Abstract
Some photosynthetic organisms live in extremely low light environments. Light limitation is associated with selective forces as well as reduced exposure to mutagens, and over evolutionary timescales it can leave a footprint on species’ genomes. Here, we present the chloroplast genomes of four green algae (Bryopsidales, Ulvophyceae), including the endolithic (limestone-boring) alga Ostreobium quekettii, which is a low light specialist. We use phylogenetic models and comparative genomic tools to investigate whether the chloroplast genome of Ostreobium corresponds to our expectations of how low light would affect genome evolution. Ostreobium has the smallest and most gene-dense chloroplast genome among Ulvophyceae reported to date, matching our expectation that light limitation would impose resource constraints reflected in the chloroplast genome architecture. Rates of molecular evolution are significantly slower along the phylogenetic branch leading to Ostreobium, in agreement with the expected effects of low light and energy levels on molecular evolution. We expected the ability of Ostreobium to perform photosynthesis in very low light to be associated with positive selection in genes related to the photosynthetic machinery, but instead, we observed that these genes may be under stronger purifying selection. Besides shedding light on the genome dynamics associated with a low light lifestyle, this study helps to resolve the role of environmental factors in shaping the diversity of genome architectures observed in nature.
Keywords: genome streamlining, photosynthesis, rates of evolution, boring algae, stoichiogenomics
Introduction
Light is rapidly attenuated under water, yet some photosynthetic organisms thrive in extremely low light marine habitats (Shashar and Stambler 1992; Mock and Kroon 2002; Larkum, Douglas, et al. 2003). Specialized lifestyles may leave a footprint on organisms’ genomes (Dutta and Paul 2012; Raven et al. 2013). For example, high-light and low light strains of the cyanobacterium Prochlorococcus have different genome sizes, GC contents and rates of molecular evolution, as well as other genome features that have been associated with their niche specialization (Hess et al. 2001; Rocap et al. 2003; Dufresne et al. 2005; Paul et al. 2010). Similar studies targeting the nuclear genomes of eukaryotic algae have also begun to emerge (see Raven et al. 2013 for a review). Different ecotypes of the microalga Ostreococcus, for example, show distinctive genome traits (Jancek et al. 2008), although in this case it is not clear whether low light has played a role.
The production of high-energy cofactors (ATP and NADPH) and the uptake of nitrogen are modulated by light intensity (MacIsaac and Dugdale 1972; Cochlan et al. 1991; Kirk 1994) and therefore it is logical to expect that the genome architecture of lineages living under low light conditions is influenced by resource constraints. Selection for saving resources and shortening replication times, in addition to random genetic drift, have been associated with the loss of genes, intergenic spacers and introns, a process known as genome streamlining (Giovannoni et al. 2005; Lynch 2006; Hessen et al. 2010; Wolf and Koonin 2013). Genome architecture can also be affected by limited supply of key elements such as nitrogen and phosphorus: different nucleotides and amino acids differ in their atomic composition, so molecules containing less atoms of the limiting nutrient may provide a selective advantage in certain niches (Acquisti et al. 2009; Elser et al. 2011; Raven et al. 2013). The Prochlorococcus strain with the smallest genome and highest content of nitrogen-poor molecules is found in surface waters, where irradiance is higher but nutrients are more depleted than in the habitat of the low light strain (Rocap et al. 2003; Dufresne et al. 2005). One could expect that when light is low enough to restrict growth rates and nitrogen uptake, organisms with small genomes and a high proportion of nitrogen-poor molecules may have better evolutionary fitness.
Sunlight may also leave footprints in a genome by directly or indirectly altering molecular rates of evolution (the molecular pacemaker). Light is a major contributor to environmental energy including solar radiation, thermal energy and chemical (metabolic) energy (Clarke and Gaston 2006). Environmental energy stimulates metabolism at many levels, and it is known that energy-rich habitats are often characterized by higher evolutionary rates (Davies et al. 2004; Clarke and Gaston 2006). Solar radiation, especially ultraviolet (UV), also plays a direct mutagenic role and may thus accelerate molecular evolution (Rothschild 1999; Willis et al. 2009). Thermal and chemical energy also depend on light: light incidence increases temperatures (e.g., in the tropics) and supports primary productivity (and consequently increases the energy available for metabolism and growth). Oxidative DNA damage generally occurs during metabolic reactions; therefore higher metabolic rates can lead to higher mutation rates (Gillooly et al. 2005). Generation times also play into it, being shorter and fixing mutations (on populations) more rapidly when the environmental energy is higher, which often happens when there is a combination of higher temperatures, metabolic rates and solar radiation (Rohde 1992; Wright and Rohde 2013). As a consequence of all these factors, it is reasonable to expect that organisms living in low-energy areas, like shaded habitats, have relatively slower rates of molecular evolution.
Challenging environments may impose particular selective regimes, which could leave a footprint of positive selection in genes undergoing adaptation. Changes in proteins that provide higher fitness in a given circumstance (e.g., low light) can be detected at the molecular level by an excess of nonsynonymous substitutions over synonymous ones (Yang 1998). Evidence of positive selection for example in the Rubisco gene (involved in carbon fixation) in mosses has been associated with its adaptation to the declining levels of atmospheric CO2 since their origination in the Ordovician (Raven and Colmer 2016). In cases of organisms living in extremely low light, it would be reasonable to expect positive selection in genes related to the photosynthetic machinery, reflecting adaptation to low light. To our knowledge, this idea has never been tested in eukaryotic algae.
The siphonous green alga Ostreobium is a convenient organism to investigate photosynthesis under low light conditions (Fork and Larkum 1989; Koehne et al. 1999; Wilhelm and Jakob 2006). Ostreobium has an endolithic (limestone-boring) lifestyle: it bores into carbonate substrates and populates all sorts of marine limestones worldwide, including shells and coral skeletons. Only a small portion of the available light reaches Ostreobium in its usual habitat: ∼99% of the light can be attenuated by the first millimeter of limestone (Nienow et al. 1988; Matthes et al. 2001). Other photosynthetic organisms living on the limestone substrate can further attenuate light: the living tissue of corals and their zooxanthellae, for example, absorb 95–99.9% of the available light (Halldal 1968; Schlichter et al. 1997). Even under these extreme low light conditions, Ostreobium carries out oxygenic photosynthesis (Kühl et al. 2008). Cyanobacteria coexisting with Ostreobium enhance their light interception by manufacturing far red-absorbing chlorophylls (Chl d and f; Chen and Blankenship 2011), whereas Ostreobium has a special chlorophyll antenna that allows it to harvest far red light (Magnusson et al. 2007). Ostreobium is also able to grow in quite deep waters, being abundant even at depths over 200 m where only a handful of algal species can persist (Littler et al. 1985; Dullo et al. 1995; Aponte and Ballantine 2001). Here the light is filtered strongly towards the blue end of the spectrum, with a peak at ∼470–480 nm (Larkum and Barrett 1983) and a different light harvesting strategy is employed: the carotenoid siphonaxanthin transfers light energy to chlorophyll and the reaction centers (Kageyama et al. 1977). Thus the success of Ostreobium in terms of its cosmopolitan distribution is associated not only with its efficiency in light utilization but also its ability to employ a range of light harvesting strategies (Fork and Larkum 1989; Schlichter et al. 1997; Magnusson et al. 2007; Tribollet 2008), for which the underlying genomic basis has never been explored.
