Summary
Medium‐chain fatty acids (MCFA, C6‐14 fatty acids) are an ideal feedstock for biodiesel and broader oleochemicals. In recent decades, several studies have used transgenic engineering to produce MCFA in seeds oils, although these modifications result in unbalance membrane lipid profiles that impair oil yields and agronomic performance. Given the ability to engineer nonseed organs to produce oils, we have previously demonstrated that MCFA profiles can be produced in leaves, but this also results in unbalanced membrane lipid profiles and undesirable chlorosis and cell death. Here we demonstrate that the introduction of a diacylglycerol acyltransferase from oil palm, EgDGAT1, was necessary to channel nascent MCFA directly into leaf oils and therefore bypassing MCFA residing in membrane lipids. This pathway resulted in increased flux towards MCFA rich leaf oils, reduced MCFA in leaf membrane lipids and, crucially, the alleviation of chlorosis. Deep sequencing of African oil palm (Elaeis guineensis) and coconut palm (Cocos nucifera) generated candidate genes of interest, which were then tested for their ability to improve oil accumulation. Thioesterases were explored for the production of lauric acid (C12:0) and myristic (C14:0). The thioesterases from Umbellularia californica and Cinnamomum camphora produced a total of 52% C12:0 and 40% C14:0, respectively, in transient leaf assays. This study demonstrated that the introduction of a complete acyl‐CoA‐dependent pathway for the synthesis of MFCA‐rich oils avoided disturbing membrane homoeostasis and cell death phenotypes. This study outlines a transgenic strategy for the engineering of biomass crops with high levels of MCFA rich leaf oils.
Keywords: plant biomass, plant oils, triacylglycerol, WRINKLED, Nicotiana, DGAT, GPAT9, medium chain, fatty acid
Introduction
Over recent years, the global production of vegetable oils has experienced constant growth, with over 179 million metric tons (MMT) being produced in 2015 (OECD/FAO, 2015), with the four major oil production crops being oil palm, soya bean, canola and sunflower. An important component of global oil consumption is medium‐chain fatty acids (MCFA), here defined as fatty acids in the range of 6–14 carbons in length. In addition, their application within the food industry MCFAs is an ideal source for biodiesel and also for a wide range of oleochemical feedstocks including pharmaceuticals, personal care products, lubricants and detergents (Arkcoll, 1988; Basiron and Weng, 2004). Currently, the predominant crop sources of MCFA‐enriched oils are coconut palm and oil palm (both palm oil and palm kernel oil) (Arkcoll, 1988). The production of these crops is limited to tropical and subtropical climates (Arkcoll, 1988; Basiron, 2007; Basiron and Weng, 2004; Kumar, 2011; Laureles et al., 2002). It has been projected that the global demand for vegetable oils will increase to approximately 240 MMT in 2050 (Corley, 2009), approximately 50% more than what is currently being produced. The development of new crops that can produce MCFA‐enriched oils in temperate climates has been proposed (Dehesh, 2001; Eccleston et al., 1996; Reynolds et al., 2015; Tjellstrom et al., 2013; Voelker et al., 1992; Wiberg et al., 2000) as a way to meet the growing global demand for MCFA in oleochemical production, pharmaceutical applications and personal care products.
Many studies have investigated the modification of seed oils to contain increased MCFA content, predominantly focused on the engineering of lauric acid (C12:0) (Eccleston and Ohlrogge, 1998; Knutzon et al., 1999; Voelker et al., 1992). In oilseeds, the engineered pathway begins with the overexpression of a specialized thioesterase (FATB) that prematurely truncates the standard fatty acid elongation cycle within the plastid allowing export into the cytoplasm. The MCFA in the cytoplasm is available for incorporation into triacylglycerols (TAG) via the endogenous oilseed pathways, with the acyl‐CoA‐dependent reactions of the Kennedy pathway (glycerol‐3‐phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAAT) and diacylglycerol acyltransferase (DGAT)) being the preferred route to avoid incorporation onto the membrane bound lipid head groups, such as phosphatidylcholine (PC). Previous studies have investigated the incorporation of MCFA into seed oils following the coordinated over‐expression of FATB and LPAAT (Figure 1a), achieving up to 67% of seed oil (Knutzon et al., 1999). More recently, transcriptomic analyses have enabled the identification of new FATB and LPAAT genes from many Cuphea species, which have been used to both modify the fatty acid profiles and improve the incorporation of MCFA into the TAG, respectively, of transgenic Camelina sativa seeds (Kim et al., 2015a,b).
Figure 1.

Graphical summary of the simplified TAG assembly pathway depicting the genes that have previously been used in combination for the increased production of oils (a). The oil sample represents the oil extracted from Nicotiana tabacum leaves with increased oil content (Vanhercke et al., 2014a,b), comprised of common fatty acids. The same gene families have been targets for the enrichment of the medium‐chain fatty acid (MCFA) content of oil in both previous studies (*) and in this study, denoted by MCFA (b). The oil sample (right) represents the oil extracted from vacuum infiltrated Nicotiana benthamiana leaves with an increased MCFA content as highlighted by the gas chromatography trace. The extracted MCFA oil remained solid until 21°C. WRI1 = WRINKLED1; FAS = fatty acid synthase; FAT = fatty acid thioesterase; GPAT = glycerol‐3‐phosphate acyltransferase; LPAAT = lysophosphatidic acid acyltransferase; DGAT = diacylglycerol acyltransferase; TAG = triacylglycerol.
Evidence, although, has found that endogenous TAG synthesis pathways in developing oilseeds are not ideal for incorporating MCFA into TAG (Wiberg et al., 1997, 2000) and that newly synthesized MCFA become incorporated into membrane bound lipids, which impedes lipid flux, agronomic performance and can even result in cell death through chlorosis (Bates et al., 2014; Voelker et al., 1996). Therefore, it would seem that although MCFA can be produced in plant cells, there is a poor pathway for incorporation into seed TAG. It has also been recognized that the accumulation of unusual fatty acids in PC appears to be a bottleneck for their enriched incorporation into TAG (Bates and Browse, 2011; Reynolds et al., 2015). In the example of engineering ricinoleic acid into oilseeds, it has been demonstrated that the endogenous pathways need to be removed in conjunction with the ectopic expression of the specialized pathway counterpart (Adhikari et al., 2016; Bates and Browse, 2011; Burgal et al., 2008; Chen et al., 2016; van Erp et al., 2011, 2015).
Recent work has demonstrated that engineering high oil levels in plant biomass is a realistic proposition (Vanhercke et al., 2013, 2014a,b) with the accumulation of significant levels of TAG in Nicotiana tabacum leaves (up to 15%) being attained by the coordinated transgenic expression of genes normally involved in oil production in seeds (Vanhercke et al., 2014b). Such approaches have uncovered a synergism involving an increase the production of fatty acids in the plastid (WRINKLED1 (WRI1)), improving the assembly of fatty acids into leaf oils (DGAT) and slowing the catabolism of these oils (OLEOSIN, OLE1 (Fan et al., 2013; Winichayakul et al., 2013)); and sugar‐dependent‐1, SDP1 (Fan et al., 2014; Kelly et al., 2013; Kim et al., 2014; Vanhercke et al., 2014a). Although the production of TAG in biomass offers a new source of common vegetable oils, these new expression platforms could also be adapted to produce high levels of novel fatty acids, such as MCFA (Reynolds et al., 2015; Wood, 2014). Our first steps in this direction involved the overexpression of thioesterases from Umbellularia californica, Cinnamomum camphora and Cocos nucifera which resulted in the production of MCFA in leaf tissues (Reynolds et al., 2015). However, these metabolic pathways also resulted in high levels of MCFA in PC resulting in severe chlorosis and cell death (Bates et al., 2014; Wiberg et al., 2000), similar to conclusions drawn from oilseed engineering. The incorporation of MCFA into the membrane lipids of vegetative tissues is hence particularly problematic.
