This paper describes a method that employs difference electron density to identify DNA purines modeled in the incorrect syn/anti conformation. Different classes of mistakes that were identified by using this method to survey all PDB models containing DNA are highlighted.
Keywords: Hoogsteen base pair, DNA structure, purine, syn/anti flip
Abstract
Hoogsteen base pairs are seen in DNA crystal structures, but only rarely. This study tests whether Hoogsteens or other syn purines are either under-modeled or over-modeled, which are known problems for rare conformations. Candidate purines needing a syn/anti 180° flip were identified by diagnostic patterns of difference electron-density peaks. Manual inspection narrowed 105 flip candidates to 20 convincing cases, all at ≤2.7 Å resolution. Rebuilding and refinement confirmed that 14 of these were authentic purine flips. Seven examples are modeled as Watson–Crick base pairs but should be Hoogsteens (commonest at duplex termini), and three had the opposite issue. Syn/anti flips were also needed for some single-stranded purines. Five of the 20 convincing cases arose from an unmodeled alternate duplex running in the opposite direction. These are in semi-palindromic DNA sequences bound by a homodimeric protein and show flipped-purine-like difference peaks at residues where the palindrome is imperfect. This study documents types of incorrect modeling which are worth avoiding. However, the primary conclusions are that such mistakes are infrequent, the bias towards fitting anti purines is very slight, and the occurrence rate of Hoogsteen base pairs in DNA crystal structures remains unchanged from earlier estimates at ∼0.3%.
1. Introduction
As the carrier of genetic information, DNA plays arguably the most crucial molecular-level role in all forms of life, and great understanding has come from simply knowing what DNA looks like. Since its initial description, the Watson–Crick base-paired, B-form double-helix structure has been presented in textbooks as the self-evident truth. However, the actual history of DNA structural biology and the structures themselves are somewhat more complex and interesting.
The double-helix structure of DNA was correctly proposed by Watson and Crick in 1953 (Watson & Crick, 1953b ▸), with the critical clue coming from an X-ray photograph produced by Raymond Gosling and Rosalind Franklin (Franklin & Gosling, 1953 ▸). Watson and Crick’s model of DNA immediately provided a clear explanatory mechanism for replication (Watson & Crick, 1953a ▸), and thus for faithful genetic reproduction. With these implications, geneticists quickly adopted the structural model of DNA. Many structural biologists remained skeptical, however, especially regarding the proposed conformation of the base pairs, known as Watson–Crick (WC) base pairs (Fig. 1 ▸). The experimental data did not provide the resolution required to discern the conformation of the base pairs. In his autobiography, Maurice Wilkins, who worked with Franklin on the DNA project, indicated that Franklin was not interested in modeling for this very reason: the experimental data had not yet revealed enough detail (Wilkins, 2003 ▸). Even in their publication, Watson and Crick admitted that their structure ‘must be regarded as unproved until it has been checked against more exact results’ (Watson & Crick, 1953b ▸).
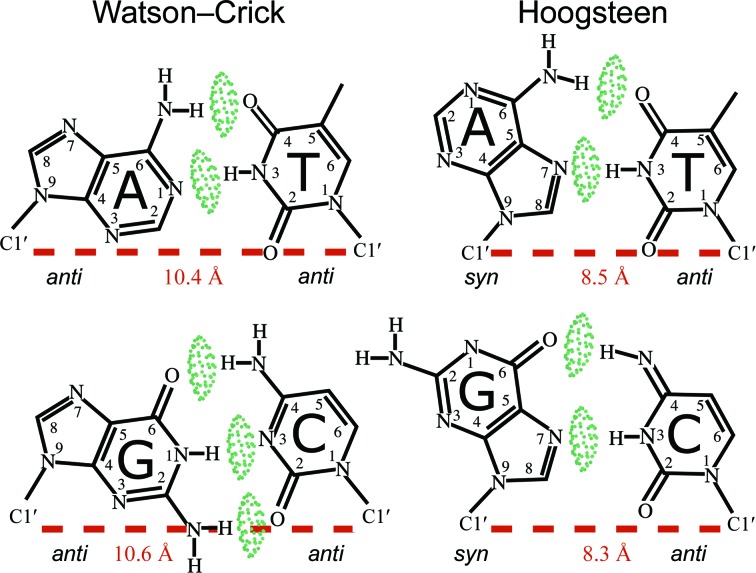
Figure 1.
WC and HG base-pair conformations. The green pillows represent hydrogen bonds. The C1′–C1′ distances for each conformation are shown in red.
In 1959, Karst Hoogsteen published the crystal structure of 1-methylthymine 9-methyladenine, which adopted a different base-pairing conformation than WC (Hoogsteen, 1959 ▸). The conformation had the purine flipped 180° and has come to be known as the Hoogsteen (HG) base pair (Fig. 1 ▸). Relative to the anti purine in WC, in an HG base pair the purine base (A or G) flips its orientation by 180°, adopting a syn conformation, different atoms hydrogen-bond and the C1′–C1′ distance across the pair is shorter. A G·C HG is less favorable than an A·T as it requires either tautomerization or protonation of N3 on dC and forms only two hydrogen bonds rather than the three seen in the WC conformation. In contrast, an A·T transition from WC to HG maintains two hydrogen bonds and does not require nonstandard protonation, giving a rationale for their much higher experimental occurrence (Zhou et al., 2015 ▸).
