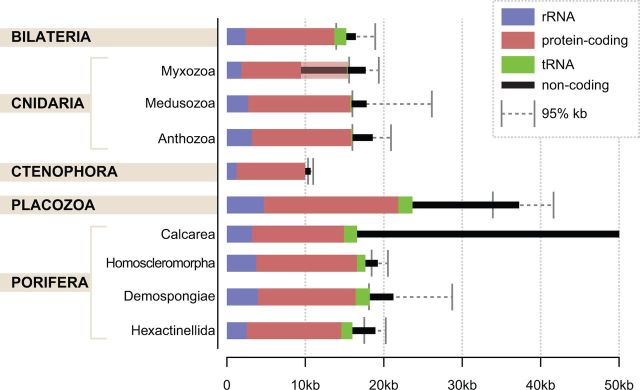
Fig. 4.—
Size and coding content of animal mitochondrial DNA. Colored bars indicate the average number of nucleotides coding for ribosomal RNA (blue), protein (red), transfer RNA (green), or noncoding (black) computed using all available complete mt-genome sequences published in the NCBI RefSeq database as of April 9, 2016. Gray error bars indicate the upper and lower boundaries of the 95% interquantile range for genome sizes (2.5th and 97.5th percentiles). The shaded portion of the red bar in Myxozoa indicates additional protein-coding genes likely present in these genomes, but not recognizable due to an extreme rate of sequence evolution.
Most of the size variation among animal mt-genomes is due to differences in the number of noncoding nucleotides, which are also distributed unevenly among intergenic regions. Bilaterian mtDNA usually contains a single large noncoding segment, also known as the “control” region, or as the “D-loop” region in mammals (Sbisa et al. 1997). In a few species, the control region was shown to contain signals for initiation and termination of replication and transcription (Goddard and Wolstenholme 1980; Clayton 1982). Among nonbilaterian animals, a single large noncoding region is present only in the mtDNA of glass sponges and perhaps ctenophores. In other nonbilaterian taxa, noncoding nucleotides are distributed more evenly among the intergenic regions. The absence of an identifiable control region in many nonbilaterians suggests that their mechanisms of replication and transcription are different from those of bilaterian animals. Furthermore, the “tRNA punctuation” model described in some bilaterian animals for processing of polycistronic mtDNA transcripts (Ojala et al. 1981) is not applicable to many nonbilaterian animals because of the presence of intergenic nucleotides and/or the scarcity or the absence of tRNA genes (see “Loss and gain of mitochondrial tRNA genes”).
More Variation in Protein-Coding Gene Content of Nonbilaterian mtDNA
Mitochondrial protein-coding gene content shows more variation in nonbilaterian compared with bilaterian animals (fig. 5). First, several additional protein-coding genes have been identified. These include atp9 for subunit 9 (subunit c) of mitochondrial F0-ATP synthase in all four classes of sponges (Lavrov et al. 2005, 2013; Wang and Lavrov 2008; Haen et al. 2014), tatC for the twin-arginine translocase subunit C in the homoscleromorph sponge family Oscarellidae (Wang and Lavrov 2007; Pett and Lavrov 2013), mutS for a putative mismatch repair protein in Octocorallia (Pont-Kingdon et al. 1995; Bilewitch and Degnan 2011), polB for the DNA dependent DNA polymerase in Medusozoa (Shao et al. 2006; Kayal et al. 2012) and one placozoan (Signorovitch et al. 2007), and intron-associated homing endonucleases, reverse transcriptases, and maturases in several lineages with mitochondrial introns. Among the genes listed above, both atp9 and tatC are also present in the mt-genome of the choanoflagellate Monosiga brevicollis (Burger et al. 2003) suggesting that these genes were inherited vertically in sponges, and lost in other animals (Wang and Lavrov 2007; Pett and Lavrov 2013). However, phylogenetic analysis does not rule out the possibility that tatC was acquired in Oscarellidae by horizontal transfer from the mtDNA of another eukaryote (Pett and Lavrov 2013). In contrast, cnidarian mutS and polB were likely acquired by horizontal gene transfer from prokaryotes or viruses (Bilewitch and Degnan 2011).
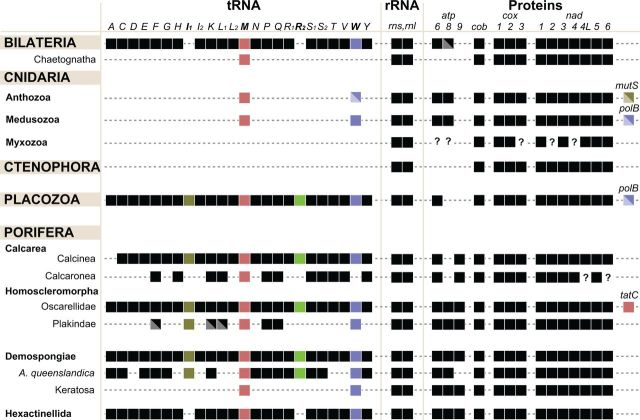
Fig. 5.—
Variation in RNA and protein-coding gene content in animal mitochondrial DNA. Left: taxonomic positions of groups with variable gene content. Central: variation in tRNA gene content. Middle: the presence of rRNA genes. Right: the presence/absence of protein-coding genes. Colored boxes highlight the identities of specific genes discussed in the text. Diagonally split boxes indicate genes that are absent in some lineages of the indicated group. Question marks denote genes for which presence or absence has not been confidently determined. Abbreviations: atp6, atp8–9: subunits 6, 8, and 9 of F0 adenosine triphosphatase (ATP) synthase; cob: apocytochrome b; cox1–3: cytochrome c oxidase subunits 1–3; nad1–6 and nad4L: NADH dehydrogenase subunits 1–6 and 4L; polB: DNA polymerase β; tatC: twin-arginine translocase component C; rns and rnl: SSU and LSU rRNAs. The tRNA genes are identified by the one-letter code for their corresponding amino acid; subscripts denote different genes for isoacceptor tRNAs, where I1 = trnI(CAU), I2 = trnI(GAU), L1 = trnL(UAG), L2 = trnL(UAA), R1 = trnR(UCG), R2 = trnR(UCU), S1 = trnS(UCN), S2 = trnS(UGA).
Second, several protein-coding genes have been lost from some nonbilaterian mt-genomes. In two ctenophores, atp6 has been transferred to the nucleus, while atp8 was not identified in either the mitochondrial or the nuclear genomes (Pett et al. 2011; Kohn et al. 2012). Similarly, atp8 appears to be lost from the mt-genomes of placozoans (Signorovitch et al. 2007; Burger et al. 2009) and calcareous sponges (Lavrov et al. 2013, 2016), while atp9 has been transferred to the nucleus in several demosponges (Erpenbeck et al. 2007; Lavrov, unpublished data). It has been thought that atp8 is also absent from mitochondrial genomes of glass sponges (Haen et al. 2007; Rosengarten et al. 2008; Haen et al. 2014). However, a plausible candidate has been recently found in Oopsacas minuta (Jourda et al. 2015) with homologous sequences identifiable in other species (unpublished data).
The most unusual mitochondrial gene content has been reported for the myxozoan genus Kudoa, where only eight protein-coding genes and some unidentified ORFs were found (Takeuchi et al. 2015). However, because of the extremely high rate of sequence evolution in myxozoan mtDNA, it is probable that some unidentified ORFs represent standard mitochondrial genes that have evolved beyond recognition (fig. 4, shaded red bar). This inference is supported by the observation that only five out of the eight identified genes have retained significant sequence similarity (E < 0.001) with their homologues outside of Myxozoa.
Loss and Gain of Mitochondrial tRNA Genes
Besides variation in protein-coding gene content, animal mt-genomes encode a variable number of transfer RNAs (mt-tRNAs) (fig. 5). Most demosponge and placozoan mt-genomes contain 24–25 tRNA genes, including trnI(CAU) and trnR(UCU). The latter two genes were lost early in the evolution of bilaterian animals in conjunction with modifications to the mitochondrial genetic code, but their products are needed for mitochondrial protein synthesis in many nonbilaterian animals, which utilize a minimally modified genetic code (see “Changes in the genetic code”). In addition, a second trnM(CAU) is present in some sponge mt-genomes, apparently coding for a separate elongator .
