Abstract
A convergent strategy was developed for the first-time synthesis of biotin-labeled GPI core glycans. These GPI conjugates are useful for various biological studies showcased by their application to the scrutiny of pore-forming bacterial toxin-GPI interaction, revealing that the phosphate group at the GPI inositol 1-O-position had a significant impact on GPI-toxin binding.
Graphical Abstract
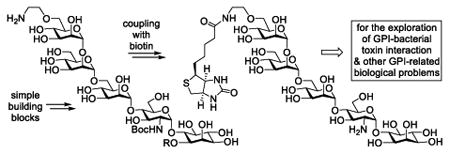
A concise and convergent synthesis of biotin-GPI glycan conjugates that were utilized to explore GPI-bacterial toxin interaction.
Post-translational protein modification by glycosylphosphotidyl-inositols (GPIs), a class of complex glycolipids, is ubiquitous in eukaryotes.1-4 One of the essential functions of GPIs is to anchor surface proteins onto the extracellular membrane.3-6 GPIs and GPI-anchored proteins play a critical role in various biological and pathological events, such as cell recognition and adhesion, signal transduction, enzymatic reaction, host defence, and viral and bacterial infections.7-9
Many GPIs have been isolated and characterized from various sources, and all of them share the conserved core structure 1 (Figure 1).1,5 GPIs insert their lipids into the cell membrane to attach GPI-anchored proteins to the cell surface. The structural diversity of natural GPIs is attributed to variations in the lipid structure and the presence of additional sugar chains or other functional groups on the mannose (Man) residues. Another conserved structural motif of GPI-anchored proteins is that all proteins are linked to the phosphoethanolamine group at the Man-III 6-O-position via their polypeptide C-terminus.
Figure 1. The conserved core structure 1 of GPIs, GPI anchoring of surface proteins onto the cell membrane, and releasing GPI-anchored proteins from the cell membrane via PI-PLC and PI-PLD-catalysed hydrolysis.
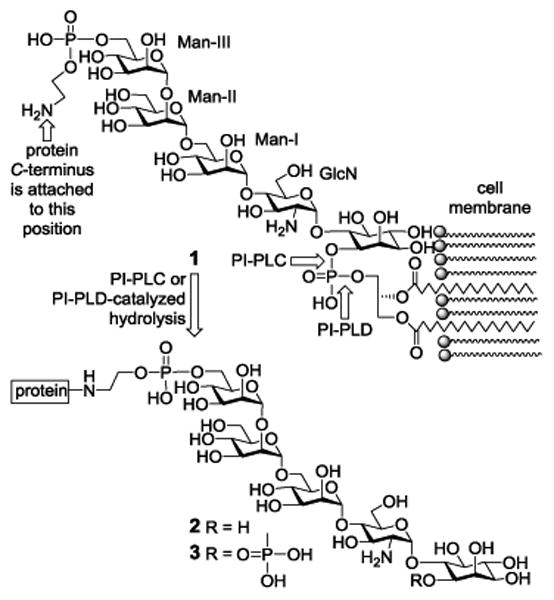
However, the amphiphilic and heterogenic properties of GPIs in nature render the isolation and analysis of GPI-linked proteins very difficult. As a result, GPI-anchored proteins are frequently studied in lipid-free forms after their lipid chains are removed either via regioselective chemical reactions, e.g., reaction with HNO2, or via enzymatic hydrolysis.1 In the latter case, two GPI-specific phospholipases, i.e., phosphoinositide phospholipase C and D (PI-PLC and PI-PLD), are useful.1,10 These enzymes, which cleave different bonds of the GPI phosphotidylinositol moiety, can be utilized to release GPI-anchored proteins from cells to afford water-soluble GPI glycan-linked proteins 2 and 3 (Figure 1). This would not only simplify the isolation of GPI-anchored proteins by using the highly conserved GPI glycan, especially its invariable glucosamine (GlcN)-inositol motif, but also facilitate in-depth analysis of these important molecules. For example, GPI-binding molecules, such as certain pore-forming bacterial toxins,11-14 can be used to pull down GPI-anchored proteins for the study of GPI-anchored proteomics.15
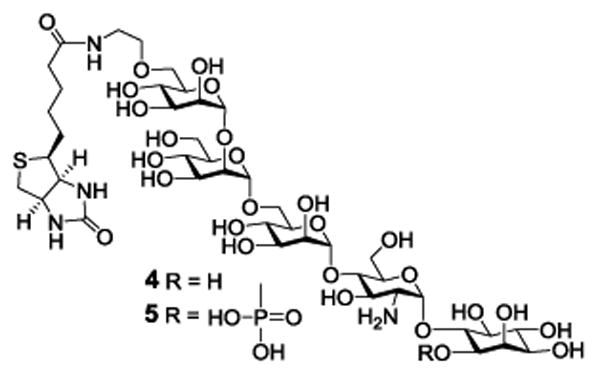
Therefore, the GPI glycans in 2 and 3, and related derivatives, should be useful biological tools. Accordingly, we explored here the synthesis of 4 and 5 (Figure 2), biotinylated derivatives of the conserved GPI glycans having the biotin molecule linked to the Man III 6-O-position. Biotin can serve as an affinity tag to enable various biological studies. As demonstrated, 4 and 5 were used to study and gain more insights into the interactions between GPIs and GPI-binding pore-forming bacterial toxins such as CAMP factor.