In 1963, crystal structures of the G·C base pair were solved, revealing only the WC conformation (Sobell et al., 1963 ▸). Although the ribose C1′ atoms are ∼2 Å closer in the HG base pair relative to WC, their experimental observation and the lack of a WC A·T base-pair structure still cast some doubt on the WC base pairing. In 1973, Alex Rich and coworkers reported the X-ray structure of A·U and G·C nucleoside phosphates (Rosenberg et al., 1973 ▸; Day et al., 1973 ▸). Both base pairs were in the WC conformation, and it was the first experimental observation of adenine involved in a WC base pair. Then, in 1980, Drew and Dickerson solved the structure of a synthetic DNA dodecamer sequence featuring a right-handed helix and WC base pairs for both G·C and A·T (Wing et al., 1980 ▸). This finally put the controversy to rest, and WC became established as the standard pairing conformation in duplex DNA for both pairs.
While WC base pairs are indeed the overwhelmingly dominant conformation in B-form DNA and A-form RNA helices, HG pairs are not absent. They are common in base triples and other complex regions of RNA tertiary structure. In DNA, HG are seen as a ubiquitous, low-level, short-lived population by dynamic NMR (Nikolova et al., 2011 ▸) and are occasionally seen in large DNA crystal structures (Zhou et al., 2015 ▸). The preference for anti over syn base orientation central to WC versus HG base pairing was recognized very early on (Haschemeyer & Rich, 1967 ▸; Olson, 1973 ▸; Davies, 1978 ▸). It has been extensively studied and upheld since then, recently for detailed χ angular preferences as a function of sugar conformation and of purine versus pyrimidine by ab initio quantum mechanics (Foloppe et al., 2002 ▸).
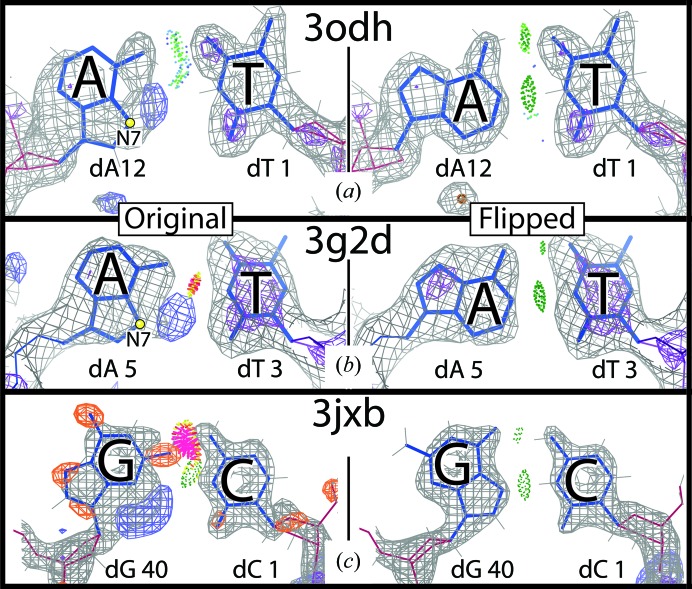
Recently, in collaboration with Hashim Al-Hashimi’s laboratory, we conducted a survey of the Protein Data Bank (PDB; Berman et al., 2000 ▸), where we identified several DNA X-ray structures containing HG pair conformations (Zhou et al., 2015 ▸). Along with reliably modeled HG base pairs, our survey uncovered a number of apparent HG base pairs with dubious conformations. While most of these have insufficiently clear electron density to identify the correct conformation, a few have unambiguous density indicating that the pair is incorrectly modeled and is really in the canonical WC conformation (e.g. Figs. 2 ▸ a and 2 ▸ b). In the opposite direction, we noticed a dubiously modeled WC G·C in PDB entry 3jxb (Watkins et al., 2010 ▸). A steric clash and the electron density, especially the difference density, indicate that the pair might be better modeled as a HG pair. A rebuild of the pair and structure refinement eliminated the difference density, clearly showing an HG conformation (Fig. 2 ▸ c).
Figure 2.
Incorrectly modeled base pairs. A·T pairs from (a) PDB entry 3odh (Vanamee et al., 2010 ▸) and (b) PDB entry 3g2d (Lakomek et al., 2010 ▸) originally modeled the adenines in the syn conformation, but the electron density clearly shows that the models are incorrect. Rebuilding the adenines in the anti conformation and refining the structures confirms the correct WC conformations in (a) and (b). (c) A WC G·C in a dubious conformation from PDB entry 3jxb (Watkins et al., 2010 ▸). Flipping the guanine and refining the structure clearly shows the correct HG conformation. Gray and purple meshes are 2mF o− DF c densities at 1.2 and 3.0σ respectively. Blue and orange mesh are mF o − DF c difference densities at 3.5 and −3.5σ, respectively.
Seeing the incorrectly modeled base pairs led us to ask how we might identify more such cases. We are specifically interested in identifying base pairs modeled as WC (or approximately WC) but which really should be HG, such as the example in PDB entry 3jxb. Here, we describe an automated method for identifying incorrectly modeled purines by searching for difference (mF o − DF c) electron-density features around purine bases. We identify a modest but significant number of purines modeled in the incorrect syn/anti conformation and show their correction. We also describe cases of apparent HG base pairs which are actually artifacts caused by the difficulties of modeling and expressing the coordinates for entire DNA helices with alternate conformations that run in opposite directions. The major aims of this paper are to solidify the HG occurrence levels reported previously and to raise awareness, among structural biologists and among structure end-users, of HG base pairs, purine syn/anti heterogeneity and alternate duplex binding.
