The highest resolution structure of bacterial type II NADH:quinone oxidoreductase was determined at 2.15 Å resolution. This structure was used for in silico quinone substrate-docking studies to investigate the binding poses of menadione and ubiquinone molecules.
Keywords: type II NADH:quinone oxidoreductase, NDH-2, respiratory enzymes, membrane proteins, quinone binding
Abstract
Type II NADH:quinone oxidoreductase (NDH-2) is a respiratory enzyme found in the electron-transport chain of many species, with the exception of mammals. It is a 40–70 kDa single-subunit monotopic membrane protein that catalyses the oxidation of NADH and the reduction of quinone molecules via the cofactor FAD. NDH-2 is a promising new target for drug development given its essential role in many bacterial species and intracellular parasites. Only two bacterial NDH-2 structures have been reported and these structures are at moderate resolution (2.3–2.5 Å). In this communication, a new crystallization platform is reported that produced high-quality NDH-2 crystals that diffracted to high resolution (2.15 Å). The high-resolution NDH-2 structure was used for in silico quinone substrate-docking studies to investigate the binding poses of menadione and ubiquinone molecules. These studies revealed that a very limited number of molecular interactions occur at the quinone-binding site of NDH-2. Given that the conformation of the active site is well defined, this high-resolution structure is potentially suitable for in silico inhibitor-compound screening and ligand-docking applications.
1. Introduction
NADH:quinone oxidoreductase plays a central role in the transfer of electrons into the electron-transport chain for the generation of ATP, and in the maintenance of the cellular NAD+/NADH redox balance. Three classes of respiratory NADH:quinone oxidoreductases, namely proton-pumping complex I, sodium-pumping NADH:quinone oxidoreductase and non-proton-translocating type II NADH:quinone oxidoreductase (NDH-2), have been identified in the respiratory chains of bacteria (Melo et al., 2004 ▸; Kerscher et al., 2008 ▸). NDH-2 is found in bacteria, fungi, yeast, protists and plants, but is absent in mammals (Melo et al., 2004 ▸). NDH-2 is essential for the growth and survival of many human pathogens, including Mycobacterium tuberculosis, Plasmodium falciparum and Toxoplasma gondii, and has attracted attention for the development of new antitubercular and antiprotazoal agents (Biagini et al., 2006 ▸; Saleh et al., 2007 ▸; Griffin et al., 2011 ▸; Weinstein et al., 2005 ▸).
Both complex I and sodium-pumping NADH:quinone oxidoreductase form large multi-subunit membrane-protein complexes. They also have the ability to generate a membrane potential by translocating either protons or sodium ions across the membrane (Kerscher et al., 2008 ▸; Melo et al., 2004 ▸; Hirst, 2013 ▸). In contrast, NDH-2 is a 40–70 kDa monotopic membrane protein that contains the cofactor FAD or FMN, and provides exergonic oxidation of NADH and reduction of quinone at two distinct substrate-binding sites. These binding sites are located on the re and si faces of the FAD isoalloxazine (Feng et al., 2012 ▸; Sena et al., 2015 ▸; Blaza et al., 2017 ▸; Heikal et al., 2014 ▸). NDH-2 does not have a proton-pumping function and may not directly support ATP synthesis by generating a proton-motive force. However, it supplies reduced quinones to terminal cytochrome oxidases and indirectly contributes to the generation of ATP.
To date, crystal structures of NDH-2 have been determined for only the following four species: Saccharomyces cerevisiae (Feng et al., 2012 ▸; Iwata et al., 2012 ▸), Caldalkalibacillus thermarum (Heikal et al., 2014 ▸), Staphylococcus aureus (Sena et al., 2015 ▸) and P. falciparum (Yang et al., 2017 ▸). NDH-2 consists of three domains. Two are Rossmann folds, one holding the cofactor FAD and the other binding NADH. The third domain is a C-terminal membrane-anchoring domain that is enriched with both hydrophobic and positively charged amino-acid residues and binds NDH-2 to the phospholipid bilayer (Iwata et al., 2012 ▸; Feng et al., 2012 ▸; Heikal et al., 2014 ▸). The quinone-binding site (Q-site) is located in this C-terminal domain. Some NDH-2 enzymes, such as that from P. falciparum, have a small additional domain (for example an EF-hand) between the second Rossmann fold and the membrane-anchoring domain, but the physiological function of this additional domain remains unknown (Yang et al., 2017 ▸; Melo et al., 2004 ▸). All of the reported NDH-2 structures have a homodimeric structure, but the functional significance of this remains unknown.