Representatives of three out of the four main lineages of nonbilaterian animals have experienced significant losses of tRNA genes. First, all mt-tRNA genes have been lost in Ctenophora (Pett et al. 2011) [the two mt-tRNA genes reported for the ctenophore Pleurobrachia bachei (Kohn et al. 2012) are annotation artifacts (Pett and Lavrov 2015)]. Second, the mt-genomes of nonmyxozoan cnidarians have lost all but one or two tRNA genes (Kayal et al. 2013), while no mt-tRNA genes have been found in two sampled species in the myxozoan genus Kudoa (Takeuchi et al. 2015). Although four mt-tRNAs have been reported from the third representative of this genus, they are also likely annotation artifacts. Third, mt-tRNA loss has occurred repeatedly in sponges, including several lineages of demosponges, the homoscleromorph family Plakinidae, and possibly some calcaronean sponges. Interestingly, one group of demosponges (Keratosa) has a pattern of mt-tRNA gene loss identical to that in most Cnidaria, encoding only genes for tryptophanyl and methioninyl mt-tRNA (Wang and Lavrov 2008).
The loss of mt-tRNA genes is commonly associated with other changes in the nuclear genome. In particular, the loss of mt-tRNAs may render some components of the mitochondrial translation system redundant, leading to the loss of their genes in the nuclear genome. For example, the maturation of (the product of the trnI(CAU) gene) involves a post-transcriptional modification of the cytosine in the anticodon (position 34) to lysidine (2-lysyl-cytidine) performed by tRNAIle-lysidine synthetase (TilS) (Soma et al. 2003), an enzyme that is not involved in maturation of cytosolic isoleucine tRNAs (Marck and Grosjean 2002). Because of the dispensable nature of this function for cytosolic protein synthesis, the loss of mitochondrial trnI(CAU) that occurred in some nonbilaterian groups and all bilaterians, was followed by the loss of the nuclear-encoded TilS gene, rendering a reacquisition of trnI(CAU) practically impossible (Pett and Lavrov 2015). Our previous research also uncovered a correlation between the loss of mt-tRNAs and the loss of nuclear-encoded mitochondrial aminoacyl-tRNA synthetases (Haen et al. 2010; Pett et al. 2011), as well as several other RNA-modifying and aminoacyl-tRNA modifying enzymes (Pett and Lavrov 2015).
Finally, some mt-tRNA genes can be gained via a process of “remolding” or “recruitment”, in which a copy of a gene is recruited from one isoaccepting group to another by a point mutation that changes the tRNA’s amino acid identity and its mRNA coupling capacity. This process has been extensively documented in both bilaterian and nonbilaterian animal mtDNA (Lavrov and Lang 2005; Wang and Lavrov 2011; Sahyoun et al. 2015).
Changes in the Genetic Code
Close unicellular relatives of animals use a nonstandard “minimally modified” genetic code for mitochondrial protein synthesis, where the only deviation from the standard genetic code is the reassignment of the UGA codon from termination to tryptophan (fig. 3). This mitochondrial genetic code was likely the ancestral condition for Metazoa (Knight et al. 2001) and is still used by most nonbilaterian animals. Early in the evolution of bilaterian animals, the meanings of two mitochondrial codons were changed, with AUA reassigned from isoleucine to methionine, and AGR from arginine to serine (Knight et al. 2001), followed by further modifications in some lineages (Watanabe and Yokobori 2011).
Several modifications to the mitochondrial genetic code have also occurred among nonbilaterian animals, some of them in parallel with those in bilaterian animals (fig. 3). Like in bilaterian animals, the identity of AGR codons has been changed from Arginine to Serine in glass sponges (Haen et al. 2007, 2014) and to either Serine or Glycine in the calcaronean sponges Sycon ciliatum, S. coactum, and P. massiliana (Lavrov et al. 2016). Furthermore, in P. massiliana the AUA codon has been reassigned from Isoleucine to Methionine. More unusual changes have occurred in calcinean sponges C. clathrus and C. aurea, where UAG has been reassigned from termination to Tyrosine and CGN codons from Arginine to Glycine (Lavrov et al. 2013), and in the calcaronean sponge L. complicata, where AGR codons have been reassigned to Methionine. In addition, at least some species of octocorals no longer use UAG in their mitochondrial coding sequences (effectively reverting to the standard genetic code), while in some ctenophores this codon has been reassigned to Serine (Pett and Lavrov 2015).
Mitochondrial Introns and Fragmented Genes
Several cases of fragmented mitochondrial genes have been reported in nonbilaterian animals, and the majority of these cases involve mitochondrial introns. Introns in nonbilaterian animal mtDNA were first found in anthozoan cnidarians (Beagley et al. 1996) and subsequently in demosponges (Rot et al. 2006), placozoans (Dellaporta et al. 2006), and homoscleromorph sponges (Wang and Lavrov 2008). It has been suggested that the more common presence of mitochondrial introns in nonbilaterian compared with bilaterian animals [they have been found only in a few annelid species among the latter (Valles et al. 2008; Richter et al. 2015)] may be due either to the lack of early separation between germ and soma, or to the low mitochondrial substitution rates observed in many of nonbilaterian species (Hausner 2012).
As is the case with organellar introns in other eukaryotes, animal mitochondrial introns are homologous to either group I or group II self-splicing introns found in bacteria and viruses (Martínez‐Abarca and Toro 2000; Hausner et al. 2014) rather than to either the spliceosomal introns or tRNA introns present in nuclear genomes (Lang et al. 2007). All hexacoral mt-genomes contain a group I intron inside nad5 and often another intron inside cox1 (Beagley et al. 1996; Goddard et al. 2006; Medina et al. 2006; Fukami et al. 2007). The intron within nad5 is highly unusual because it encompasses at least 2 and as many as 15 standard mitochondrial genes, and sometimes additional ORFs (Emblem et al. 2014). One or two group I introns are also present in cox1 in some members of the homoscleromorph sponge family Plakinidae (Gazave et al. 2010), and three different group I introns have been described in cox1 from several groups of demosponges (Rot et al. 2006; Szitenberg et al. 2010; Erpenbeck et al. 2015; Huchon et al. 2015). Finally, all placozoan mt-genomes contain six to seven group I introns (Signorovitch et al. 2007). The structure of cox1 in these species is most unusual in that it consists of seven to eight exons contained in three segments with different transcriptional polarities, which are joined together by group I intron trans-splicing, one of few reports of such a process in vivo (Burger et al. 2009). Homing endonucleases of the LAGLIDADG type can often be found encoded in cox1 group I introns, and the patchy phylogenetic distribution of these proteins within these groups is consistent with the typical life cycle of homing endonucleases, which involves recurrent horizontal transfer among closely related species (Goddard and Burt 1999).
Group II introns are less common in animal mitochondria, but as many as four different forms have been identified in placozoan mt-genomes (Signorovitch et al. 2007; Burger et al. 2009). In addition, two group II introns have recently been found in demosponges (Huchon et al. 2015). Phylogenetic analysis of the reverse transcriptase encoded by the latter introns suggests that they may have originated by horizontal transfer from a red algal donor (Huchon et al. 2015). Surprisingly, group II mitochondrial introns have also been discovered in several annelid species (Valles et al. 2008; Richter et al. 2015), the only example of introns in bilaterian animals.
No introns have been found in glass sponges, calcareous sponges, ctenophores, medusozoan cnidarians, and myxozoans, or most bilaterian animals, groups in which accelerated rates of mt-sequence evolution have been observed. This lack of introns in fast-evolving mtDNA is consistent with observations from other groups of eukaryotes, and can potentially be explained by the mutational hazard hypothesis (Lynch et al. 2006), which asserts that strong mutation pressure, resulting from a high mutation rate, suppresses the proliferation of noncoding elements, including introns. At the same time, no mitochondrial introns have been found in either octocoral cnidarians or the homoscleromorph sponge family Oscarellidae, two groups with exceptionally low rates of mitochondrial sequence evolution (Gazave et al. 2010; McFadden et al. 2011), indicating that other factors are also at play.
In some cases, not only mitochondrial genes but also their products can be fragmented. For example, in the calcareous sponges C. clathrus and C. aurea, both rRNA genes are discontinuous and located on different chromosomes (Lavrov et al. 2013, unpublished data), but encode conserved rRNA structures when pieced together. Fragmented rRNA genes have also been found in the oyster genus Crassostrea (Milbury and Gaffney 2005; Milbury et al. 2010), and in Placozoa, where large subunit rRNA is encoded in two pieces (Signorovitch et al. 2007). Typically, rRNA fragments assemble as separate molecules, but an example of trans-splicing has been recently reported in the Euglenozoan flagellate Diplonema (Valach et al. 2014). There is no evidence of rRNA fragment trans-splicing in animal mitochondria.