2. Methods
The primary difference between WC and HG base pairs is the conformation of the purine base: syn versus anti χ orientation around the glycosidic bond joining the base to the deoxy-ribose sugar. Thus, to find incorrectly modeled DNA base pairs, we aimed to identify purine decoys, i.e. a purine modeled as syn but which really should be anti or vice versa. To do this, we created a general pattern of difference density using the mismodeled purines shown in Fig. 2 ▸ as a reference. This pattern is the basis of the method described below.
2.1. The automated program find_purine_decoys
We wrote an automated program called find_purine_decoys which identifies difference density peaks in predicted locations of potential problems on and around the purine. The program takes a PDB code, downloads the coordinates and structure factors, and returns a list of potential purine decoys, if any are present in the structure. The program requires deposited structure factors for the given PDB entry in order to run. For each purine in the structure, the program uses four steps to identify whether or not the purine is a potential decoy.
(i) Create a search region in real space around the purine.
(ii) Grid the search region.
(iii) Calculate difference values at the grid points.
(iv) Test for difference peaks at predicted locations.
2.2. Create a search region
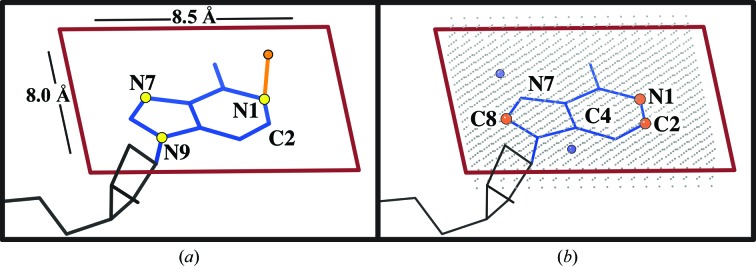
The search region is constructed by drawing a box around the purine (Fig. 3 ▸) in a coordinate frame defined from the plane of the base. The vector between N7 and N1 defines the X direction. The cross-product of the N1–N7 and N1–N9 vectors defines the Z direction, represented by the orange vector in Fig. 3 ▸(a). The origin (the lower left corner of the rectangle, near the ribose) is defined as the point 6 Å left in X and 4 Å down in Y from the N1 atom. The full rectangle spans 8.5 Å in X and 8.0 Å in Y (red outline in Fig. 3 ▸ a).
Figure 3.
(a) A visual explanation of constructing the search region around a purine. The atoms marked with yellow balls were used to define the base plane. The orange vector defines a normal to this plane. (b) A visual representation of the constructed five-layer search grid (gray) where difference (mF o − DF c) values are calculated. The blue and orange balls mark the search regions for positive and negative difference density, respectively.
2.3. Grid the search regions
Within the constructed rectangle (Fig. 3 ▸), a two-dimensional grid is created on the base plane, with the crystallographic standard spacing of 1/4 of the resolution. Identical grids are placed 0.35 and 0.7 Å above and below the base plane, limited to avoid peaks much out of the base plane. This creates a three-dimensional grid of points, five planes and 1.4 Å deep, with the central plane being the base plane (Fig. 3 ▸ b).
2.4. Identify difference peaks
A σA-weighted difference (mF o − DF c) map is calculated, and the difference density value at each grid point is estimated using eight-point interpolation. Difference peaks are identified by searching for multiple adjacent grid points above a positive σ threshold or below a negative σ threshold. The number of adjacent points required and the positive and negative σ density thresholds can be changed. The defaults, used in this study, for a peak to be assigned are ≥4 adjacent points with density in the appropriate direction and with absolute value above the crystallographic standard default of ±3σ.
2.5. Determine potential
The incorrectly modeled base pairs that were identified during our survey exhibited general patterns of difference density (Fig. 2 ▸). Based on this, we looked for negative peaks at the Watson–Crick edge near N1 or C2, or at the sugar edge near C8 (orange balls in Fig. 3 ▸ b). We also looked for positive peaks out from N7 or C4 (blue balls in Fig. 3 ▸ b). These two test points are at box coordinates 3.5, 1.25 near C4 and 1.75, 5.5 near N7. The program reports three levels of decoy candidates from very strong to weak. Very strong decoy candidates have difference density peaks near all five test points. Strong decoy candidates have difference peaks near any three of the five test points or near both positive test points. Weak decoy candidates have difference peaks near any two of the five test points.
2.6. Identifying purine decoys in the PDB
In June 2014 all crystal structures containing DNA but excluding RNA, and with structure-factor data, were downloaded from the PDB. Potential decoys were identified by running find_purine_decoys and selecting those in the strong and very strong categories as described above. Owing to variations in data quality, manual inspection was required to select those purines that truly looked like convincing candidate decoys. For these candidates, manual rebuilding and refinement with PHENIX (Adams et al., 2010 ▸) were performed to test whether the purine was indeed a decoy.
3. Results
Our program, find_purine_decoys, identified 105 potential purine decoys in the strong and very strong categories. Manual inspection of the electron density, especially peak shape and local noise level, was performed to confirm the convincing decoys, i.e. purines which showed convincing evidence that they were modeled in the incorrect anti/syn conformation (qualitative notes on these are given in Supplementary Table S1). A total of 20 purines were identified as convincing decoys and are listed in Table 1 ▸. Further inspection, rebuilding and refinement confirmed 14 flips, five unmodeled alternates and one case with density that was too ambiguous to discern the correct conformation.
Table 1. Manually identified plausible purine decoys from the list returned by find_purine_decoys .