Over 4000 membrane-protein structures are available in the Protein Data Bank, but less than 22% of these structures have been determined at resolutions with numerical values lower than 2.0 Å (http://www.rcsb.org). High-resolution structures have been determined for the eukaryotic NDH-2 yeast Ndi1 (PDB entry 4g6h) at 2.26 Å resolution (Feng et al., 2012 ▸) and P. falciparum PfNDH-2 (PDB entry 5jwc) at 2.05 Å resolution (Yang et al., 2017 ▸). Reported bacterial NDH-2 structures are in the 2.3–2.5 Å resolution range (Sousa et al., 2017 ▸; Heikal et al., 2014 ▸). In silico compound screening and docking experiments are relatively easy to set up and do not require a large investment compared with conventional biochemical drug screening (Lagarde et al., 2015 ▸). Notwithstanding this, successful application relies heavily on the quality of the scaffold protein structure (Lagarde et al., 2015 ▸; Warren et al., 2012 ▸). The crystal structure of a protein is not error-free, and coordination errors largely correlate with the resolution at which the structure is determined (Warren et al., 2012 ▸). Ideally, the structure will have been determined with a high-quality data set (i.e. high resolution).
Previously, we reported a 2.5 Å resolution crystal structure of NDH-2 from the thermoalkaliphilic bacterium C. thermarum (Heikal et al., 2014 ▸). In this study, we report a new crystallization platform that produced high-quality NDH-2 crystals resulting in a high-resolution (2.15 Å) bacterial NDH-2 structure. The high-resolution NDH-2 structure was used for in silico quinone substrate-docking studies to investigate the binding poses of menadione and ubiquinone molecules, which were predicted to adopt similar binding poses at the Q-site. The quinone head group is clamped by Glu317 and Ile379, and only one hydrogen bond forms to the N3 atom of the FAD isoalloxazine.
2. Materials and methods
2.1. NDH-2 protein expression and purification
NDH-2 from C. thermarum was expressed and purified as described previously (Heikal et al., 2014 ▸).
2.2. Crystallization
The NDH-2 crystallization was performed using an established method (Heikal et al., 2014 ▸), with modification of the conditions. We altered the original Morpheus crystallization buffer conditions (Gorrec, 2009 ▸), used a higher concentration of NDH-2 and used microseeding (Luft & DeTitta, 1999 ▸). The original buffer was 0.1 M Bicine/Tris pH 8.5 containing a mixture of 1,6-hexanediol, 1-butanol, 1,2-propanediol, 2-propanol, 1,4-butanediol and 1,3-propanediol (the final concentration of each component was 0.02 M), 10%(v/v) glycerol and 20%(v/v) polyethylene glycol 4000. The new buffer was 0.1 M Bicine/Tris buffer pH 8.5 containing 10%(v/v) polyethylene glycol 4000 and 25%(v/v) ethylene glycol. d,l-Lysine (30–150 mM) was added to the new buffer when required to slow the formation of crystals and to aid the growth of larger crystals. Dimethyl sulfoxide [DMSO; 1–7%(v/v)] was added when hydrophobic ligands were co-crystallized with NDH-2. For the crystal seeding stock, crystals were harvested in a Hampton Research seeding tube containing 100 µl of the new crystallization buffer excluding lysine. Stocks were prepared according to the manufacturer’s instructions (Hampton Research, Aliso Viejo, California, USA). An NDH-2 sample with a concentration of 30 mg ml−1 was used for crystallization. Crystallization was performed using the hanging-drop vapour-diffusion method in 24-well plates, in which a drop containing 1 µl protein solution mixed with 1 µl precipitant buffer solution was set up against 1 ml reservoir solution. An aliquot (0.2 µl) of an appropriately diluted seeding stock was added immediately after mixing the protein and precipitant buffer solutions. Plates were incubated at 18°C and crystals formed immediately after seeding. The crystals were not harvested until day 4 to allow sufficient crystal growth to obtain high-quality crystals. Crystal quality rapidly deteriorated after day 7. It was serendipitously discovered that high-quality crystals that diffracted to beyond 2.3 Å resolution were produced with the addition of 1–10 mM menadione (MD) to the new buffer system. No additional cryoprotectant was required, and crystals were flash-cooled in liquid nitrogen for data collection.