The light-driven genomic traits of Ostreobium can only be investigated in a comparative framework. While algal nuclear genome sequences are still scarce, chloroplast genomes are better sampled and constitute a powerful tool for molecular evolutionary studies (Lemieux et al. 2014). Ostreobium belongs to the Bryopsidales (Ulvophyceae), a diverse order of seaweeds for which only a handful of chloroplast genomes are available (Leliaert and Lopez-Bautista 2015). Additional chloroplast genomes of species from this order can help us investigate genomic traits correlated to low light in Ostreobium.
The goal of this study is to evaluate the evolutionary dynamics of the chloroplast genome of the low light alga Ostreobium using comparative and phylogenetic methods. Because comparative analyses in a phylogenetic context require a sufficiently large sample of genomes, we present the chloroplast genomes of four green algae, including Ostreobium quekettii and members from three other families in the same order, all previously uncharacterized. We used a combination of stoichiogenomics (the study of elemental composition of macromolecules; Elser et al. 2011) and models of molecular rate variation to investigate our expectations for a lineage adapted to low light conditions. Our first expectation related to light-dependent resources limitation: if the Ostreobium lineage has evolved in low-energy and low-nutrient conditions, its chloroplast genome can be expected to be smaller, more compact (i.e., with less intergenic spacers, introns and repeats) and contain less nitrogen than the chloroplast genomes of related algae. Our second expectation was that the phylogenetic branch leading to Ostreobium has slower rates of molecular evolution (i.e., mutation rates) than other branches in the phylogeny due to fewer mutations induced by UV and slower generation times often associated with low energy niches. Lastly, we would expect genes related to its photosynthetic machinery to have experienced positive selection and enabled Ostreobium’s highly efficient light utilization.
Materials and Methods
Sequencing, Assembly and Annotation
Total genomic DNA of Ostreobium quekettii, Halimeda discoidea, Derbesia sp. and Caulerpa cliftonii were extracted using a modified cetyl trimethylammonium bromide (CTAB) method described in Cremen et al. (2016) and sequenced on an Illumina platform. The collection sites and library preparation details are described in the Supplementary Materials. Sequences were submitted to European Nucleotide Archive and GenBank (accession numbers LT593849, KX808496, KX808497 and KX808498).
Sequences were assembled using CLC Genomics Workbench 7.5.1 (http://www.clcbio.com). Circularity and scaffold regions were resolved by comparing the CLC assembly with assemblies generated independently with MEGAHIT (Li et al. 2015), SOAPdenovo2 (Luo et al. 2012) and SPADES (Nurk et al. 2013). Details about the assembly settings and quality checks are reported in the Supplementary Materials. A combination of automated pipelines and manual editing was used to annotate the chloroplast genomes, which is also described in the Supplementary Materials.
Comparative Analysis
In order to compare Ostreobium with other Ulvophyceae, the chloroplast genomes of Bryopsis plumosa (NC_026795), Tydemania expeditions (NC_026796), Ulva sp. (KP720616), Pseudendoclonium akinetum (AY835431) and Oltmannsiellopsis viridis (NC_008099), available in GenBank, were included in our comparative analysis. Genome features were extracted with Geneious 9.0.4 (Kearse et al. 2012). Hypothetical ORFs with <300 bp were excluded and the tilS gene was re-annotated as a pseudogene (not a CDS) in Tydemania and Bryopsis, where it has a frame shift or a stop codon in the middle of the gene. The number of repeats, including tandem and palindromic repeats, were calculated with the Geneious implementation of Phobos v.3.3.11 (Mayer 2007) and with the Emboss suite (http://www.bioinformatics.nl/emboss-explorer/); see Supplementary Materials for details.
Nitrogen (N) content quantification was based on the counts of N atoms per nucleotide or amino acid using the formula described in Acquisti et al. (2009): where is the number of N atoms in the i-th base and is the proportion of each base in the chloroplast genome. For the nucleotide counts we used nC = nG = 4 and nA = nT = 3.5 (Acquisti et al. 2009). For the coding DNA sequences (exons) we used nA = 5, nT = 2, nG = 5, and nC = 3. For amino acid counts (the theoretical proteome) we used n = 2 for asparagine, glutamine, lysine and tryptophan; n = 3 for histidine; n = 4 for arginine; and n = 1 for other amino acids. Copy number and expression levels play a major role in N utilization, but neither qPCR nor transcriptome analysis could be carried out because our source materials were of different developmental stages and environmental conditions. Instead, we investigated N-content in coding sequences and amino acids on a gene by gene basis, in addition to doing so at the whole chloroplast genome level. Assuming that expression levels of genes correlate among species, the gene by gene approach should reduce the problem of differential expression between genes and make for more realistic among-species comparisons. Finally, we also evaluated whether the average length of coding sequences is smaller in Ostreobium, as gene size reduction has been observed in some endosymbionts with reduced genome sizes (Charles et al. 1999).
Phylogeny, Rates of Evolution and Selection Analysis
The coding sequences of all species were aligned at the amino acid level using a locally installed version of MAFFT v7.215 (Katoh et al. 2002), with multithreading and default parameters, and then the aligned amino acid sequences were converted back to nucleotides using RevTrans (Wernersson and Pedersen 2003). The ftsH, rpoB, rpoC1, rpoC2 and ycf1 genes could not be reliably aligned (according to a visual assessment) and were excluded along with the tilS pseudogene from downstream analyses. A maximum likelihood phylogeny was built using RAxML (Stamatakis 2006) with a GTR + Γ model, a partitioning strategy separating 1st, 2nd and 3rd codon positions, and a rapid bootstrap search of 500 replicates. Oltmannsiellopsis viridis, Pseudendoclonium akinetum and Ulva sp. were used as outgroups.
In order to test whether DNA mutation (substitution) rates were slower in the Ostreobium lineage, we studied lineage-specific rates of molecular evolution using the baseml program from the PAML v.4.7 package (Yang 2007). We chose to perform this test at the nucleotide (rather than at the amino acid) level because we expect environmental energy to affect rates of molecular evolution at the nucleotide level (i.e., regardless whether the mutations are synonymous or nonsynonymous). We compared the fit of a model with unique rates of evolution across all branches (global clock) to a model with a different rate for the Ostreobium lineage (local clock) using the Akaike Information Criterion (AIC). Because rates of molecular evolution inherently vary among species, a model with two rates is likely to better fit the data than a single-rate model. While this is taken into consideration when calculating model fit (AIC penalizes parameter-rich models), we also verified the rates of molecular evolution under a relaxed clock model, whereby rates are free to vary on all branches of the phylogeny (see Supplementary Materials).
To evaluate whether photosynthetic genes have been under positive selection in the Ostreobium lineage, we excluded gene alignments containing less than four species and grouped (concatenated) genes into 15 gene classes (cf. Wicke et al. 2011). We analyzed this data set using the branch model implemented in PAML (Yang 1998, 2007) and the random effects branch-site model (branch-site REL) implemented in HyPhy (Kosakovsky Pond et al. 2005, 2011). The branch model was run with the codeml program, using the F3 × 4 codon model (Goldman and Yang 1994; Yang 2007). We compared the fit of a model with differential dN/dS ratio (ω) for Ostreobium and the background lineages, to a model with a universal ω for all branches (the null hypothesis) using the Akaike information criterion (AIC). This approach directly tests our hypothesis, but has a risk of returning a good fit for poor models because the null hypothesis (universal ω) may be overly simple (see Kosakovsky Pond et al. 2011). Therefore we also used the branch-site REL, which allows detecting positive selection in all branches of the phylogeny and the proportion of sites under selection (Kosakovsky Pond et al. 2005, 2011). We evaluated whether positive selection had occurred in the Ostreobium lineage with the likelihood ratio test and P values (with the Holm correction procedure) implemented in the branch-site REL method in HyPhy (Kosakovsky Pond et al. 2011).