All of these examples were rebuilt and refined. In the ‘Confirm’ column ‘Yes’ confirms that the flip was appropriate, ‘UMA’ indicates that unmodeled alternates explain the difference density and ‘?’ indicates ambiguous density.
| PDB code | Resolution (Å) | Chain | No. | Alternative | Type | Confirm | Structural context |
|---|---|---|---|---|---|---|---|
| 1jkr | 2.27 | B | 16 | A | dA | Yes | Sticky end |
| 2xm3 | 2.30 | Q | 12 | dG | Yes | Sticky end −1 | |
| 2xo6 | 1.90 | E | 12 | dG | Yes | Sticky end −1 | |
| 3brf | 2.47 | C | 2 | dA | Yes | Sticky end −1 | |
| 3v6t | 1.85 | H | 2 | dA | Yes | Blunt end | |
| 3hxo | 2.40 | B | 23 | dG | Yes | DNA aptamer | |
| 2wq7 | 2.00 | D | 8 | dG | Yes | Single strand | |
| 3v9w | 1.70 | G | 7 | dA | Yes | Single strand | |
| 4dtn | 1.96 | T | 3 | dA | Yes | Single strand | |
| 3v6j | 2.30 | B | 7 | dA | Yes | DNA Pol Dpo4 n − 2 | |
| 1s97 | 2.40 | J | 7 | dA | Yes | DNA Pol Dpo4 n − 2 | |
| 3gx4 | 2.70 | Z | 219 | dA | Yes | In helix, HG > WC | |
| 4i2o | 1.77 | X | 5 | dA | Yes | In helix, HG > WC | |
| 3odh | 2.30 | G | 12 | dA | Yes | Blunt end, HG > WC | |
| 4l0z | 2.70 | C | 1 | dG | ? | Sticky end | |
| 3g99 | 1.80 | C | 1 | dA | UMA | Sticky end | |
| 3g9i | 1.85 | D | 8 | dA | UMA | In helix | |
| 3g9o | 1.65 | D | 1 | dA | UMA | Sticky end | |
| 3g9p | 1.65 | D | 1 | dA | UMA | Sticky end | |
| 3p57 | 2.19 | G | 1 | dA | UMA | Sticky end |
Of the 14 purines modeled in the incorrect anti/syn conformation, two were at the n − 2 position in DNA polymerase Dpo4, near the active site. Three were modeled as Hoogsteen but were confirmed to be WC. Four are outside a canonical helix, in either a single-stranded region or another tertiary context. Five were at an oligonucleotide terminus (or −1 to the terminus): four of these were at sticky ends and one was at a blunt end. This preference for ends corroborates our Hoogsteen survey findings, where we observed HG base pairs enriched in strands at duplex termini (Zhou et al., 2015 ▸). The following subsections highlight the details of selected examples, both confirmed flips and unmodeled alternates.
3.1. Terminal ends
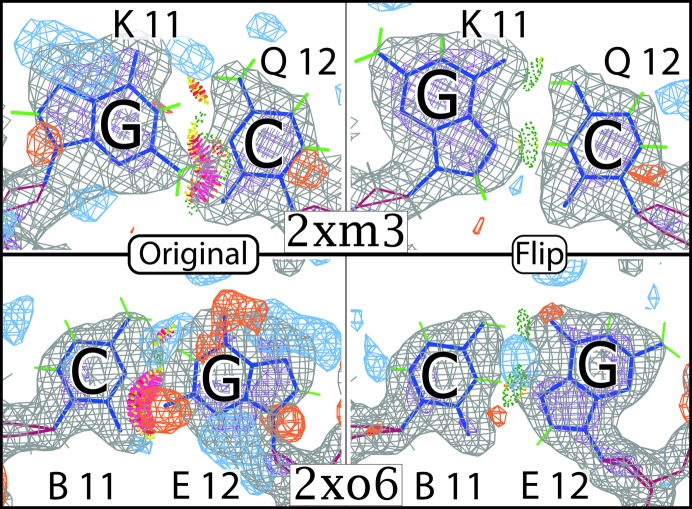
Crystals are made up of repeating unit cells related by translational symmetry. When double-helical DNA is included, the unit cells often line up in a way that allows the DNA ends in each unit cell to stack with one another. This creates semi-continuous DNA helices throughout the whole crystal. Crystallographers often take advantage of this property and design oligonucleotides that have complementary sticky ends, in the hope of promoting stacking of the oligonucleotides and thereby crystallization. Our survey identified five confirmed purine flips at double-helix ends: four at sticky ends (Supplementary Figs. S1, S4, S5 and S6) and one at a blunt end (Supplementary Fig. S11).
Two of these examples come from two structures of insertion sequence Dra2 transposase from Deinococcus radiodurans (ISDra2; Hickman et al., 2010 ▸). To see both bases in the pair one must view symmetry-related contacts, as the pyrimidine and purine are sticky-end overlaps of the helix. Both base pairs have obvious steric clashes between the guanine and cytosine bases. After flipping the purines and refining the structures, the clashes disappear, two HG hydrogen bonds form and the fit to the density improves substantially (Fig. 4 ▸), confirming the correct fit. These pairs exemplify the fact that the syn conformation is much more likely at terminal positions. where the purine has more freedom to move than in the tighter confines of a regular B-form helix interior.
Figure 4.
Mismodeled G·C pairs in ISDra2. These both occur at the oligonucleotide terminus in sticky ends. This means that the purine and pyrimidine are in different asymmetric units. Both pairs fit substantially better in the flipped conformation as syn and HG. The pyrimidines are modeled in their rare tautomer to satisfy the hydrogen bond to N7 on the purine.