2.3. Data collection and processing
The X-ray diffraction data were collected using a microfocus beam at the Australian Synchrotron MX2 beamline. The detector distance was set to 300 mm. Each diffraction image was collected with 30% beam attenuation and a 1° oscillation angle. Three data sets spanning 60, 90 and 60° were collected from a single crystal with 1.5, 2.0 and 2.5 s exposures, respectively. The data-collection point was moved each time to avoid radiation damage. All data sets were processed using the XDS package (Kabsch, 2010 ▸), and were merged and scaled using AIMLESS (Evans & Murshudov, 2013 ▸) in the CCP4 suite (Winn et al., 2011 ▸). To minimize the introduction of model bias into the new structure, a polyalanine model was generated from chain B of PDB entry 4nwz (Heikal et al., 2014 ▸) using CHAINSAW (Stein, 2008 ▸). Molecular replacement was performed using the Phaser crystallographic software with a polyalanine model (McCoy et al., 2007 ▸). Structure refinement and manual modelling were carried out using PHENIX (Adams et al., 2009 ▸) and Coot (Emsley et al., 2010 ▸). PyMOL (Schrödinger) was used to create the figures. Data-collection and processing statistics are given in Table 1 ▸.
Table 1. Data-collection and refinement statistics for the high-resolution NDH-2 structure and comparison to the previously determined structure.
Values in parentheses are for the highest resolution shell.
| Higher resolution | Previous structure | |
|---|---|---|
| Data-collection statistics | ||
| Wavelength (Å) | 0.954 | 0.954 |
| Resolution (Å) | 48.9–2.15 (2.19–2.15) | 65.3–2.50 (2.64–2.50) |
| Space group | P21 | P21 |
| Unit-cell parameters | ||
| a (Å) | 72.8 | 72.8 |
| b (Å) | 113.6 | 114.5 |
| c (Å) | 129.8 | 130.6 |
| β (°) | 91.0 | 92.0 |
| R merge | 0.092 (0.696) | 0.060 (1.320) |
| R p.i.m. | 0.075 (0.610) | 0.035 (0.761) |
| Mean I/σ(I) | 17.3 (2.2) | 15.8 (1.1) |
| Completeness (%) | 91.9 (98.0) | 99.6 (100.0) |
| Multiplicity | 4.1 (3.1) | 3.9 (4.0) |
| Total No. of reflections | 428866 (17441) | 290728 (43141) |
| No. of unique reflections | 105403 (5541) | 73752 (10759) |
| CC1/2 | 0.982 (0.566) | 0.999 (0.394) |
| Wilson B factor (Å2) | 38.6 | 76.0 |
| Refinement statistics | ||
| Resolution (Å) | 48.9–2.15 (2.17–2.15) | 64.6–2.50 (2.56–2.50) |
| R work | 0.208 (0.309) | 0.217 (0.307) |
| R free | 0.238 (0.324) | 0.269 (0.370) |
| R.m.s.d.s | ||
| Bonds (Å) | 0.003 | 0.010 |
| Angles (°) | 0.716 | 1.144 |
| Chiral volume (Å3) | 0.003 | 0.07 |
| No. of atoms | ||
| Protein | 11721 | 12029 |
| Water | 502 | 213 |
| FAD | 212 | 212 |
| Average B factors (Å2) | ||
| Main chain | 53.6 | 77.2 |
| Side chain | 56.0 | 83.7 |
| Water | 50.2 | 58.4 |
| FAD | 40.9 | 59.7 |
| Ramachandran plot statistics (%) | ||
| Favoured regions | 98.1 | 96.8 |
| Allowed regions | 1.9 | 3.2 |
| Outliers | 0 | 0 |
| PDB entry | 5wed | 4nwz |
2.4. Molecular modelling of ubiquinone and MD in the Q-site