Figure 2. Structures of the designed GPI core glycan-biotin conjugates 4 and 5.
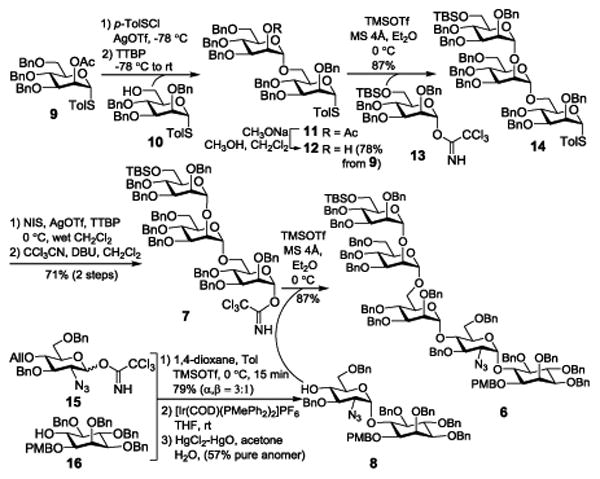
Our synthesis commenced with the assembly of the conserved GPI core glycan 6 by a convergent [3 + 2] strategy from glycosyl donor 7 and acceptor 8 (Scheme 1), followed by regioselective manipulation of temporary protections and functionalization. In turn, 7 was prepared from 9, 10 and 13, while 8 was prepared from 15 and 16. First, pre-activation-based glycosylation16 of 9 with 10 using para-toluenesulfenyl triflate (TolSOTf), prepared in situ from para-toluenesulfenyl chloride (TolSCl) and silver triflate (AgOTf), as the promoter and 2,4,6-tri-t-butylpyrimidine (TTBP) as the acid scavenge afforded α-linked disaccharide 11 stereoselectively. The α-configuration of the newly generated glycosidic bond in 11 was confirmed by its anomeric 1H and 13C NMR coupling constant (JH1′-C1′: 173 Hz).17 NaOMe-promoted deacetylation of 11 and glycosylation of the resulting 12 with 13 under the influence of trimethylsilyl triflate (TMSOTf) provided trisaccharide 14 (JH1″-C1″: 176 Hz) stereoselectively. Although 14 could be used as a glycosyl donor for coupling with 8, a similar reaction in the literature18 gave a relatively low yield (51%). To get improved efficiency for the key [3 + 2] coupling, we probed alternative glycosylation methods. Therefore, 14 was converted into trichloroacetimidate 7, a more reactive glycosyl donor,19 on N-iodosuccinimide (NIS)-mediated thioglycoside hydrolysis and then reaction of resulting hemiacetal with trichloroacetonitrile and 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU). In the meantime, 8 was synthesized by reaction of 16 with 15 in 1,4-dioxane and toluene (1:1), which produced an inseparable mixture of α,β-anomers (3:1), and then [Ir(COD)(PMePh2)2]PF6-mediated two-step one-pot removal of the allyl (All) group. Compound 8 was easily separated from the β-isomer by column chromatography to give a good overall yield (57%). Finally, glycosylation of 8 with 7 under the promotion of TMSOTf went smoothly to afford an excellent yield (87%). The α-configuration of the newly formed mannosyl linkage in 6 was again verified by its anomeric 1H-13C coupling constant (JH1′-C1′: 175 Hz).