In this report, we extend the MCFA metabolic pathway by combining a series of previously published gene ensembles with three different DGAT1‐like genes isolated from the kernel of Elaeis guineensis (African oil palm). We also identified a functional GPAT9 from C. nucifera that was included in the metabolic pathway for improving the incorporation of MCFA into seed oils (Figure 1b). In this study, we demonstrate an improvement in MCFA utilization, which results in more efficient sequestering of MCFA in TAG, while also effectively limiting the accumulation of MCFA in membrane lipids. This nonseed approach also allows metabolic engineers to avoid some of the bottlenecks associated with oilseed engineering where the more common fatty acids are rapidly and specifically incorporated into seed oils.
Results
Identification of functional GPAT9 from Cocos nucifera (coconut)
AtGPAT9 has recently been identified as being responsible for the acylation of acyl‐CoAs to the sn‐1 position of the glycerol‐3‐phosphate (G3P) backbone, which also demonstrated the ability to increase TAG production (Shockey et al., 2016; Singer et al., 2016). Based on the fatty acid composition of oil extracted from C. nucifera (coconut), it was hypothesized that a GPAT9 from coconut may assist in boosting the MCFA content of transgenic oils. A homologous gene was identified by BLAST searching an assembled coconut endosperm transcriptome. Following isolation and sequencing of the full‐length transcript of interest, the open reading frame for the predicted CnGPAT9 was identified. The open reading frame was used for subsequent sequence alignment with the AtGPAT9 homolog, revealing that the sequences are ≈78% identical (Figure S1). Following multiple sequence alignment with other annotated GPAT sequences (Figure S2), it was confirmed that the isolated CnGPAT9 transcript clustered with other identified and predicted GPAT9 sequences.
Following identification of the full‐length sequence, the candidate CnGPAT9 was cloned into pJP3343 for testing functionality. Initially, the CnGPAT9 was tested for functionality using the transient N. benthamiana infiltration assay, by testing its ability to increase TAG content, through comparison against the AtGPAT9 as a positive control. Total lipids were extracted and analysed to determine the effect of GPAT9 expression on TAG content (Figure 2). From comparison with the p19 alone samples (0.1 ± 0.0%), it was determined that the expression of either AtGPAT9 (P = 0.05) and CnGPAT9 (P = 0.01) was sufficient in leading to significant increases in the TAG content in the leaf being 0.5 ± 0.2% and 0.7 ± 0.1%, respectively. However, there was no significant differences in the increased TAG effect between either GPAT9 candidates (P = 0.18). Collectively, these data illustrated that the isolated candidate sequence from coconut was a functional GPAT9.
Figure 2.

Testing the effect of glycerol‐3‐phosphate acyltransferase 9 (GPAT9) expression on triacylglycerol (TAG) content, comparing genes from Arabidopsis thaliana (AtGPAT9) and Cocos nucifera (CnGPAT9), determined by application of the transient Nicotiana benthamiana leaf expression assay (n = 4).
DGAT1 from oil palm promotes production of MCFA‐enriched oils
It has been previously demonstrated that MCFA‐containing oils could be produced in the leaves of N. benthamiana (Reynolds et al., 2015). Chlorosis was observed in some gene combinations following the high proportion of MCFA accumulating in the membrane lipids, especially PC. It was therefore hypothesized that the introduction of a DGAT capable of utilizing MCFA for esterification into TAG may increase the MCFA content while alleviating the chlorosis phenotype, while also increasing TAG production.
The publication of the Elaeis guineensis (African oil palm) transcriptome (Dussert et al., 2013) revealed gene candidates that may be involved in lipid synthesis pathways. These data included sequences for three predicted DGAT1s, which were codon optimized for expression in N. tabacum and synthesized by GeneArt. Although these candidates had not yet been characterized in terms of their substrate specificity, the fatty acid profile of the oils from oil palm (palm oil and palm kernel oil) (Edem, 2002) suggested that these DGAT1 candidates may exhibit specificity for MCFA substrates. These EgDGAT1 candidates were infiltrated in combination with AtWRI1 (Arabidopsis thaliana WRINKLED1) and CnLPAAT, both with and without the co‐expression of CcTE (thioesterase from Cinnamomum camphora), to determine their ability to both mediate TAG production and the incorporation of MCFA into TAG. Following 5 days of expression, observations of the leaves revealed that the negative chlorosis phenotype was associated with several gene combinations (Figure S3), but correlated with the expression of the CcTE in particular. Further analysis revealed that the chlorosis phenotype was alleviated by the addition of EgDGAT1.1 (a gene which is expressed in the palm kernel) when compared with AtDGAT1. It was hypothesized that the phenotypic improvement was due to the improved capacity of the EgDGAT1.1 to sequester elevated levels of MCFA into TAG. Analysis of the predicted EgDGAT1 genes via multiple alignments of the translated proteins (Figure S4) revealed that both the EgDGAT1.2 and EgDGAT1.3 isoforms may be nonfunctional due to the absence of highly conserved C‐ and N‐terminal motifs (Cao, 2011), which are responsible for the catalytic and regulatory activities of the DGAT1 enzyme, respectively (Liu et al., 2012; Xu et al., 2008).
Total lipids were extracted and analysed to better understand the relationship between chlorosis and particular gene combinations. The total fatty acid profile (Figure 3; blue) revealed that in the absence of CcTE expression, the TFA content was similar following the addition of either AtDGAT1 or EgDGAT1.1 (P = 0.23). Following the introduction of the CcTE, the TFA content was significantly higher for treatments including EgDGAT1.1 than compared to AtDGAT1 (P = 0.015). The same correlation was observed for TAG content (Figure 3; red). Although the TAG content was similar for AtWRI1 + AtDGAT1 and AtWRI1 + EgDGAT1.1 samples (P = 0.45), following the addition of CcTE the TAG content was significantly increased for samples expressing EgDGAT1.1, compared to samples expressing the AtDGAT1 (P = 0.027). This result suggested that following CcTE expression, in the presence of AtDGAT1, fatty acid synthesis (FAS) was inhibited due to inefficient assembly of the MCFA into glycerolipids. Conversely, there appeared to be no inhibition of FAS following the addition of EgDGAT1.1 (here within referred to as EgDGAT1) highlighted by increases in both the TFA and TAG content, implying improved incorporation efficiency for MCFAs.
Figure 3.
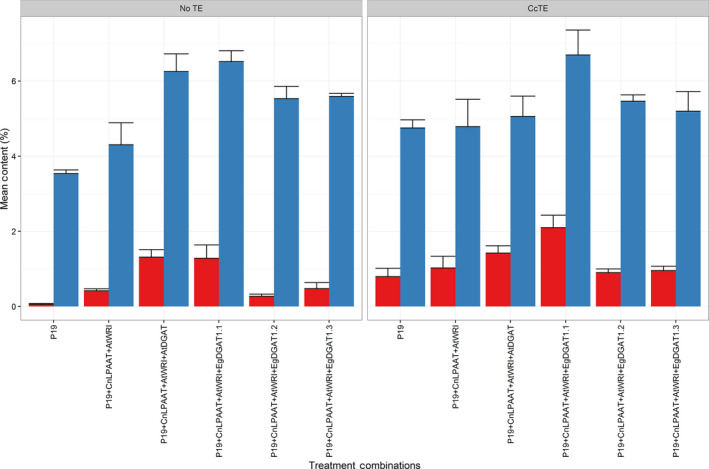
Total fatty acid (TFA; blue) and triacylglycerol (TAG; red) contents of Nicotiana benthamiana leaf infiltration samples (n = 3) were quantified. ‘CcTE’ represents the expression of the thioesterase from Cinnamomum camphora, whereas ‘No TE’ represents samples where CcTE expression was absent. CnLPAAT = Cocos nucifera lysophosphatidic acid acyltransferase; AtWRI = Arabidopsis thaliana WRINKLED1; AtDGAT = A. thaliana diacylglycerol acyltransferase 1; EgDGAT = Elaeis guineensis diacylglycerol acyltransferase.
The fatty acid composition of the phospholipid fraction was also analysed (Figure 4). Total phospholipids were fractionated by TLC and prepared for analysis by the preparation of FAME. Analysis of the fatty acid composition of the phospholipids revealed a significant reduction in the accumulation of MCFA, particularly C14:0 and C16:0, following the over‐expression of the EgDGAT1 (P = 0.008), than compared to the expression of AtDGAT1. This suggested that the reduced accumulation of MCFA into membrane lipids assisted in the improvement of the chlorosis phenotype.