3.2. Wishful Hoogsteens
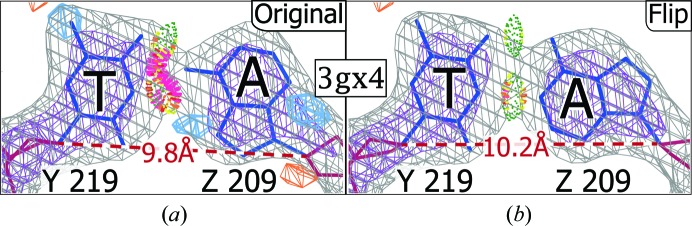
The goal at the onset of this project was to identify HG base pairs incorrectly modeled as WC. In the course of our survey, we also identified three base pairs with the opposite problem, i.e. modeled as HG (or HG-like) but which should really be WC. Two had serious steric clashes, while the third had the bases too far apart to make the canonical HG hydrogen bonds. An important consideration when determining whether to model an HG is the separation of the ribose sugars. On average, the WC and HG C1′–C1′ distances are 10.5 and 8.4 Å, respectively. For an A·T in PDB entry 3gx4, as modeled, the syn conformation brings the C1′ atoms closer to each other than in WC base pairs, but not close enough to form good HG hydrogen bonds (Fig. 5 ▸ a). Flipping and refining improves the model and the fit to density, confirming the WC conformation (Fig. 5 ▸ b).
Figure 5.
A wishful Hoogsteen. (a) This example from PDB entry 3gx4 was originally modeled as an HG but created serious clashes and difference density peaks. (b) Flipping the purine, and making a WC base pair, fits the density better and creates two strong hydrogen bonds.
3.3. Hoogsteens in DNA polymerase Dpo4
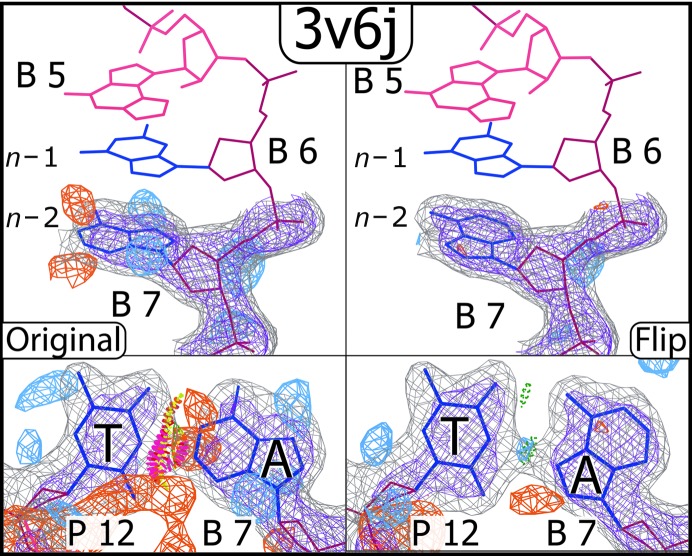
Dpo4 from Sulfolobus solfataricus is a member of the Y-family of DNA polymerases known for its ability to bypass lesions and its high error rate (Boudsocq et al., 2001 ▸). In two separate structures of Dpo4, we identified purines in the n − 2 position (two base pairs from the insertion site) that were modeled in the incorrect syn/anti conformation (Supplementary Figs. S2 and S10). Originally modeled as WC, these base pairs are genuine HG pairs. It is of interest that these rare conformations are being observed in a polymerase whose role is to bypass lesions. While it is difficult to ascertain the true reason for the observed conformation, inspection of the local environment provides some hints.
In PDB entry 1s97, the adjacent base pair in the n − 1 position is a G·T mismatch wherein the guanine is flipped to form a reverse wobble conformation, with hydrogen bonds between dG O6 and dT N3 and between dG N1 and dT O4 (Trincao et al., 2004 ▸). The unusual conformation of n − 1 may have something to do with the HG conformation seen at the n − 2 position. In PDB entry 3v6j (Zhao et al., 2012 ▸) the n − 1 base pair is correctly modeled as an HG, perhaps as a result of the bulky triple-ring base (N 2,3-ethenoguanine) in the insertion site, which makes more van der Waals contacts with the n − 1 guanine syn conformation than it would with the anti conformation. Again, these adjacent conformations provide a possible influence on the preference for syn at n − 2 (Fig. 6 ▸).
Figure 6.
An example of a flip from a Dpo4 polymerase structure (PDB entry 3v6j). The bulky three-ring base in the insertion site may be influencing the preference for the syn conformation seen at the n − 1 and n − 2 positions.
3.4. Other purine flips
Our survey included four authentic purine flips outside the canonical helical context. A dG at a strand-switch site in a structure of the DNA aptamer ARC1172 (PDB entry 3hxo) was incorrectly modeled in the syn conformation (Huang et al., 2009 ▸). The dG should really be anti and makes one strong hydrogen bond to its dT pair (Supplementary Fig. S8). The only single-stranded purine where we identified syn as the correct conformation was in DNA photolyase (PDB entry 2wq7; Glas et al., 2010 ▸). The dG, originally modeled incorrectly as anti (Supplementary Fig. S3), is in the middle of the duplex but is single-stranded since the intended base-pair position is the lesion bound by the protein. The flip with the clearest density is from a structure of RNase T bound to a three-nucleotide product (PDB entry 3v9v; Hsiao et al., 2012 ▸). The dA at the 3′ end of the short strand stacks with two phenylalanines. The modeled syn conformation is clearly wrong, and the density patterning makes the flip to anti correction fairly obvious (Supplementary Fig. S12).