The structures of ubiquinone and MD were built in Maestro (Schrödinger) and then prepared for docking using LigPrep (Schrödinger). The Q-site in chain B of the new NDH-2 crystal structure was used for docking (Sastry et al., 2013 ▸). The structure of NDH-2 was prepared using Protein Preparation Wizard (Schrödinger). Both ubiquinone and MD were then modelled in the Q-site using the induced-fit docking (Farid et al., 2006 ▸; Sherman, Beard et al., 2006 ▸; Sherman, Day et al., 2006 ▸) protocol in the Schrödinger Suite (Schrödinger). The centre of the grid was defined as the centroid of residues 13, 44, 46, 47, 316, 317, 320, 347, 348, 349, 350, 376, 379, 380, 382 and 383. The side chains of residues 13, 347, 382 and 383 were trimmed off for initial docking. The van der Waals radii of the ligand and receptor atoms were scaled by a factor of 0.5. The top 20 poses from the initial docking were retained for optimization of the receptor residue conformations. Residues within a 5 Å distance of the respective docked ligand, with the exception of Glu317 and Glu321, were refined. The ligand was then re-docked into the top 20 newly generated receptor conformations that are within 30 kcal mol−1 of the best structure generated after refinement, using the extra-precision (XP) mode of Glide (Schrödinger).
3. Results and discussion
3.1. Production of high-quality NDH-2 crystals for improved resolution
We successfully established a new crystallization platform that consistently generated high-quality NDH-2 crystals that diffracted to beyond 2.5 Å resolution (20 out of 47 crystals tested). The original crystallization conditions used a mixture of volatile alcohols, which produced crystals that were difficult to handle. Consequently, the NDH-2 crystals were loosely packed and were sensitive to changes in the buffer conditions during crystal transfer. Most of the crystals obtained using the old buffer system diffracted to a resolution lower than 3.0 Å. Fortunately, we obtained one crystal that diffracted to 2.5 Å resolution in an earlier study (Heikal et al., 2014 ▸). The fragility of the NDH-2 crystals was also evident in the number of failed ligand-soaking experiments that occurred using both the old and the new crystallization systems.
Compared with the old system, the new system was better for co-crystallization with both hydrophilic and hydrophobic ligands. Using this system, we previously determined the structure of an NAD+–NDH-2 complex (Blaza et al., 2017 ▸). Under the original conditions, no crystals formed in the presence of DMSO, which was added to solubilize highly hydrophobic NDH-2 substrates (e.g. quinones) and inhibitors. In contrast, with the new system we could use up to 7%(v/v) DMSO and still obtain NDH-2 crystals. In this study, we attempted to co-crystallize NDH-2 in the presence of 1,4-naphthoquinone, MD and several NDH-2 inhibitors (e.g. phenothiazines) that presumably bind at the Q-site. Although a number of crystals were obtained from the co-crystallization trials, none of the co-crystals had electron densities corresponding to these ligands either at the Q-site or at any other sites in the structure. However, we discovered that the inclusion of 1–10 mM MD in the crystallization buffer produced many crystals that diffracted to beyond 2.3 Å resolution.