Scheme 1. Synthesis of orthogonally protected GPI core glycan 6.
With 6 in hand, we attempted first its coupling with biotin via a succinyl group (Scheme 2). Consequently, 6 was treated with Et3N·3HF to remove the t-butyldimethylsilyl (TBS) group, which was followed by acylation of the free Man-III 6-OH group with succinic anhydride and condensation of the resulting acid with an ethylenediamine derivative 18 of biotin in the presence of 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyri-dinium 3-oxid hexafluorophosphate (HATU). The fully protected biotin-GPI glycan conjugate 19 was obtained in a good overall yield (56%) from 6. However, debenzylation of 19 via catalytic hydrogenolysis in a mixture of dichloromethane, methanol and water was unexpectedly slow, possibly due to the interference of biotin. After 10 days of reaction, all benzyl groups in 19 were removed finally, but partial hydrolysis of the ester linkage was also noticed and the two products were inseparable.
Scheme 2. Attempted synthesis of an ester-linked biotin-GPI glycan conjugate.
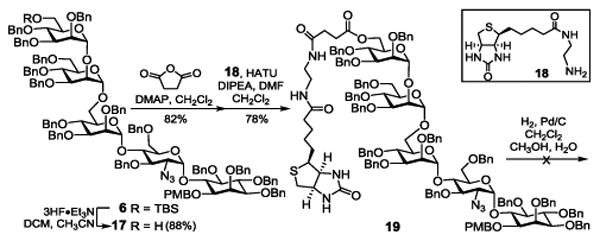
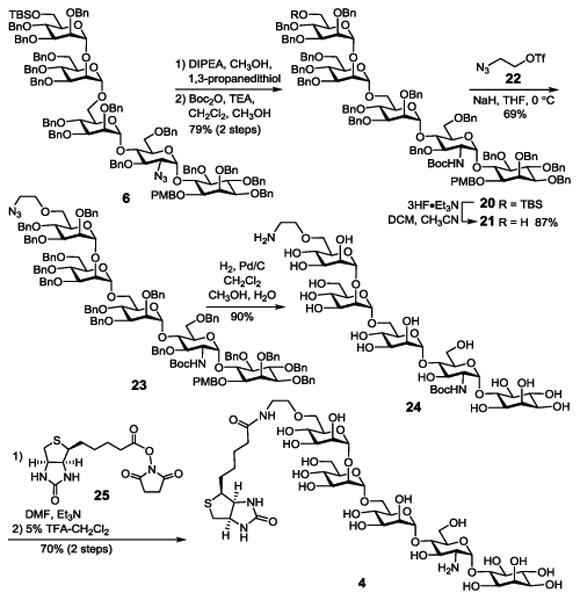
Because of the instability of an ester bond at the Man-III 6-O-position to hydrolysis, which would render such GPI conjugates nearly useless, we decided to probe alternative linkers for GPI-biotin conjugation. For this purpose, we became interested in ethanolamine, because as shown in 4 and 5, it can be attached to GPI glycans via an ether linkage while its other terminus can couple with biotin via a stable amide bond. In the synthesis of 4 (Scheme 3), the ethanolamine moiety was first introduced as ethanolazide. To differentiate this azido group from the azido group in GlcN and enable regioselective modifications later on, 6 was converted into 20 with the amino group of GlcN protected with a tert-butyloxycarbonyl (Boc) group, achieved by reduction of the azide with thiol and then reaction of the resulting amine with Boc anhydride. The N-Boc group was expected to be stable in remaining synthetic manoeuvring but easily removable under mildly acid conditions in the final deprotection step. Thereafter, 20 was treated with 3HF·Et3N to expose the Man-III 6-OH group, which was then alkylated with triflate 22 to introduce the linker. Reductive debenzylation of 23 with concomitant reduction of the azide under an H2 atmosphere using 10% Pd as the catalyst afforded deprotected glycan 24. Regioselective coupling of 24 carrying a free amino group with an active ester 25 of biotin went smoothly as verified by MS analysis, which was followed by removal of the Boc group with 5% trifluoroacetic acid (TFA) to eventually generate 4 (70% for two steps). Its structure was confirmed by NMR and MS data.
Scheme 3. Synthesis of biotin-GPI glycan conjugate 4.