A very interesting case in our survey is a modeled dA (chain T residue 3 in PDB entry 4dtn) in the single-stranded region, before incorporation, in DNA polymerase from bacteriophage RB69 (Xia et al., 2012 ▸). The modeled syn conformation is obviously incorrect, and the 1.96 Å resolution electron density clearly shows the anti conformation to be correct (Supplementary Fig. S13). In the publication for this structure (Xia et al., 2012 ▸) the authors solved a total of nine polymerase structures and reported the identity of the purine of interest as dA. In three of these structures, including PDB entry 4dtn, the highlighted purine is modeled as dA and stacks with tryptophan 547 on the polymerase. However, in the six remaining structures this purine does not interact with the tryptophan and is modeled as dG, with unambiguous electron-density support. After flipping the dA to anti in PDB entry 4dtn, the density strongly suggests that the correct base identity is dG (Supplementary Fig. S13). In combination with the evidence of the six other structures in this study, there is little doubt that this example needs a flip to anti and also a change of base identity, despite the publication.
3.5. Unmodeled alternates
Our survey uncovered a high number of potential purine flips in several crystal structures of the glucocorticoid receptor (GR) bound to DNA determined by Meijsing and coworkers (Meijsing et al., 2009 ▸). Further investigation of these structures identified potential flips in adenine tracts. We were initially excited by this finding as the flips looked to be genuine and at high resolution. An example is PDB entry 3g9p at 1.65 Å resolution. We tested our hypothesis by rebuilding the model with the purine flips and refining the entire structure. The flipped purines fitted the 2F o − DF c density much better; however, there were new large positive difference density peaks between the purines and pyrimidines in the flipped pairs. Also, the well resolved ribose sugars were at a distance that was inconsistent with a Hoogsteen base pair and suggested WC pairing (Supplementary Fig. S15). There had to be another answer.
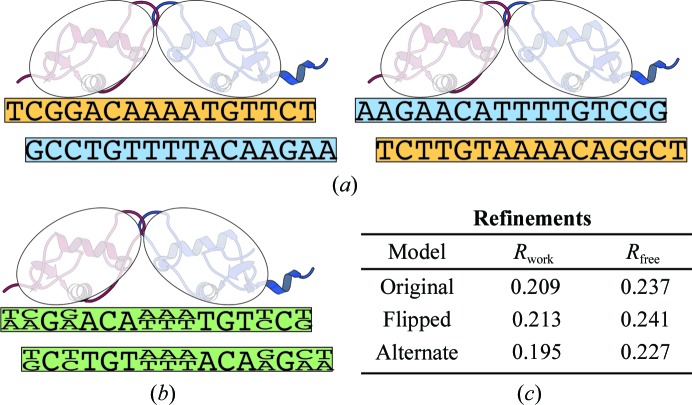
That answer turns out to lie in the fact that GR binds to semi-palindromic sequences (Strähle et al., 1987 ▸), as seen for the bound oligonucleotides in each of the structures. This means that there are two possible binding modes, as shown in Fig. 7 ▸(a). Within the crystal structure, both binding modes are present at about equal levels in different unit cells, creating alternate identities at bases where the palindrome is imperfect, as shown in Fig. 7 ▸(b) and Supplementary Fig. S15. In several of the GR structures the density for these base pairs indeed suggests alternates. For PDB entry 3g9p, after building both whole-helix alternates and refining (including occupancies), the two-alternate model was confirmed. While the purine-flipped model increased the R free by 0.4%, the two-alternate model decreased the R free by 1% (Fig. 7 ▸ c).
Figure 7.
A simplified two-dimensional representation of GR binding one of its target semi-palindromic sequences (from PDB entry 3g9o). This figure does not attempt to represent the complexities seen in the full three-dimensional crystal. (a) Since the palindrome is imperfect, GR has two binding modes. (b) In the asymmetric unit, the two binding modes are effectively superimposed, causing alternate base identities at locations where the palindrome is imperfect. (c) R factors for refinements of PDB entry 3g9p. Examples of individual base pairs with electron density are shown in Supplementary Fig. S15.
Another whole-helix alternate case was found in the transcription factor FixK2 from Bradyrhizobium japonicum (PDB entry 4i2o; Bonnet et al., 2013 ▸; Supplementary Fig. S16). Originally modeled in the syn conformation, adenine X5 took part in an A·T HG-like pairing, but the sugar C1 atoms were too far apart for canonical HG hydrogen bonds. The flip to anti was confirmed by a better density fit and better sterics, with strong WC hydrogen bonds (Supplementary Fig. S17, top). However, reminiscent of the glucocorticoid receptor, the DNA bound to the FixK2 homodimer is semi-palindromic (just six base pairs at the specific binding sites are identical; see Supplementary Fig. S16), and density anomalies were suggestive of an unmodeled T·A/C·G alternate at dA X5. Most tellingly, if the helix bound in both orientations then the two central base pairs would contain purine/pyrimidine alternates. Their very well ordered electron density indeed showed evidence of such alternates (Supplementary Fig. S17). Fitting and refining a second, reversed copy of the helix, both separately (‘Model B’) and as alternates (‘Model A/B’), confirmed this hypothesis: the whole-helix alternate model fitted the density best at all five purine/pyrimidine sites as well as at dA X5, and it decreased the R free by >1% (Supplementary Fig. S17). The case of PDB entry 4i2o thus contains both a purine flip and a whole-helix unmodeled alternate.