RNA Editing and Translational Frameshifting
Some animal mt-genomes encode aberrant tRNAs and/or mRNAs that are corrected by RNA editing during or after transcription (Gott and Emeson 2000). Mitochondrial RNA editing in bilaterian animals involves primarily tRNAs (Börner et al. 1997). As an extreme example, nearly half of nucleotides in the mt-tRNAs of onychophorans are added post-transcriptionally (Segovia et al. 2011). A few putative cases of mRNA editing have also been reported in bilaterian animal mitochondria, most of which manifest as variation in the length of polythymidine poly(T) tracts (Vanfleteren and Vierstraete 1999; Riepsamen et al. 2008; Denoeud et al. 2010; but see Riepsamen et al. 2011). A few RNA-DNA differences have recently been reported in human mtDNA (Bar-Yaacov et al. 2013).
In nonbilaterian animals, both tRNA and mRNA editing has been found. The mtDNA of C. clathrus encodes tRNA molecules with multiple mismatches in the aminoacyl acceptor stem, which undergo post-transcriptional RNA editing (Lavrov et al. 2013). In Placozoa, a conserved uracil (U) position in cox1 is required for proper splicing of a group I intron, but a cytosine (C) is required at the same site for a functional COX protein subunit. This apparent conflict is resolved by converting U to C by an RNA editing mechanism that takes place after splicing is completed (Burger et al. 2009). Recently, widespread and persistent mt-mRNA editing was discovered in calcaronean sponges (Lavrov et al. 2016), consisting of single or double U insertions in pre-existing poly(U) tracts. A total of 225, 435, and 451 nucleotides were inserted in mt-mRNAs identified in three analyzed species (Lavrov et al. 2016).
In glass sponges, several single nucleotide insertions in otherwise conserved mitochondrial coding sequences are retained in mRNA and have been interpreted as +1 translational frameshifting sites (Haen et al. 2007; Rosengarten et al. 2008). Most of these insertions occur immediately upstream of conserved glycine GGA codons and result in the formation of in-frame TGG or CGG codons, which are otherwise not used or very rarely used in glass sponge mitochondrial coding sequences (Haen et al. 2014). These unusual sequence features, along with some unusual changes in the corresponding glycine tRNAs, suggest an “out-of-frame pairing” model of translational frameshifting, in which the extra nucleotide is excluded from pairing at the A-site of the ribosome (Buchan and Stansfield 2007). Our recent work revealed that frameshifting sites have evolved repeatedly in glass sponge mtDNA, and can persist for hundreds of millions of years (Haen et al. 2014). Among bilaterian animals, translational frameshifting has been reported in birds and turtles (Harlid et al. 1997; Mindell et al. 1998; Zardoya and Meyer 1998; Parham et al. 2006), Polyrhachis ants (Beckenbach et al. 2005) and the eastern oyster (Milbury and Gaffney 2005). Interestingly, AGR codons in human mtDNA, originally inferred to code for termination, have been recently reinterpreted as mitochondrial frameshifting sites (Temperley et al. 2010).
Variable Rates of Sequence Evolution
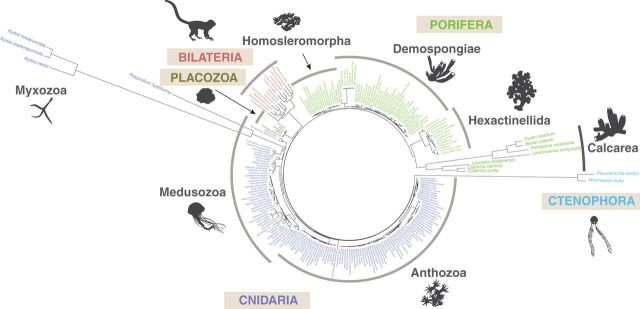
It was observed early in the study of animal mtDNA that the rate of nucleotide substitutions was an order of magnitude higher in mitochondrial compared with nuclear genes (Brown et al. 1979). Subsequently, a wide range of mitochondrial substitution rates have been estimated within vertebrate lineages, including mammals, where the rates can vary 100-fold and birds, where the rates can vary 30-fold (Nabholz et al. 2008; Nabholz et al. 2009). Analysis of mtDNA sequences from nonbilaterian animals adds another dimension to these observations by showing persistent differences in rates among major animal groups (fig. 6). In particular, demosponges, homoscleromorphs, and most cnidarians have relatively low rates of mitochondrial substitutions, glass sponges, and bilaterian animals have intermediate rates, while ctenophores, calcareous sponges, and myxozoans have high to extremely high rates (fig. 6).
Fig. 6.—
mtDNA-based animal phylogeny showing variable rates of mt-sequence evolution. Complete or nearly complete mt-genomes from all available nonbilaterian animals and a few selected bilaterian animals were downloaded from the GenBank. In addition, mitochondrial genomes of Periphylla periphylla and Polypodium hydriforme, were assembled from high throughput transcriptomic and genomic data (SRX956805 and SRX687102, respectively). Inferred amino acid sequences from nine mitochondrial genes (cob, cox1-3, nad1-5) were aligned with MAFFT v7.215 (Katoh and Standley 2013). Conserved blocks within the alignments were selected with Gblocks 0.91 b (Talavera and Castresana 2007) using relaxed parameters (parameters 1 and 2 = ½, parameter 3 = 8, parameter 4 = 5, all gap positions in parameter 5). Cleaned alignments were concatenated in a dataset 2,228 positions in length. Bilateria, Cnidaria, Ctenophora, Placozoa, and Porifera were constrained as monophyletic and the best constrained topology was identified using RAxML with the MTREV + GAMMA + F substitution model and 32 initial tree searches. Subsequently, branch lengths were re-estimated with the CAT + GTR + Γ4 model in PhyloBayes MPI 1.4e (Lartillot et al. 2013).
An increased substitution rate may be caused by an elevated mutation rate (which determines the rate of neutral evolution) and/or an increased probability of fixation of non-neutral mutations (Kimura 1993; reviewed in Baer et al. 2007). A high mutation rate in bilaterian mtDNA, first inferred from phylogenetic comparisons, was later corroborated and revised upward by pedigree analyses and mutation accumulation experiments (Parsons et al. 1997; Denver et al. 2000; Haag-Liautard et al. 2008; Xu et al. 2012). The high mutation rate in animal mtDNA is often attributed to damage from reactive oxygen species (ROS) released as byproducts during oxidative phosphorylation (Richter et al. 1988; Yakes and Van Houten 1997) and has been proposed to be a function of the metabolic rate (Martin and Palumbi 1993; Gillooly et al. 2005). However, no support for this “metabolic rate hypothesis” has been found in subsequent studies (Lanfear et al. 2007). Furthermore, recent studies that have utilized ultra-sensitive sequencing of somatic mtDNA revealed that most mutagenic events are not G -> T transversions expected as the result of ROS damage but transitions, likely caused by errors made by DNA polymerase and/or spontaneous deamination of cytidine and adenosine during replication (reviewed in Larsson 2010; Ameur et al. 2011; Kennedy et al. 2013; Itsara et al. 2014). Thus, changes in mitochondrial mutation rates are more likely the result of changes in nuclear-encoded proteins responsible for the maintenance and replication of mtDNA rather than different levels of ROS production. Because of the lower mutation load expected for small genomes, natural selection for mtDNA replication fidelity should be relaxed compared with the nuclear genome, resulting in a higher mutation rate (Lynch 2010). It has also been suggested that the elevated mutation rate in animal mtDNA may be driven by positive selection for a greater supply of adaptive mutations (Wallace 2007), but we view this proposition as highly unlikely. Although alleles causing a high mutation rate (mutator alleles) do exist in natural populations and can become fixed by hitchhiking with linked beneficial mutations (Taddei et al. 1997), this is not expected in mtDNA, which is unlinked from its nuclear-encoded replication machinery (Lynch 2011). Even in asexual bacterial populations, hitchhiker mutator alleles are generally short-lived and/or exist at low frequencies due to the inevitable long-term deleterious mutation load associated with a high mutation rate (Giraud et al. 2001). Experimental increase of mutation rate in mammalian mtDNA lead to aggravated aging, impaired brain development, and reduced lifespan (Ross et al. 2013, 2014).