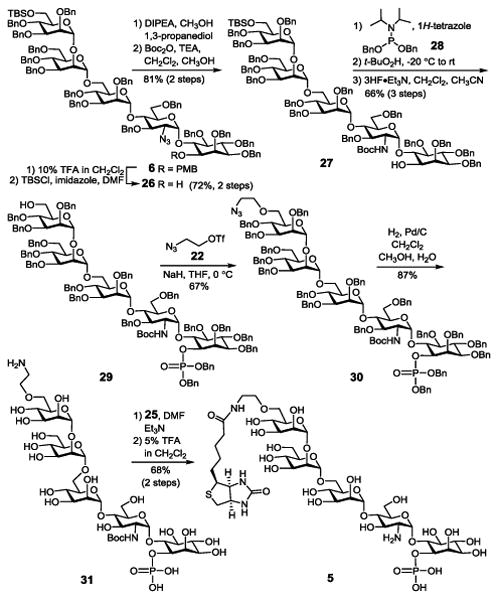
The synthesis of 5, which had a phosphate group at the inositol 1-O-position, from 6 (Scheme 4) started with removing the p-methoxybenzyl (PMB) group. Since it was previously found that ceric ammonium nitrate (CAN)-mediated PMB removal from a GPI was complex and incomplete,18 we probed the reaction of 6 with 2,3-dichloro-5,6-dicyanobenzoquinone (DDQ), but it also provided a complex result. Consequently, we chose to remove PMB using 10% TFA in DCM, which was fast and complete but the TBS group was partly removed. Thus, the product mixture was treated with TBSCl and imidazole to selectively protect the primary hydroxyl group again. Next, the azido group in 26 was reduced and the resultant amine was protected with the Boc group as described above to give 27. Phosphorylation of 27 with phosphoramidite 28 by a two-step one-pot protocol produced the fully protected phosphate in situ, which was treated with 3HF·Et3N to remove the TBS group to generate 29 (66%). It is worth noting that an attempt to reduce the azide after inositol 1-O-phosphorylation was unsuccessful possibly because of the interactions between the reducing reagent and the phosphate group. Next, linker introduction to 29, reductive debenzylation of the resultant 30, coupling 31 with biotin, and removal of the N-Boc group under the above-described conditions afforded the synthetic target 5, which was confirmed by NMR and MS data.
Scheme 4. Synthesis of biotin-GPI glycan conjugate 5 with a phosphate group at the inositol 1-O-position.
To showcase the application potential of biotin-labelled GPIs, 4 and 5 were utilized to study the interactions between GPI and GPI-binding pore-forming bacterial toxin, such as CAMP factor from Streptococcus agalactiae.14 Many bacteria secrete toxic proteins, known as toxins, as their major virulent factors which can bind to certain components on the host cell, form porous oligomers, and insert the pores into the host cell membrane to cause damage.20 It has been revealed that some of these pore-forming bacterial toxins, such as CAMP factor, use GPI as their binding target to facilitate the pore-forming process.12,21,22 Our previous studies utilizing synthetic GPIs and related derivatives suggested that the conserved GlcN-Inositol moiety of GPIs was crucial for GPI-CAMP factor binding and a synthetic GPI analog could inhibit CAMP factor-mediated toxicity to the host cell.23,24 However, all GPIs and GPI derivatives used in previous studies contained lipids, thus whether free GPI glycans, e.g., that in 2 or 3, can bind to pore-forming toxins and how the phosphate group may affect the binding are not clear.
To answer the above questions and gain insights into GPI-toxin interactions, we applied 4 and 5 to analysing CAMP factor-GPI glycan binding by enzyme-linked immunosorbent assay (ELISA), which was made possible by the biotin tag. In the experiments, ELISA plates were first coated with CAMP factor and then with a phosphate buffered saline Tween-20 (PBST) solution of bovine serum albumin (BSA) to block any nonspecific GPI-binding sites. Subsequently, serially diluted solutions of 4 and 5 (0.024∼200 μg/mL, ca. 0.021∼180 μM) in phosphate buffered saline (PBS) were added in the plates. The plates were washed with PBST and incubated with a solution of horseradish peroxidase (HRP)-streptavidin conjugate. After washed again with PBST, the plates were developed with 3,3′,5,5′-tetramethylbenzidine (TMB). The enzymatic reaction product was assessed by measuring the light absorption at 450 nm wavelength. The observed optical density (OD) value should be proportional to the concentration of the enzymatic reaction product - a measure of the HRP-streptavidin conjugate adhered to the plates, which was in turn determined by the amounts of 4 and 5 captured by CAMP factor coated on the plates. Negative controls for these experiments were those with CAMP factor replaced by BSA for plate coating.