4. Discussion
Our survey identified just 14 purines that, with confidence, require flipping in DNA-containing X-ray PDB entries as of June 2014 with deposited diffraction data (as required since February 2008). A limitation of this, or any other method, is that the correctness of a specific local conformation cannot be reliably assessed without considering the fit to the experimental data as well as the geometry and sterics (clashes and hydrogen bonds). Just half of the 14 flip examples became authentic HG base pairs, with five of those seven at oligonucleotide duplex termini. Three other flip examples were originally modeled as HG pairs but are actually WC base pairs. The rest are single-stranded, near nonstandard bulky bases in DNA polymerase or in a DNA aptamer.
These findings confirm the views of Hoogsteen base pairs for DNA in solution versus in crystals. NMR sees very few stable HG base pairs but does observe low populations (∼0.08–2.73%) of rapidly transient HG at essentially every site in DNA (Alvey et al., 2014 ▸). Crystal structures cannot see such low-population alternative conformations, but rather see individual, stabilized HG base pairs at a similar overall frequency (0.3%; Zhou et al., 2015 ▸). We have now verified that potential mismodeling in either direction has not distorted this frequency, as has occurred for some other very rare conformations, such as cis-nonproline peptides (Croll, 2015 ▸; Williams & Richardson, 2015 ▸).
Our method looked for difference density patterns around DNA purine bases to identify potential flips. This demonstrates that, while noisy, difference density can add important information to the consideration of 2mF o − DF c density, model quality and sterics. In this survey we identified 20 plausible purine decoys, but only 14 of them turned out to be authentic flips. The other examples revealed a second distinct class of problem: unmodeled strand alternates. The patterns of difference density peaks for these two classes have a similar appearance, and only at mid to high resolution with good local 2mF o − DF c density can they be distinguished.
To discern the correct class of conformational problem when the difference density suggests an incorrect model, one must consider the position of the purine in the sequence, the oligonucleotide sequence and the relative orientation of the bases (if in a base pair). If an anti purine is in a base pair but at an oligonucleotide terminus or near a distorted section of helix, a flip to syn is plausible, either at full occupancy or as a mixture. If, on the other hand, the purine is in a semi-palindromic duplex, an unmodeled alternate conformation of the entire helix in the opposite orientation is probably the answer. This should especially be considered when the oligonucleotide is bound to a homodimer. If this is the case, the positions of predicted purine/pyrimidine alternates will have density suggestive of alternate identities (in well ordered regions).
Another indication is the C1′–C1′ distance in the base pair. If the overall positions of the two sugars are clear and the C1′–C1′ distance is about 10 Å or greater, then an HG conformation is not appropriate. A syn/anti flip within a base pair always involves movement of one or both sugar rings and a slight adjustment of the adjacent backbone, making post-flip refinement a necessity. At resolutions better than about 2 Å, one can often see an improved fit of the sugar, although there are usually no difference peaks. Less well resolved electron density, however, allows the sugar to change position or conformation substantially without affecting the fit to a diagnostically useful degree. Unfortunately, at resolutions worse than about 2.5 Å it is very difficult, and often impossible, to discern the correct conformation. When the local choice of conformation is truly ambiguous, it is most appropriate to model WC, which has three orders of magnitude higher prior probability in duplex DNA.
This survey had no resolution cutoff, i.e. PDB entries of all resolutions were run through find_purine_decoys. We felt that a resolution cutoff was unnecessary because meaningful difference density at low resolution is not prevalent; we were also curious about what the program would identify at all resolutions. The resolution range of the 105 putative purine-decoy examples identified by find_purine_decoys was 0.98–3.2 Å. The 20 deemed to be plausible flips were in the resolution range 1.7–2.7 Å. Most of the rest were judged to be correct as deposited, while 24 examples had electron density that was too ambiguous to discern the correct conformation. These spanned resolutions from 1.6 to 3.2 Å. These facts illustrate that confidently assigning the correct syn/anti conformation depends on having both mid- to high-resolution density and also local, well resolved density. Some regions of electron density are disordered or otherwise ambiguous, even within high-resolution structures.
Overall, both mismodeled ‘hidden’ Hoogsteens and unjustified syn purines do occur. However, crystallographers are generally doing a good job in modeling purines. They are neither under-modeling nor over-modeling frequently enough to significantly change the HG occurrence statistics from the 0.3% that we found in our previous collaborative HG survey (Zhou et al., 2015 ▸). Also confirmed here are the HG preferences for A·T over G·C and for duplex termini.
Supplementary Material
Supplementary Material.. DOI: 10.1107/S2059798317013729/cb5098sup1.pdf
Acknowledgments
We would like to thank Michael Prisant for managing the database, web and compute servers used in this work. We would also like to thank Hashim Al-Hashimi and his laboratory members, especially Allison Stelling, for useful discussion during this study and manuscript preparation.
Funding Statement
This work was funded by National Institutes of Health, National Institute of General Medical Sciences grants R01-GM073919 and PO1-GM063210. Agilent Foundation grant 3216.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Alvey, H. S., Gottardo, F. L., Nikolova, E. N. & Al-Hashimi, H. M. (2014). Nature Commun. 5, 4786. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bonnet, M., Kurz, M., Mesa, S., Briand, C., Hennecke, H. & Grütter, M. G. (2013). J. Biol. Chem. 288, 14238–14246. [DOI] [PMC free article] [PubMed]
- Boudsocq, F., Iwai, S., Hanaoka, F. & Woodgate, R. (2001). Nucleic Acids Res. 29, 4607–4616. [DOI] [PMC free article] [PubMed]
- Croll, T. I. (2015). Acta Cryst. D71, 706–709. [DOI] [PubMed]
- Davies, D. B. (1978). Prog. Nucl. Magn. Reson. Spectrosc. 12, 135–225.
