Abstract
HIV entry inhibitors are highly effective in controlling virus replication. We have developed a lentiviral vector that expresses a secreted entry inhibitor, soluble CD4 (sCD4), which binds to the HIV envelope glycoproteins and inactivates the virus. We have shown that sCD4 was secreted from gene-modified CD4+ T cells, as well as from human umbilical cord blood-derived CD34+ hematopoietic stem/progenitor cells (HSPCs), and protected unmodified HIV target cells from infection in vitro. To investigate the in vivo application of our approach, we injected gene-modified HSPCs into NOD/SCID/γcnull (NSG) mice. NSG hosts supported multi-lineage differentiation of human gene-modified HSPCs. Upon challenge with HIV, humanized mice capable of secreting sCD4 demonstrated a reduction of viral load over time compared to control humanized mice. In contrast to gene therapy approaches that render only gene-modified HIV target cells resistant to infection, our approach also showed protection of unmodified CD4+ T cells in the peripheral blood and tissues. Our findings provide support for the continuous delivery of secreted entry inhibitors via gene therapy as an alternative to oral administration of antiretroviral drugs or injection of antiretroviral proteins, including antibodies.
Keywords: HIV, gene therapy, NSG mouse model, entry inhibitor, soluble CD4, hematopoietic stem cells, T cells, HIV entry, promoter, humanized mice
Introduction
Despite major advances in drug design, infections with HIV remain incurable, and the number of individuals living with HIV continues to rise. Two recent approaches used to treat three HIV-infected patients have given novel insights into the requirements for controlling HIV replication in the absence of antiretroviral drug therapy (ART). The Berlin patient was transplanted with allogeneic hematopoietic stem cells (HSCs) that were resistant to HIV due to a rare naturally occurring deletion in the C-C motif chemokine receptor 5 (CCR5) gene.1 The two Boston patients received allogeneic HSCs without the CCR5 deletion, but remained on ART during the procedure.2 All three patients showed complete donor chimerism and were free of detectable levels of HIV after the therapy. However, upon cessation of ART, viral rebound was detected in the Boston patients, but not in the Berlin patient. These studies provide proof of principle that replacing a patient’s immune system with genetically resistant cells can lead to a functional cure. We and others have employed a vast array of gene therapy approaches to render autologous cells resistant to HIV infection.3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Contrary to what was observed in the Berlin patient, the majority of HIV target cells remain unmodified and susceptible to HIV in a gene therapy setting. In an ideal scenario, the gene-modified HIV target cells would have a survival advantage over unmodified cells and replace the susceptible cells over time. However, clinical trials have revealed that gene-modified cells fail to expand in patients,18, 19, 20 probably because of indirect cytopathic effects of HIV replication in the unmodified target cell population.21 A decrease in viral load was observed in a pre-clinical macaque model in which chemotherapeutic agents were used to select in vivo for gene-modified cells that contained a drug resistance gene in addition to a gene conferring HIV resistance.22 Nevertheless, unmodified cells recovered and gene-modified cells decreased to pre-infection levels as soon as the viral loads decreased, thus effectively replenishing the susceptible HIV target cell population.22 Therefore, it is imperative to protect both the unmodified and gene-modified HIV target cell populations for the long-term control of HIV infection.
In contrast to conventional HIV gene therapy strategies that render HIV target cells resistant to infection, modifying cells to produce a secreted antiviral protein would lead to a systemic protective effect. Intuitively, cells of the immune system are best suited to produce antiviral proteins, but only a limited number of studies examined secretion of entry inhibitors from HIV target cells.23, 24 We have previously designed a lentiviral vector for the secretion of a single-chain variable fragment targeting CCR5 (scFvPRO140) and have shown that gene-modified HIV target cells and neighboring unmodified target cells are protected from infection.25 Nevertheless, a major limitation of CCR5-targeting strategies is their ineffectiveness against C-X-C motif chemokine receptor 4 (CXCR4)-tropic HIV. Unlike CCR5, CXCR4 is not dispensable for the host26, 27 and close to 50% of treatment-experienced patients harbor HIV that can utilize CXCR4.28 Furthermore, a shift to CXCR4-tropic HIV was observed in a patient who received a treatment similar to the Berlin patient.29 Therefore, a therapy should be effective against both CCR5- and CXCR4-tropic HIV. The monoclonal antibody VRC01 partially mimics the interaction of the HIV envelope glycoprotein 120 (gp120) with the CD4 receptor and inhibits infection irrespective of co-receptor tropism.30 While VRC01 is effective against a broad range of HIV isolates, the virus rapidly develops resistance to the antibody without impairing the ability of gp120 to bind to the CD4 receptor.31, 32 Soluble CD4 (sCD4) is a truncated version of the CD4 receptor that contains the gp120 binding site. Contrary to VRC01, gp120 mutations that affect binding to sCD4 inevitably reduce the ability of the virus to bind to the CD4 receptor and compromise the replicative fitness of the virus.32 An initial clinical trial based on the administration of recombinant sCD4 proved to be disappointing because only modest reductions in viral load were observed upon administration of sCD4.33 Follow-up in vitro studies showed that some patient isolates required significantly higher concentrations of sCD4 for inhibition than initially anticipated.34 Based on these findings, twice-daily administration of sCD4 for 4 weeks was examined in a clinical trial. In the highest dose group, two out of three patients achieved and maintained complete neutralization of cell-free virus during the treatment period.35, 36 All clinical trials were safe, and no sCD4-mediated enhancement of infection was observed. However, frequent injections of high concentrations of recombinant sCD4 were not deemed feasible for the long-term treatment of patients.
Upon expression in human cells, sCD4 is secreted into the surrounding media. We have previously shown that adding gene-modified cells expressing sCD4 to infected primary blood lymphocytes suppressed viral spread in co-culture experiments.37 Here, we extend these studies and show that gene-modified primary CD4+ T cells and CD34+ hematopoietic stem/progenitor cells (HSPCs) secreted significant quantities of sCD4 that conferred protection to gene-modified and unmodified HIV target cells in vitro. NOD/SCID/γcnull (NSG) mice supported multi-lineage differentiation of human gene-modified HSPCs. Upon infection of the NSG hosts, sCD4 secreted from HSPCs and their progeny inhibited HIV replication and protected CD4+ T cells from infection. Our findings provide in vivo evidence for the safety and feasibility of a gene therapy strategy based on the secretion of an entry inhibitor from gene-modified HSPCs and their progeny cells.
Results
Promoter Analysis
Lentiviral vectors integrate into the genome of host cells and are the vectors of choice for the stable expression of transgenes. Self-inactivating (SIN) lentiviral vectors were designed to prevent transactivation of neighboring genes, vector mobilization, and transcriptional interference with the internal promoters used to express transgenes.38, 39 We chose a bicistronic SIN vector design for the simultaneous expression of the codon-optimized sCD4 gene and the fluorescent reporter ZsGreen1 gene in the modified cells.
To optimize expression in hematopoietic cell lineages, we tested SIN vectors containing different internal promoters (Figure 1A). The human cytomegalovirus (CMV) immediate-early promoter/enhancer was used because it allows high levels of transgene expression in several cell types, including HSPCs.40 We also examined the activity of the CMV promoter in conjunction with a ubiquitous chromatin opening element (UCOE; UCMV promoter) that has previously been shown to prevent silencing of a viral promoter in the context of a lentiviral vector.41 In contrast to viral promoters, housekeeping genes should not be prone to silencing and might also be safer than a viral promoter, particularly for long-lived transduced human cell applications. Therefore, we included the human elongation factor 1-alpha (EF1α) promoter in our analysis. The vectors LV-CMV-sCD4, LV-UCMV-sCD4, and LV-EF1α-sCD4 were used to transduce a human embryonic kidney cell line (293T), a myeloid cell line (K562), B lymphoid cell lines (Bjab and Raji), and T lymphoid cell lines (Jurkat and Pm1). The gene-modified cells were cultured for 2 weeks and analyzed for ZsGreen1 fluorescence by flow cytometry (Figure 1B). All promoters were active in the 293T cell line, but CMV promoter-mediated expression varied in the other tested cell lines and was especially weak in the two T cell lines. In comparison to the CMV promoter, the UCMV promoter improved expression in both T cell lines, but not in other cell types. The EF1α promoter was active in myeloid as well as B and T lymphoid cell lines and mediated significantly higher expression than the CMV or UCMV promoter in T lymphoid cell lines.
Figure 1.