3.2. High-resolution structure of NDH-2
The 2.15 Å resolution structure of NDH-2 from C. thermarum was determined in space group P21, with unit-cell parameters a = 72.8, b = 113.6, c = 129.8 Å, β = 91°. These parameters were almost unchanged from those of the original 2.5 Å resolution structure (Table 1 ▸). After molecular replacement, four molecules were found in an asymmetric unit with well resolved electron density, allowing residues 3–396 to be built in all four chains. This is in contrast to the previous NDH-2 structure, in which the D chain was disordered compared with the other chains and some residues (83–90, 333–337 and 359–365) were missing (Heikal et al., 2014 ▸). As described earlier, we did not observe additional electron density corresponding to MD in the structure.
The new structure improved the overall quality of the NDH-2 model. In the current model, more residues (98.1%) were built in the favoured regions of the Ramachandran plot than in the previous structure. The number of water molecules found in the new structure increased to approximately 500, which was 2.5 times that in the previous structure. The improved resolution was also evident in the lower average Wilson B factor and average B factor of the refined macromolecules and FAD ligands in the new structure, respectively (Table 1 ▸). In particular, the average Wilson B factor of the new data set decreased to 38.6 Å2, which is approximately half that of the previously determined structure.
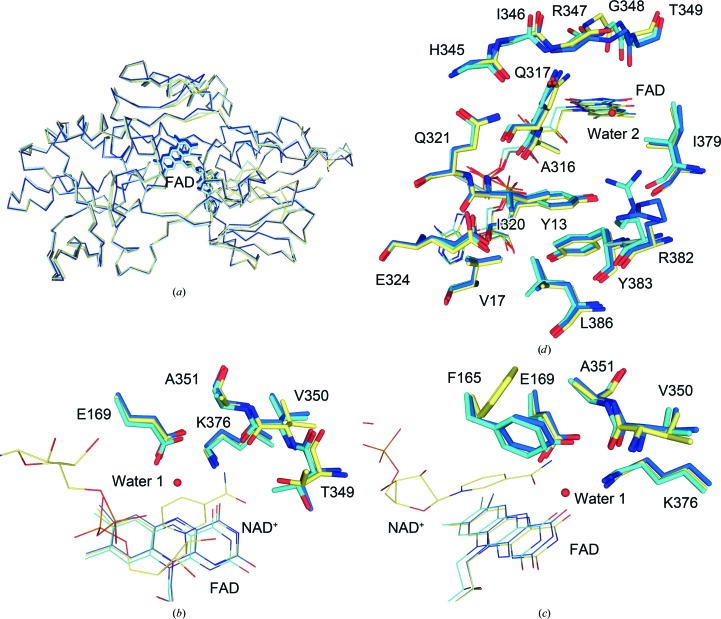
Among the four molecules in the asymmetric unit, the chain B molecule had the lowest average B factor (50.0 Å2 for all atoms). Therefore, it was selected for subsequent structural analysis. For the same reason, chains A and B were used for a homodimer comparison. Overall, the wild-type structures from this study and our previous study did not differ significantly, with an r.m.s.d. of 0.526 Å over 392 Cα atoms (Fig. 1 ▸ a). Their homodimeric structures were also comparable, with an r.m.s.d. of 0.766 Å over 784 Cα atoms. No noticeable structural differences were observed for the residues involved in the binding sites for NADH and quinone (Figs. 1 ▸ b, 1 ▸ c and 1 ▸ d). Next, the high-resolution structure was compared with the structure of the NDH-2–NAD+ complex (PDB entry 5kms; Blaza et al., 2017 ▸). Again, overall, the two structures were comparable, with an r.m.s.d. of 0.407 over 394 Cα atoms. Only a couple of minor differences were noted at the nicotinamide binding site (Figs. 1 ▸ b and 1 ▸ c; Blaza et al., 2017 ▸). These included minor structural rearrangements of the side chain of Thr349, with hydrogen-bond formation between its hydroxyl group and the nicotinamide O atom, and repositioning of the Phe165 side chain upon NADH binding. As expected, the two homodimers were very similar, with an r.m.s.d. of 0.440 Å over 788 Cα atoms, reconfirming that NAD(H) binding does not induce structural rearrangement in NDH-2.
Figure 1.