Figure 4.
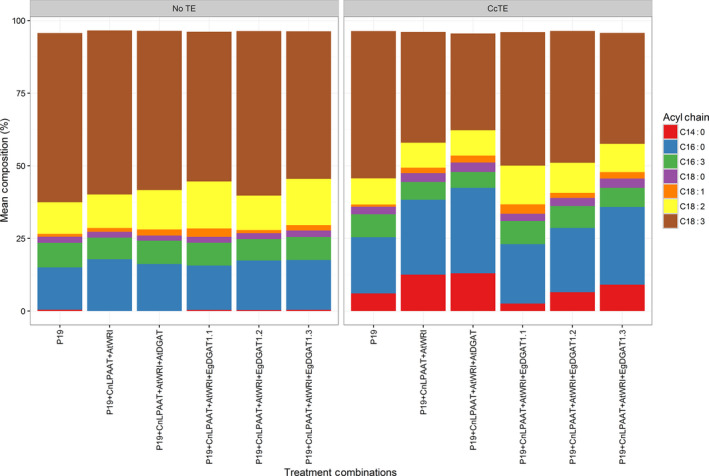
Investigating the effects of DGAT1 co‐expression on fatty acid profile changes in the membrane lipids (phospholipids) (n = 3). Results in the figure represent each fatty acid as a percentage of total fatty acid methyl esters (FAME) of the phospholipid fraction only. ‘CcTE’ represents the expression of the thioesterase from Cinnamomum camphora. CnLPAAT = Cocos nucifera lysophosphatidic acid acyltransferase; AtWRI = Arabidopsis thaliana WRINKLED1; AtDGAT = A. thaliana diacylglycerol acyltransferase 1; EgDGAT = Elaeis guineensis diacylglycerol acyltransferase.
The reconfiguration of a Kennedy pathway for efficient MCFA accumulation
Following confirmation of the CnGPAT9, the GPAT9 was used for subsequent studies that involved investigating its capability to use MCFA acyl‐CoAs as substrates for TAG assembly. The CnGPAT9 was tested through the sequential reconfiguration of the Kennedy pathway, rebuilding the pathway for improving MCFA composition, with all infiltrations performed using AtWRI1 to elevate fatty acid synthesis. After 5 days of gene expression, photographs of the leaves (Figure S5) were taken before the infiltrated zones were harvested and processed for the extraction of total lipids as previously described (Wood et al., 2009).
Both the fatty acid composition of TAG and the TAG content were determined by GC‐FID (Figures 5 and 6, respectively). With the co‐expression of UcTE, the sequential addition of each acyltransferase resulted in both significantly increased total TAG content (P ≤ 0.05), and a significantly increased accumulation of C12:0 in the TAG profile (P ≤ 0.02). The highest level of C12:0 accumulation achieved was 51.6 ± 2.0%, which was associated with the combined expression of UcTE + AtWRI1 + CnGPAT9 + CnLPAAT + EgDGAT1, with a total TAG content of 2.4 ± 0.7%. It was also observed that this combination was associated with the recovery of the chlorosis phenotype, predicted to be a result of efficient sequestering of laurate into TAG, and hence the exclusion from membrane lipids. Similar results were observed with the associated co‐expression of the CcTE. The highest level of accumulation of C14:0 was 40.3 ± 1.2%, with the combination of CcTE + AtWRI1 + CnGPAT9 + CnLPAAT, although there was no significant difference in the TAG content compared to the addition of the CnGPAT9 only (P = 0.08). The greatest TAG production was achieved following the further addition of the EgDGAT1, with a total TAG content of 2.8 ± 0.2%. However, the fatty acid composition of TAG was altered following the additional expression of EgDGAT1, with both a significant reduction in C14:0 (P = 0.015) and a significant increase in C16:0 (P = 0.013) content. This shift in profile suggests that EgDGAT1 exhibits a stronger substrate preference for C16:0 compared to C14:0. Consistent with the UcTE scenario, a significant improvement in the chlorotic phenotype was observed following the addition of the EgDGAT1. However, in the scenario of samples associated with the expression of CnTE2, the sequential addition of the acyltransferases did not result in any significant differences in either the fatty acid profile of TAG, or the total TAG content. This may be due to the native acyltransferases' ability to efficiently utilize the increased flux of C16:0 acyl‐CoA associated with the activity of CnTE2. However, the observed chlorotic phenotype suggests that efficient assembly of TAG was not occurring. Consequently, further investigations into the composition of other lipid classes, particularly the membrane lipids, were essential in understanding the occurrence of chlorosis.
Figure 5.

Fatty acid composition analysis of triacylglycerol (TAG), determined by the analysis of fatty acid methyl esters (FAME) via gas chromatography–flame ionization detection (GC‐FID) (n = 3). Each thioesterase was infiltrated in combination with CnGPAT9 alone, CnGPAT9 + CnLPAAT or CnGPAT9 + CnLPAAT + EgDGAT1, with all treatments including expression of AtWRI1 (Arabidopsis thaliana WRINKLED1). CcTE = Cinnamomum camphora thioesterase; CnTE2 = Cocos nucifera thioesterase; UcTE = Umbellularia californica thioesterase; CnGPAT9 = C. nucifera glycerol‐3‐phosphate acyltransferase 9; CnLPAAT = C. nucifera lysophosphatidic acid acyltransferase; EgDGAT1 = Elaeis guineensis diacylglycerol acyltransferase.
Figure 6.

Quantified TAG content, determined by the analysis of fatty acid methyl esters (FAME) via gas chromatography–flame ionization detection (GC‐FID) (n = 3). The results were grouped by thioesterase expression including from Cinnamomum camphora (CcTE), Cocos nucifera (CnTE2) or Umbellularia californica (UcTE). All treatments were also co‐infiltrated with WRI1 from Arabidopsis thaliana (AtWRI1). CnGPAT9 = C. nucifera glycerol‐3‐phosphate acyltransferase 9; CnLPAAT = C. nucifera lysophosphatidic acid acyltransferase; EgDGAT1 = Elaeis guineensis diacylglycerol acyltransferase 1.
Further investigations into the effects of the sequential addition of acyltransferases on the utilization of acyl‐CoAs for the assembly of MCFA‐enriched glycerolipids were performed using QQQ‐LCMS, to reveal any differences in MCFA assembly and distribution. The integrated analysis including DAG (Figure S6), PC and TAG reveals an extensive amount of information in respect to the assembly process of lipids. From the expression of CnGPAT9, in the background of UcTE + AtWRI1 (Figure 7), it was observed that CnGPAT9 appears to utilize C12:0 for assembly, based on the presence of PC 30:3 (C12:0 plus C18:3). It was reasoned that the sn‐2 position is most likely occupied by the C18:3, due to either the esterification of C12:0 to the sn‐1 position via CnGPAT9 or from the absence of CnLPAAT. The presence of some TAG 42:3 suggests that the native DGATs exhibit some capability of utilizing C12:0 for TAG assembly (12:0/18:3/12:0). Following the subsequent addition of CnLPAAT, a significant amount of PC 24:0 (di‐C12:0) was being produced, indicative that C12:0 was being efficiently esterified to both the sn‐1 and sn‐2 positions of the G3P backbone. However, without a DGAT that exhibits strong substrate preference for C12:0, most of the produced laurate remains sequestered in membrane lipids. With the further addition of the EgDGAT1, previously shown to be capable of utilizing MCFA for lipid assembly, a shift in laurate accumulation was observed. This shift involved the reduction MCFAs accumulating in PC, and a correlating increase production of MCFA‐enriched TAG. Most notable was the shift from PC 24:0 (without EgDGAT1) to the accumulation of TAG 36:0 (tri‐C12:0) (with EgDGAT1), highlighting that laurate was being efficiently incorporated into all three position of the G3P backbone. Significant increases were also observed for other MCFA‐enriched TAG species including TAG 38:0, TAG 40:0 and TAG 42:0. These results confirm that the expression of an appropriate DGAT1 is essential for the efficient incorporation of the unusual fatty acids of interest (in this scenario, C12:0) into TAG. Conclusively, these results highlighted that the expression of the EgDGAT1 effectively relieves the accumulation of laurate in PC and promotes efficient production of laurate‐enriched TAG in plant leaf lipids.