Promoter Analysis
(A) Overview of the SIN lentiviral vectors with different internal promoters. (B) Flow cytometric analysis of transduced cell lines and primary cells. Cells were transduced with LV-CMV-sCD4, LV-UCMV-sCD4, or LV-EF1α-sCD4 as described in the Materials and Methods. HSPCs were analyzed 4 days post-transduction. All other cell types were analyzed 14 days post-transduction. The percentage of gene-modified cells is indicated, and the median fluorescence intensity is shown in parentheses. (C) Western blot analysis for the presence of sCD4 in culture media of 293T or Jurkat cells transduced with the LV-CMV-sCD4 (CMV), LV-UCMV-sCD4 (UCMV), or LV-EF1α-sCD4 (EF1α) at 2–3 weeks post-transduction. (D) Signal peptide analysis. The native CD4 signal peptide was substituted by the alpha-1 antitrypsin (AAT) signal peptide sequence. Jurkat cells were transduced with LV-EF1α-sCD4 (CD4) or LV-EF1α-AAT-sCD4 (AAT), and culture media were analyzed by anti-His-tag ELISA. A two-tailed unpaired t test was used to determine statistical significance; *p < 0.05. Data are means and SEM representative of two independent experiments performed in duplicates. Ψ, packaging signal; cPPT, central polypurine tract; IRES, internal ribosome entry site; RRE, Rev response element; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; ΔU3, deletion in the U3 promoter.
The three vectors with the different promoters were subsequently used to transduce primary CD4+ T cells and HSPCs (Figure 1B). Similar to data obtained from T lymphoid cell lines, the UCMV promoter was more active than the CMV promoter in both primary cell types. Nevertheless, the vector containing the EF1α promoter mediated the highest expression of the ZsGreen1 gene in CD4+ T cells and CD34+ HSPCs.
Western blot analyses showed that sCD4 was present in the culture supernatants of transduced 293T cell lines, irrespective of the vector used (Figure 1C). In contrast, sCD4 was only detectable in the supernatants of Jurkat cells transduced with the vector containing the EF1α promoter (Figure 1C). We and others have previously shown that utilizing the human alpha-1 antitrypsin (AAT) signal peptide can increase the secretion of recombinant proteins in human cells.42, 43 LV-EF1α-sCD4 was modified to contain the sCD4 gene with the AAT signal peptide to generate LV-EF1α-AAT-sCD4. Substituting the native CD4 signal peptide with the AAT signal peptide resulted in an sCD4 increase of 50% in the culture supernatants of gene-modified Jurkat cells as determined by ELISA (Figure 1D). Based on these results, LV-EF1α-AAT-sCD4 was used in subsequent experiments, unless indicated otherwise.
Differentiation Potential of Gene-Modified HSPCs
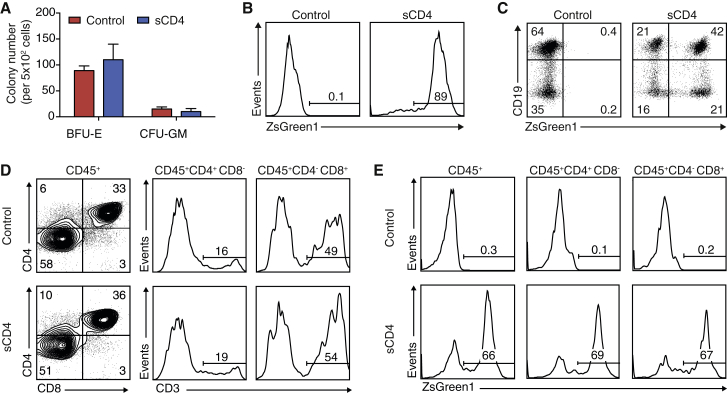
To analyze whether sCD4 interferes with the differentiation potential of HSPCs, we transduced HSPCs with LV-EF1α-AAT-sCD4. Gene-modified HSPCs (CD34+ZsGreen1+) were sorted 24 hr post-transduction and plated in methylcellulose, co-cultured with OP9 cells or co-cultured with OP9-DL4 cells to analyze myeloid progenitor cell, B cell, or T cell development, respectively.44, 45, 46, 47, 48 Gene-modified HSPCs formed burst-forming units erythrocytes (BFU-E) and colony-forming units granulocyte-macrophage (CFU-GM) in methylcellulose cultures (Figure 2A). Flow cytometry analysis of the cells from these cultures revealed that the EF1α promoter remained active in the myeloid progenitor cells after 2 weeks of culture (Figure 2B). After 2–3 weeks of co-culture of gene-modified HSPCs with OP9 cells, the presence of gene-modified B cells (CD19+) was evident (Figure 2C). Following co-culture with OP9-DL4 cells for 53 days, dual-positive (CD4+CD8+) and single-positive (CD4+CD8−CD3high or CD4−CD8+CD3high) cells emerged (Figure 2D). Flow cytometry analysis of CD4+CD8− and CD4−CD8+ cells showed that the EF1α promoter remained active and allowed similar levels of expression in these cell subsets (Figure 2E). Although in vitro-differentiated cultures do not fully recapitulate events of in vivo differentiation, the results indicate that gene-modified HSPCs expressing sCD4 have multi-lineage myeloid cell, B cell, and T cell potential.
Figure 2.
Gene-Modified HSPCs Are Capable of Multilineage Differentiation In Vitro
(A) 5 × 102 gene-modified CD34+ZsGreen1+ HSPCs (sCD4) or unmodified HSPCs (control) were placed in methylcellulose, and cultures were scored for BFU-E and CFU-GM after 14 days. The data are means and SEM representative of two independent experiments performed in duplicate. (B) Representative flow cytometric analysis for promoter activity in methylcellulose. All colonies from the methylcellulose culture were collected, stained with anti-CD33 antibodies and DAPI, and analyzed for ZsGreen1. (C) Gene-modified or control HSPCs were co-cultured with OP9 cells for 2–3 weeks and analyzed by flow cytometry for the presence of CD19+ B cells and promoter activity. Plots are representative of two independent experiments. (D) Control HSPCs or HSPCs expressing sCD4 were co-cultured with OP9-DL4 cells for 53 days and analyzed by flow cytometry for the presence of CD4+CD8− and CD4−CD8+ that co-express CD3. Representative plots from three similar experiments are shown. (E) Flow cytometry analysis of EF1α promoter activity in OP9-DL4 culture-derived lymphoid cells pre-gated on CD45+ cells, CD45+CD4+CD8− or CD45+CD4−CD8+ populations.
Antiviral Effect of sCD4
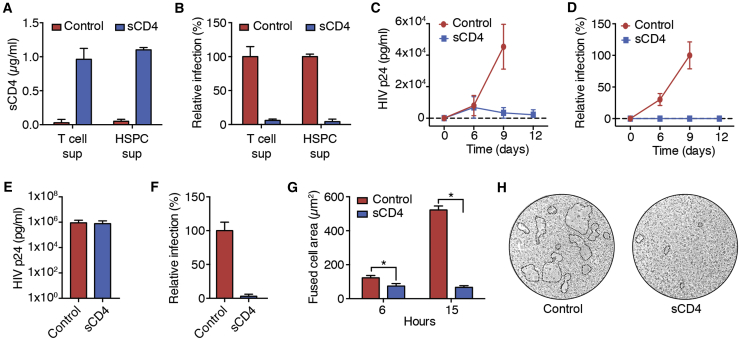
CD4+ T cells isolated from PBMCs and HSPCs enriched from human umbilical cord blood (UCB) were transduced with LV-EF1α-AAT-sCD4 (∼60% and ∼30% gene marking, respectively). Culture supernatants from gene-modified T cells and HSPCs contained ∼1 μg/mL sCD4 (Figure 3A), which was further shown to mediate >95% inhibition of replication-incompetent HIVJRFL entry in single-round infection assays (Figure 3B).
Figure 3.
Antiviral Effect of sCD4
(A and B) Primary CD4+ T cells or HSPCs were transduced with LV-EF1α-AAT-sCD4. Unmodified cells served as a control. 5 × 105 cells/mL were cultured for 4 days, and the culture supernatants (sups) were harvested. (A) T cell and HSPC culture supernatants were analyzed by His-tag ELISA for the presence of sCD4. (B) Single-round infection assays with HIVJRFL were performed in the presence of T cell and HSPC culture supernatants. The number of infected TZM-bl cells was determined as described in the Materials and Methods. Data are means and SEM from two independent experiments performed in duplicate. (C and D) Primary CD4+ T cells were transduced with LV-EF1α (control) or LV-EF1α-AAT-sCD4 (∼60% gene modification) and infected with HIVIIIB. (C) Culture supernatants from infected T cell cultures were analyzed by p24 ELISA. (D) Culture supernatants from infected T cell cultures were used to infect TZM-bl cells in single-round infection assays. Data are means and SEM from two independent experiments performed in duplicate. (E and F) 293T cells were transduced with LV-CMV-AAT-CD4. Unmodified 293T cells (control) or gene-modified 293T cells (sCD4) were transfected with plasmids for the production of replication-incompetent HIVJRFL. (E) Culture supernatants from transfected 293T cells were analyzed by p24 ELISA. (F) Culture supernatants from transfected 293T cells were used to infect TZM-bl cells in single-round infection assays. Data are means and SEM from three independent experiments. (G) TZM-bl cells were transduced with LV-EF1α-AAT-sCD4. Unmodified (control) or gene-modified TZM-bl cells (sCD4) were co-cultured with 293T cells transiently expressing HIVJRFL Env. Syncytia formation was analyzed by light microscopy, and the surface area covered by fused cells was determined. A two-tailed unpaired t test was used to determine statistical significance; *p < 0.05. Data are means and SEM from two independent experiments performed in duplicates. (H) Representative microscope images of fused cells from the 15-h time point are shown. The dotted lines indicate the area of fused cells.