Results
Four New Chloroplast Genomes of Bryopsidales
The sequence data of Ostreobium queketti, Halimeda discoidea, Derbesia sp. and Caulerpa cliftonii were assembled into complete (circular mapping) chloroplast genomes (fig. 1 andsupplementary fig. S1, Supplementary Material online). The mean coverage was 235× for Ostreobium, 3,983× for Halimeda, 1,116× for Caulerpa and 469× for Derbesia (supplementary fig. S2, Supplementary Material online). Two gapped scaffold regions in Halimeda seem to have a (possibly polymorphic) number of repeats. One of these gaps was closed with an alternative assembler software (SPADES, Nurk et al. 2013) and the other was coded as stretch of Ns (see supplementary fig. S2, Supplementary Material online). The main genome features including their sizes are shown in table 1.
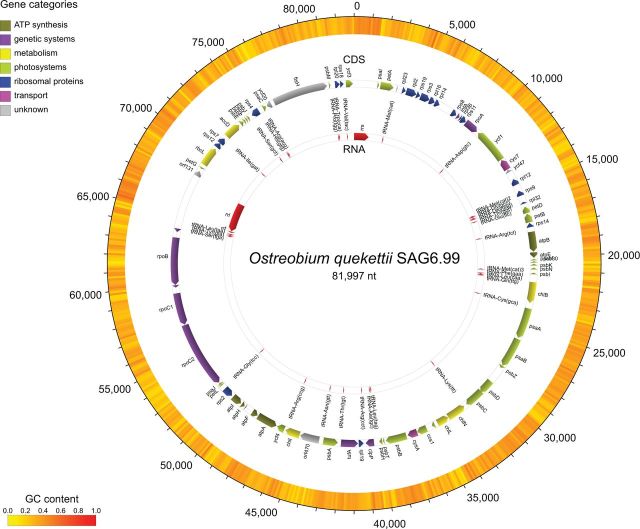
Fig. 1.—
Gene map of the Ostreobium quekettii chloroplast genome. Genes are colored by their known function.
Table 1.
Summary of the Chloroplast Genome Features of Ostreobium quekettii and Comparison with Other Ulvophyceae Chloroplast Genomes
| Species | Genome size (bp) | N content genome | N content coding DNA | N content proteome | GC content (%) | Introns | Repeats (50 bp+) | Tandem repeatsa | Palind. seqs | Int. spacers (%)b | Accession number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oltmannsiellopsis viridis | 151,933 | 3.702 | 3.698 | 1.361 | 40.5 | 10 | 84 | 5 | 652 | 39.57 | NC_008099 |
| Pseudendoclonium akinetum | 195,867 | 3.657 | 3.699 | 1.379 | 31.5 | 28 | 100 | 22 | 418 | 37.46 | AY835431 |
| Ulva sp. | 99,983 | 3.626 | 3.669 | 1.366 | 25.3 | 5 | 12 | 2 | 410 | 22.67 | KP720616 |
| Ostreobium quekettii | 81,997 | 3.656 | 3.692 | 1.369 | 31.9 | 6 | 8 | 1 | 100 | 11.96 | LT593849 |
| Bryopsis plumosa | 106,859 | 3.650 | 3.692 | 1.359 | 30.8 | 13 | 12 | 1 | 161 | 20.40 | NC_026795 |
| Derbesia sp. | 115,765 | 3.644 | 3.685 | 1.374 | 29.7 | 12 | 8 | 5 | 146 | 19.09 | KX808497 |
| Caulerpa cliftonii | 131,135 | 3.688 | 3.675 | 1.378 | 37.6 | 11 | 15 | 7 | 115 | 25.74 | KX808498 |
| Halimeda discoidea c | 122,075 | 3.653 | 3.681 | 1.363 | 32.2 | 14 | 19 | 11 | 112 | 19.96 | KX808496 |
| Tydemania expeditionis | 105,200 | 3.668 | 3.656 | 1.377 | 32.8 | 11 | 7 | 1 | 72 | 18.73 | NC_026796 |
Note.—Nitrogen (N) content in Genome and Coding DNA based on nucleotides, N content in Proteome based on amino acids counts.
Only tandem repeats with 15–1,000 bp were included in the count.
Excluding ORFs < 300 bp.
Halimeda has one scaffold with a unknown number of repeats annotated with 100 Ns.
Palind seqs, Palindromic repeats; Int. spacers, Intergenic spacers.
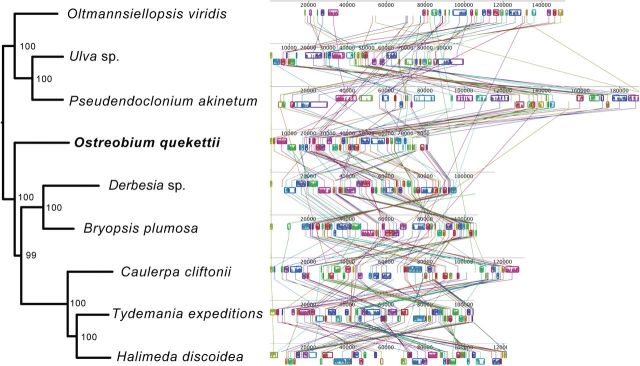
Gene content of Ostreobium is similar to related algae but it lacks the chloroplast envelope membrane protein gene (cemA) that is present in all other Ulvophyceae sequenced to date (supplementary table S1, Supplementary Material online). The tRNA(Ile)-lysidine synthase gene (tilS) seems to be a pseudogene in Ostreobium, Halimeda and Derbesia as it contains multiple in-frame stop codons. We could not identify it at all in Caulerpa cliftonii (i.e., no tBLASTx hits with e-values < 0.001 and identity > 50%, using Bryopsis plumosa as reference), although this pseudogene has been found in another Caulerpa species (Zuccarello et al. 2009). None of our chloroplast genomes have the organelle division inhibitor factor gene (minD), supporting the notion that this gene has been lost from the chloroplasts of Bryopsidales (Leliaert and Lopez-Bautista 2015). Like Ulva, Bryopsis and Tydemania, the chloroplast genomes sequenced here do not have the quadripartite architecture often found in green algae and land plants (Lemieux et al. 2000; Pombert 2005). Despite an overall highly conserved gene content, the Ulvophyceae genomes have multiple rearrangements as indicated in the Mauve alignment (fig. 2).
Fig. 2.—
Mauve alignment of chloroplast genomes available for algae of the class Ulvophyceae, including the endolithic alga Ostreobium quekettii and the three seaweeds sequenced in this study. Colored boxes indicate regions of synteny (collinear blocks, identified by the Progressive Mauve algorithm). The species are sorted according to a Maximum Likelihood phylogeny based on a concatenated alignment of the coding sequences of the chloroplast genomes; bootstrap values are indicated near branch nodes.