While the rate of fixation for neutral mutations is equal to the mutation rate, that of non-neutral mutations also depends on their fitness effect (expressed as selection coefficient) and effective population size (Kimura 1993). Both theoretical considerations (Kimura 1968) and analyses of empirical data (Weinreich and Rand 2000; Nielsen and Yang 2003; Popadin et al. 2007; Lartillot and Poujol 2011) suggest that the majority of mtDNA mutations are deleterious and thus should be eliminated by selection. Although positive selection has also been inferred in the mtDNA of some lineages, usually in association with postulated higher demand for energy production [e.g., in anthropoid primates with an expanded neocortical brain (Grossman et al. 2004), bats (Shen et al. 2010), and mammals at high altitudes (Hassanin et al. 2009)] demonstrating the occurrence of adaptive substitutions in mtDNA evolution can be difficult (Meiklejohn et al. 2007; Nei et al. 2010). Nevertheless, it has been estimated that up to a quarter of nonsynonymous substitutions in various animal species may be fixed by adaptive evolution (James et al. 2016).
The strength of genetic drift, which is inversely proportional to the effective population size, will also influence the probability of fixation of non-neutral mutations, with higher rates of deleterious substitutions in smaller populations, and higher rates of beneficial substitutions in larger populations (Ohta 1973). The effect of genetic drift on mtDNA is amplified by several features of mitochondrial biology, including its uniparental (maternal) inheritance, effective haploidy, and lack (or scarcity) of recombination (Ballard and Whitlock 2004; Aanen et al. 2014). A commonly observed outcome of stronger genetic drift in mtDNA is the more rapid fixation of mildly deleterious mutations (Lynch and Blanchard 1998) as reported for large versus small mammals (Popadin et al. 2007). It has also been inferred that selective sweeps occur predominantly in invertebrate rather than vertebrate animals, due to their larger population sizes (Bazin et al. 2006; Meiklejohn et al. 2007). Finally, genetic drift should influence the mitochondrial mutation rate itself due to accumulation of mildly deleterious mutations in nuclear-encoded mitochondrial replication proteins (Lynch 2010; Sung et al. 2012).
Given the impact that reduced effective population size can have on substitution rates by increasing both the mutation rate and the probability of fixation of slightly deleterious mutations, life history features may help explain the accelerated rates of mitochondrial sequence evolution observed in some nonbilaterian animals. For example, the fastest evolving animal mtDNA belongs to Myxozoa, one of very few examples of parasitic nonbilaterian animals [the others being a related cnidarian endoparasite Polypodium hydriforme (Evans et al. 2008), and Eulampetia pancerina, a ctenophore with a parasitic larval stage]. As a result of their parasitic lifestyle, there are recurrent bottlenecks in myxozoan populations as their hosts are infected by a small number of spores (Kent et al. 2001). In addition, sexual reproduction usually takes place between descendants of a single founder actinospore, leading to inbreeding and further reduction in the effective population size of mtDNA. Ctenophores, another group with highly accelerated mitochondrial substitution rates, also have an unusual biology, characterized by frequent selfing, hermaphroditism, and large fluctuations in population size (Baker and Reeve 1974; Pianka 1974).
The effect of smaller population size is often most visible in changes (likely deleterious) that accumulate in tRNA and rRNA structures (Lynch 1996). The mt-genome of the ctenophore M. leidyi, a selfing hermaphrodite (Baker and Reeve 1974; Pianka 1974), represents one of the most extreme examples, where the sizes of the small and large subunit rRNAs have been reduced to no more than 368 and 878 bp, respectively, with only a few identifiable conserved secondary structures (Pett et al. 2011). Interestingly, in the calcinean sponge C. clathrus mt-rRNA structures are relatively well conserved despite high substitution rates in mitochondrial coding sequences.
General Trends in the Evolution of Animal mtDNA
Thirty-five years after the first animal mtDNA sequence was determined (Anderson et al. 1981), we are finally attaining a comprehensive view of animal mt-genome diversity. The emerging picture reveals more variation in animal mtDNA organization and evolution than previously appreciated, much of which is found among nonbilaterian phyla. Conversely, many of the genetic novelties thought to be associated with animal mtDNA (Wolstenholme 1992b) appear to be restricted to Bilateria. The richness of mitochondrial features in nonbilaterian animals is particularly striking in the light of the relatively small number nonbilaterian species with complete mt-genomes (around 200), which is an order of magnitude smaller than the number for Bilateria (over 6000) (fig. 1).
In part, the greater mtDNA diversity in nonbilaterian animals can be explained by the deeper divergences among nonbilaterian comparing to bilaterian phyla (Dos Reis et al. 2015). However, mtDNA diversity does not appear to be simply a function of time, as most major changes map to early branches in animal evolution (fig. 1). This can be seen, for example, in the remarkable uniformity of mtDNA in most bilaterian animals, which suggests that characteristic features of bilaterian mtDNA emerged after the split of Bilateria and Cnidaria in the late Precambrian but prior to the diversification of bilaterian animals in the early Cambrian. Similarly, a distinct organization of glass sponge mtDNA was established prior to the divergence of crown lineages in the Cambrian and has been maintained thereafter (Haen et al. 2014). Comparable patterns of mtDNA evolution are also found within ctenophores and placozoans (Signorovitch et al. 2007; Pett et al. 2011; Kohn et al. 2012), although it is less clear when these patterns were established.
It is not immediately clear why these early patterns should have been so widely conserved, but an intriguing possibility is that early changes in the evolution of certain groups canalized the future course of evolutionary events in mtDNA. For example, early changes in the bilaterian mt-tRNA and/or mt-protein import machinery may have prevented the replacement of mt-tRNAs with their nuclear counterparts, a process that is quite common outside of Bilateria (Schneider 2011; Pett and Lavrov 2015), but very rare among bilaterian animals [the exception of chaetognaths (Faure and Casanova 2006) deserves further investigation (e.g. see Barthelemy and Seligmann 2016)].
As with other biological systems, the evolution of animal mtDNA is also marked by convergence. One of the most striking examples is the parallel evolution of glass sponge and bilaterian mtDNA. Both groups of animals share a similar mtDNA organization with a single large noncoding region, an identical change in the genetic code, a modification in the structure of tRNA(Ser), as well as a particular bias in nucleotide composition (Haen et al. 2007). Other examples of convergent mitochondrial evolution in animals include transitions to a multipartite, linear genome organization in calcareous sponges and the cnidarian class Cubozoa and the loss of the same set of tRNA genes in Cnidaria and the demosponge lineage Keratosa. Interestingly, parallel evolution in mtDNA can precipitate parallel changes in the nuclear genomes, as has been shown by the loss of nuclear encoded mt-aminoacyl-tRNA synthetases in association with the loss of mt-tRNAs. It remains to be seen whether other changes in the content and structure of mt-genomes can lead to convergent evolution in other nuclear genes and molecular pathways.
Future Directions
Faced with the abundance of bilaterian mt-genome sequences, it has been argued that further sampling of animal mt-genomes may be of limited use for studying mtDNA evolution (Smith 2015). This may be true for some groups. However, current mtDNA sampling for nonbilaterian animals remains poor in comparison with their considerable phylogenetic diversity, and further studies in these groups have the potential to significantly expand our understanding of mitochondrial evolution.
In particular, there are several nonbilaterian groups, such as ctenophores, calcareous sponges, and myxozoan cnidarians, where sampling is clearly inadequate. Probably not coincidentally, these are also the groups that have either unusual mt-genome organization or extremely high rates of mt-sequence evolution, or both. For all these groups, mtDNA sequences became available only with the advent of next generation DNA sequencing technologies, which made it possible and cost-effective to sequence total cellular DNA from targeted species and bioinformatically assemble mtDNA. Interestingly, unusual mt-genomes may remain undiscovered in otherwise well-sampled groups of animals because traditional methods have failed to obtain positive results. For example, preliminary data suggest that multipartite mtDNA has evolved in some anthozoan cnidarians (order Ceriantharia) (Brugler and France 2007; Kayal et al. 2013) and demosponges (genus Mycale) (unpublished data) but has not been fully described due to sequencing/assembly difficulties. The discoveries of unusual mt-genomes open new questions in the study of animal mtDNA, such as the formation, maintenance, and expression of multipartite genomes, which in the absence of mitosis should represent a formidable task. Studying such genomes could also provide alternative explanations for unusual experimental results in other supposedly more familiar taxa. Indeed, several recent studies have forced us to rethink mtDNA biology in even the most familiar “model” animal species (e.g. Guo et al. 2003; Temperley et al. 2010; Lewis et al. 2015).
Finally, in many groups our knowledge of mitochondrial biology is limited to the mtDNA sequence itself, while the vast majority of proteins functioning in mitochondria are encoded by nuclear genomes. Recent explosive growth in the quantity of nuclear genomic and transcriptomic data from nonmodel organisms, including many nonbilaterian animals, will provide a fruitful avenue for understanding mitochondrial physiology and evolution in animals.