The gene-modified CD4+ T cells were subsequently infected with HIVIIIB. On day 6 post-infection, production of HIV p24 was evident in control cells transduced with an empty vector and cells transduced with LV-EF1α-AAT-sCD4 (Figure 3C), indicating that at least some cells were infected despite sCD4 being present in the culture media. However, virus production increased over time from cells transduced with the control vector, whereas it declined from cells expressing sCD4. Across all time points, virus present in the culture supernatants from cells expressing sCD4 was non-infectious when tested in a single-round infection assay (Figure 3D).
To further examine the antiviral effect, 293T cells expressing sCD4 were transfected with plasmids for the production of replication-incompetent HIVJRFL. Culture supernatants from unmodified control cells and gene-modified cells expressing sCD4 contained comparable amounts of HIV p24 (Figure 3E), suggesting that virus release is not affected by sCD4 expression or its presence in the culture media. Similar to the results from primary CD4+ T cells, the virus produced in gene-modified 293T cells failed to infect HIV target cells in a single-round infection assay (Figure 3F).
To determine whether sCD4 expressed/secreted from the gene-modified cells reduces HIV infectivity by neutralizing envelope proteins present on the cell surface, we co-cultured TZM-bl cells expressing sCD4 with 293T cells expressing the HIVJRFL envelope protein. In this cell fusion assay, interaction of cell-surface-associated gp120 with the CD4 receptor on uninfected HIV target cells is marked by the formation of syncytia consisting of large multinucleated cells. In co-cultures with unmodified control cells, syncytia formation increased over time, and severe cytopathic effects were visible after 15 hr (Figure 3G). In contrast, syncytia formation remained low in co-cultures with HIV target cells that expressed sCD4 (Figure 3G). Representative microscope images are shown in Figure 3H. Overall, these results suggest that sCD4 secreted from the gene-modified cells can protect HIV target cells by neutralizing cell-free virions as well as cell-surface-associated HIV Env.
In Vivo Differentiation of Gene-Modified HSPCs
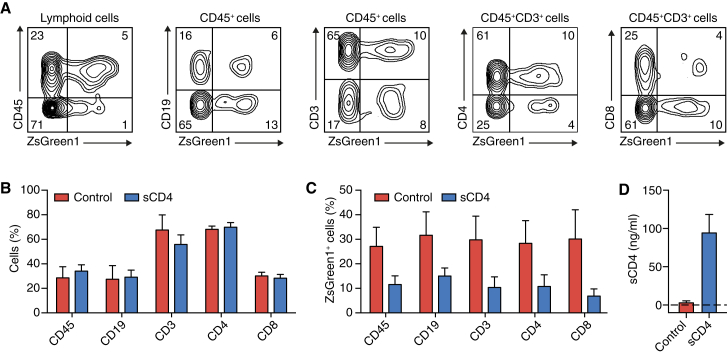
HSPCs were transduced with LV-EF1α-AAT-sCD4 or with an identical LV-EF1α control vector that lacked the sCD4 gene. The cells were analyzed by flow cytometry after 24 hr. We observed generally higher transduction efficacies with the control vector than with the vector encoding sCD4 (Table 1). The HSPCs were injected into NSG mice, and the peripheral blood was analyzed for the presence of human cells 13–19 weeks post-injection. Representative flow cytometry plots for a mouse from the sCD4 group are shown in Figure 4A. The blood of mice from the control group and the sCD4 group contained comparable proportions of human leukocytes (CD45+), B cells (CD19+), and T cells (CD3+) as shown in Figure 4B. Control and sCD4 mice also showed a similar ratio of CD4+ and CD8+ cells within the CD3+ T cell population (Figure 4B). The overall percentage of gene-modified cells was reduced in control and sCD4 mice (Figure 4C) in comparison to the level of gene modification achieved pre-injection (Table 1), which is likely due to transient expression of the ZsGreen1 gene from unintegrated vector that was present directly after the transduction and was lost upon cell differentiation.49 However, consistent with the initial in vitro transduction, control mice had higher percentages of gene-modified cells than mice of the sCD4 group (Figure 4C). The level of gene modification in mice was equal throughout the different cell populations within the control group and the sCD4 group (Figure 4C). Development of myeloid cells was minimal as expected (data not shown) because of lack of human species-specific cytokine support in NSG mice.50 As observed previously in immunocompromised mouse strains, engraftment between individual mice was heterogeneous,51 but no correlation between the level of gene modification and engraftment was evident (Table 2). Plasma samples of mice injected with HSPCs expressing sCD4 contained 0.1 μg/mL sCD4 (Figure 4D).
Table 1.
Gene Modification of Human HSPCs
| Transduction | Control |
sCD4 |
||
|---|---|---|---|---|
| CD34+ Cells (%) | ZsGreen1+ Cells (%) | CD34+ Cells (%) | ZsGreen1+ Cells (%) | |
| 1 | 94 | 43 | 90 | 23 |
| 2 | 97 | 41 | 95 | 33 |
| 3 | 93 | 28 | 94 | 13 |
| Mean ± SEM | 95 ± 1 | 37 ± 5 | 93 ± 2 | 23 ± 6 |
HSPCs isolated from human UCB were transduced with LV-EF1α (control) or LV-EF1α-AAT-CD4 (sCD4). The cells were analyzed for the presence of CD34 and the percentage of ZsGreen1+ cells within the CD34+ cell population on day 1 post-transduction.
Figure 4.
Generation of Humanized Mice Capable of Expressing sCD4
NSG mice were engrafted with HSPCs transduced with LV-EF1α (control) or LV-EF1α-AAT-sCD4 (sCD4). (A–C) 13–19 weeks post-injection, peripheral blood was analyzed for the presence of human CD45+, CD19+, CD3+, CD4+, and CD8+ cells and the presence of gene-modified cells within the same cell populations. CD19+ and CD3+ cells were pre-gated for CD45. CD4+ and CD8+ cells were pre-gated for CD45 and CD3. (A) Representative flow cytometry images for a mouse from the sCD4 group. (B) Human cell population in humanized mice. (C) Gene marking across cell populations in humanized mice. (D) The concentration of sCD4 in the peripheral blood of humanized mice was analyzed by ELISA. All data are expressed as means and SEM (n = 9 for the control group; n = 12 for the sCD4 group).
Table 2.
Engraftment in NSG Mice
| Mouse No. | Control |
sCD4 |
||
|---|---|---|---|---|
| CD45+ Cells (%) | ZsGreen1+ Cells (%) | CD45+ Cells (%) | ZsGreen1+ Cells (%) | |
| 1 | 62 | 25 | 48 | 4 |
| 2 | 8 | 53 | 55 | 5 |
| 3 | 20 | 34 | 47 | 14 |
| 4 | 2 | 7 | 47 | 6 |
| 5 | 6 | 73 | 22 | 25 |
| 6 | 23 | 28 | 31 | 18 |
| 7 | 42 | 10 | 2 | 12 |
| 8 | 14 | 9 | 25 | 43 |
| 9 | 80 | 5 | 21 | 5 |
| 10 | – | – | 54 | 4 |
| 11 | – | – | 46 | 2 |
| 12 | – | – | 11 | 1 |
| Arithmetic mean | 29 | 27 | 34 | 12 |
| Geometric mean | 17 | 19 | 26 | 7 |
| Median | 20 | 25 | 39 | 6 |
Mice were injected with HSPCs transduced with LV-EF1α (control) or LV-EF1α-AAT-sCD4 (sCD4). The peripheral blood of mice was analyzed for the presence of human CD45+ cells and for the percentage of ZsGreen1+ cells within the CD45+ cell population 13–19 weeks post-injection of HSPCs.
In Vivo Inhibition of HIV Infection
The mice were challenged with HIVBaL 5 months post-injection of HSPCs, and the viral load in the peripheral blood was measured by qRT-PCR every 2 weeks up to 8 weeks post-infection (Figure 5A). The viral load between individual mice was heterogeneous but increased over time in control mice. In contrast, viral load continuously decreased in sCD4 mice. In comparison to control mice, sCD4 mice showed a 14-fold difference in viral load 8 weeks post-infection (Figure 5A). The two sCD4 mice with the lowest levels of gene marking (1% and 2%) showed a change in viral load similar to control mice and were therefore grouped separately (sCD4-lowGM; Figure 5A). Details of the different mouse groups are provided in Table 3.
Figure 5.