Figure 7.
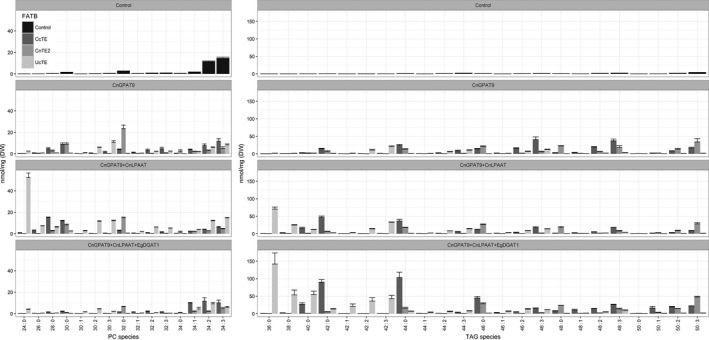
Comparative analysis of phosphatidylcholine (PC) and triacylglycerol (TAG) lipid species, associated with the expression of thioesterases from either Cinnamomum camphora (CcTE), Cocos nucifera (CnTE2) or Umbellularia californica (UcTE) (n = 3). ‘Control’ refers to samples that were infiltrated with p19 only. All other samples were co‐infiltrated with both p19 and WRINKLED1 from Arabidopsis thaliana (AtWRI1). CnGPAT9 = C. nucifera glycerol‐3‐phosphate acyltransferase 9; CnLPAAT = C. nucifera lysophosphatidic acid acyltransferase; EgDGAT1 = Elaeis guineensis diacylglycerol acyltransferase.
A similar pattern was also observed in the case study involving the over‐expression of CcTE (Figure 7). From the expression of CnGPAT9, in the background of CcTE + AtWRI1, it was observed that CnGPAT9 appears to utilize C14:0 for assembly, based on the accumulation of PC 28:0 (di‐C14:0) and PC 30:0 (C14:0 plus C16:0). It appears that the native LPAAT genes are somewhat capable of utilizing C14:0 acyl‐CoA for lipid assembly based on the presence of PC 28:0, indicating that C14:0 was being esterified to both the sn‐1 and sn‐2 positions. Similarly, the native DGATs also appear capable of utilizing C14:0 for TAG assembly, based on the production of TAG 42:0 (tri‐C14:0). However, the subsequent addition of CnLPAAT to the system increased utilization of C14:0 acyl‐CoA evident from the significantly increased abundance of PC 28:0, which indicated an increased efficiency of esterification to the sn‐2 position of the G3P backbone. This increased accumulation of MCFA was also correlated with a more severe chlorosis phenotype then compared to the CnGPAT9 alone, most likely attributed to the increased saturation of the membrane lipids. The further addition of the EgDGAT1 resulted in almost the complete removal of MCFA accumulation in PC. This was associated with an increased production of MCFA‐enriched TAG species, particularly TAG 40:0, TAG 42:0, TAG 44:0 and TAG 46:0, all of which include the incorporation of C14:0. It was suspected that the EgDGAT1 exhibits a stronger preference for C16:0 than compared to C14:0, based on the higher abundance of TAG 44:0 (14:0/14:0/16:0) than TAG 42:0 (tri‐C14:0). This is supported by the similar trend seen in the changes in total fatty acid composition of TAG (Figure 5).
However, in the case study involving the over‐expression of CnTE2 (Figure 7), the pattern was not as consistent as previously seen. From the expression of CnGPAT9, in the background of CnTE2 + AtWRI1, it was observed that CnGPAT9 also utilizes C16:0 for assembly, based on the accumulation of PC 32:0 (di‐C16:0). Based on the fatty acid profile of N. benthamiana leaves, it was expected that the native LPAATs and DGATs exhibit substrate specificity for the incorporation of C16:0 acyl‐CoAs into glycerolipids, evidenced from the increased production of C16:0‐enriched TAG species, through simply over‐expressing a thioesterase with C16:0 specificity. Although the subsequent additions of the CnLPAAT and EgDGAT1 did not appear to significantly affect the overall TAG composition, there was significant reduction in the total MCFA accumulation in PC lipids. Importantly, the addition of the EgDGAT1 was associated with a reduction in the degree of leaf chlorosis, although not completely recovered.
Through analysing the effect that CnGPAT9 expression has on the fatty acid profile of both the PC and TAG lipid fractions, it was concluded that CnGPAT9 is an important factor in contributing towards both MCFA accumulation and increasing the total production of TAG in plant leaves. In all thioesterase scenarios, the low abundance of MCFA‐containing DAG species suggests that they are actively converted to PC via the activities of either PDCT or CPT (Bates and Browse, 2011, 2012; Bates et al., 2012), in the absence of a MCFA‐specific DGAT. The addition of EgDGAT1 changes the metabolic flux of the system, pushing MCFA towards TAG accumulation via the Kennedy pathway, and hence away from incorporation into membrane lipids via avoiding the conversion of DAG to PC.
Modification of TAG profile through thioesterase co‐expression
With the knowledge that fatty acid thioesterases are essential factors in the fatty acid profile determination of plant seed oils (Harwood, 1996), it seems reasonable to suggest that further modifications of the fatty acid profile could be achieved through the combinatorial expression of different thioesterases. The thioesterases (UcTE, CcTE and CnTE2) were tested in combinations with each other using the transient N. benthamiana infiltration system. The thioesterases were either infiltrated alone, in pairs or in trio, with all thioesterase combinations being tested with and without the high oil background, consisting of AtWRI1 + CnLPAAT + EgDGAT1.
To understand the contribution of each thioesterase on the assembly of TAG, the TAG composition and fatty acid profile was analysed via LC‐MS. Initially TAG was characterized by its fatty acid profile (Figure 8). With the expression of UcTE in the high oil background, C12:0 accounted for 48% of the TAG profile. However, significant decreases in the C12:0 content were observed, associated with significant increases in C14:0 and C16:0 following the addition of CcTE or CnTE2, respectively. Conversely, a similar pattern was observed in the scenario of CnTE2 expression, which results in the accumulation of 58% C16:0 in TAG when expressed alone. The content of C16:0 was decreased following co‐expression of another thioesterase, correlated with increased proportions of C12:0 and C14:0 with the co‐expression of UcTE or CcTE, respectively. When the thioesterases were co‐expressed, the total MCFA content accounted for approximately 75% of the total fatty acid profile. Interestingly, this co‐ordinated expression of all three thioesterases produced a TAG profile which was similar in proportions of C12:0 (24%), C14:0 (16%) and C16:0 (34%).
Figure 8.

Investigating the effects of combined thioesterase expression on triacylglycerol (TAG) fatty acid composition. Results are presented as the percentage for each fatty acid, determined by GC‐FID (n = 4). The ‘Control’ refers to the infiltration of p19 alone. The label ‘Thioesterase Only’ refers to the expression of p19 with the respective thioesterase combination. AtWRI1 = Arabidopsis thaliana WRINKLED1; CnLPAAT = C. nucifera lysophosphatidic acid acyltransferase; EgDGAT1 = Elaeis guineensis diacylglycerol acyltransferase; CcTE = Cinnamomum camphora thioesterase; CnTE = Cocos nucifera thioesterase; UcTE = Umbellularia californica thioesterase.