Acknowledgments
We thank Daniel Sloan, Jonathan Wendel, Kateryna Makova—our associate editor and two anonymous reviewers for their insightful comments on earlier versions of this article, and Paul Veach for his help with illustrations. A large part of the research reviewed here was supported by a grant from the National Science Foundation to DVL (DEB-0829783). W.P. was supported in part by the French National Research Agency (ANR) grant Ancestrome (ANR-10-BINF-01-01).
Literature Cited
- Aanen D, Spelbrink J, Beekman M. 2014. What cost mitochondria? The maintenance of functional mitochondrial DNA within and across generations. Philos Trans R Soc Lond B Biol Sci. 369:20130438.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abascal F, Posada D, Zardoya R. 2012. The evolution of the mitochondrial genetic code in arthropods revisited. Mitochondrial DNA 23:84–91. [DOI] [PubMed] [Google Scholar]
- Ameur A, et al. 2011. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 7:e1002028.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290:457–465. [DOI] [PubMed] [Google Scholar]
- Armstrong MR, Block VC, Phillips MS. 2000. A multipartite mitochondrial genome in the potato cyst nematode Globodera pallida. Genetics 154:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ax P. 1996. Multicellular animals: a new approach to the phylogenetic order in nature. Berlin (Germany): Springer. [Google Scholar]
- Baer C, Miyamoto M, Denver D. 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 8:619–631. [DOI] [PubMed] [Google Scholar]
- Baker L, Reeve M. 1974. Laboratory culture of the lobate ctenophore Mnemiopsis mccradyi with notes on feeding and fecundity. Mar Biol. 26:57–62. [Google Scholar]
- Ballard J, Whitlock M. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744. [DOI] [PubMed] [Google Scholar]
- Bar-Yaacov D, et al. 2013. RNA-DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Res. 23:1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemy R, Seligmann H. 2016. Cryptic tRNAs in chaetognath mitochondrial genomes. Comput Biol Chem. 62:119–132. [DOI] [PubMed] [Google Scholar]
- Bazin E, Glemin S, Galtier N. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science. 312:570–572. [DOI] [PubMed] [Google Scholar]
- Beagley CT, et al. 1995. Mitochondrial genomes of Anthozoa (Cnidaria). Progress Cell Res. 5:149–153. [Google Scholar]
- Beagley CT, Okada NA, Wolstenholme DR. 1996. Two mitochondrial group I introns in a metazoan, the sea anemone Metridium senile: one intron contains genes for subunits 1 and 3 of NADH dehydrogenase. Proc Natl Acad Sci U S A. 93:5619–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckenbach A, Robson S, Crozier R. 2005. Single nucleotide +1 frameshifts in an apparently functional mitochondrial cytochrome b gene in ants of the genus Polyrhachis. J Mol Evol. 60:141–152. [DOI] [PubMed] [Google Scholar]
- Bernt M, et al. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69:352–364. [DOI] [PubMed] [Google Scholar]
- Berthier F, Renaud M, Alziari S, Durand R. 1986. RNA mapping on Drosophila mitochondrial DNA: precursors and template strands. Nucleic Acids Res. 14:4519–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilewitch J, Degnan S. 2011. A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol Biol. 11:228.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, et al. 2015. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Yokobori S, Mörl M, Dörner M, Pääbo S. 1997. RNA editing in metazoan mitochondria: staying fit without sex. FEBS Lett. 409:320–324. [DOI] [PubMed] [Google Scholar]
- Borst P, Kroon A. 1969. Mitochondrial DNA: physicochemical properties, replication, and genetic function. Int Rev Cytol. 26:107–190. [DOI] [PubMed] [Google Scholar]
- Breton S, et al. 2014. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 30:555–564. [DOI] [PubMed] [Google Scholar]
- Breton S, et al. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 28:1645–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge D, Cunningham CW, Schierwater B, Desalle R, Buss LW. 1992. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci U S A. 89:8750–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman S, Mcfadden C. 2012. The mitochondrial genome of Paraminabea aldersladei (Cnidaria: Anthozoa: Octocorallia) supports intramolecular recombination as the primary mechanism of gene rearrangement in octocoral mitochondrial genomes. Genome Biol Evol. 4:994–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, George M, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci. 76:1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugler M, France S. 2007. The complete mitochondrial genome of the black coral Chrysopathes formosa (Cnidaria:Anthozoa:Antipatharia) supports classification of antipatharians within the subclass Hexacorallia. Mol Phylogenet Evol. 42:776–788. [DOI] [PubMed] [Google Scholar]
- Buchan J, Stansfield I. 2007. Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell 99:475–487. [DOI] [PubMed] [Google Scholar]
- Burger G, Forget L, Zhu Y, Gray MW, Lang BF. 2003. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc Natl Acad Sci U S A. 100:892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray M, Forget L, Lang B. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5:418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Yan Y, Javadi P, Lang B. 2009. Group I-intron trans-splicing and mRNA editing in the mitochondria of placozoan animals. Trends Genet. 25:381–386. [DOI] [PubMed] [Google Scholar]
- Cameron S, Yoshizawa K, Mizukoshi A, Whiting M, Johnson K. 2011. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera). BMC Genomics 12:394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capt C, Passamonti M, Breton S. 2015. The human mitochondrial genome may code for more than 13 proteins. Mitochondrial DNA 1–4. [DOI] [PubMed] [Google Scholar]
- Chandel NS. 2014. Mitochondria as signaling organelles. BMC Biol. 12:34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. 1984. The Drosophila mitochondrial genome. Oxf Surv Eukaryot Genes 1:1–35. [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22:252–271. [DOI] [PubMed] [Google Scholar]
- Clayton DA. 1982. Replication of animal mitochondrial DNA. Cell 28:693.. [DOI] [PubMed] [Google Scholar]
- Collins AG. 2002. Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J Evol Biol. 15:418–432. [Google Scholar]
- Daly M, et al. 2007. The phylum Cnidaria: a review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 127–182. [Google Scholar]
- Dellaporta S, et al. 2006. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci U S A. 103:8751–8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, et al. 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330:1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D, Morris K, Lynch M, Vassilieva L, Thomas W. 2000. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science 289:2342–2344. [DOI] [PubMed] [Google Scholar]
- Dickey A, et al. 2015. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics 16:439.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann M, Haen KM, Lavrov DV, Wörheide G. 2012. Molecular phylogeny of glass sponges (Porifera, Hexactinellida): increased taxon sampling and inclusion of the mitochondrial protein-coding gene, cytochrome oxidase subunit I. Hydrobiologia 687:11–20. [Google Scholar]
- Dong W, Song S, Jin D, Guo X, Shao R. 2014. Fragmented mitochondrial genomes of the rat lice, Polyplax asiatica and Polyplax spinulosa: intra-genus variation in fragmentation pattern and a possible link between the extent of fragmentation and the length of life cycle. BMC Genomics 15:44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Reis M, et al. 2015. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 25:2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Beaupré H, et al. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol BIol 10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C, Ryan J. 2015. The evolution of animal genomes. Curr Opin Genet Dev. 35:25–32. [DOI] [PubMed] [Google Scholar]
- Eitel M, Osigus H, Desalle R, Schierwater B. 2013. Global diversity of the placozoa. PLoS One 8:e57131.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emblem A, et al. 2014. Sea anemones possess dynamic mitogenome structures. Mol Phylogenet Evol. 75:184–193. [DOI] [PubMed] [Google Scholar]
- Ender A, Schierwater B. 2003. Placozoa are not derived cnidarians: evidence from molecular morphology. Mol Biol Evol. 20:130–134. [DOI] [PubMed] [Google Scholar]
- Erpenbeck D, Aryasari R, Hooper J, Worheide G. 2015. A mitochondrial intron in a verongid sponge. J Mol Evol. 80:13–17. [DOI] [PubMed] [Google Scholar]
- Erpenbeck D, et al. 2007. Mitochondrial diversity of early-branching metazoa is revealed by the complete mt genome of a haplosclerid demosponge. Mol Biol Evol. 24(1):19–22. [DOI] [PubMed] [Google Scholar]
- Evans N, Lindner A, Raikova E, Collins A, Cartwright P. 2008. Phylogenetic placement of the enigmatic parasite, Polypodium hydriforme, within the Phylum Cnidaria. BMC Evol Biol. 8:139.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E, Casanova J. 2006. Comparison of chaetognath mitochondrial genomes and phylogenetical implications. Mitochondrion 6:258–262. [DOI] [PubMed] [Google Scholar]
- Faure E, et al. 2011. Probable presence of an ubiquitous cryptic mitochondrial gene on the antisense strand of the cytochrome oxidase I gene. Biol Direct. 6:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE. 1992. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol Biochem Parasitol. 52:145.. [DOI] [PubMed] [Google Scholar]