In Vivo Inhibition of HIV Infection
NSG mice were injected with HSPCs transduced with LV-EF1α (control) or LV-EF1α-AAT-sCD4 (sCD4). sCD4-lowGM denotes two mice of the sCD4 group with 1% and 2% gene marking. Details of the different mouse groups are shown in Table 3. Five months post-injection, the mice were infected with HIVBaL. (A) HIV RNA copies in the peripheral blood were determined at the indicated time points post-infection. The fold change of HIV RNA is shown for all groups. The viral load was ∼5 × 104 copies/mL for the control group, ∼1.5 × 105 copies/mL for the sCD4 group, and ∼5 × 104 copies/mL for the sCD4-lowGM group at 2 weeks post-infection. For sCD4 versus control, a two-way repeated measures ANOVA showed a significant effect (f(3, 48) = 3.191, p < 0.05) with the Bonferroni multiple comparisons test revealing a significant difference at 8 weeks post-infection (∗p < 0.05). (B) Peripheral blood was analyzed for the percentage of human CD4+ T cells at the indicated time points. Lymphoid cells were pre-gated on CD45+CD3+ cells. For sCD4 versus control, a two-way repeated measures ANOVA showed a significant effect (f(3, 27) = 4.843, p < 0.01) with the Bonferroni multiple comparisons test revealing a significant difference at 4, 6, and 8 weeks post-infection (∗p < 0.05). (C) Representative flow cytometry plots from week 6 post-infection are shown. (D) Peripheral blood was analyzed for the presence of ZsGreen1+ T cells. Statistical significance was determined using a paired t test (*p < 0.05); pre-infection = 13–19 weeks after injection of HSPCs; post-infection = 8 weeks after injection of HIVBaL. All data are expressed as means and SEM. The viral load data are representative of three independent experiments. Cell analyses were performed for two of the experiments.
Table 3.
Overview of the Different Mouse Groups
| Analysis | Control |
sCD4 |
sCD4-lowGM |
|||
|---|---|---|---|---|---|---|
| Viral Load | T Cells | Viral Load | T Cells | Viral Load | T Cells | |
| GM (%) | 22.1 | 17.4 | 13.6 | 17.8 | 1.5 | 1.5 |
| n | 8 | 5 | 10 | 6 | 2 | 2 |
| Mouse No. | 1–8 | 4–8 | 1–10 | 5–10 | 11–12 | 11–12 |
The average gene marking (GM), number of mice (n), and which mice were analyzed are shown for each group and analysis.
Over the same time period, the percentage of human CD4+ and CD8+ T cells in the peripheral blood of infected mice was examined by flow cytometry. We observed a decrease in CD4+ T cells in control and sCD4-lowGM mice (Figure 5B). In comparison to control mice, sCD4 mice had higher percentages of human CD4+ T cells 4, 6, and 8 weeks post-infection (Figure 5B). Representative flow cytometry plots for the time point 6 weeks post-infection are shown in Figure 5C. We also observed an increase in the percentage of gene-modified T cells in the peripheral blood of sCD4 and sCD4-lowGM mice at 8 weeks post-infection in comparison to pre-infection levels (Figure 5D).
The control and sCD4 mice were sacrificed 9–10 weeks post-infection, and the percentage of human CD4+ T cells was analyzed in the thymus, spleen, and bone marrow. Representative flow cytometry plots are shown in Figure 6A. The analyses showed that higher levels of CD4+ T cells were present in the thymus, spleen, and bone marrow in sCD4 mice in comparison to control mice (Figures 6B), although the effect did not formally reach statistical significance for the thymus.
Figure 6.
Tissue Analysis of HIV-Infected Humanized Mice Lacking or Expressing sCD4
Nine weeks post-infection, tissues were harvested and analyzed for the presence of CD4+ and CD8+ T cells. (A) Flow cytometry analysis of cells isolated from the thymus, spleen, and bone marrow of infected mice. (B) CD4+/CD8+ T cell ratio in the thymus, spleen, and bone marrow. Lymphoid cells were gated on CD45+CD3+ cells. The median for each group and tissue is shown. One outlier (CD4/CD8 ratio of 16) is not shown in the control group of the thymus analysis. Statistical significance was determined with a Mann-Whitney U test; n = 5 for control (control mice 4–8) and n = 6 for sCD4 (sCD4 mice 5–10).
Discussion
The success of gene therapy approaches that render HIV target cells non-permissive to infection is limited because of ongoing HIV replication in the unmodified HIV target cell population, which impairs the expansion of gene-modified cells. In contrast, secretion of entry inhibitors can result in a systemic control of HIV by preventing infection of gene-modified and unmodified HIV target cells. We have shown that gene-modified CD4+ T cells and HSPCs produce significant quantities of the entry inhibitor sCD4. Gene-modified HSPCs expressing sCD4 were shown to have multi-lineage differentiation potential in vitro and in vivo. Upon infection of humanized mice generated with gene-modified HSPCs expressing sCD4, we observed a reduction in viral load and protection of the CD4+ T cell population in comparison to control mice. Our study provides proof of principle for the safety and efficacy of secreting an sCD4-based entry inhibitor from gene-modified HSPCs and their progeny in a pre-clinical model of HIV infection.
We evaluated lentiviral vectors containing the CMV, UCMV, or EF1α promoter in different cell lines, primary CD4+ T cells, and HSPCs. The UCMV promoter was significantly more active than the CMV promoter in primary CD4+ T cells and to a lesser extent in T lymphoid cell lines and HSPCs. However, the EF1α promoter was highly active in all tested cell types and performed significantly better in HSPCs than either the CMV or UCMV promoter. Importantly, the EF1α promoter remained active upon in vitro differentiation of gene-modified HSPCs into myeloid progenitor cells, B cells, and T cells. In NSG mice injected with gene-modified HSPCs, we observed equal percentages of gene-modified cells across different cell subsets. Therefore, our results provide further evidence for using the EF1α promoter when transgene expression across different hematopoietic cell lineages is desired.
Two applications of a monocistronic lentiviral vector at an MOI of 100 have previously been used to transduce HSPCs in pre-clinical and clinical studies for the treatment of Wiskott-Aldrich syndrome (WAS).52, 53 We performed two transductions with a bicistronic lentiviral vector at an MOI of 50 to generate gene-modified HSPCs. In comparison to the WAS studies, we observed lower levels of overall gene marking, which was likely due to bicistronic lentiviral vectors being less efficient in transducing HSPCs than monocistronic vectors.49 We also observed higher transduction efficacies for the control vector than for the vector encoding sCD4. Similar findings have previously been reported.18 LV-EF1α and LV-EF1α-AAT-sCD4 contained an internal ribosome entry site (IRES) element to initiate the translation of the fluorescent marker, but the control vector lacked the sCD4-encoding sequence. The smaller size of LV-EF1α in comparison to LV-EF1α-AAT-sCD4 could have resulted in increased transduction. Additionally, the second open reading frame in bicistronic vectors is often less effectively translated than the first open reading frame, which could have contributed to the observed discrepancy.54
Injecting gene-modified HSPCs into mice after 4 days of in vitro culture resulted in similar levels of engraftment in the control and sCD4 groups. Although the overall engraftment was lower than what has been reported for mice that were injected with fresh HSPCs or HSPCs cultured for 1 day,55 comparable levels of engraftment have previously been reported for gene-modified HSPCs that were cultured for 3 days prior to injection.56 Gene modification and expression of sCD4 did not affect the engraftment of HSPCs because there were mice with higher levels of gene modification and higher percentages of CD45+ cells in both the control and sCD4 groups.
Although sCD4 is generally more effective against CXCR4-tropic strains, we used CCR5-tropic HIVBaL to infect mice because the majority of circulating patient isolates utilize CCR5 as a co-receptor. Following the infection of the humanized mice, mice of the control and sCD4 groups showed signs of infection 2 weeks after HIV injection. However, the viral load in mice of the sCD4 group decreased over time. We obtained similar results when gene-modified CD4+ T cells were infected with HIV. Our in vitro results suggest that sCD4 present in culture supernatants renders virus particles non-infectious by neutralizing HIV Env on cell-free virions and on the surface of infected cells. Indeed, it has recently been shown that the entry inhibitors maraviroc and FIT20 cause an accumulation of non-infectious virus in the culture supernatants of infected cells.57 However, qRT-PCR-based methods are unable to distinguish between infectious and non-infectious virus. Therefore, viral load measurements in our study were likely overestimated by qRT-PCR, and a greater protective effect could be observed by looking at the percentage of infectious virus particles or infected cells.
Nevertheless, we observed protection of CD4+ T cells from HIV-mediated depletion and an increase of gene-modified T cells in mice expressing sCD4. Continued production of sCD4 may lead to high local concentrations of sCD4, especially at the sites of HIV replication. If the extracellular concentration of sCD4 is sufficiently high to neutralize the majority of HIV, gene-modified and unmodified HIV target cells should have been equally protected from infection. The observed increase in ZsGreen1+ cells in the sCD4 group may indicate that gene-modified T cells have a survival advantage, potentially because the local concentration of sCD4 directly around the gene-modified cells is higher.