- Day, R. O., Seeman, N. C., Rosenberg, J. M. & Rich, A. (1973). Proc. Natl Acad. Sci. USA, 70, 849–853. [DOI] [PMC free article] [PubMed]
- Foloppe, N., Hartmann, B., Nilsson, L. & MacKerell, A. D. (2002). Biophys. J. 82, 1554–1569. [DOI] [PMC free article] [PubMed]
- Franklin, R. E. & Gosling, R. G. (1953). Nature (London), 171, 740–741. [DOI] [PubMed]
- Glas, A. F., Kaya, E., Schneider, S., Heil, K., Fazio, D., Maul, M. J. & Carell, T. (2010). J. Am. Chem. Soc. 132, 3254–3255. [DOI] [PubMed]
- Haschemeyer, A. & Rich, A. (1967). J. Mol. Biol. 27, 369–384. [DOI] [PubMed]
- Hickman, A. B., James, J. A., Barabas, O., Pasternak, C., Ton-Hoang, B., Chandler, M., Sommer, S. & Dyda, F. (2010). EMBO J. 29, 3840–3852. [DOI] [PMC free article] [PubMed]
- Hoogsteen, K. (1959). Acta Ctyst. 12, 822–823.
- Hsiao, Y.-Y., Duh, Y., Chen, Y.-P., Wang, Y.-T. & Yuan, H. S. (2012). Nucleic Acids Res. 40, 8144–8154. [DOI] [PMC free article] [PubMed]
- Huang, R.-H., Fremont, D. H., Diener, J. L., Schaub, R. G. & Sadler, J. E. (2009). Structure, 17, 1476–1484. [DOI] [PMC free article] [PubMed]
- Lakomek, K., Dickmanns, A., Ciirdaeva, E., Schomacher, L. & Ficner, R. (2010). J. Mol. Biol. 399, 604–617. [DOI] [PubMed]
- Meijsing, S. H., Pufall, M. A., So, A. Y., Bates, D. L., Chen, L. & Yamamoto, K. R. (2009). Science, 324, 407–410. [DOI] [PMC free article] [PubMed]
- Nikolova, E. N., Kim, E., Wise, A. A., O’Brien, P. J., Andricioaei, I. & Al-Hashimi, H. M. (2011). Nature (London), 470, 498–502. [DOI] [PMC free article] [PubMed]
- Olson, W. K. (1973). Biopolymers, 12, 1787–1814. [DOI] [PubMed]
- Rosenberg, J. M., Seeman, N. C., Kim, J. J. P., Suddath, F. L., Nicholas, H. B. & Rich, A. (1973). Nature (London), 243, 150–154. [DOI] [PubMed]
- Sobell, H. M., Tomita, K. I. & Rich, A. (1963). Proc. Natl Acad. Sci. USA, 49, 885–892. [DOI] [PMC free article] [PubMed]
- Strähle, U., Klock, G. & Schütz, G. (1987). Proc. Natl Acad. Sci. USA 84, 7871–7875. [DOI] [PMC free article] [PubMed]
- Trincao, J., Johnson, R. E., Wolfle, W. T., Escalante, C. R., Prakash, S., Prakash, L. & Aggarwal, A. K. (2004). Nature Struct. Mol. Biol. 11, 457–462. [DOI] [PubMed]
- Vanamee, É. S., Viadiu, H., Chan, S.-H., Ummat, A., Hartline, A. M., Xu, S.-Y. & Aggarwal, A. K. (2010). Nucleic Acids Res. 39, 712–719. [DOI] [PMC free article] [PubMed]
- Watkins, D., Mohan, S., Koudelka, G. B. & Williams, L. D. (2010). J. Mol. Biol. 396, 1145–1164. [DOI] [PubMed]
- Watson, J. D. & Crick, F. H. (1953a). Nature (London), 171, 964–967. [DOI] [PubMed]
- Watson, J. D. & Crick, F. H. (1953b). Nature (London), 171, 737–738. [DOI] [PubMed]
- Wilkins, M. (2003). The Third Man of the Double Helix: The Autobiography of Maurice Wilkins. Oxford University Press.
- Williams, C. & Richardson, J. (2015). Comput. Crystallogr. Newsl. 6, 2–6. https://www.phenix-online.org/newsletter/CCN_2015_01.pdf.
- Wing, R., Drew, H., Takano, T., Broka, C., Tanaka, S., Itakura, K. & Dickerson, R. E. (1980). Nature (London), 287, 755–758. [DOI] [PubMed]
- Xia, S., Vashishtha, A., Bulkley, D., Eom, S. H., Wang, J. & Konigsberg, W. H. (2012). Biochemistry, 51, 4922–4931. [DOI] [PMC free article] [PubMed]
- Zhao, L., Christov, P. P., Kozekov, I. D., Pence, M. G., Pallan, P. S., Rizzo, C. J., Egli, M. & Guengerich, F. P. (2012). Angew. Chem. Int. Ed. 51, 5466–5469. [DOI] [PMC free article] [PubMed]
- Zhou, H., Hintze, B. J., Kimsey, I. J., Sathyamoorthy, B., Yang, S., Richardson, J. S. & Al-Hashimi, H. M. (2015). Nucleic Acids Res. 43, 3420–3433. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material.. DOI: 10.1107/S2059798317013729/cb5098sup1.pdf