Genome Economics
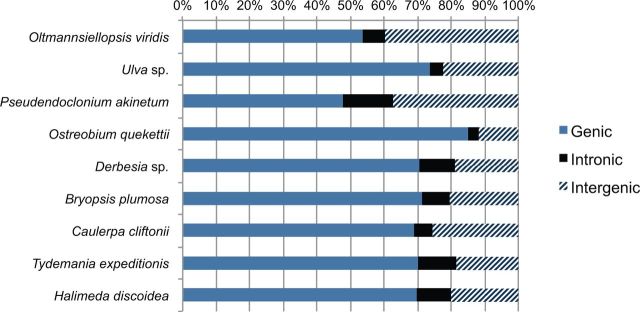
In order to evaluate some of our expectations regarding light-driven resource limitations on chloroplast genomes, we compared the chloroplast genome of Ostreobium with those of the eight other algae from the class Ulvophyceae in terms of size, compactness (gene-density) and nitrogen content. With 81,997 bp, Ostreobium has the smallest and most gene-dense chloroplast genome of all Ulvophyceae sequenced to date (table 1 and fig. 3). The size reduction in the Ostreobium chloroplast genome is not caused by gene loss (78 of 79 common plastid genes are present, supplementary table S1, Supplementary Material online) but by a reduction of intergenic spacers, introns and repeats (table 1 andfig. 3). Intergenic spacers compose only 11.9% of the Ostreobium chloroplast genome, compared with an average of 25.4% (std 8.4%) in other Ulvophyceae. Ostreobium also has a small number of introns, missing even the highly conserved tRNA-Leu (uaa) group I intron (Simon et al. 2003) that is present in other Bryopsidales chloroplast genomes (Leliaert and Lopez-Bautista 2015; this study). Nitrogen utilization in Ostreobium did not differ substantially from other algae, either in the nucleotide composition of the complete chloroplast DNA, the coding regions, or the amino acids of predicted proteins (table 1). Likewise, the N counts on a gene by gene basis did not reveal any obvious pattern (supplementary table S2, Supplementary Material online). The average gene length in Ostreobium was found to be similar to related algae (median difference of gene sizes = 0, supplementary table S3, Supplementary Material online).
Fig. 3.—
Proportion of genes, introns and intergenic spacers in the chloroplast genomes of algae of the class Ulvophyceae. Only ORFs >300 bp were included in the count. The percentage of intronic regions includes the intronic ORFs present in some species.
Rates of Evolution
To investigate whether the molecular pacemaker along the branch leading to Ostreobium is slower than in the remainder of the tree, we constructed a Maximum Likelihood (ML) phylogeny from the chloroplast genomes (71 genes concatenated, 47,559 bp, fig. 2) and fitted two models of molecular evolution to the same data set. We found that a model with differential rates of evolution for the branch leading to Ostreobium and the remaining branches of the phylogeny fits the data much better (ΔAIC = 92) than a model with a homogeneous rate across the entire tree. The branch rate parameter values estimated by ML are 0.81 for the Ostreobium branch versus 1.00 for the remainder of the tree. In other words, the relative rate of molecular evolution along the Ostreobium branch is 19% slower than along the other branches of the phylogeny.
A similar result was obtained by calculating the rates of molecular evolution with a relaxed molecular clock, but some branches other than the Ostreobium branch also had slower rates of molecular evolution (supplementary fig. S3, Supplementary Material online). Except for Bryopsis and Ostreobium, all other Bryopsidales showed a relatively fast rate. The rate estimated for the Ostreobium branch corresponds to 65% of the rates averaged across all other branches of the phylogeny.
Selection on Genes Related to Photosynthesis
Our third expectation was that genes related to the photosynthetic pathway have experienced positive selection in the lineage leading to Ostreobium. We concatenated genes encoding different subunits of the same protein to improve signal from short gene alignments. Using the branch model of Yang (1998), we tested whether the ω ratio (dN/dS) of the branch leading to Ostreobium differs from the background ω for other lineages in the phylogeny. If they do differ significantly, and if ω is >1, then positive selection could be inferred (Yang 1998). However, we found no indication that genes in the branch leading to Ostreobium have been under positive selection (table 2 and supplementary table S4, Supplementary Material online). Instead, we observed that most of the proteins related to the photosynthetic machinery have a stronger signature of purifying selection in the Ostreobium lineage than in other branches of the phylogeny (ΔAIC > 4, table 2). We note though that these results should be interpreted with prudence given the methods’ susceptibility of returning a good fit for poor models (see Kosakovsky Pond et al. 2011). In order to verify these results in light of an alternative method, we performed a second analysis using the random effects branch-site model.
Table 2.
Omega Values (dN/dS) for the Different Gene Classes in the Chloroplast Genomes of Ulvophycean Algae
| Single-ω model |
Two-ω model |
ΔAIC | ||
|---|---|---|---|---|
| ω global | ω background | ω Ostreobium | ||
| Photosynthetic light reactions | ||||
| atp | 0.025 | 0.028 | 0.009 | 25.043 |
| pet | 0.032 | 0.034 | 0.016 | 3.625 |
| psa | 0.019 | 0.021 | 0.007 | 29.851 |
| psb | 0.029 | 0.031 | 0.013 | 40.538 |
| Photosynthetic dark reactions | ||||
| chl | 0.022 | 0.024 | 0.012 | 4.749 |
| ccsA | 0.025 | 0.025 | 0.032 | −1.938 |
| rbcL | 0.020 | 0.023 | 0.007 | 13.922 |
| Translation and protein-modifying enzymes | ||||
| clp | 0.014 | 0.017 | 0.004 | 2.825 |
| infA | 0.039 | 0.044 | 0.020 | 0.660 |
| rpl | 0.039 | 0.040 | 0.029 | −0.643 |
| rps | 0.035 | 0.036 | 0.022 | 1.179 |
| tufA | 0.023 | 0.025 | 0.011 | 1.516 |
| Proteins not related to photosynthesis | ||||
| accD | 0.036 | 0.041 | 0.017 | 1.379 |
| cys | 0.027 | 0.027 | 6.291 | −2.000 |
| rpo | 0.003 | 0.003 | 0.002 | −1.749 |
Note.—Two models were tested a single ω for all lineages and a model with different ω values for Ostreobium and all other species. The goodness of fit of the two-ω over the single-ω model is given by ΔAIC.
The second analysis with the branch-site REL model, which can detect selection in all branches of the tree and parts of the alignment without having to specify lineages of interest a priori (Kosakovsky Pond et al. 2011), confirmed that there are no signatures of positive selection along the Ostreobium lineage (supplementary table S5, Supplementary Material online). As in the previous analysis, the branch-site REL model suggests that several gene classes have experienced stronger purifying selection in the lineage leading to Ostreobium: many of the genes show smaller ω values in the Ostreobium lineage (both mean ω and ω1—representing purifying selection) and a higher proportion of sites under the purifying selective regime when compared with the average values obtained for all other branches (supplementary table S5, Supplementary Material online).