A unique element of our approach is the secretion of antiviral proteins from all hematopoietic cell lineages, which increases the ratio of producer cells to HIV target cells. For example, we observed a B cell/T cell ratio of 1:2 and a CD4+ T cell/CD8+ T cell ratio of 2:1 in our humanized NSG mice. Therefore, mice with 10% overall gene marking would have at least 22.5 gene-modified producer cells per 100 HIV target cells. However, an important consideration worth noting is that myeloid engraftment in NSG mice is limited. Because approximately 60% of CD45+ cells in human blood are CD33+ myeloid cells,58, 59 they would likely contribute significantly to the production of sCD4. Even though the HSPC transplantation into the NSG model reconstitutes most human immune system lineages, there is a clear need for an improved model that would truly test the full benefit of secretion from multipotent HSPCs. Another disadvantage of the NSG mouse model is the inability to mount an immune response against HIV.60 sCD4 has the unique ability to induce conformational changes exposing conserved epitopes in the HIV envelope proteins.61, 62 Antibodies against conserved CD4-induced epitopes are readily formed in patients,63 but exhibit limited antiviral effects because their epitopes are inaccessible in the absence of virus binding to CD4.61 sCD4 could enable pre-existing antibodies targeted against the CD4-induced epitopes to bind HIV Env on virus-producing cells and destroy infected cells via antibody-dependent cellular cytotoxicity.64 More recently developed mouse strains, e.g., MISTRG mice, or macaques, that better simulate the human immune system may be used in the future.59, 65, 66
sCD4 has been shown to enhance infection at subinhibitory concentrations in vitro. However, administration of sCD4 was safe in clinical trials.33, 35, 36 Therefore, it is unclear whether sCD4-mediated enhancement of infection plays a role in vivo. The in vitro enhancing effects of sCD4 are apparent within days of culture.62, 67, 68, 69 Additionally, emergence of resistant virus was reported ∼1 month after administration of the antibodies VRC01 or 3BNC117 in recent clinical trials.31, 32 We have not observed an enhancement of infection or a viral load rebound, but long-term testing in mice may be necessary in order to exclude with certainty that HIV escapes inhibition by sCD4. We and other groups have shown that sCD4 can be covalently linked to synergistic inhibitors, such as sCD4-induced epitope-targeting antibody fragments (e.g., sCD4-scFv17b), CCR5-mimicking peptides (e.g., eCD4-immunoglobulin [Ig]), or fusion inhibitors (e.g., 2DLT or sCD4-FIT45).37, 68, 69, 70, 71 The resulting bifunctional proteins drastically improved the antiviral effect of sCD4 without enabling infection of CD4-negative cells. Furthermore, bifunctional inhibitors should also minimize the emergence of escape mutants. We are currently investigating the use of secreted bifunctional proteins to inhibit CCR5-tropic and the more aggressively replicating CXCR4-tropic strains in humanized mice.
Gardner et al.69 have shown that adeno-associated virus (AAV)-mediated secretion of eCD4-Ig by muscle cells can protect macaques from HIV infection. We are currently exploring AAV-mediated secretion of sCD4-based inhibitors. Although promising, the feasibility of using AAV to mediate secretion of therapeutic proteins in high quantities in humans still needs to be determined. The benefit of an HSPC-based approach is that high quantities of inhibitors are likely not necessary because secretion occurs directly at the sites of viral replication. Human trials would be necessary to further evaluate the safety and efficacy of expressing sCD4-based inhibitors using HSPCs. Alternatively, autologous CD4+ and CD8+ T cells secreting entry inhibitors could be engineered and used to augment the efficacy of existing therapies.
After more than two decades, gene therapy is emerging as a powerful tool to treat diseases that were deemed incurable. Although the initial cost of gene therapy is high, a one-time treatment with gene-modified HSPCs or T cells is more cost-effective than lifelong ART.72 With the recent successes,52, 73, 74, 75, 76, 77, 78 it is foreseeable that technical and financial barriers will be further reduced, and gene therapy could become a treatment option for a larger population of persons living with HIV.
Materials and Methods
Cells
HEK293T cells were a gift from Dr. Jason Moffat (University of Toronto, Toronto, ON, Canada). The B lymphoid Bjab and Raji cell lines were received from Dr. Lori Frappier (University of Toronto). Bone marrow-derived OP9 cells expressing Notch ligand Delta-like 4 (OP9-DL4) cells were previously described.44 T lymphoid Jurkat cells and myeloid K562 cells were provided by Dr. Donald Branch (University of Toronto). The following cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH: T lymphoid Pm1 cells from Dr. Marvin Reitz,79 and CD4+CCR5+CXCR4+ TZM-bl from Drs. John C. Kappes and Xiaoyun Wu and Tranzyme.80, 81, 82, 83, 84 PBMCs from healthy donors were received from Dr. Mario Ostrowski (University of Toronto). Primary CD4+ T cells were isolated from PBMCs via negative selection using magnetic beads conjugated to antibodies that target CD4− cells according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Human UCB samples were obtained from consenting mothers following delivery in accordance with the approved guidelines established by the research ethics board of Sunnybrook Health Sciences Centre (Toronto, ON, Canada). UCB samples were collected, and HSPC-containing fractions were purified from these samples as previously described.46
Plasmids
pLV-CMV was generated by replacing the PstI-to-NheI fragment of pLVX-IRES-ZsGreen1 (Clontech, Mountain View, CA, USA) with the compatible NsiI-to-AvrII fragment from pLJM1-EGFP.85 The UCOE (nt 1,397–2,942 of the HNRPA2B1 gene) along with a ClaI site at the 5′ end and nt 2,184–2,378 from pLV-CMV at the 3′ end were synthesized by Genscript (Piscataway, NJ, USA). pLV-UCMV was generated by inserting the UCOE as a ClaI-to-NdeI fragment into the same sites of pLV-CMV. Cloning the BssHII-to-BamHI fragment of pHIV-ZsGreen186 into the corresponding sites of pLV-CMV resulted in pLV-EF1α. The gene encoding sCD4 (amino acids 1–209 of CD4 followed by GGGSGAGCCPGCCHHHHHH) and the nucleotide sequence encoding the signal peptide of human AAT (amino acids 1–24) with an EcoRI site at the 5′ end and nt 76–218 of the open reading frame of sCD4 were synthesized by Genscript. The sCD4 gene was flanked by a 5′ EcoRI site and a 3′ NotI site and was inserted into the same sites of pLV-CMV to generate pLV-CMV-sCD4. The EcoRI-to-ApaI fragment containing the AAT sequence was used in a tripartite ligation reaction with the ApaI-to-BamHI and BamHI-to-EcoRI fragments of pLV-CMV-sCD4 to generate pLV-CMV-AAT-sCD4 with the AAT signal peptide. The gene encoding sCD4 was transferred as an EcoRI-to-AvrII fragment into the same restriction sites of pLV-UCMV or pLV-EF1α to generate pLV-UCMV-sCD4 or pLV-EF1α-sCD4, respectively. pLV-UCMV-AAT-sCD4 and pLV-EF1α-AAT-sCD4 were generated by transferring sCD4 with the AAT signal peptide as an EcoRI-to-AvrII fragment from pLV-CMV-AAT-sCD4 into the same restriction sites in pLV-UCMV-sCD4 or pLV-EF1α-sCD4, respectively. pJRFL-Env was a kind gift from Dr. Branch. The following plasmids were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: psPAX2 and pMD2.G from Dr. Didier Trono, and pSG3ΔEnv from Drs. Kappes and Wu.82, 87
Production of Vectors and Viruses
Lentiviral vectors were produced and concentrated by ultracentrifugation as previously described.88 The titer of vector preparations was determined by measuring ZsGreen1 fluorescence in 293T cells.37 Replication-incompetent HIV was generated as described previously by transfecting 293T cells with 10.5 μg of pSG3ΔEnv and 10.5 μg of pJRFL-Env encoding HIV gp160.42 The 50% tissue culture infectious dose (TCID50) of replication-incompetent HIV was determined using TZM-bl cells.42, 89 HIVIIIB and HIVBaL were kind gifts from Drs. Branch and Ostrowski, respectively.
Transductions
For the transduction of cell lines, 1 × 105 cells were mixed with the lentiviral vector and 8 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO, USA). 293T cells were transduced at an MOI of 2. All other cell lines were transduced at an MOI of 10. Bjab, Raji, K562, Pm1, and Jurkat cells were centrifuged with the vectors at 2,000 × g in swinging bucket rotors at 32°C for 1–2 hr. Primary CD4+ T cells were activated with anti-CD28/anti-CD3 antibodies conjugated to magnetic beads (Thermo Fisher Scientific) and cultured in AIM-V serum-free medium (Thermo Fisher Scientific) supplemented with 10% human serum AB (Sigma-Aldrich) and 200 inducing units/mL interleukin-2 (IL-2; BioShop, Burlington, ON, Canada) for 24 hr. 1 × 105 cells were spinoculated with vectors at an MOI of 50 in the presence of 5 μg/mL polybrene at 2,000 × g at 32°C for 1–2 hr. The cells were resuspended and incubated for 16 hr. The media were replaced by fresh media, and the cells were incubated for 6 hr, followed by another vector application. HSPCs were activated in X-VIVO 10 serum-free hematopoietic cell media (Lonza, Basel, Switzerland) containing thrombopoietin (TPO; 10 ng/mL), FMS-like tyrosine kinase 3 ligand (Flt3L; 100 ng/mL), stem cell factor (SCF; 100 ng/mL), and IL-3 (30 ng/mL). Twenty-four hours post-activation, 1 × 105 HSPCs and lentiviral vector at an MOI of 50 were added to Retronectin (20 μg/mL; Clontech)-coated plates and incubated for 24 hr, followed by another vector application.