The TAG profile was further analysed through determining the abundance of individual TAG species (Figure S7), which reveals more information in respect to TAG assembly and the co‐ordinated effects of the studied thioesterases on TAG composition. When UcTE was expressed alone, the most abundant TAG species was TAG 36:0 (tri‐laurate), with TAG 38:0, TAG 40:0, TAG 42:0, TAG 42:1, TAG 42:2 and TAG 42:3 also being highly abundant. Following the co‐expression of CcTE and CnTE2, there were significant decreases in the abundance of TAG 36:0, TAG 38:0 and unsaturated TAG 42 species. This correlated with significant increases for TAG 42:0, TAG 44:0 and TAG 46:0. The increased abundance of larger TAG species can be justified by the increased availability of C14:0 and C16:0 for incorporation into TAG, following the expression of CcTE and CnTE2. When CcTE was expressed alone, the most abundant TAG species was TAG 44:0, with TAG 42:0, TAG 48:3, TAG 50:2 and TAG 50:3 being almost equally abundant. With the addition of UcTE, increases were observed for the lower molecular weight TAG species particularly TAG 36:0, TAG 38:0 and TAG 40:0, due to the increased availability of C12:0 for TAG assembly. Alternatively, when combined with CnTE2, increases were observed for higher molecular weight TAG species particularly TAG 46:0, TAG 48:0 and TAG 50:3, indicative of increased availability of C16:0. When only the CnTE2 was expressed, the most abundant TAG species were TAG 46:0, TAG 48:0, TAG 50:2 and TAG 50:3. Similar to the CcTE scenario, the co‐expression with UcTE led to an increase in the abundance of many lower molecular weight TAG species including TAG 36:0, TAG 38:0, TAG 40:0, TAG 42:0 and TAG 44:0. The addition of the CcTE also led to changes in the TAG composition, primarily increased abundance of TAG 40:0, TAG 42:0, TAG 44:0 and TAG 46:0. From the combinatorial expression of all three thioesterases, there was a broad range in the TAG composition, highlighting the increased availability of C12:0, C14:0 and C16:0 for the assembly of glycerolipids.
Discussion
In the seeds of native plants, the incorporation of unusual fatty acids is almost exclusively confined to TAG and typically excluded from membrane lipids, most likely because they interfere with proper membrane functions and are often deleterious to the plant cells (Millar et al., 2000). A different scenario has been observed in transgenic plants that have attempted to modify the oil fatty acid profiles, such as increasing the lauric acid content (Knutzon et al., 1999). Although high levels of laurate accumulation in plant oils have been achieved in the seeds of transgenic canola, at ≈67%, there was a significant level of laurate being sequestered in PC during seed development (Wiberg et al., 1997). In this earlier transgenic canola work, de novo DAG containing laurate is not being efficiently converted to TAG by the resident DGAT but is instead being converted to the membrane lipid PC. The native canola LPCAT lacks the capability to handle MCFAs (Zhang et al., 2015), so the route to PC could be via PDCT or CPT exchange. Consequently, this inefficient utilization of laurate for TAG synthesis was also associated with a negatively correlated penalty in total oil yields (Knutzon et al., 1999).
Similar to the expression of MCFA in seed oil, the over‐expression of MCFA in the leaf high oil model (Vanhercke et al., 2013) with the co‐expression of CnGPAT9 and CnLPAAT there is a metabolic bottleneck identified with the sequestering of MCFA in PC. It appeared that the CnGPAT9 does not exhibit substrate specificities, but rather actively incorporates acyl‐CoAs based on their availability, although this requires further investigation. The low abundance of MCFA‐containing DAG species suggests that de novo DAG is quickly converted to PC through the activities of PDCT or CPT, due to the absence of a substrate‐specific DGAT capable of using the MCFA‐containing DAG for TAG assembly. It was hypothesized that the addition of a DGAT1 with substrate specificity for MCFA is essential to relieve this bottleneck and hence promote synthesis of MCFA‐enriched TAG. Previously, it has been demonstrated that PDAT is also involved in the maintenance of membrane homoeostasis, through the removal of damaged or unusual fatty acids from the membrane lipids and sequestering them into TAG (Fan et al., 2013, 2014). This suggests that the expression of a substrate‐specific PDAT may have assisted with the recovery of the observed phenotype. This study demonstrated that the over‐expression of the DGAT1 from E. guineensis (EgDGAT1) was sufficient enough in restoring membrane homoeostasis by avoiding the initial accumulation of MCFA in PC. The expression of EgDGAT1 proved that a substrate‐specific DGAT is essential for the efficient assembly of TAG, through the relationship of decreased PC species and increased TAG species containing MCFA.
The addition of EgDGAT1 plays an important role in the assembly of TAG, particularly those containing C12:0 and C14:0, associated with expression of UcTE and CcTE, respectively. The increased amounts of tri‐MCFA TAG species, compared to the samples expressing only the CnGPAT9 and CnLPAAT, highlights the importance of expressing an appropriate DGAT for efficient TAG assembly. Collectively, these results highlight an efficient assembly pathway for the incorporation of MCFA into plant oils, while also efficiently avoiding accumulation in membrane lipids. We hypothesized that through the relief of the metabolic bottleneck of PC accumulation, the complete assembly pathway assists in promoting higher oil yields. This was supported by the observed increases in both TFA and TAG contents following EgDGAT1 expression, and hence achieved via preventing inhibition of the FAS cycle previously observed with an incomplete pathway for the utilization of MCFA‐CoA (Knutzon et al., 1999; Wiberg et al., 1997). This is also supported by the results of previous studies that have demonstrated that the inefficient utilization of unusual fatty acids for the assembly of glycerolipids is correlated with both their accumulation in phospholipids (Knutzon et al., 1999) and the inhibition of fatty acid synthesis (Bates et al., 2014).
It is suspected that the concept of substrate‐specific DGATs could be further extended into the work of improving the accumulation of other unusual fatty acids, such as short‐chain, epoxy and hydroxyl fatty acids, in both seed and plant biomass platforms. The identification of a substrate‐specific DGAT followed by its over‐expression in the system of interest should assist in the incorporation of the respective fatty acid into TAG. Another alternative for improving the accumulation of unusual fatty acids could be the modification of the active site through methods such as site‐directed mutagenesis (Liu et al., 2010; Morand et al., 1998) and hence modify or improve the substrate specificity of acyltransferases. For example, the EgDGAT1 exhibited some specificity for C14:0, but preferred C12:0 and C16:0. It may be possible to modify the active site of the EgDGAT1 to explicitly utilize C14:0 for incorporation into TAG. Although in this study significant levels of C14:0 incorporation into TAG was achieved, with ≈40%, the addition of an engineered DGAT with explicit substrate specificity for C14:0 should contribute to increasing the myristate content of TAG.
It has been well established that thioesterases are an important factor in the determination of the fatty acid composition of TAG (Bates et al., 2013; Dehesh et al., 1996a; Harwood, 1996). Historically, transgenic work has been associated with the over‐expression of single thioesterases (Dehesh et al., 1996b; Dörmann et al., 2000; Eccleston et al., 1996; Leonard et al., 1998), for the modification of TAG composition. In this study, different combinations of thioesterases were studied to investigate the potential shift in the MCFA composition of TAG and also highlight the contribution that thioesterases have towards determining and modifying the oil composition of both the native species and transgenic plants, respectively. It was demonstrated that the TAG fatty acid composition can be significantly altered through the co‐expression of thioesterases with different substrate specificities, where they work together to determine the final lipid profile. This occurrence could be explained through an increased variety of acyl‐CoAs that are available for TAG assembly and hence demonstrating that thioesterases are an integral factor in the determination of the fatty acid profile of plant oils. To efficiently utilize the increased variety within the acyl‐CoA pool for TAG assembly, it may also be necessary to over‐express various DGATs with suitable substrate specificity.
It has long been considered that the expression of a DGAT1 plays a significant role in determining plant oil compositions (Bates and Browse, 2012). Importantly, the results we present indicate that the expression of a substrate‐specific DGAT1 is essential in the avoidance of bottleneck accumulation of MCFA in PC and hence directing the flux of MCFA to TAG production. This is achieved through the efficient utilization of the MCFA‐CoAs, produced by the termination of fatty acid synthesis by thioesterase expression, for the synthesis of TAG from de novo DAG. This reconfigured Kennedy pathway for improving MCFA incorporation into TAG has the potential to be further explored in both seed and leaf oil contexts and may potentially be applied for improving the accumulation of other unusual fatty acids. Of particular importance, the identification of an appropriate DGAT for the fatty acid profile of interest (MCFA or other unusual fatty acids) is essential for achieving increased oil production with specifically tailored fatty acid compositions.