Comparison of the new (blue), old (PDB entry 4nwz; cyan) and NAD+-bound (PDB entry 5kms; yellow) NDH-2 structures. (a) Superposition of the three structures. (b) Comparison with the nicotinamide-binding site. The identification codes for the NAD+ molecule and the water molecule are PDB entries 5kms and 5wed, respectively. Phe165 is omitted for clarity. (c) A view of (b) rotated by 60° clockwise. Thr349 is omitted for clarity. (d) Comparison with the quinone-binding site. Side chains are omitted for residues 345–349 for clarity.
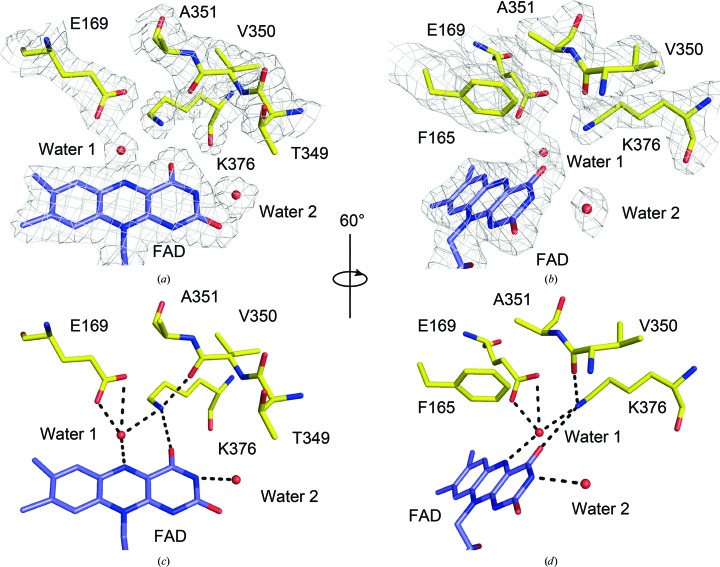
3.3. Chemical geometries of the two substrate-binding sites
At 2.15 Å resolution, the electron density around the NDH-2 catalytic centre is well defined (Figs. 2 ▸ a and 2 ▸ b), providing a clear picture of atomic interactions at the active site of bacterial NDH-2. Residues Phe165, Glu169, Thr349–Ala351 and Lys376 are at the heart of the nicotinamide binding site on the re face of the FAD isoalloxazine (Figs. 2 ▸ c and 2 ▸ d). Previously, it has been shown that residues Phe165, Thr349 and Val350 are directly involved in accommodating the nicotinamide group of NADH (Blaza et al., 2017 ▸). Next, we looked at the Glu169 residue, which is a highly conserved residue in the NDH-2 family that potentially plays an important catalytic role in NADH oxidation in yeast Ndi1 (Feng et al., 2012 ▸) and S. aureus NDH-2 (Marreiros et al., 2017 ▸). In our structure, this residue firmly secured a water molecule (water 1) that resides next to the FAD ring (Fig. 2 ▸ c). A relatively tight hydrogen bond formed between this water and the N5 atom of the isoalloxazine ring with a distance of 2.8 Å. Notably, in the previous structure this water was present in only two out of four chains and was loosely bonded to FAD (3.1–3.2 Å; Heikal et al., 2014 ▸). An equivalent water molecule has been identified in the structure of yeast Ndi1 (PDB entry 4g6g; Feng et al., 2012 ▸) but not in that of P. falciparum PfNDH-2 (PDB entry 5jwa; Yang et al., 2017 ▸). Thus, the involvement of this water in catalysis remains unclear. It has been reported that a conserved mutation to an aspartate at the Glu242 residue of Ndi1, which is equivalent to Glu169 in NDH-2 from C. thermarum, causes a significant growth defect in S. cerevisiae (Feng et al., 2012 ▸). This suggests that maintenance of the chemical environment, including the orientation of the glutamic acid side chain and water 1 at the re face of the FAD isoalloxazine, are important for NADH oxidation. The glutamic acid residue is highly conserved among all FAD-dependent reductases that use NADH or NADPH (Lee et al., 2014 ▸; Marreiros et al., 2017 ▸), and the orientation of the glutamate side chain and water 1 is likely to be a major determinant of whether the pK a is optimal for NADH oxidation and FAD reduction. Finally, the new structure suggests that Lys376 plays an important role in stabilizing both the short linker separating the binding sites for NADH and quinone and the orientation of the FAD isoalloxazine. The ∊-amino group of Lys376 hydrogen-bonds to a carbonyl O atom of Val350 and to the O4 atom of isoalloxazine. The B factor of the ∊-amino N atom is 41.0 Å2, which is the lowest among those of the residues shown in Figs. 2 ▸(b) and 2 ▸(c), which supports the proposed stabilization. This residue also coordinates to a water 1 molecule. In yeast Ndi1 (PDB entry 4g6g; Feng et al., 2012 ▸) and P. falciparum PfNDH-2 (PDB entry 5jwa; Yang et al., 2017 ▸) Lys376 is substituted by a tyrosine residue (Tyr482 and Tyr504) but plays the same role.