Detection of sCD4 in Culture Media
Western blot analysis was performed as previously described.25, 37 His-tag ELISA (Genscript) was performed according to the manufacturer’s instructions. The concentration of sCD4 in culture media was determined in relation to purified His-tagged sCD4 as described previously.42
Single-Round Infection Assays
Single-round infection assays were essentially performed as described previously.42 Briefly, replication-incompetent HIVJRFL at an MOI of 0.05 was incubated with sCD4-containing or mock culture supernatants for 20 min. The mixture was added to 5 × 104 TZM-bl cells in the presence of 8 μg/mL polybrene. Alternatively, 100 μL of virus-containing culture supernatants was used to infect TZM-bl cells. The cells were incubated for 2 days, fixed with 1% formaldehyde, and stained overnight in a staining solution (0.4 mM potassium ferricyanide, 0.4 mM potassium ferrocyanide, 20 mM magnesium chloride, 0.4 μg/mL X-Gal). Three light microscope images were taken for each well, and the number of infected (β-galactosidase+) cells was determined to score infection.
Inhibition of Virus Release
293T cells were transduced with LV-CMV-AAT-CD4. Replication-incompetent HIVJRFL was produced from gene-modified and unmodified 293T cells as described above. Virus in the culture supernatants was quantified by p24 ELISA (XpressBio, Frederick, MD, USA), and 100 ng of p24-equivalent was tested for infectivity in single-round infection assays.
Inhibition of Cell Fusion
TZM-bl cells were transduced with LV-EF1α-AAT-sCD4. For the expression of HIV envelope proteins, 293T cells were transfected with 10.5 μg of psPAX2 and 10.5 μg of pJRFL-Env using the calcium phosphate method as previously described.37 After 24 hr, the media were exchanged and the cells were incubated for 1 additional day. Prior to the assay, the cells were detached using PBS containing 1 mM EDTA and washed twice with PBS supplemented with 1% fetal bovine serum. 1 × 105 transfected 293T cells were incubated with 2 × 105 gene-modified or unmodified TZM-bl cells. At the indicated time points, the cells were fixed with 1% formaldehyde and stained with Giemsa. Three light microscope images per well were taken, and the surface area of fused cells was determined using Adobe Photoshop.
Infection of CD4+ T Cells
CD4+ T cells were transduced with LV-EF1α or LV-EF1α-AAT-sCD4. Four days post-transduction, 5 × 105 gene-modified cells in 100 μL culture media were mixed with 100 μL media containing 18 ng of p24-equivalent of HIVIIIB (MOI ∼0.4). The mixture was incubated for 4–5 hr in order to allow virus adsorption. The cells were washed to remove unbound virus and cultured in AIM-V medium with 10% human serum and 200 U/mL IL-2. Virus production was determined by p24 ELISA (XpressBio). Progeny virus infectivity was determined by performing single-round infection assays using 100 μL of culture supernatants from infected cell cultures.
Induction of T Cell Development
Gene-modified HSPCs were co-cultured with OP9-DL4 cells as previously described for unmodified HSPCs.44, 45, 46
Induction of B Cell Development
Gene-modified HSPCs were co-cultured with OP9 cells and 50 ng/mL IL-7, 5 ng/mL Flt3L, and 30 ng/mL SCF similar to what was previously described for unmodified HSPCs.44, 45, 46, 47, 48
Methylcellulose Assay
Analysis of hematopoietic progenitor potential was performed by plating 5 × 102 sorted gene-modified HSPCs (CD34+ZsGreen1+) or unmodified HSPCs in methylcellulose H4435 (Stem Cell Technologies). Colonies containing erythroid and myeloid cells were scored after 14 days based on morphologic characteristics. For promoter analysis, colonies were washed in PBS to remove methylcellulose. For each analysis, all colonies were collected from one plate and stained with DAPI and an anti-CD33 antibody for flow cytometry.
Infection of Humanized Mice
NOD.cg-PrkdcscidIL2rgtm/Wjl/Sz (NSG) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and housed and bred in a pathogen-free facility. Following transduction, 5 × 105 HSPCs were washed and intrahepatically injected into 4-day-old NSG mice irradiated with 150 rad. Mice were infected 20 weeks post-injection with 10,000 TCID50 of HIVBaL in 100 μL of PBS by intraperitoneal injection. Blood samples were obtained by saphenous bleeds at the indicated time points. Plasma was analyzed for HIV RNA copy numbers by qRT-PCR at Department of Microbiology, Mount Sinai Hospital (Toronto, ON, Canada). Cells isolated from the blood, thymus, spleen, and bone marrow were analyzed by flow cytometry. All animal procedures were approved by the Sunnybrook Health Science Centre and University of Toronto Animal Care Committees.
Flow Cytometry
Cells were stained with standard antibody panels (BD Bioscience, Mississauga, ON, Canada, and eBioscience, San Diego, CA, USA) as previously described.46 The following fluorophore-conjugated antibodies were used in this study: anti-CD19-phycoerythrin (PE), anti-CD8-PE, anti-CD34-allophycocyanin (APC), anti-CD34-APC, anti-mouse CD45-APC, anti-CD4-APC, anti-CD33-Alexa Fluor 700 (AF700), anti-CD4-AF700, anti-CD45-APC/Cy7, and anti-CD3-PE/Cy7. Stained cells were analyzed with a FACSCalibur or an LSR-II cytometer (BD Biosciences). Some data were acquired with an Epics XL MCL (Beckman Coulter, Brea, CA, USA). Data analyses were performed using FlowJo (Tree Star, Ashland, OR, USA).
Statistics
The data are presented as mean ± SEM unless otherwise indicated. To determine statistical significance, we used a two-tailed unpaired t test. A two-way repeated measures ANOVA, a paired t test, and Mann-Whitney U test were performed where indicated. Results with a p value <0.05 were considered statistically significant and marked with an asterisk.
Author Contributions
A.F. and J.S. designed and performed the experiments, analyzed the data, and wrote the manuscript; S.A. assisted with some of the flow cytometry analyses; D.L. assisted with HIV testing in humanized mice; S.R. participated in the design of the study and edited the manuscript; J.C.Z.-P. and S.J. designed the study, analyzed the data, and edited the manuscript; S.J. conceived the idea and initiated the study.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This research was funded by the Canadian Institutes of Health Research and the Canadian Foundation for AIDS Research. J.C.Z.-P. is supported by a Canada Research Chair in Developmental Immunology and by The Krembil Foundation.