- Feng J, et al. 2014. New phylogenomic and comparative analyses provide corroborating evidence that Myxozoa is Cnidaria. Mol Phylogenet Evol. 81:10–18. [DOI] [PubMed] [Google Scholar]
- Fiala I, Bartošová-Sojková P, Whipps CM. 2015. Classification and phylogenetics of Myxozoa In: Okamura B, Gruhl A, Bartholomew JL, editors. Myxozoan evolution, ecology and development. Cham: Springer; p 85–110. [Google Scholar]
- Figueroa D, Baco A. 2015. Octocoral mitochondrial genomes provide insights into the phylogenetic history of gene order rearrangements, order reversals, and cnidarian phylogenetics. Genome Biol Evol. 7:391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegontov P, et al. 2015. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol Biol Evol. 32:1115–1131. [DOI] [PubMed] [Google Scholar]
- Foox J, Siddall M. 2015. The road to Cnidaria: history of phylogeny of the Myxozoa. J Parasitol. 101:269–274. [DOI] [PubMed] [Google Scholar]
- Fricova D, et al. 2010. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5' termini. Microbiology 156:2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami H, Chen C, Chiou C, Knowlton N. 2007. Novel group I introns encoding a putative homing endonuclease in the mitochondrial cox1 gene of scleractinian corals. J Mol Evol. 64:591–600. [DOI] [PubMed] [Google Scholar]
- Gazave E, et al. 2010. Molecular phylogeny restores the supra-generic subdivision of homoscleromorph sponges (Porifera, Homoscleromorpha). PLoS One 5:e14290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, et al. 2011. No longer Demospongiae: Homoscleromorpha formal nomination as a fourth class of Porifera. Hydrobiologia 1–8. [Google Scholar]
- Gibson T, Blok V, Dowton M. 2007. Sequence and characterization of six mitochondrial subgenomes from Globodera rostochiensis: multipartite structure is conserved among close nematode relatives. J Mol Evol. 65:308–315. [DOI] [PubMed] [Google Scholar]
- GIGA Community of Scientists 2014. The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes. J. Heredity 105:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly JF, Allen AP, West GB, Brown JH. 2005. The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc Natl Acad Sci U S A. 102:140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud A, et al. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608. [DOI] [PubMed] [Google Scholar]
- Gissi C, et al. 2010. Hypervariability of ascidian mitochondrial gene order: exposing the myth of deuterostome organelle genome stability. Mol Biol Evol. 27:211–215. [DOI] [PubMed] [Google Scholar]
- Goddard J, Wolstenholme D. 1980. Origin and direction of replication in mitochondrial DNA molecules from the genus Drosophila. Nucleic Acids Res. 8:741–757. [PMC free article] [PubMed] [Google Scholar]
- Goddard M, Burt A. 1999. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci U S A. 96:13880–13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M, Leigh J, Roger A, Pemberton A. 2006. Invasion and persistence of a selfish gene in the Cnidaria. PLoS One 1:e3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. 2000. Functions and mechanisms of RNA editing. Annu Rev Genet. 34:499–531. [DOI] [PubMed] [Google Scholar]
- Gray M, Burger G, Lang B. 1999. Mitochondrial evolution. Science 283:1476–1481. [DOI] [PubMed] [Google Scholar]
- Gray M, Lang B, Burger G. 2004. Mitochondria of protists. Annu Rev Genet. 38:477–524. [DOI] [PubMed] [Google Scholar]
- Grossman L, Wildman D, Schmidt T, Goodman M. 2004. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 20:578–585. [DOI] [PubMed] [Google Scholar]
- Gualberto J, et al. 2014. The plant mitochondrial genome: dynamics and maintenance. Biochimie. 100:107–120. [DOI] [PubMed] [Google Scholar]
- Guo B, et al. 2003. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 423:456–461. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. 2008. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 6:e204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haen K, Lang B, Pomponi S, Lavrov D. 2007. Glass sponges and bilaterian animals share derived mitochondrial genomic features: a common ancestry or parallel evolution? Mol Biol Evol. 24:1518–1527. [DOI] [PubMed] [Google Scholar]
- Haen K, Pett W, Lavrov D. 2010. Parallel loss of nuclear-encoded mitochondrial aminoacyl-tRNA synthetases and mtDNA-encoded tRNAs in Cnidaria. Mol Biol Evol. 27:2216–2219. [DOI] [PubMed] [Google Scholar]
- Haen K, Pett W, Lavrov D. 2014. Eight new mtDNA sequences of glass sponges reveal an extensive usage of + 1 frameshifting in mitochondrial translation. Gene 535:336–344. [DOI] [PubMed] [Google Scholar]
- Harlid A, Janke A, Arnason U. 1997. The mtDNA sequence of the ostrich and the divergence between paleognathous and neognathous birds. Mol Biol Evol. 14:754–761. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Ropiquet A, Couloux A, Cruaud C. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol. 68:293–310. [DOI] [PubMed] [Google Scholar]
- Hausner G. 2012. Introns, Mobile Elements, and Plasmids In: Bullerwell CE, editor. Organelle Genetics. Berlin, Heidelberg (Germany: ): Springer; p 329–357. [Google Scholar]
- Hausner G, Hafez M, Edgell DR. 2014. Bacterial group I introns: mobile RNA catalysts. Mobile DNA 5:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfenbein K, Fourcade H, Vanjani R, Boore J. 2004. The mitochondrial genome of Paraspadella gotoi is highly reduced and reveals that chaetognaths are a sister group to protostomes. Proc Natl Acad Sci U S A. 101:10639–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchon D, Szitenberg A, Shefer S, Ilan M, Feldstein T. 2015. Mitochondrial group I and group II introns in the sponge orders Agelasida and Axinellida. BMC Evol Biol. 15:278.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, et al. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae). Mitochondrial DNA 25:29–30. [DOI] [PubMed] [Google Scholar]
- Itsara L, et al. 2014. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 10:e1003974.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J, Piganeau G, Eyre-Walker A. 2016. The rate of adaptive evolution in animal mitochondria. Mol Ecol. 25:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Barker S, Shao R. 2013. Substantial variation in the extent of mitochondrial genome fragmentation among blood-sucking lice of mammals. Genome Biol Evol. 5:1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourda C, Santini S, Rocher C, Le Bivic A, Claverie J. 2015. Mitochondrial genome sequence of the glass sponge Oopsacas minuta. Genome Announc. 3:e00823–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnkowska A, et al. 2016. A Eukaryote without a mitochondrial organelle. Curr Biol. 26:1274–1284. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, et al. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, et al. 2012. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol. 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Lavrov D. 2008. The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene 410:177–186. [DOI] [PubMed] [Google Scholar]
- Kayal E, Roure B, Philippe H, Collins A, Lavrov D. 2013. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol Biol. 13:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Salk J, Schmitt M, Loeb L. 2013. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9:e1003794.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent M, et al. 2001. Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol. 48:395–413. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1993. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217:624–626. [DOI] [PubMed] [Google Scholar]
- Klassen R, Meinhardt F. 2007. Linear protein-primed replicating plasmids in eukaryotic microbes In: editors. Microbial Linear Plasmids. Berlin (Germany): Springer; p 187–226. [Google Scholar]
- Knight R, Freeland S, Landweber L. 2001. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2:49–58. [DOI] [PubMed] [Google Scholar]
- Kohn A, et al. 2012. Rapid evolution of the compact and unusual mitochondrial genome in the ctenophore, Pleurobrachia bachei. Mol Phylogenet Evol. 63:203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Thomas JA, Welch JJ, Brey T, Bromham L. 2007. Metabolic rate does not calibrate the molecular clock. Proc Natl Acad Sci. 104:15388–15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B, Laforest M, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet. 23:119–125. [DOI] [PubMed] [Google Scholar]
- Larsson N. 2010. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 79:683–706. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Poujol R. 2011. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol Biol Evol. 28:729–744. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Lavrov D, Lang B. 2005. Transfer RNA gene recruitment in mitochondrial DNA. Trends Genet. 21:129–133. [DOI] [PubMed] [Google Scholar]
- Lavrov D, Adamski M, Chevaldonné P, Adamska M. 2016. Extensive mitochondrial mRNA editing and unusual mitochondrial genome organization in calcaronean sponges. Curr Biol. 26:86–92. [DOI] [PubMed] [Google Scholar]
- Lavrov D, Forget L, Kelly M, Lang B. 2005. Mitochondrial genomes of two demosponges provide insights into an early stage of animal evolution. Mol Biol Evol. 22:1231–1239. [DOI] [PubMed] [Google Scholar]