Discussion
An Economical Genome
The endolithic alga Ostreobium has a remarkably small and compact chloroplast genome (figs. 1 and 3; table 1). We found that the economic nature of the Ostreobium chloroplast genome is not accomplished by a replacement of expensive nucleotides or amino acids (i.e., containing more N atoms) by more economic ones, but by an overall reduction of intergenic regions (fig. 3). Energy limitation resulting from the low light niche that this alga occupies may have contributed to an evolutionary reduction of the genome size. Due to the limited light available for photosynthesis, saving energy in any aspect of its cell biology including genome replication and transcription would result in a selective advantage. Introns significantly increase the costs of transcription (Lehninger et al. 1993; Castillo-Davis et al. 2002). Likewise, repeats and intergenic spacers consume resources, so these can be under selection towards reduction in energy-poor environments (Dufresne et al. 2005; Giovannoni et al. 2005).
Besides natural selection, neutral factors as random genetic drift and population sizes can also shape genome architecture (Lynch 2006; Lynch et al. 2006) and may have contributed to the chloroplast genome streamlining in Ostreobium. Genome reduction resulting from neutral evolution (or from a relaxation of purifying selection) is typically observed in obligate parasitic or symbiotic species, which tend to have small effective population sizes and therefore a higher influence of genetic drift (Mira et al. 2001; Wolf and Koonin 2013). Genes that are no longer essential to survival are under nearly neutral evolution, so the reduced genomes of parasitic species typically show substantial gene loss (e.g., loss of genes involved in photosynthesis) and, sometimes, an accumulation of pseudogenes (Mira et al. 2001; de Koning and Keeling 2006; McNeal et al. 2007; Wicke et al. 2013; Yan et al. 2015). Ostreobium, in contrast, has a tightly packed chloroplast genome, virtually no gene loss (except for cemA) and no sign of gene size reduction, supporting a considerable role of adaptive processes on its genome streamlining.
Evidently, not all photosynthetic organisms living in low light environments have reduced genome sizes. Acaryochloris marina is a shade specialist with an 8.3 Mb genome, which is large for a cyanobacteria (Swingley et al. 2008; Larsson et al. 2011). In this case, a different mechanism can be speculated on: by producing chlorophyll d, Acaryochloris may not experience the same resource constraints that Ostreobium does, and as it occupies a relatively uncompetitive niche, this cyanobacterium could be under relaxed purifying selection which might culminate in genome expansion (see Swingley et al. 2008; Larsson et al. 2011). Wolf and Koonin (2013) proposed the existence of two phases in genome evolution: an explosive innovation phase that leads to an increase in genome complexity followed by a longer reductive phase. It is possible that the Acaryochloris genome size reflects its recent innovation/adaptive phase while the Ostreobium lineage, which has occupied an endolithic low light lifestyle for more than 500 million years (Vogel and Brett 2009; Marcelino and Verbruggen 2016), possibly has been in a reductive stage over a longer timespan. The genus Acaryochloris, however, is much older than 500 Ma (Sánchez-Baracaldo 2015), and in order to test this hypothesis it would be necessary to know when the genus acquired chlorophyll d and when it transitioned to a shaded lifestyle. Naturally, algae living in well-lit habitats can also have small chloroplast genomes. Small cells tend to have small genomes, and small chloroplast genomes have been observed in picoplanktonic species like Ostreococcus and other Prasinophyceae species (Derelle et al. 2006; Lemieux et al. 2014). Small genomes in bloom forming species, as Ostreococcus, could be a selective advantage given their reduced replication times (see Cavalier-Smith 2005), however, at least for prokaryotes, no correlation between duplication times and genome sizes has been observed (Mira et al. 2001). The effects of random genetic drift and population sizes likely play a major role in shaping genome sizes in these cases (Lynch 2006).
Interestingly, small chloroplast genomes have also been observed in other organisms commonly inhabiting resource-poor niches. Plants in the Gnetophytes have a reduced chloroplast genome associated with a reduced number of introns and intergenic regions, in addition to some gene loss, which the authors suggest to be an adaptation to the resource-constrained habitats that these plants occupy (Wu et al. 2009). The recently published chloroplast genome of the palmophyllalean alga Verdigellas peltata, which typically occurs in deep waters and other shaded environments, is also small (79,444 bp long), compact and intronless (Leliaert et al. 2016). These observations support our hypothesis on the effects of low light and resource constraints on chloroplast genome size, though more extremophile species need to be sequenced in order to verify whether this pattern sustains.
Our results are restricted to the chloroplast genome. Based on microspectrophotometry estimates, Ostreobium’s nuclear genome (2C ≈ 0.5 pg) is on the smaller side of the genome size range among Ulvophyceae (0.1–6.1 pg) (Kapraun 2007). Because no nuclear genomes of Ulvophyceae have been sequenced to date, it is not currently feasible to analyze associations between low light and nuclear genome evolution. We anticipate that when more complete nuclear genomes are sequenced, equivalent analyses of the impact of resource constraints on nuclear genome evolution will follow.
Regarding gene loss, the cemA gene, involved in the uptake of inorganic carbon into chloroplasts (Rolland 1997), is the only gene absent from Ostreobium but present in all other Ulvophyceae (supplementary table S1, Supplementary Material online). Knock-out experiments in Chlamydomonas have shown that cemA is not essential for life or photosynthesis, but that its disruption drastically increases light sensitivity: mutants lacking a functional cemA have a lower threshold level of light perceived as excessive, so they accumulate large amounts of zeaxanthin, which is a pigment that dissipates excess light as heat (Rolland 1997). Consequently, mutants are only able to grow (photoautotrophically) under low light conditions (Rolland 1997). Although the possibility that this gene has been transferred to the nucleus in Ostreobium cannot be completely ruled out, cemA was neither found in other assembled contigs nor in a recently sequenced Ostreobium transcriptome (see section “Searching for cemA” in the Supplementary Materials). Once the capacity to tolerate high light is lost, there would be strong constraints on subsequent transitions to higher-light habitats, providing a plausible explanation for why Ostreobium lineages have diversified abundantly within the endolithic niche (Marcelino and Verbruggen 2016; Sauvage et al. 2016) but are not known to have diversified out of it (i.e., given origin to nonendolithic species). Endolithic algal species are often light saturated at low light intensities but some experimental studies show that they are able to photoacclimate to light levels approaching full solar irradiance (see Tribollet 2008 for a review). There are high levels of cryptic diversity within endolithic green algae (Marcelino and Verbruggen 2016; Sauvage et al. 2016) and it is not known which species are able to cope with higher levels of light, raising the question of whether cemA has been lost in other lineages of Ostreobium and whether they acquired other mechanisms to tolerate high light.
We expected to observe a larger proportion of nitrogen-poor molecules in the Ostreobium chloroplast genome for several reasons. First, low light irradiance limits the uptake of nitrogen (MacIsaac and Dugdale 1972; Cochlan et al. 1991) and it has been empirically demonstrated that Ostreobium growth is limited by nitrogen and phosphorous in naturally occurring concentrations (Carreiro-Silva et al. 2012). Second, absorption of nutrients may be difficult in endolithic environments due to limited circulation and thicker diffusive boundary layers (see Larkum, Koch, et al. 2003). However, our results indicate that the nitrogen content in the Ostreobium chloroplast genome (and predicted proteome) is similar to those of other algae in the same class (table 1 and supplementary tables S2 and S3, Supplementary Material online).