References
- 1.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 2.Henrich T.J., Hu Z., Li J.Z., Sciaranghella G., Busch M.P., Keating S.M., Gallien S., Lin N.H., Giguel F.F., Lavoie L. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J. Infect. Dis. 2013;207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi S., Van Brunschot A., Asad S., van der Elst I., Read S.E., Bernstein A. Inhibition of human immunodeficiency virus type 1 multiplication by antisense and sense RNA expression. J. Virol. 1991;65:5524–5530. doi: 10.1128/jvi.65.10.5524-5530.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weerasinghe M., Liem S.E., Asad S., Read S.E., Joshi S. Resistance to human immunodeficiency virus type 1 (HIV-1) infection in human CD4+ lymphocyte-derived cell lines conferred by using retroviral vectors expressing an HIV-1 RNA-specific ribozyme. J. Virol. 1991;65:5531–5534. doi: 10.1128/jvi.65.10.5531-5534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramezani A., Joshi S. Comparative analysis of five highly conserved target sites within the HIV-1 RNA for their susceptibility to hammerhead ribozyme-mediated cleavage in vitro and in vivo. Antisense Nucleic Acid Drug Dev. 1996;6:229–235. doi: 10.1089/oli.1.1996.6.229. [DOI] [PubMed] [Google Scholar]
- 6.Lamothe B., Joshi S. Current developments and future prospects for HIV gene therapy using interfering RNA-based strategies. Front. Biosci. 2000;5:D527–D555. doi: 10.2741/lamothe. [DOI] [PubMed] [Google Scholar]
- 7.Ding S.F., Lombardi R., Nazari R., Joshi S. A combination anti-HIV-1 gene therapy approach using a single transcription unit that expresses antisense, decoy, and sense RNAs, and trans-dominant negative mutant Gag and Env proteins. Front. Biosci. 2002;7:a15–a28. doi: 10.2741/ding. [DOI] [PubMed] [Google Scholar]
- 8.Ramezani A., Ma X.Z., Nazari R., Joshi S. Development and testing of retroviral vectors expressing multimeric hammerhead ribozymes targeted against all major clades of HIV-1. Front. Biosci. 2002;7:a29–a36. doi: 10.2741/ramezani. [DOI] [PubMed] [Google Scholar]
- 9.Nazari R., Ma X.Z., Joshi S. Inhibition of human immunodeficiency virus-1 entry using vectors expressing a multimeric hammerhead ribozyme targeting the CCR5 mRNA. J. Gen. Virol. 2008;89:2252–2261. doi: 10.1099/vir.0.2008/001222-0. [DOI] [PubMed] [Google Scholar]
- 10.Nazari R., Joshi S. Exploring the potential of group II introns to inactivate human immunodeficiency virus type 1. J. Gen. Virol. 2008;89:2605–2610. doi: 10.1099/vir.0.2008/004333-0. [DOI] [PubMed] [Google Scholar]
- 11.Liem S.E., Ramezani A., Li X., Joshi S. The development and testing of retroviral vectors expressing trans-dominant mutants of HIV-1 proteins to confer anti-HIV-1 resistance. Hum. Gene Ther. 1993;4:625–634. doi: 10.1089/hum.1993.4.5-625. [DOI] [PubMed] [Google Scholar]
- 12.Melekhovets Y.F., Joshi S. Fusion with an RNA binding domain to confer target RNA specificity to an RNase: design and engineering of Tat-RNase H that specifically recognizes and cleaves HIV-1 RNA in vitro. Nucleic Acids Res. 1996;24:1908–1912. doi: 10.1093/nar/24.10.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singwi S., Ramezani A., Ding S.F., Joshi S. Targeted RNases: a feasibility study for use in HIV gene therapy. Gene Ther. 1999;6:913–921. doi: 10.1038/sj.gt.3300884. [DOI] [PubMed] [Google Scholar]
- 14.Singwi S., Joshi S. Potential nuclease-based strategies for HIV gene therapy. Front. Biosci. 2000;5:D556–D579. doi: 10.2741/singwi. [DOI] [PubMed] [Google Scholar]
- 15.Holt N., Wang J., Kim K., Friedman G., Wang X., Taupin V., Crooks G.M., Kohn D.B., Gregory P.D., Holmes M.C., Cannon P.M. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiGiusto D.L., Krishnan A., Li L., Li H., Li S., Rao A., Mi S., Yam P., Stinson S., Kalos M. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burke B.P., Levin B.R., Zhang J., Sahakyan A., Boyer J., Carroll M.V., Colón J.C., Keech N., Rezek V., Bristol G. Engineering cellular resistance to HIV-1 infection in vivo using a dual therapeutic lentiviral vector. Mol. Ther. Nucleic Acids. 2015;4:e236. doi: 10.1038/mtna.2015.10. [DOI] [PubMed] [Google Scholar]
- 18.Woffendin C., Ranga U., Yang Z., Xu L., Nabel G.J. Expression of a protective gene-prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. USA. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Lunzen J., Glaunsinger T., Stahmer I., von Baehr V., Baum C., Schilz A., Kuehlcke K., Naundorf S., Martinius H., Hermann F. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol. Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- 20.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matrajt L., Younan P.M., Kiem H.P., Schiffer J.T. The majority of CD4+ T-cell depletion during acute simian-human immunodeficiency virus SHIV89.6P infection occurs in uninfected cells. J. Virol. 2014;88:3202–3212. doi: 10.1128/JVI.03428-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younan P.M., Polacino P., Kowalski J.P., Peterson C.W., Maurice N.J., Williams N.P., Ho O., Trobridge G.D., Von Laer D., Prlic M. Positive selection of mC46-expressing CD4+ T cells and maintenance of virus specific immunity in a primate AIDS model. Blood. 2013;122:179–187. doi: 10.1182/blood-2013-01-482224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egerer L., Volk A., Kahle J., Kimpel J., Brauer F., Hermann F.G., von Laer D. Secreted antiviral entry inhibitory (SAVE) peptides for gene therapy of HIV infection. Mol. Ther. 2011;19:1236–1244. doi: 10.1038/mt.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph A., Zheng J.H., Chen K., Dutta M., Chen C., Stiegler G., Kunert R., Follenzi A., Goldstein H. Inhibition of in vivo HIV infection in humanized mice by gene therapy of human hematopoietic stem cells with a lentiviral vector encoding a broadly neutralizing anti-HIV antibody. J. Virol. 2010;84:6645–6653. doi: 10.1128/JVI.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falkenhagen A., Ameli M., Asad S., Read S.E., Joshi S. Gene therapy using a secreted single chain variable fragment targeting CCR5 to inhibit HIV infection. J. Antivir. Antiretrovir. 2013;5:085–091. [Google Scholar]
- 26.Eash K.J., Means J.M., White D.W., Link D.C. CXCR4 is a key regulator of neutrophil release from the bone marrow under basal and stress granulopoiesis conditions. Blood. 2009;113:4711–4719. doi: 10.1182/blood-2008-09-177287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaix J., Nish S.A., Lin W.H., Rothman N.J., Ding L., Wherry E.J., Reiner S.L. Cutting edge: CXCR4 is critical for CD8+ memory T cell homeostatic self-renewal but not rechallenge self-renewal. J. Immunol. 2014;193:1013–1016. doi: 10.4049/jimmunol.1400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann C. The epidemiology of HIV coreceptor tropism. Eur. J. Med. Res. 2007;12:385–390. [PubMed] [Google Scholar]
- 29.Kordelas L., Verheyen J., Beelen D.W., Horn P.A., Heinold A., Kaiser R., Trenschel R., Schadendorf D., Dittmer U., Esser S., Essen HIV AlloSCT Group Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N. Engl. J. Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 30.Lin R.Z., Dreyzin A., Aamodt K., Li D., Jaminet S.C., Dudley A.C., Melero-Martin J.M. Induction of erythropoiesis using human vascular networks genetically engineered for controlled erythropoietin release. Blood. 2011;118:5420–5428. doi: 10.1182/blood-2011-08-372946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch R.M., Boritz E., Coates E.E., DeZure A., Madden P., Costner P., Enama M.E., Plummer S., Holman L., Hendel C.S., VRC 601 Study Team Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch R.M., Wong P., Tran L., O’Dell S., Nason M.C., Li Y., Wu X., Mascola J.R. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J. Virol. 2015;89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schooley R.T., Merigan T.C., Gaut P., Hirsch M.S., Holodniy M., Flynn T., Liu S., Byington R.E., Henochowicz S., Gubish E. Recombinant soluble CD4 therapy in patients with the acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. A phase I-II escalating dosage trial. Ann. Intern. Med. 1990;112:247–253. doi: 10.7326/0003-4819-112-4-247. [DOI] [PubMed] [Google Scholar]
- 34.Daar E.S., Ho D.D. Relative resistance of primary HIV-1 isolates to neutralization by soluble CD4. Am. J. Med. 1991;90(4A):22S–26S. doi: 10.1016/0002-9343(91)90407-o. [DOI] [PubMed] [Google Scholar]
- 35.Schacker T., Coombs R.W., Collier A.C., Zeh J.E., Fox I., Alam J., Nelson K., Eggert E., Corey L. The effects of high-dose recombinant soluble CD4 on human immunodeficiency virus type 1 viremia. J. Infect. Dis. 1994;169:37–40. doi: 10.1093/infdis/169.1.37. [DOI] [PubMed] [Google Scholar]
- 36.Schacker T., Collier A.C., Coombs R., Unadkat J.D., Fox I., Alam J., Wang J.P., Eggert E., Corey L. Phase I study of high-dose, intravenous rsCD4 in subjects with advanced HIV-1 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;9:145–152. [PubMed] [Google Scholar]