- Lavrov D, et al. 2013. Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol Biol Evol. 30:865–880. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Brown WM. 2001. Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8, normally-structured tRNAs, and has a gene arrangement relatable to those of coelomate metazoans. Genetics 157:621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV, Lang BF. 2014. Mitochondrial genomes in unicellular relatives of animals In: Wells RD,, Bond JS, Klinman J, Masters BSS, Bell E, editors. Molecular Life Sciences. New York (NY): Springer, pp. 1–4. [Google Scholar]
- Le T, et al. 2000. Phylogenies inferred from mitochondrial gene orders—a cautionary tale from the parasitic flatworms. Mol Biol Evol. 17:1123–1125. [DOI] [PubMed] [Google Scholar]
- Lewis S, et al. 2015. A rolling circle replication mechanism produces multimeric lariats of mitochondrial DNA in Caenorhabditis elegans. PLoS Genet. 11:e1004985.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane A, Marzuki S, Ozawa T, Tanaka M. 1989. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet 1:642–645. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kurokawa T, Sekino M, Tanabe T, Watanabe K. 2013. Complete mitochondrial DNA sequence of the ark shell Scapharca broughtonii: an ultra-large metazoan mitochondrial genome. Comp Biochem Physiol Part D Genomics Proteomics 8:72–81. [DOI] [PubMed] [Google Scholar]
- Lynch M. 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller's ratchet in mitochondrial genomes. Mol Biol Evol. 13:209–220. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2010. Evolution of the mutation rate. Trends Genet. 26:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2011. The lower bound to the evolution of mutation rates. Genome Biol Evol. 3:1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Blanchard JL. 1998. Deleterious mutation accumulation in organelle genomes. Genetica 102–103:29–39. [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311:1727–1730. [DOI] [PubMed] [Google Scholar]
- Manuel M. 2006. Phylogeny and evolution of calcareous sponges. Can J Zool. 84:225–241. [Google Scholar]
- Marcadé I, et al. 2007. Structure and evolution of the atypical mitochondrial genome of Armadillidium vulgare (Isopoda, Crustacea). J Mol Evol. 65:651–659. [DOI] [PubMed] [Google Scholar]
- Marck C, Grosjean H. 2002. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. rna 8:1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Palumbi SR. 1993. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci U S A. 90: 4087–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abarca F, Toro N. 2000. Group II introns in the bacterial world. Mol Microbiol. 38:917–926. [DOI] [PubMed] [Google Scholar]
- Mcbride H, Neuspiel M, Wasiak S. 2006. Mitochondria: more than just a powerhouse. Curr Biol. 16:R551–R560. [DOI] [PubMed] [Google Scholar]
- Mcfadden C, et al. 2011. Limitations of mitochondrial gene barcoding in Octocorallia. Mol Ecol Resour. 11:19–31. [DOI] [PubMed] [Google Scholar]
- Medina M, Collins A, Takaoka T, Kuehl J, Boore J. 2006. Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci U S A. 103:9096–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C, Montooth K, Rand D. 2007. Positive and negative selection on the mitochondrial genome. Trends Genet. 23:259–263. [DOI] [PubMed] [Google Scholar]
- Milbury C, Gaffney P. 2005. Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Mar Biotechnol. (NY) 7:697–712. [DOI] [PubMed] [Google Scholar]
- Milbury C, Lee J, Cannone J, Gaffney P, Gutell R. 2010. Fragmentation of the large subunit ribosomal RNA gene in oyster mitochondrial genomes. BMC Genomics 11:485.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills CE. 2014. Phylum Ctenophora: list of all valid species names. Electronic internet document available at http://faculty.washington.edu/cemills/Ctenolist.html. Published by the author, web page established March 1998, last updated 16 March 2014.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15:1568–1571. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Machida R, Nishida S. 2010. Complete mitochondrial genome sequences of the three pelagic chaetognaths Sagitta nagae, Sagitta decipiens and Sagitta enflata. Comp Biochem Physiol Part D Genomics Proteomics 5:65–72. [DOI] [PubMed] [Google Scholar]
- Miyazawa H, Yoshida M, Tsuneki K, Furuya H. 2012. Mitochondrial genome of a Japanese placozoan. Zool Sci. 29:223–228. [DOI] [PubMed] [Google Scholar]
- Moroz L, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C, Cardenas P. 2015. Proposal for a revised classification of the Demospongiae (Porifera). Front Zool. 12:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 76:444–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. 2008. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol. 25:120–130. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. 2009. The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol. 9:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Suzuki Y, Nozawa M. 2010. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 11:265–289. [DOI] [PubMed] [Google Scholar]
- Nesnidal M, Helmkampf M, Bruchhaus I, El-Matbouli M, Hausdorf B. 2013. Agent of whirling disease meets orphan worm: phylogenomic analyses firmly place Myxozoa in Cnidaria. PLoS One 8:e54576.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. 2003. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol Biol Evol. 20:1231–1239. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature 246:96–98. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474. [DOI] [PubMed] [Google Scholar]
- Okimoto R, Macfarlane J, Clary D, Wolstenholme D. 1992. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 130:471–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osigus H, Eitel M, Bernt M, Donath A, Schierwater B. 2013. Mitogenomics at the base of Metazoa. Mol Phylogenet Evol. 69:339–351. [DOI] [PubMed] [Google Scholar]
- Pagliarini D, Rutter J. 2013. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 27:2615–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, et al. 2013. The complete mitochondrial genome of Hydra vulgaris (Hydroida: Hydridae). Mitochondrial DNA 25:418–419. [DOI] [PubMed] [Google Scholar]
- Papillon D, Perez Y, Caubit X, Le Parco Y. 2004. Identification of chaetognaths as protostomes is supported by the analysis of their mitochondrial genome. Mol Biol Evol. 21:2122–2129. [DOI] [PubMed] [Google Scholar]
- Parham J, et al. 2006. The phylogeny of Mediterranean tortoises and their close relatives based on complete mitochondrial genome sequences from museum specimens. Mol Phylogenet Evol. 38:50–64. [DOI] [PubMed] [Google Scholar]
- Parsons T, et al. 1997. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 15:363–368. [DOI] [PubMed] [Google Scholar]
- Petersen J, et al. 2014. Chromera velia, endosymbioses and the rhodoplex hypothesis–plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol Evol. 6:666–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett W, Lavrov D. 2013. The twin-arginine subunit C in Oscarella: origin, evolution, and potential functional significance. Integr Comp Biol. 53:495–502. [DOI] [PubMed] [Google Scholar]
- Pett W, Lavrov D. 2015. Cytonuclear interactions in the evolution of animal mitochondrial tRNA metabolism. Genome Biol Evol. 7:2089–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett W, et al. 2011. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: insight from mtDNA and the nuclear genome. Mitochondrial DNA 22:130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianka ND. 1974. Ctenophora In: Giese AC, Pearse JS, editors. Reproduction of marine invertebrates. New York (NY): Academic Press. [Google Scholar]
- Pisani D, et al. 2015. Genomic data do not support comb jellies as the sister group to all other animals. Proc Natl Acad Sci U S A. 112:15402–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Haddock S, Sogin M, Harbison G. 2001. A molecular phylogenetic framework for the phylum Ctenophora using 18S rRNA genes. Mol Phylogenet Evol. 21:218–230. [DOI] [PubMed] [Google Scholar]
- Pont-Kingdon GA, et al. 1995. A coral mitochondrial mutS gene. Nature 375(6527):109–111. [DOI] [PubMed] [Google Scholar]
- Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K. 2007. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci U S A. 104:13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race HL, Herrmann RG, Martin W. 1999. Why have organelles retained genomes? Trends Genet. 15:364–370. [DOI] [PubMed] [Google Scholar]
- Richter C, Park J, Ames B. 1988. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 85:6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Schwarz F, Hering L, Boggemann M, Bleidorn C. 2015. The utility of genome skimming for phylogenomic analyses as demonstrated for glycerid relationships (Annelida, Glyceridae). Genome Biol Evol. 7:3443–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riepsamen A, Blok V, Phillips M, Gibson T, Dowton M. 2008. Poly(T) variation within mitochondrial protein-coding genes in Globodera (Nematoda: Heteroderidae). J Mol Evol. 66:197–209. [DOI] [PubMed] [Google Scholar]
- Riepsamen A, et al. 2011. Poly(T) variation in heteroderid nematode mitochondrial genomes is predominantly an artefact of amplification. J Mol Evol. 72:182–192. [DOI] [PubMed] [Google Scholar]