Several potential explanations can be raised. First, seaweeds in general may naturally be under nitrogen limitation (Vitousek and Howarth 1991; Harrison and Hurd 2001), resulting in all of the examined genomes having similar nitrogen content. Alternatively, genome replication and DNA repair may be less frequent in Ostreobium as a consequence of the reduced environmental energy, slow metabolism and growth, therefore a slower rate of nitrogen intake may be required and this economic aspect of Ostreobium is not reflected in its genome. Sample size could also be an issue: previous studies on N bias used nuclear genomes (Acquisti et al. 2009) and patterns may not be visible in the smaller chloroplast genome. Nitrogen limitation may also lead to overall genome reduction (Kang et al. 2015) rather than biases in nucleotide and amino acid composition. Finally, nitrogen utilization is largely dependent on the number of copies of the chloroplast genome and expression levels, which cannot be detected in our analyses. If fresh DNA extractions from algae growing in their natural conditions and belonging to the same developmental stage were available, would be interesting to perform comparative qPCR and transcriptome analyses to test whether this is the case.
In Prochlorococcus, it is the high-light strain that contains less nitrogen in its genome, although it is not substantially different from one of the low light strains (Dufresne et al. 2005). In this case, the nitrogen availability in the water column seems to play a more important role than a restricted nitrogen-uptake ability due to light limitation. Heterotrophic pathways have been observed in the genome of Prochlorococcus, especially in the low light strains, suggesting that they might use other sources of energy in addition to light (García-Fernández and Diez 2004). This potentially mitigates the effects of low irradiance on nitrogen uptake in this organism, which would explain why low light Prochlorococcus strains have more nitrogen in their genomes. A recent review (Raven et al. 2013) suggests a theoretical association between AT/GC ratios in genomes (which could culminate in nitrogen bias) and UV irradiation, but notes that this is not commonly observed in nature because multiple other factors influencing genome content may play a more significant role than light alone.
Slow Rates of Evolution
The results of two independent tests show that Ostreobium has a relatively slow rate of molecular evolution than closely related lineages. Other ulvophytes also seem to have slow rates of molecular evolution, which might be related to other species traits not analyzed here, but in Ostreobium, the most reasonable explanations relate to the effects of the low light niche that this endolithic alga occupies. Sunlight, including UV radiation, induces DNA damage, mutations and rearrangements (Ries et al. 2000; Raven et al. 2013; Kumar et al. 2014). While these changes often get repaired (see Boesch et al. 2011 for mechanisms), the frequency with which remaining mutations are passed through generations dictates the molecular pacemaker (Baer et al. 2007). Following this logic, low light lineages will likely have slower rates of molecular evolution than lineages living in high light conditions, as observed in Ostreobium and in low light strains of Prochlorococcus (Dufresne et al. 2005). In Prochlorococcus, it is likely that the loss of DNA repair genes also contributes to an increase in mutation rates in high light strains (Dufresne et al. 2005).
Sunlight also shapes evolutionary rates through environmental energy—it sustains primary productivity and ambient temperature. Energy-rich habitats are the epicenter of evolutionary change worldwide (Davies et al. 2004; Jetz and Fine 2012; Wright and Rohde 2013). This environmental energy is positively correlated to metabolic rates in many organisms (Allen et al. 2002) and the by-products of metabolic reactions (e.g., reactive oxygen and nitrogen species) are another major source of mutations (Gillooly et al. 2005; Boesch et al. 2011). It has been proposed that more solar radiation and higher temperatures increase metabolism and growth rates, shortening generation times and increasing mutation rates (Rohde 1992). Shorter generations lead to more mutations accumulated per unit of time, so species living in high-energy habitats tend to have faster rates of molecular evolution (Bromham 2011). One could speculate that the low energy niche that Ostreobium occupies results in slow metabolic rates and generation times (although they are unknown for this alga), culminating in a slow molecular pacemaker. Longer generation times have been associated with slow rates of molecular evolution in tree ferns (Zhong et al. 2014), which are also shade plants (Page 2002).
Selection in the Ostreobium Chloroplast Genome
We did not find evidence for positive selection on genes related to photosynthesis in the lineage leading to Ostreobium (table 2 and supplementary table S5, Supplementary Material online). On the contrary, we observed some signs, though weak, of stronger purifying selection in this lineage.
Ostreobium is known to have several features that facilitate low light photosynthesis. It is able to produce red-shifted chlorophylls and uses an uncommon uphill energy transfer from these chlorophylls to photosystem II (Koehne et al. 1999; Wilhelm and Jakob 2006). The photosynthesis-related proteins that are more likely to be affected by low light (e.g., the light harvesting complex superfamily and the pigments involved in light capture) are encoded in the nucleus (Green and Parson 2003), and so innovations in these genes would not be detected in our analysis. The recently sequenced nuclear genome of the seagrass Zostera marina revealed an expanded number of light harvesting complex B genes (Olsen et al. 2016). Like Ostreobium, Zostera is adapted to a light depleted (aquatic) niche when compared with its land plant relatives. We expect that interesting findings will result for Ostreobium with the analysis of transcriptome and nuclear genome data.
Another scenario that may have contributed to not observing selection is that the lineage leading to Ostreobium could have experienced an early burst of positive selection followed by purifying selection, and such a history may go undetected in analyses. If innovations related to low light adaptation appeared early in Ostreobium evolution and increased its fitness, it is expected that they would be immediately followed by purifying selection—especially if the loss of the cemA gene caused intolerance to high-light and confined the ancestral endolithic lineage to shaded habitats (where any mutation decreasing photosynthesis performance is likely to lead to decreased fitness). This scenario provides a plausible explanation for the stronger purifying selection on photosynthesis-related genes in the branch leading to Ostreobium when compared with other branches in the phylogeny. The available tools may not have enough power to detect faint episodes of selection, particularly if the data are saturated with synonymous substitutions or if selection occurred at deep internal branches (Kosakovsky Pond et al. 2011; Gharib and Robinson-Rechavi 2013). Although both analyses show some sign of a stronger purifying selection in the Ostreobium lineage, these results should be interpreted with caution as the phylogeny contains long branches (implying long periods of time: Ostreobium, for example, diverged 500 Ma ago), therefore substitutions may have saturated the data to a point where evolution cannot be reliably characterized by the models. Simulations mimicking the evolution of algal chloroplast genomes may help to characterize those methodological limitations. Finally, the power of these analyses will certainly increase as more genomic data of high and low light-adapted lineages become available.
Conclusion
We present the chloroplast genomes of four green algae (Bryopsidales) and investigate the genomic footprints of a low light lifestyle in the endolithic Ostreobium quekettii. This alga has the smallest and most gene-packed chloroplast genome among Ulvophyceae, which is a possible adaptation to light-related resources constraints. The molecular pacemaker is significantly slower in the phylogenetic branch leading to Ostreobium, consistent with a scenario where low energy levels reduce rates of molecular evolution. Unexpectedly, we observed some signs of higher levels of purifying selection in the photosynthesis-related genes in Ostreobium when compared with other algae. It is still unclear whether this result is allied to an early episodic positive selection followed by a strong purifying selection or to a methodological limitation, as the current methods may not have the power to detect selection in deep-branching lineages, especially if the data are saturated with substitutions. Sequencing additional chloroplast and nuclear genomes of different Ostreobium lineages and other low light adapted species will help to further clarify the genomic correlates of low light adaptations.