Methods
Gene selection
Thioesterases were identified from previous publications, with codon optimized gene coding sequences being synthesized (Geneart, Regensburg, Germany) for Cinnamomum camphora 14:0‐ACP thioesterase (referred to as ‘CcTE’, Q39473.1 (Yuan et al., 1995)), Umbellularia californica 12:0‐ACP thioesterase (UcTE, Q41635.1 (Voelker et al., 1992), Cocos nucifera acyl‐ACP thioesterase FatB2 (CnTE2, AEM72520.1 (Jing et al., 2011)). The C. nucifera LPAAT (CnLPAAT, Q42670.1 (Knutzon et al., 1995)) was also synthesized. WRI1 and DGAT1 expression vectors were produced as previously described by Vanhercke et al. (2013).
From a recently published transcriptome of African oil palm (Elaeis guineensis) (Dussert et al., 2013), three DGAT candidates were chosen for testing their capability of efficiently utilizing MCFA for the assembly of leaf lipids. The DGATs from oil palm were selected based on the fatty acid compositions of palm oil and palm kernel oil (Edem, 2002), being high in MCFA content.
The glycerol‐3‐phosphate acyltransferase 9 from C. nucifera (CnGPAT9) was identified from a recently constructed transcriptome, constructed from RNA isolated from developing coconut endosperm. Using the recently identified and confirmed Arabidopsis thaliana GPAT9 (AtGPAT9) (Shockey et al., 2016) as the BLAST query, a good candidate for GPAT9 from coconut was identified. The CnGPAT9 (NCBI accession number: KX235871) was identified based on sequence homology with AtGPAT9. Using the synthesized coconut cDNA as template, Phusion high fidelity (New England Biolabs, Hitchin, England) PCR was performed (according to manufacturer's instructions) to attempt the amplification of the full length CnGPAT9 sequence.
Construct assembly
Each gene was cloned into the EcoRI site of a binary vector, pJP3343, which already contained a 35S promoter with duplicated enhancer region (Vanhercke et al., 2013). Agrobacterium tumefaciens strain AGL1 was transformed with each of the constructs.
Nicotiana benthamiana transient assay
Transient expression in N. benthamiana leaves was performed as previously described (Wood et al., 2009), with some minor modifications. A. tumefaciens cultures containing the gene coding for the p19 viral suppressor protein and the chimeric gene(s) of interest were mixed such that the final OD600 of each culture was equal to 0.1 prior to infiltrations. Each complete experiment was repeated five times to confirm reproducibility, although the data were presented for only a single experiment. For fatty acid profile analyses, a total of 16 leaves from six plants were infiltrated with the different gene combinations. The samples being compared were randomly located, with a p19 control infiltrated for each plant. After infiltrations, the N. benthamiana plants were grown for a further 5 days before leaf discs were harvested, freeze‐dried, weighed and stored at −80 °C.
Total lipid extraction and fatty acid profile analysis
Total lipids were extracted from freeze‐dried N. benthamiana leaves. During the extraction of total lipids, TAG 51:0 (tri‐C17:0) was added as the internal standard for the quantification of both the TAG and total fatty acid (TFA) contents. Freeze‐dried leaf tissue was ground to powder in a microcentrifuge tube containing a metallic ball using Reicht tissue lyser (Qiagen) for 3 min. at 20 frequency/s. Chloroform:methanol (2:1, v/v) was added and mixed for a further 3 min. on the tissue lyser before the addition of 1:3 (v/v) of 0.1 m KCl. The sample was then mixed for a further 3 min. before centrifugation (5 min. at 14 000 g ), after which the lower lipid phase was collected. The remaining phase was washed once with chloroform, and the lower phase extracted and pooled with the earlier extract. Lipid phase solvent was then evaporated completely using N2 gas flow and the lipids resuspended in 5 μL chloroform per mg of original dry leaf weight.
Fatty acid methyl esters (FAMEs) of total lipids (equivalent to 10 mg dry weight) were produced by incubating extracted lipid in 1 N methanolic HCl (Supelco, Bellefonte, PA) at 80 °C for 3 h. FAMEs were analysed by an Agilent 7890A gas chromatograph coupled with flame ionization detector (GC‐FID, Agilent Technologies, Palo Alto, CA), on a BPX70 column (30 m, 0.25 mm inner diameter, 0.25 μm film thickness, SGE) essentially as described previously (Zhou et al., 2011), except the column temperature programme. The column temperature was programmed as an initial temperature at 100 °C holding for 3 min, ramping to 240 °C at a rate of 7 °C/min and holding for 1 min. NuChek GLC‐426 was used as the external reference standard. Peaks were integrated with Agilent Technologies ChemStation software (Rev B.04.03 (16)).
TLC analysis
From the total lipid extracts (equivalent to 10 mg dry weight), TAG and polar lipids were fractionated by TLC (Silica gel 60, MERCK) in hexane:diethylether:acetic acid (70:30:1 v/v/v) and visualized by spraying Primuline (Sigma, 5 mg/100 mL acetone:water (80:20 v/v)) and exposing plate under UV. TLC analysis was primarily used for the identification of fatty acid composition of TAG and phospholipids from lipid extraction samples. This also enabled the determination of the total TAG content for each sample. The TAG and phospholipid fractions were scraped from the TLC plates and methylated according to the FAME preparation protocol described previously.
LC‐MS analysis
Lipids extracted from 1 mg dry leaf weight were resuspended and diluted to 1 mg/mL in mL butanol:methanol (1:1, v/v) and analysed by liquid chromatography–mass spectrometry (LC‐MS), based on previously described methods (Petrie et al., 2012). Briefly, lipids were chromatographically separated using a Waters BEH C8 (100 × 2.1 mm, 2.7 μm) fitted to an Agilent 1290 series LC and 6490 triple quadrupole LC‐MS with Jet Stream ionization with a binary gradient flow rate of 0.2 mL/min. The mobile phases were as follows: A. H2O:acetonitrile (10:90, v/v) with 10 mm ammonium formate and 0.2% acetic acid; B. H2O:acetonitrile:isopropanol (5:15:80, v/v) with 10 mm ammonium formate and 0.2% acetic acid. For the phosphatidylcholine (PC) and lysophosphatidylcholine (LPC), species hydrogen adducts were quantified by the characteristic 184 m/z phosphatidyl head group ion under positive ionization mode. The ammonium adducts of monogalactosyl diacylglycerol (MGDG), digalactosyl diacylglycerol (DGDG), diacylglycerol (DAG) and TAG lipid species were analysed by the neutral loss of singular fatty acids C12 to C18. Multiple reaction monitoring (MRM) lists were based on the following major fatty acids: 12:0, 14:0, 16:0, 16:3, 18:0, 18:1, 18:2, 18:3, using a collision energy of 28 V for all lipid classes except for DAG where a collision energy of 14 V was used. Individual MRM TAG was identified based on ammoniated precursor ion and product ion from neutral loss.
Statistical analysis
To determine whether there was any significant differences between samples, statistical analysis was performed using either the data analysis package for Excel 2013 or the dplyr, ggplot2 packages from R using RStudio (Horton and Kleinman, 2015). Statistical analyses were performed using students T‐test, for two samples assuming unequal variances. The experiments were designed with either three or four replicates, to enable accurate analysis of any significant differences.
Conflict of interest
We confirm that this work is original and has not been published elsewhere nor is there any associated conflict of interest.
Supporting information
Figure S1. Complete protein sequence alignment of GPAT9 from Arabidopsis thaliana (AtGPAT9) (Shockey et al., 2016) and Cocos nucifera (CnGPAT9) identified from transcriptome.
Figure S2. Phylogenetic relationship of glycerol‐3‐phosphate acyltransferase (GPAT) genes from various species.
Figure S3. Investigating the accumulation and assembly of MCFA in plant leaf lipids, testing different DGAT1 candidates, with representative images shown for each treatment (n = 3).
Figure S4. Analysis of diacylglycerol acyltransferase 1 (DGAT1) protein sequences by multiple sequence alignment, investigating differences in Arabidopsis thaliana (AtDGAT1; NP_179535) and predicted DGAT1 isoforms from Elaeis guineensis (Dussert et al., 2013).