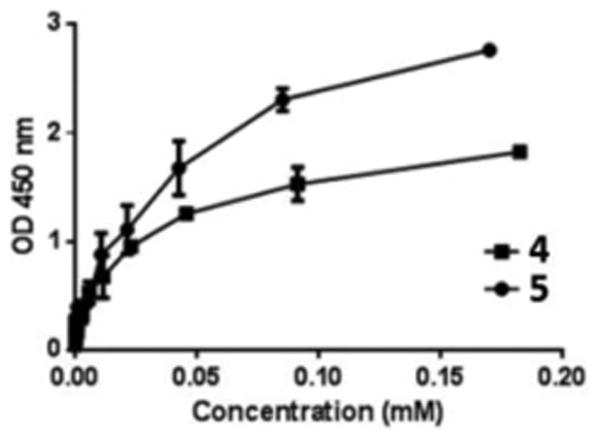
The ELISA results depicted in Figure 3 revealed clearly the close correlation between the OD450 values and the concentration of 4 and 5 and that at low μM concentrations 4 and 5 already had substantial binding to CAMP factor. The results demonstrated unambiguously the high affinity binding between CAMP factor on the plate and GPI glycans in solution. Under the experimental condition, the OD450 values for both 4 and 5 started to level off and reach the maxima beyond 200 μM. Importantly, the OD450 values of 5 were significantly higher than that of 4 for all tested concentrations. The result has disclosed that GPI glycan with a phosphate group at the inositol 1-O-position had higher binding affinity to CAMP factor than the glycan without the phosphate, suggesting the functional significance of this phosphate group in the GPI anchor-CAMP factor binding process.
Figure 3. ELISA results of CAMP factor binding with 4 and 5.
In summary, biotin was successfully coupled with GPI glycans to afford structurally defined GPI glycan-biotin conjugates. Although there have been a number of GPI and GPI conjugate syntheses presented in the literature,25,26 such as recent report to prepare the Toxoplasma gondii GPI,27 to the best of our knowledge, the current work represents the first chemical synthesis of biotin-labeled GPI glycans. The synthesis is highlighted by convergent assembly of orthogonally protected GPI core glycan 6, followed by manipulation of the orthogonal protections and regiospecific phosphorylation and biotinylation. Biotin was linked to the GPI Man-III 6-O-position via a bifunctional group by relatively stable ether and amide bonds. Regiospecific biotinylation was achieved with free GPI glycans. This synthetic strategy is anticipated to be broadly useful for preparing other biotinylated GPI derivatives.
Although a metabolic engineering strategy for the biotinylation GPI anchors on live cells was developed, its main utility was to facilitate on-cell GPI and GPI-anchored protein labelling and isolation of GPI-linked proteins for GPI-anchored proteomics analysis.28 Biotin-labeled GPIs synthesized here should be very useful for many other studies of GPIs. For example, conjugates 4 and 5 were employed to study GPI-CAMP factor interaction by ELISA semi-quantitatively, which was enabled by the biotin tag. Our preliminary results have verified not only the specific and strong binding between GPI glycan and CAMP factor but also the biological significance of the phosphate group at the inositol 1-O-position. Whereas the results are useful for gaining insights into GPI binding with pore-forming bacterial toxins, detailed and quantitative analysis of this process employing 4 and 5 as tools is undergoing in our lab. Nonetheless, the current work showcased the value of biotin-labeled GPI glycans.
Supplementary Material
Acknowledgments
National Institute of General Medical Sciences (NIGMS) and National Institutes of Health (NIH) are acknowledged for their generous support (R01 GM090270) of this project. We thank Prof. M. Palmer at Waterloo University for providing CAMP factor used in this research.