Supplementary Material
Acknowledgments
This work was supported by the Australian Biological Resources Study (RFL213-08), the Australian Research Council (FT110100585, DP150100705), the Holsworth Wildlife Research Endowment and the Sapere Aude Advanced grant from the Danish Council for Independent Research for the Natural Sciences. The Sophie Ducker Postgraduate Scholarship supported the publication fee. V.R.M. and M.C.M.C. receive a University of Melbourne scholarship. The Caulerpa sample was collected under the DEC Flora permit 10006072. We thank John West for providing the Derbesia strain and Claude Payri for facilitating field work in PNG. We thank four anonymous reviewers for their helpful comments. We are thankful to Karolina Fucikova, John Raven and the members of the Verbruggen lab for valuable insights during the execution of this study and preparation of the article.
Literature Cited
- Acquisti C, Elser JJ, Kumar S. 2009. Ecological nitrogen limitation shapes the DNA composition of plant genomes. Mol Biol Evol. 26:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AP, Brown JH, Gillooly JF. 2002. Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science 297:1545–1548. [DOI] [PubMed] [Google Scholar]
- Aponte NE, Ballantine DL. 2001. Depth distribution of algal species on the deep insular fore reef at Lee Stocking Island, Bahamas. Deep Sea Res I Oceanogr Res Pap. 48:2185–2194. [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 8:619–631. [DOI] [PubMed] [Google Scholar]
- Boesch P, et al. 2011. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta Mol Cell Res. 1813:186–200. [DOI] [PubMed] [Google Scholar]
- Bromham L. 2011. The genome as a life-history character: why rate of molecular evolution varies between mammal species. Philos Trans R Soc B Biol Sci. 366:2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreiro-Silva M, Kiene WE, Golubic S, McClanahan TR. 2012. Phosphorus and nitrogen effects on microbial euendolithic communities and their bioerosion rates. Mar Pollut Bull. 64:602–613. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. 2002. Selection for short introns in highly expressed genes. Nat Genet. 31:415–418. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot. 95:147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles H, Mouchiroud D, Lobry J, Gonçalves I, Rahbe Y. 1999. Gene size reduction in the bacterial aphid endosymbiont Buchnera. Mol Biol Evol. 16:1820–1822. [DOI] [PubMed] [Google Scholar]
- Chen M, Blankenship RE. 2011. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 16:427–431. [DOI] [PubMed] [Google Scholar]
- Clarke A, Gaston KJ. 2006. Climate, energy and diversity. Proc Biol Sci. 273:2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochlan WP, Price NM, Harrison PJ. 1991. Effects of irradiance on nitrogen uptake by phytoplankton: comparison of frontal and stratified communities. Mar Ecol Prog Ser. 69:103–116. [Google Scholar]
- Cremen MCM, Huisman JM, Marcelino VR, Verbruggen H. 2016. Taxonomic revision of Halimeda (Bryopsidales, Chlorophyta) in southwestern Australia. Aust Syst Bot. 29:41–54. [Google Scholar]
- Davies TJ, Savolainen V, Chase MW, Moat J, Barraclough TG. 2004. Environmental energy and evolutionary rates in flowering plants. Proc R Soc B Biol Sci. 271:2195–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Keeling PJ. 2006. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 103:11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. 2005. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dullo W-C, et al. 1995. Factors controlling holocene reef growth: an interdisciplinary approach. Facies 32:145–188. [Google Scholar]
- Dutta C, Paul S. 2012. Microbial lifestyle and genome signatures. Curr Genomics 13:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser JJ, Acquisti C, Kumar S. 2011. Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends Ecol Evol. 26:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fork DC, Larkum AWD. 1989. Light harvesting in the green alga Ostreobium sp., a coral symbiont adapted to extreme shade. Mar Biol. 103:381–385. [Google Scholar]
- García-Fernández JM, Diez J. 2004. Adaptive mechanisms of nitrogen and carbon assimilatory pathways in the marine cyanobacteria Prochlorococcus. Res Microbiol. 155:795–802. [DOI] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M. 2013. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol Biol Evol. 30:1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly JF, Allen AP, West GB, Brown JH. 2005. The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc Natl Acad Sci. 102:140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Green BR, Parson WW, editors. 2003. Light-harvesting antennas in photosynthesis. Dordrecht: Springer Netherlands. [Google Scholar]
- Halldal P. 1968. Photosynthetic capacities and photosynthetic action spectra of endozoic algae of the massive coral Favia. Biol Bull. 134:411–424. [Google Scholar]
- Harrison PJ, Hurd CL. 2001. Nutrient physiology of seaweeds: application of concepts to aquaculture. Cah Biol Mar. 42:71–82. [Google Scholar]
- Hess WR, et al. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth Res. 70:53–71. [DOI] [PubMed] [Google Scholar]
- Hessen DO, Jeyasingh PD, Neiman M, Weider LJ. 2010. Genome streamlining and the elemental costs of growth. Trends Ecol Evol. 25:75–80. [DOI] [PubMed] [Google Scholar]
- Jancek S, Gourbière S, Moreau H, Piganeau G. 2008. Clues about the genetic basis of adaptation emerge from comparing the proteomes of two Ostreococcus ecotypes (Chlorophyta, Prasinophyceae). Mol Biol Evol. 25:2293–2300. [DOI] [PubMed] [Google Scholar]
- Jetz W, Fine PVA. 2012. Global gradients in vertebrate diversity predicted by historical area-productivity dynamics and contemporary environment. PLoS Biol. 10:e1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama A, Yokohama Y, Shimura S, Ikawa T. 1977. An efficient excitation energy transfer from a carotenoid, siphonaxanthin to chlorophyll a observed in a deep-water species of chlorophycean seaweed. Plant Cell Physiol. 18:477–480. [Google Scholar]
- Kang M, Wang J, Huang H. 2015. Nitrogen limitation as a driver of genome size evolution in a group of karst plants. Sci Rep. 5:11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF. 2007. Nuclear DNA content estimates in green algal lineages: Chlorophyta and Streptophyta. Ann Bot. 99:677–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JTO. 1994. Light and photosynthesis in aquatic ecosystems. Cambridge: Cambridge University Press. [Google Scholar]
- Koehne B, Elli G, Jennings RC, Wilhelm C, Trissl HW. 1999. Spectroscopic and molecular characterization of a long wavelength absorbing antenna of Ostreobium sp. Biochim Biophys Acta Bioenerg. 1412: 94–107. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, et al. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 28:3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Kühl M, Holst G, Larkum AWD, Ralph PJ. 2008. Imaging of oxygen dynamics within the endolithic algal community of the massive coral Porites lobata. J Phycol. 44:541–550. [DOI] [PubMed] [Google Scholar]
- Kumar RA, Oldenburg DJ, Bendich AJ. 2014. Changes in DNA damage, molecular integrity, and copy number for plastid DNA and mitochondrial DNA during maize development. J Exp Bot. 65:6425–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum AWD, Barrett J. 1983. Light-harvesting processes in algae. Adv Bot Res. 10:1–219. [Google Scholar]
- Larkum AWD, Douglas SE, Raven JA, editors. 2003. Photosynthesis in algae. Dordrecht: Springer Netherlands. [Google Scholar]
- Larkum AWD, Koch EW, Kühl M. 2003. Diffusive boundary layers and photosynthesis of the epilithic algal community of coral reefs. Mar Biol. 142:1073–1082. [Google Scholar]
- Larsson J, Nylander JA, Bergman B. 2011. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol. 11:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. 1993. Principles of biochemistry. 2nd edn New York; Worth Publishers. [Google Scholar]
- Leliaert F, et al. 2016. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci Rep. 6:25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, Lopez-Bautista JM. 2015. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics 16:204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M. 2000. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 403:649–652. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M. 2014. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genomics 15:857.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. [DOI] [PubMed] [Google Scholar]
- Littler MM, Littler DS, Blair SM, Norris JN. 1985. Deepest known plant life discovered on an uncharted seamount. Science 227:57–59. [DOI] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2006. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol. 60:327–349. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311:1727–1730. [DOI] [PubMed] [Google Scholar]
- MacIsaac JJ, Dugdale RC. 1972. Interactions of light and inorganic nitrogen in controlling nitrogen uptake in the sea. Deep Sea Res Oceanogr Abstr. 19:209–232. [Google Scholar]
- Magnusson SH, Fine M, Kühl M. 2007. Light microclimate of endolithic phototrophs in the scleractinian corals Montipora monasteriata and Porites cylindrica. Mar Ecol Prog Ser. 332:119–128. [Google Scholar]
- Marcelino V, Verbruggen H. 2016. Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci Rep. 6: 31508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes U, Turner SJ, Larson DW. 2001. Light attenuation by limestone rock and its constraint on the depth distribution of endolithic algae and cyanobacteria. Int J Plant Sci. 162:263–270. [Google Scholar]
- Mayer C. 2007. Phobos: a tandem repeat search tool. [cited 2016 Sep2 ]. Available from: http://www.geneious.com/plugins/phobos-plugin.