Figure S5. Photos of Nicotiana benthamiana transient study, investigating the reconstruction of the Kennedy pathway tailored for improving the accumulation of MCFA, with representative images shown for each treatment (n = 3).
Figure S6. Analysis of diacylglycerol (DAG) lipid species, associated with the expression of thioesterase from Umbellularia californica (UcTE), Cinnamomum camphora (CcTE) or Cocos nucifera (CnTE2) (n = 3). Control refers to samples that were p19 only infiltrations. CnGPAT9 = C. nucifera glycerol‐3‐phosphate acyltransferase 9; CnLPAAT = C. nucifera lysophosphatidic acid acyltransferase; EgDGAT1 = Elaeis guineensis diacylglycerol acyltransferase.
Figure S7. Following Nicotiana benthamiana infiltrations testing different combinations of thioesterases, the triacylglycerol (TAG) species profile was investigated via LC‐MS.
References
- Adhikari, N.D. , Bates, P.D. and Browse, J. (2016) WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase‐expressing seeds. Plant Physiol. 171, 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkcoll, D. (1988) Lauric oil resources. Econ. Bot. 42, 195–205. [Google Scholar]
- Basiron, Y. (2007) Palm oil production through sustainable plantations. Eur. J. Lipid Sci. Technol. 109, 289–295. [Google Scholar]
- Basiron, Y. and Weng, C.K. (2004) The oil palm and its sustainability. J. Oil Palm Res. 16, 1–10. [Google Scholar]
- Bates, P.D. and Browse, J. (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J. 68, 387–399. [DOI] [PubMed] [Google Scholar]
- Bates, P.D. and Browse, J. (2012) The significance of different diacylglycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 3, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, P.D. , Fatihi, A. , Snapp, A.R. , Carlsson, A.S. , Browse, J. and Lu, C. (2012) Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 160, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, P.D. , Stymne, S. and Ohlrogge, J. (2013) Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. [DOI] [PubMed] [Google Scholar]
- Bates, P.D. , Johnson, S.R. , Cao, X. , Li, J. , Nam, J.W. , Jaworski, J.G. , Ohlrogge, J.B. et al. (2014) Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc. Natl Acad. Sci. USA, 111, 1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgal, J. , Shockey, J. , Lu, C. , Dyer, J. , Larson, T. , Graham, I. and Browse, J. (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol. J. 6, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. (2011) Structure‐function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Res. Notes, 4, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.Q. , van Erp, H. , Martin‐Moreno, J. , Johnson, K. , Morales, E. , Browse, J. , Eastmond, P.J. et al. (2016) Expression of castor LPAT2 enhances ricinoleic acid content at the sn‐2 position of triacylglycerols in lesquerella seed. Int. J. Mol. Sci. 17, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley, R.H.V. (2009) How much palm oil do we need? Environ. Sci. Policy, 12, 134–139. [Google Scholar]
- Dehesh, K. (2001) How can we genetically engineer oilseed crops to produce high levels of medium‐chain fatty acids? Eur. J. Lipid Sci. Technol. 103, 688–697. [Google Scholar]
- Dehesh, K. , Edwards, P. , Hayes, T. , Cranmer, A.M. and Fillatti, J. (1996a) Two novel thioesterases are key determinants of the bimodal distribution of acyl chain length of cuphea palustris seed oil. Plant Physiol. 110, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh, K. , Jones, A. , Knutzon, D.S. and Voelker, T.A. (1996b) Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of Ch FatB2, a thioesterase cDNA from Cuphea hookeriana. Plant J. 9, 167–172. [DOI] [PubMed] [Google Scholar]
- Dörmann, P. , Voelker, T.A. and Ohlrogge, J.B. (2000) Accumulation of palmitate in arabidopsis mediated by the acyl‐acyl carrier protein thioesterase FATB1. Plant Physiol. 123, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussert, S. , Guerin, C. , Andersson, M. , Joet, T. , Tranbarger, T.J. , Pizot, M. , Sarah, G. et al. (2013) Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 162, 1337–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston, V.S. and Ohlrogge, J.B. (1998) Expression of lauroyl‐acyl carrier protein thioesterase in brassica napus seeds induces pathways for both fatty acid oxidation and biosynthesis and implies a set point for triacylglycerol accumulation. Plant Cell Online, 10, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston, V. , Cranmer, A. , Voelker, T. and Ohlrogge, J. (1996) Medium‐chain fatty acid biosynthesis and utilization in Brassica napus plants expressing lauroyl‐acyl carrier protein thioesterase. Planta, 198, 46–53. [Google Scholar]
- Edem, D.O. (2002) Palm oil: biochemical, physiological, nutritional, hematological and toxicological aspects: A review. Plant Foods Hum. Nutr. 57, 319–341. [DOI] [PubMed] [Google Scholar]
- van Erp, H. , Bates, P.D. , Burgal, J. , Shockey, J. and Browse, J. (2011) Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol. 155, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp, H. , Shockey, J. , Zhang, M. , Adhikari, N.D. and Browse, J. (2015) Reducing isozyme competition increases target fatty acid accumulation in seed triacylglycerols of transgenic Arabidopsis. Plant Physiol. 168, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Yan, C. , Zhang, X. and Xu, C. (2013) Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in arabidopsis leaves. Plant Cell Online, 25, 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Yan, C. , Roston, R. , Shanklin, J. and Xu, C. (2014) Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward beta‐oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell, 26, 4119–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood, J.L. (1996) Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta BBA ‐ Lipids Lipid Metab. 1301, 7–56. [DOI] [PubMed] [Google Scholar]
- Horton, N.J. and Kleinman, K. (2015) Using R and RStudio for Data Management, Statistical Analysis, and Graphics. Boca Raton, Florida, USA: CRC Press. [Google Scholar]
- Jing, F. , Cantu, D. , Tvaruzkova, J. , Chipman, J. , Nikolau, B. , Yandeau‐Nelson, M. and Reilly, P. (2011) Phylogenetic and experimental characterization of an acyl‐ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 12, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A.A. , van Erp, H. , Quettier, A.L. , Shaw, E. , Menard, G. , Kurup, S. and Eastmond, P.J. (2013) The sugar‐dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 162, 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.J. , Yang, S.W. , Mao, H.‐Z. , Veena, S.P. , Yin, J.‐L. and Chua, N.‐H. (2014) Gene silencing of Sugar‐dependent 1 (JcSDP1), encoding a patatin‐domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol. Biofuels, 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J. , Silva, J.E. , Iskandarov, U. , Andersson, M. , Cahoon, R.E. , Mockaitis, K. and Cahoon, E.B. (2015a) Structurally divergent lysophosphatidic acid acyltransferases with high selectivity for saturated medium chain fatty acids from Cuphea seeds. Plant J. 84, 1021–1033. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Silva, J.E. , Vu, H.S. , Mockaitis, K. , Nam, J.‐W. and Cahoon, E.B. (2015b) Toward production of jet fuel functionality in oilseeds: identification of FatB acyl‐acyl carrier protein thioesterases and evaluation of combinatorial expression strategies in Camelina seeds. J. Exp. Bot. 66, 4251–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon, D.S. , Lardizabal, K.D. , Nelsen, J.S. , Bleibaum, J.L. , Davies, H.M. and Metz, J.G. (1995) Cloning of a coconut endosperm cDNA encoding a 1‐acyl‐sn‐glycerol‐3‐phosphate acyltransferase that accepts medium‐chain‐length substrates. Plant Physiol. 109, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon, D.S. , Hayes, T.R. , Wyrick, A. , Xiong, H. , Maelor Davies, H. and Voelker, T.A. (1999) Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn‐2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant Physiol. 120, 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S.N. (2011) Variability in coconut (Cocos nucifera L.) germplasm and hybrids for fatty acid profile of oil. J. Agric. Food Chem. 59, 13050–13058. [DOI] [PubMed] [Google Scholar]