Figure 2.
Nicotinamide-binding site of C. thermarum NDH-2. (a) A top view of the re face of the FAD isoalloxazine. The positions of both an FAD molecule (purple) and residues (yellow) involved in the nicotinamide-binding site are supported by an electron-density map shown as a grey mesh (2F o − F c map contoured at 1σ). (b) A view of (a) rotated by 60° clockwise. (c) The same view as in (a) but without the electron-density map. (d) The same view as in (b) but without the electron-density map. Phe165 is omitted in (a) and (c) for clarity. Potential hydrogen-bonding interactions are shown by dashed lines in (c) and (d).
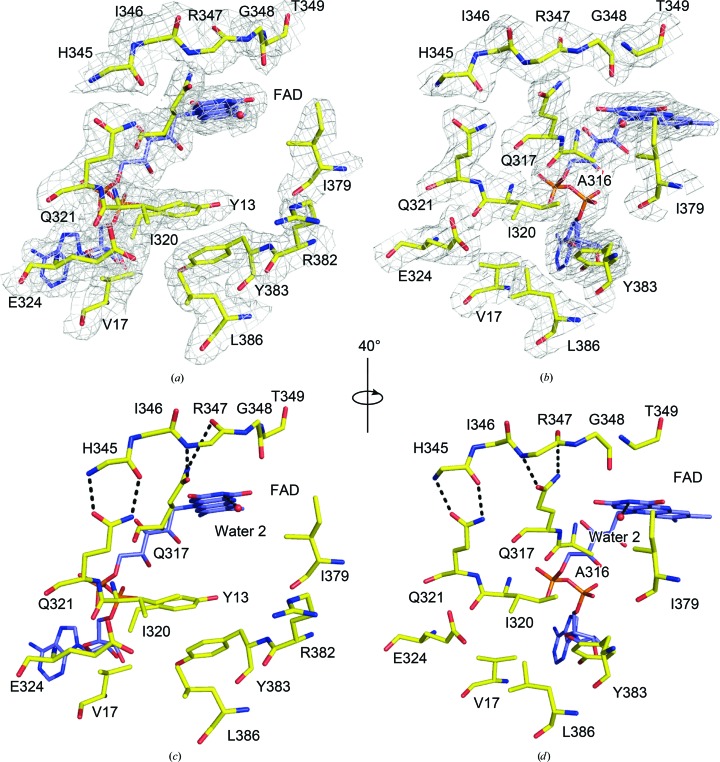
The electron density at the slot-shaped Q-site is also well defined. All residues and their side chains were unambiguously modelled (Figs. 3 ▸ a and 3 ▸ b). In the present structure, the previously proposed contacts between the Q-site motif AQXAXQ (Heikal et al., 2014 ▸; Marreiros et al., 2016 ▸) and the short linker separating the two substrate-binding sites were confirmed with greater confidence. Hydrogen-bonding interactions between the main-chain atoms in the short linker and the two glutamine side chains were clearly supported by the electron-density maps (Figs. 3 ▸ a and 3 ▸ b). The B factors of the atoms involved in hydrogen bonding were lower than those of other atoms at the Q-site. The average B factor of the two O and N atoms of the two glutamate side chains was 50.4 Å2, whereas the average B factor for the carboxyl O atoms and N atoms of the Glu324 and Arg382 side chains was 77.2 Å2. The low B factor for the O and N atoms is further evidence of the hydrogen-bonding interactions. A well defined second water molecule (water 2) directly hydrogen-bonding to an N3 atom of the isoalloxazine was found at the Q-site (Figs. 2 ▸ and 3 ▸). In a similar manner to water 1, this water was present in only one chain in the previous structure but was present in all chains in the present structure. Interestingly, in the in silico quinone substrate modelling the positions of this water and the quinone carbonyl O atom overlapped (see §3.4).