- Rosengarten R, Sperling E, Moreno M, Leys S, Dellaporta S. 2008. The mitochondrial genome of the hexactinellid sponge Aphrocallistes vastus: evidence for programmed translational frameshifting. BMC Genomics 9:33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Coppotelli G, Hoffer B, Olson L. 2014. Maternally transmitted mitochondrial DNA mutations can reduce lifespan. Sci Rep. 4:6569.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, et al. 2013. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature 501:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rot C, Goldfarb I, Ilan M, Huchon D. 2006. Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol Biol. 6:71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein ND, et al. 2013. Deep sequencing of mixed total DNA without barcodes allows efficient assembly of highly plastic ascidian mitochondrial genomes. Genome Biol Evol. 5:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone C, et al. 2002. Mitochondrial DNA in metazoa: degree of freedom in a frozen event. Gene 286:3–12. [DOI] [PubMed] [Google Scholar]
- Sahyoun A, et al. 2015. Towards a comprehensive picture of alloacceptor tRNA remolding in metazoan mitochondrial genomes. Nucleic Acids Res. 43:8044–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K. 1990. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol Rev. 54:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbisa E, Tanzariello F, Reyes A, Pesole G, Saccone C. 1997. Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene 205:125–140. [DOI] [PubMed] [Google Scholar]
- Schneider A. 2011. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem. 80:1033–1053. [DOI] [PubMed] [Google Scholar]
- Schulz F, et al. 2016. A Rickettsiales symbiont of amoebae with ancient features. Environ Microbiol. 18:2326–2342. [DOI] [PubMed] [Google Scholar]
- Segovia R, Pett W, Trewick S, Lavrov D. 2011. Extensive and evolutionarily persistent mitochondrial tRNA editing in velvet worms (phylum Onychophora). Mol Biol Evol. 28:2873–2881. [DOI] [PubMed] [Google Scholar]
- Shao Z, Graf S, Chaga O, Lavrov D. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 381:92–101. [DOI] [PubMed] [Google Scholar]
- Shen Y, et al. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci U S A. 107:8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorovitch A, Buss L, Dellaporta S. 2007. Comparative genomics of large mitochondria in Placozoans. PLoS Genet. 3:e13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simion P, Bekkouche N, Jager M, Queinnec E, Manuel M. 2015. Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora. Zoology (Jena) 118:102–114. [DOI] [PubMed] [Google Scholar]
- Singh T, et al. 2009. Tunicate mitogenomics and phylogenetics: peculiarities of the Herdmania momus mitochondrial genome and support for the new chordate phylogeny. BMC Genomics 10:534.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. 2015. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics. 151):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, et al. 2012. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol. 4:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Keeling P. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 112:10177–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola E, et al. 2015. Evolutionary analysis of mitogenomes from parasitic and free-living flatworms. PLoS One 10:e0120081.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, et al. 2003. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell 12:689–698. [DOI] [PubMed] [Google Scholar]
- Stampar S, Maronna M, Kitahara M, Reimer J, Morandini A. 2014. Fast-evolving mitochondrial DNA in Ceriantharia: a reflection of hexacorallia paraphyly? PLoS One 9:e86612.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25:1129–1137. [DOI] [PubMed] [Google Scholar]
- Sultana T, et al. 2013. Comparative analysis of complete mitochondrial genome sequences confirms independent origins of plant-parasitic nematodes. BMC Evol Biol. 13:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W, Ackerman M, Miller S, Doak T, Lynch M. 2012. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci U S A. 109:18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szitenberg A, Rot C, Ilan M, Huchon D. 2010. Diversity of sponge mitochondrial introns revealed by cox 1 sequences of Tetillidae. BMC Evol Biol. 10:288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, et al. 1997. Role of mutator alleles in adaptive evolution. Nature 387:700–702. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, et al. 2015. The mitochondrial genomes of a myxozoan genus Kudoa are extremely divergent in Metazoa. PLoS One 10:e0132030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Temperley R, Richter R, Dennerlein S, Lightowlers R, Chrzanowska-Lightowlers ZM. 2010. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science 327:301.. [DOI] [PubMed] [Google Scholar]
- Valach M, Moreira S, Kiethega G, Burger G. 2014. Trans-splicing and RNA editing of LSU rRNA in Diplonema mitochondria. Nucleic Acids Res. 42:2660–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles Y, Halanych K, Boore J. 2008. Group II introns break new boundaries: presence in a bilaterian's genome. PLoS One 3:e1488.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren J, Vierstraete A. 1999. Insertional RNA editing in metazoan mitochondria: the cytochrome b gene in the nematode Teratocephalus lirellus. RNA 5:622–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt O, Erpenbeck D, Wörheide G. 2008. A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics 9:350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt O, Wülfing E, Wörheide G. 2012. Molecular phylogenetic evaluation of classification and scenarios of character evolution in calcareous sponges (Porifera, class Calcarea). PLoS One 7:e33417.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Nickisch-Rosenegk M, Brown WM, Boore JL. 2001. Complete sequence of the mitochondrial genome of the tapeworm Hymenolepis diminuta: gene arrangements indicate that Platyhelminths are Eutrochozoans. Mol Biol Evol. 18:721–730. [DOI] [PubMed] [Google Scholar]
- Wallace D. 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 76:781–821. [DOI] [PubMed] [Google Scholar]
- Wallace DC. 1992. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science 256:628–632. [DOI] [PubMed] [Google Scholar]
- Wang X, Lavrov D. 2007. Mitochondrial genome of the homoscleromorph Oscarella carmela (Porifera, Demospongiae) reveals unexpected complexity in the common ancestor of sponges and other animals. Mol Biol Evol. 24:363–373. [DOI] [PubMed] [Google Scholar]
- Wang X, Lavrov D. 2008. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae. PLoS One 3:e2723.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lavrov D. 2011. Gene recruitment—a common mechanism in the evolution of transfer RNA gene families. Gene 475:22–29. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu M. 2015. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 5:7949.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrior R, Gall J. 1985. The mitochondrial DNA of Hydra attenuata and Hydra littoralis consists of two linear molecules. Arch Des Sci. 38:439–445. [Google Scholar]
- Watanabe K, Yokobori S. 2011. tRNA modification and genetic code variations in animal mitochondria. J Nucleic Acids 2011:623095.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Bessho Y, Kawasaki M, Hori H. 1999. Mitochondrial genes are found on minicircle DNA molecules in the mesozoan animal Dicyema. J Mol Biol. 286:645–650. [DOI] [PubMed] [Google Scholar]
- Wei D, et al. 2012. The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS One 7:e33973.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Rand DM. 2000. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics 156:385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan N, Kocot K, Moroz L, Halanych K. 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc Natl Acad Sci U S A. 112:5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme DR. 1992a. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR. 1992b. Genetic novelties in mitochondrial genomes of multicellular animals. Curr Opin Genet Dev. 2:918–925. [DOI] [PubMed] [Google Scholar]
- Wörheide G, et al. 2012. Deep phylogeny and evolution of sponges (phylum Porifera). Adv Mar Biol. 61:1–78. [DOI] [PubMed] [Google Scholar]
- WoRMS Editorial Board (2015). World Register of Marine Species. Available at: http://www.marinespecies.org [Google Scholar]
- Xu S, et al. 2012. High mutation rates in the mitochondrial genomes of Daphnia pulex. Mol Biol Evol. 29:763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes F, Van Houten B. 1997. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 94:514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata F, et al. 2015. Phylogenomic analyses support traditional relationships within Cnidaria. PLoS One 10:e0139068.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardoya R, Meyer A. 1998. Complete mitochondrial genome suggests diapsid affinities of turtles. Proc Natl Acad Sci U S A. 95:14226–14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. 2011. Animal biodiversity: an introduction to higher-level classification and taxonomic richness. Zootaxa 3148:7–12. [DOI] [PubMed] [Google Scholar]
- Zouros E. 2013. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 40:1–31. [Google Scholar]
- Zouros E, Freeman K, Ball A, Pogson G. 1992. Direct evidence for extensive paternal mitochondrial DNA inheritance in the marine mussel Mytilus. Nature 359:412–414. [DOI] [PubMed] [Google Scholar]