- 37.Falkenhagen A., Ameli M., Asad S., Read S.E., Joshi S. A novel gene therapy strategy using secreted multifunctional anti-HIV proteins to confer protection to gene-modified and unmodified target cells. Gene Ther. 2014;21:175–187. doi: 10.1038/gt.2013.70. [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi H., Blömer U., Takahashi M., Gage F.H., Verma I.M. Development of a self-inactivating lentivirus vector. J. Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varma N., Janic B., Ali M., Iskander A., Arbab A. Lentiviral based gene transduction and promoter studies in human hematopoietic stem cells (hHSCs) J. Stem Cells Regen. Med. 2011;7:41–53. doi: 10.46582/jsrm.0701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F., Frost A.R., Blundell M.P., Bales O., Antoniou M.N., Thrasher A.J. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol. Ther. 2010;18:1640–1649. doi: 10.1038/mt.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falkenhagen A., Asad S., Read S.E., Joshi S. Lentiviral expression system for the purification of secreted proteins from human cell cultures. BMC Biotechnol. 2016;16:66. doi: 10.1186/s12896-016-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun B., Zhang H., Benjamin D.K., Jr., Brown T., Bird A., Young S.P., McVie-Wylie A., Chen Y.T., Koeberl D.D. Enhanced efficacy of an AAV vector encoding chimeric, highly secreted acid alpha-glucosidase in glycogen storage disease type II. Mol. Ther. 2006;14:822–830. doi: 10.1016/j.ymthe.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohtashami M., Shah D.K., Kianizad K., Awong G., Zúñiga-Pflücker J.C. Induction of T-cell development by Delta-like 4-expressing fibroblasts. Int. Immunol. 2013;25:601–611. doi: 10.1093/intimm/dxt027. [DOI] [PubMed] [Google Scholar]
- 45.La Motte-Mohs R.N., Herer E., Zúñiga-Pflücker J.C. Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood. 2005;105:1431–1439. doi: 10.1182/blood-2004-04-1293. [DOI] [PubMed] [Google Scholar]
- 46.Awong G., Herer E., Surh C.D., Dick J.E., La Motte-Mohs R.N., Zúñiga-Pflücker J.C. Characterization in vitro and engraftment potential in vivo of human progenitor T cells generated from hematopoietic stem cells. Blood. 2009;114:972–982. doi: 10.1182/blood-2008-10-187013. [DOI] [PubMed] [Google Scholar]
- 47.Cho S.K., Webber T.D., Carlyle J.R., Nakano T., Lewis S.M., Zúñiga-Pflücker J.C. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:9797–9802. doi: 10.1073/pnas.96.17.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt T.M., Zúñiga-Pflücker J.C. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 49.Dupuy F.P., Mouly E., Mesel-Lemoine M., Morel C., Abriol J., Cherai M., Baillou C., Nègre D., Cosset F.L., Klatzmann D., Lemoine F.M. Lentiviral transduction of human hematopoietic cells by HIV-1- and SIV-based vectors containing a bicistronic cassette driven by various internal promoters. J. Gene Med. 2005;7:1158–1171. doi: 10.1002/jgm.769. [DOI] [PubMed] [Google Scholar]
- 50.Shultz L.D., Ishikawa F., Greiner D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 51.McDermott S.P., Eppert K., Lechman E.R., Doedens M., Dick J.E. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood. 2010;116:193–200. doi: 10.1182/blood-2010-02-271841. [DOI] [PubMed] [Google Scholar]
- 52.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scaramuzza S., Biasco L., Ripamonti A., Castiello M.C., Loperfido M., Draghici E., Hernandez R.J., Benedicenti F., Radrizzani M., Salomoni M. Preclinical safety and efficacy of human CD34(+) cells transduced with lentiviral vector for the treatment of Wiskott-Aldrich syndrome. Mol. Ther. 2013;21:175–184. doi: 10.1038/mt.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones S., Peng P.D., Yang S., Hsu C., Cohen C.J., Zhao Y., Abad J., Zheng Z., Rosenberg S.A., Morgan R.A. Lentiviral vector design for optimal T cell receptor gene expression in the transduction of peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Hum. Gene Ther. 2009;20:630–640. doi: 10.1089/hum.2008.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiekmeijer A.S., Pike-Overzet K., Brugman M.H., Salvatori D.C., Egeler R.M., Bredius R.G., Fibbe W.E., Staal F.J. Sustained engraftment of cryopreserved human bone marrow CD34(+) cells in young adult NSG mice. Biores. Open Access. 2014;3:110–116. doi: 10.1089/biores.2014.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kramer V.G., Schader S.M., Oliveira M., Colby-Germinario S.P., Donahue D.A., Singhroy D.N., Tressler R., Sloan R.D., Wainberg M.A. Maraviroc and other HIV-1 entry inhibitors exhibit a class-specific redistribution effect that results in increased extracellular viral load. Antimicrob. Agents Chemother. 2012;56:4154–4160. doi: 10.1128/AAC.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Table 22-1 Blood Cells. In: Alberts B., Johnson A., Lewis J., editors. Molecular Biology of the Cell. 4th edition. Garland Science; 2002. https://www.ncbi.nlm.nih.gov/books/NBK26919/table/A4143/ [Google Scholar]
- 59.Rongvaux A., Willinger T., Martinek J., Strowig T., Gearty S.V., Teichmann L.L., Saito Y., Marches F., Halene S., Palucka A.K. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brehm M.A., Shultz L.D., Luban J., Greiner D.L. Overcoming current limitations in humanized mouse research. J. Infect. Dis. 2013;208(Suppl 2):S125–S130. doi: 10.1093/infdis/jit319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salzwedel K., Smith E.D., Dey B., Berger E.A. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 2000;74:326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haim H., Si Z., Madani N., Wang L., Courter J.R., Princiotto A., Kassa A., DeGrace M., McGee-Estrada K., Mefford M. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson J.E., Elliott D.H., Martin E.A., Micken K., Rosenberg E.S. High frequencies of antibody responses to CD4 induced epitopes in HIV infected patients started on HAART during acute infection. Hum. Antibodies. 2005;14:115–121. [PubMed] [Google Scholar]
- 64.Williams K.L., Cortez V., Dingens A.S., Gach J.S., Rainwater S., Weis J.F., Chen X., Spearman P., Forthal D.N., Overbaugh J. HIV-specific CD4-induced antibodies mediate broad and potent antibody-dependent cellular cytotoxicity activity and are commonly detected in plasma from HIV-infected humans. EBioMedicine. 2015;2:1464–1477. doi: 10.1016/j.ebiom.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veselinovic M., Charlins P., Akkina R. Modeling HIV-1 mucosal transmission and prevention in humanized mice. Methods Mol. Biol. 2016;1354:203–220. doi: 10.1007/978-1-4939-3046-3_14. [DOI] [PubMed] [Google Scholar]
- 66.Peterson C.W., Wang J., Norman K.K., Norgaard Z.K., Humbert O., Tse C.K., Yan J.J., Trimble R.G., Shivak D.A., Rebar E.J. Long-term multilineage engraftment of autologous genome-edited hematopoietic stem cells in nonhuman primates. Blood. 2016;127:2416–2426. doi: 10.1182/blood-2015-09-672337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arthos J., Cicala C., Steenbeke T.D., Chun T.W., Dela Cruz C., Hanback D.B., Khazanie P., Nam D., Schuck P., Selig S.M. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J. Biol. Chem. 2002;277:11456–11464. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- 68.Lu L., Pan C., Li Y., Lu H., He W., Jiang S. A bivalent recombinant protein inactivates HIV-1 by targeting the gp41 prehairpin fusion intermediate induced by CD4 D1D2 domains. Retrovirology. 2012;9:104. doi: 10.1186/1742-4690-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardner M.R., Kattenhorn L.M., Kondur H.R., von Schaewen M., Dorfman T., Chiang J.J., Haworth K.G., Decker J.M., Alpert M.D., Bailey C.C. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature. 2015;519:87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dey B., Del Castillo C.S., Berger E.A. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J. Virol. 2003;77:2859–2865. doi: 10.1128/JVI.77.5.2859-2865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falkenhagen A., Joshi S. Further characterization of the bifunctional HIV entry inhibitor sCD4-FIT45. Mol. Ther. Nucleic Acids. 2017;7:387–395. doi: 10.1016/j.omtn.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burnett J.C., Zaia J.A., Rossi J.J. Creating genetic resistance to HIV. Curr. Opin. Immunol. 2012;24:625–632. doi: 10.1016/j.coi.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Zhang F., Adams S., Bjorkegren E., Bayford J., Brown L., Davies E.G. Hematopoietic stem cell gene therapy for adenosine deaminase-deficient severe combined immunodeficiency leads to long-term immunological recovery and metabolic correction. Sci. Transl. Med. 2011;3:97ra80. doi: 10.1126/scitranslmed.3002716. [DOI] [PubMed] [Google Scholar]
- 74.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Adams S., Howe S.J., Al Ghonaium A., Bayford J., Brown L., Davies E.G. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 75.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Bougnères P., Schmidt M., Kalle C.V., Fischer A., Cavazzana-Calvo M., Aubourg P. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol. 2012;507:187–198. doi: 10.1016/B978-0-12-386509-0.00010-7. [DOI] [PubMed] [Google Scholar]
- 76.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 78.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 79.Lusso P., Cocchi F., Balotta C., Markham P.D., Louie A., Farci P., Pal R., Gallo R.C., Reitz M.S., Jr. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Platt E.J., Wehrly K., Kuhmann S.E., Chesebro B., Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derdeyn C.A., Decker J.M., Sfakianos J.N., Wu X., O’Brien W.A., Ratner L., Kappes J.C., Shaw G.M., Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei X., Decker J.M., Liu H., Zhang Z., Arani R.B., Kilby J.M., Saag M.S., Wu X., Shaw G.M., Kappes J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takeuchi Y., McClure M.O., Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J. Virol. 2008;82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Platt E.J., Bilska M., Kozak S.L., Kabat D., Montefiori D.C. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J. Virol. 2009;83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Welm B.E., Dijkgraaf G.J., Bledau A.S., Welm A.L., Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 88.Kutner R.H., Zhang X.Y., Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 89.Vodicka M.A., Goh W.C., Wu L.I., Rogel M.E., Bartz S.R., Schweickart V.L., Raport C.J., Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]