Footnotes
Electronic Supplementary Information (ESI) available: experimental procedures for the syntheses and biological studies, NMR and MS data and spectra of all synthetic intermediates and final products. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Ferguson MAJ, Williams AF. Ann Rev Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 2.Englund PT. Ann Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 3.Cole RN, Hart GW. In: New Comprehensive Biochemistry. Montreuil J, Vliegenthart JFG, Schachter H, editors. Vol. 29. Elsevier; Amsterdam: 1997. pp. 69–88. [Google Scholar]
- 4.Hwa K. Adv Exp Med Biol. 2001;491:207–214. doi: 10.1007/978-1-4615-1267-7_15. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson MAJ. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- 6.Pittet M, Conzelmann A. Biochim Biophys Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Kasahara K, Sanai Y. Glycoconjugate J. 2000;17:153–162. doi: 10.1023/a:1026576804247. [DOI] [PubMed] [Google Scholar]
- 8.Marmor MD, Julius M. J Biol Regul Homeostatic Agents. 2000;14:99–115. [PubMed] [Google Scholar]
- 9.Paulick MG, Forstner MB, Groves JT, Bertozzi CR. PNAS. 2007;104:20332–20337. doi: 10.1073/pnas.0710139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Low MG. Biochem J. 1987;244:1–13. doi: 10.1042/bj2440001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrami L, Fivaz M, Glauser PE, Parton RG, van Der Goot FG. J Cell Biol. 1998;140:525–540. doi: 10.1083/jcb.140.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep DB, Nelson KL, Raja SM, Pleshak EN, Buckley JT. J Biol Chem. 1998;273:2355–2360. doi: 10.1074/jbc.273.4.2355. [DOI] [PubMed] [Google Scholar]
- 13.Gordon VM, Nelson KL, Buckley JT, Stevens VL, Tweten RK, Elwood PC, Leppla SH. J Biol Chem. 1999;274:27274–27280. doi: 10.1074/jbc.274.38.27274. [DOI] [PubMed] [Google Scholar]
- 14.Lang S, Palmer M. J Biol Chem. 2003;278:38167–38173. doi: 10.1074/jbc.M303544200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao P, Nairn AV, Hester S, Moremen KW, O'Regan RM, Oprea G, Wells L, Pierce M, Abbott KL. J Biol Chem. 2012;287:25230–25240. doi: 10.1074/jbc.M112.339465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Huang L, Wang H, Ye X. Angew Chem Int Ed. 2004;43:5221–5224. doi: 10.1002/anie.200460176. [DOI] [PubMed] [Google Scholar]
- 17.Podlasek CA, Wu J, Stripe WA, Bondo PB, Serianni AS. J Am Chem Soc. 1995;117:8635–8644. [Google Scholar]
- 18.Xue J, Guo Z. J Am Chem Soc. 2003;125:16334–16339. doi: 10.1021/ja0382157. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt RR. Pure & Appl Chem. 1989;61:1257. [Google Scholar]
- 20.van der Goot FG. Pore-Forming Toxins. Springer-Verlag; Berlin: 2001. [Google Scholar]
- 21.Braun V, Focareta T. Crit Rev Microbiol. 1991;18:115–158. doi: 10.3109/10408419109113511. [DOI] [PubMed] [Google Scholar]
- 22.Parker MW, van der Goot FG, Buckley T. Mol Microbiol. 1996;19:205–212. doi: 10.1046/j.1365-2958.1996.355887.x. [DOI] [PubMed] [Google Scholar]
- 23.Lang S, Xue J, Guo Z, Palmer M. Med Microbiol Immunol. 2007;196:1–10. doi: 10.1007/s00430-006-0021-2. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Shen Z, Zeng X, Lang S, Palmer M, Guo Z. Carbohydr Res. 2008;343:1718–1729. doi: 10.1016/j.carres.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Swarts BM, Guo Z. Adv Carbohydr Chem Biochem. 2012;67:137–219. doi: 10.1016/B978-0-12-396527-1.00004-8. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikolaev AV, Al-Maharik N. Nat Prod Rep. 2011;28:970–1020. doi: 10.1039/c0np00064g. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai YH, Gçtze S, Azzouz N, Hahm HK, Seeberger PH, Silva DV. Angew Chem Int Ed. 2011;50:9961–9964. doi: 10.1002/anie.201103483. [DOI] [PubMed] [Google Scholar]
- 28.Lu L, Gao J, Guo Z. Angew Chem Int Ed. 2015;54:9679–9682. doi: 10.1002/anie.201503814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