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW. 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596. [DOI] [PubMed] [Google Scholar]
- Mock T, Kroon BMA. 2002. Photosynthetic energy conversion under extreme conditions—II: the significance of lipids under light limited growth in Antarctic sea ice diatoms. Phytochemistry 61:53–60. [DOI] [PubMed] [Google Scholar]
- Nienow JA, McKay CP, Friedmann EI. 1988. The cryptoendolithic microbial environment in the Ross Desert of Antarctica: light in the photosynthetically active region. Microb Ecol. 16:271–289. [PubMed] [Google Scholar]
- Nurk S, et al. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in Computational Molecular Biology: 17th Annual International Conference, RECOMB 2013, Beijing, China, April 7-10, 2013. Proceedings. Berlin, Heidelberg: Springer. p. 158–170.
- Olsen JL, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530:331–335. [DOI] [PubMed] [Google Scholar]
- Page CN. 2002. Ecological strategies in fern evolution: a neopteridological overview. Rev Palaeobot Palynol. 119:1–33. [Google Scholar]
- Paul S, Dutta A, Bag SK, Das S, Dutta C. 2010. Distinct, ecotype-specific genome and proteome signatures in the marine cyanobacteria Prochlorococcus. BMC Genomics 11:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombert J-F. 2005. The Chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of Chlorophyte lineages. Mol Biol Evol. 22:1903–1918. [DOI] [PubMed] [Google Scholar]
- Raven JA, Beardall J, Larkum AWD, Sanchez-Baracaldo P. 2013. Interactions of photosynthesis with genome size and function. Philos Trans R Soc B Biol Sci. 368:20120264–20120264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Colmer TD. 2016. Life at the boundary: photosynthesis at the soil–fluid interface. A synthesis focusing on mosses. J Exp Bot. 67: 1613–1623. [DOI] [PubMed] [Google Scholar]
- Ries G, et al. 2000. Elevated UV-B radiation reduces genome stability in plants. Nature 406:98–101. [DOI] [PubMed] [Google Scholar]
- Rocap G, et al. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047. [DOI] [PubMed] [Google Scholar]
- Rohde K. 1992. Latitudinal gradients in species diversity: the search for the primary cause. Oikos 65:514–527. [Google Scholar]
- Rolland N. 1997. Disruption of the plastid ycf10 open reading frame affects uptake of inorganic carbon in the chloroplast of Chlamydomonas. Embo J. 16:6713–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild LJ. 1999. The influence of UV radiation on Protistan evolution. J Eukaryot Microbiol. 46:548–555. [DOI] [PubMed] [Google Scholar]
- Sánchez-Baracaldo P. 2015. Origin of marine planktonic cyanobacteria. Sci Rep. 5:17418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage T, Schmidt WE, Suda S, Fredericq S. 2016. A metabarcoding framework for facilitated survey of endolithic phototrophs with tufA. BMC Ecol. 16:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichter D, Kampmann H, Conrady S. 1997. Trophic potential and photoecology of endolithic algae living within coral skeletons. Mar Ecol. 18:299–317. [Google Scholar]
- Shashar N, Stambler N. 1992. Endolithic algae within corals – life in an extreme environment. J Exp Mar Bio Ecol. 163:277–286. [Google Scholar]
- Simon D, Fewer D, Friedl T, Bhattacharya D. 2003. Phylogeny and self-splicing ability of the plastid tRNA-Leu group I Intron. J Mol Evol. 57:710–720. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Swingley WD, et al. 2008. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci U S A. 105:2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribollet A. 2008. The boring microflora in modern coral reef ecosystems: a review of its roles In: Wisshak M, Tapanila L, editors. Current developments in bioerosion. Berlin, Heidelberg: Springer Berlin Heidelberg; p. 67–94. [Google Scholar]
- Vitousek P, Howarth R. 1991. Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry 13:87–115. [Google Scholar]
- Vogel K, Brett CE. 2009. Record of microendoliths in different facies of the Upper Ordovician in the Cincinnati Arch region USA: the early history of light-related microendolithic zonation. Palaeogeogr Palaeoclimatol Palaeoecol. 281:1–24. [Google Scholar]
- Wernersson R, Pedersen AG. 2003. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 31:3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25:3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, DePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C, Jakob T. 2006. Uphill energy transfer from long-wavelength absorbing chlorophylls to PSII in Ostreobium sp. is functional in carbon assimilation. Photosynth Res. 87:323–329. [DOI] [PubMed] [Google Scholar]
- Willis KJ, Bennett KD, Birks HJB. 2009. Variability in thermal and UV-B energy fluxes through time and their influence on plant diversity and speciation. J Biogeogr. 36:1630–1644. [Google Scholar]
- Wolf YI, Koonin EV. 2013. Genome reduction as the dominant mode of evolution. BioEssays 35:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SD, Rohde K. 2013. Energy and spatial order in niche and community. Biol J Linn Soc. 110:696–714. [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM. 2009. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet Evol. 52:115–124. [DOI] [PubMed] [Google Scholar]
- Yan D, et al. 2015. Auxenochlorella protothecoides and Prototheca wickerhamii plastid genome sequences give insight into the origins of non-photosynthetic algae. Sci Rep. 5:14465.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 15:568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhong B, Fong R, Collins LJ, McLenachan PA, Penny D. 2014. Two new fern chloroplasts and decelerated evolution linked to the long generation time in tree ferns. Genome Biol Evol. 6:1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarello GC, Price N, Verbruggen H, Leliaert F. 2009. Analysis of a plastid multigene data set and the phylogenetic position of the marine macroalga Caulerpa filiformis (Chlorophyta). J Phycol. 45:1206–1212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.