- Laureles, L.R. , Rodriguez, F.M. , Reaño, C.E. , Santos, G.A. , Laurena, A.C. and Mendoza, E.M.T. (2002) Variability in fatty acid and triacylglycerol composition of the oil of coconut (Cocos nucifera L.) hybrids and their parentals. J. Agric. Food Chem. 50, 1581–1586. [DOI] [PubMed] [Google Scholar]
- Leonard, J.M. , Knapp, S.J. and Slabaugh, M.B. (1998) A Cupheaβ‐ketoacyl‐ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases. Plant J. 13, 621–628. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Siloto, R.M. and Weselake, R.J. (2010) Role of cysteine residues in thiol modification of acyl‐CoA:diacylglycerol acyltransferase 2 from yeast. Biochemistry, 49, 3237–3245. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Siloto, R.M. , Lehner, R. , Stone, S.J. and Weselake, R.J. (2012) Acyl‐CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 51, 350–377. [DOI] [PubMed] [Google Scholar]
- Millar, A.A. , Smith, M.A. and Kunst, L. (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci. 5, 95–101. [DOI] [PubMed] [Google Scholar]
- Morand, L.Z. , Patil, S. , Quasney, M. and German, J.B. (1998) Alteration of the fatty acid substrate specificity of lysophosphatidate acyltransferase by site‐directed mutagenesis. Biochem. Biophys. Res. Commun. 244, 79–84. [DOI] [PubMed] [Google Scholar]
- OECD/FAO (2015) OECD‐FAO Agricultural Outlook (Edition 2015). Paris: OECD Publishing. [Google Scholar]
- Petrie, J.R. , Shrestha, P. , Zhou, X.‐R. , Mansour, M. , Liu, Q. , Belide, S. , Nichols, P. et al. (2012) Metabolic engineering plant seeds with fish oil‐like levels of DHA. PLoS ONE, 7, e49165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, K. , Taylor, M. , Zhou, X.‐R. , Vanhercke, T. , Wood, C. , Blanchard, C. , Singh, S. et al. (2015) Metabolic engineering of medium‐chain fatty acid biosynthesis in Nicotiana benthamiana plant leaf lipids. Front. Plant Sci. 6, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey, J. , Regmi, A. , Cotton, K. , Adhikari, N. , Browse, J. and Bates, P.D. (2016) Identification of arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 170, 163–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, S.D. , Chen, G. , Mietkiewska, E. , Tomasi, P. , Jayawardhane, K. , Dyer, J.M. and Weselake, R.J. (2016) Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids. J. Exp. Bot. 65, 4627–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellstrom, H. , Strawsine, M. , Silva, J. , Cahoon, E.B. and Ohlrogge, J.B. (2013) Disruption of plastid acyl:acyl carrier protein synthetases increases medium chain fatty acid accumulation in seeds of transgenic Arabidopsis. FEBS Lett. 587, 936–942. [DOI] [PubMed] [Google Scholar]
- Vanhercke, T. , El Tahchy, A. , Shrestha, P. , Zhou, X.‐R. , Singh, S.P. and Petrie, J.R. (2013) Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 587, 364–369. [DOI] [PubMed] [Google Scholar]
- Vanhercke, T. , El Tahchy, A. , Liu, Q. , Zhou, X.R. , Shrestha, P. , Divi, U.K. , Ral, J.P. et al. (2014a) Metabolic engineering of biomass for high energy density: oilseed‐like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 12, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke, T. , Petrie, J.R. and Singh, S.P. (2014b) Energy densification in vegetative biomass through metabolic engineering. Biocatal. Agric. Biotechnol. 3, 75–80. [Google Scholar]
- Voelker, T.A. , Worrell, A.C. , Anderson, L. , Bleibaum, J. , Fan, C. , Hawkins, D.J. , Radke, S.E. et al. (1992) Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science (New York, N.Y.), 257, 72–74. [DOI] [PubMed] [Google Scholar]
- Voelker, T.A. , Hayes, T.R. , Cranmer, A.M. , Turner, J.C. and Davies, H.M. (1996) Genetic engineering of a quantitative trait: metabolic and genetic parameters influencing the accumulation of laurate in rapeseed. Plant J. 9, 229–241. [Google Scholar]
- Wiberg, E. , Banas, A. and Stymne, S. (1997) Fatty acid distribution and lipid metabolism in developing seeds of laurate‐producing rape (Brassica napus L.). Planta, 203, 341–348. [DOI] [PubMed] [Google Scholar]
- Wiberg, E. , Edwards, P. , Byrne, J. , Stymne, S. and Dehesh, K. (2000) The distribution of caprylate, caprate and laurate in lipids from developing and mature seeds of transgenic Brassica napus L. Planta, 212, 33–40. [DOI] [PubMed] [Google Scholar]
- Winichayakul, S. , Scott, R.W. , Roldan, M. , Hatier, J.H. , Livingston, S. , Cookson, R. , Curran, A.C. et al. (2013) In vivo packaging of triacylglycerols enhances Arabidopsis leaf biomass and energy density. Plant Physiol. 162, 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, C.C. (2014) Leafy biofactories: producing industrial oils in non‐seed biomass. EMBO Rep. 15, 201–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, C.C. , Petrie, J.R. , Shrestha, P. , Mansour, M.P. , Nichols, P.D. , Green, A.G. and Singh, S.P. (2009) A leaf‐based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol. J. 7, 914–924. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Francis, T. , Mietkiewska, E. , Giblin, E.M. , Barton, D.L. , Zhang, Y. , Zhang, M. et al. (2008) Cloning and characterization of an acyl‐CoA‐dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site‐directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol. J. 6, 799–818. [DOI] [PubMed] [Google Scholar]
- Yuan, L. , Voelker, T.A. and Hawkins, D.J. (1995) Modification of the substrate specificity of an acyl‐acyl carrier protein thioesterase by protein engineering. Proc. Natl Acad. Sci. USA, 92, 10639–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Jasieniecka‐Gazarkiewicz, K. , Wan, X. , Luo, L. , Zhang, Y. , Banas, A. , Jiang, M. et al. (2015) Molecular characterization of two lysophospholipid:acyl‐CoA acyltransferases belonging to the MBOAT family in nicotiana benthamiana. PLoS ONE, 10, e0144653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.R. , Green, A.G. and Singh, S.P. (2011) Caenorhabditis elegans Delta12‐desaturase FAT‐2 is a bifunctional desaturase able to desaturate a diverse range of fatty acid substrates at the Delta12 and Delta15 positions. J. Biol. Chem. 286, 43644–43650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Complete protein sequence alignment of GPAT9 from Arabidopsis thaliana (AtGPAT9) (Shockey et al., 2016) and Cocos nucifera (CnGPAT9) identified from transcriptome.
Figure S2. Phylogenetic relationship of glycerol‐3‐phosphate acyltransferase (GPAT) genes from various species.
Figure S3. Investigating the accumulation and assembly of MCFA in plant leaf lipids, testing different DGAT1 candidates, with representative images shown for each treatment (n = 3).
Figure S4. Analysis of diacylglycerol acyltransferase 1 (DGAT1) protein sequences by multiple sequence alignment, investigating differences in Arabidopsis thaliana (AtDGAT1; NP_179535) and predicted DGAT1 isoforms from Elaeis guineensis (Dussert et al., 2013).
Figure S5. Photos of Nicotiana benthamiana transient study, investigating the reconstruction of the Kennedy pathway tailored for improving the accumulation of MCFA, with representative images shown for each treatment (n = 3).
Figure S6. Analysis of diacylglycerol (DAG) lipid species, associated with the expression of thioesterase from Umbellularia californica (UcTE), Cinnamomum camphora (CcTE) or Cocos nucifera (CnTE2) (n = 3). Control refers to samples that were p19 only infiltrations. CnGPAT9 = C. nucifera glycerol‐3‐phosphate acyltransferase 9; CnLPAAT = C. nucifera lysophosphatidic acid acyltransferase; EgDGAT1 = Elaeis guineensis diacylglycerol acyltransferase.
Figure S7. Following Nicotiana benthamiana infiltrations testing different combinations of thioesterases, the triacylglycerol (TAG) species profile was investigated via LC‐MS.