Figure 3.
Quinone-binding site of C. thermarum NDH-2. (a) A quinone-binding site viewed from the si face of the FAD flavin ring. A 2F o − F c map contoured at 1σ is shown as a grey mesh. (b) A view of (a) rotated by 40° clockwise. (c) The same view as (a) but without the electron-density map. (d) The same view as (b) but without the electron-density map. Ala316 is omitted for clarity in (a) and (c). Arg382 is omitted in (b) and (d) for clarity. Potential hydrogen-bonding interactions are shown by dashed lines in (c) and (d).
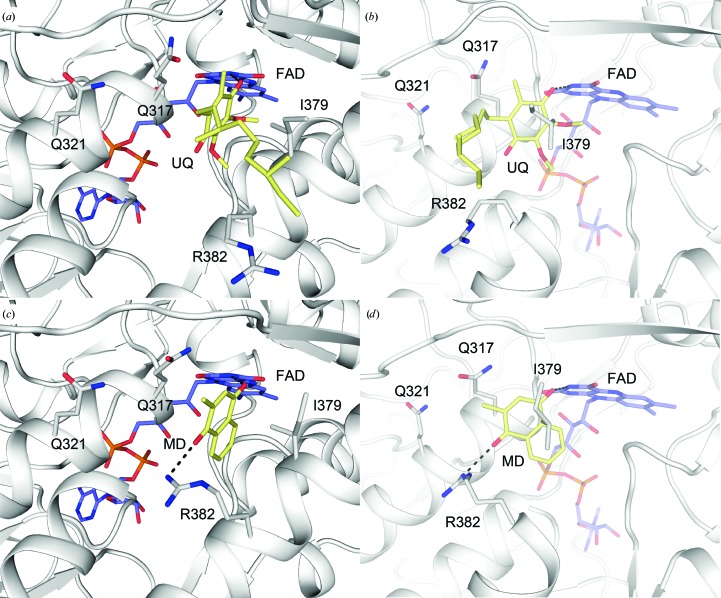
3.4. Modelling of quinone substrates at the Q-site
Previously, we determined the crystal structure of NDH-2 complexed with NAD+, but were unable to determine the structure of the quinone–substrate complex despite a number of attempts (Blaza et al., 2017 ▸). The structure of yeast Ndi1 complexed with ubiquinone is the only structure that shows a physiologically relevant substrate bound at the Q-site of NDH-2 (Feng et al., 2012 ▸). To understand how quinone molecules bind at the Q-site of the bacterial NDH-2 enzyme, we performed an in silico docking study based on the structure of the yeast Ndi1 complexed with ubiquinone (PDB entry 4g73; Feng et al., 2012 ▸) and used it to model both the physiologically relevant substrate MD and a physiologically unrelated ubiquinone.
A lack of ligand/substrate presence in the Q-site of the bacterial NDH-2 crystal structure made it difficult to predict whether the conformations adopted by some residues (Arg347 and Arg382) were blocking the ubiquinone-binding pocket. Superposition of the Q-site between the bacterial NDH-2 crystal structure and yeast Ndi1 structure showed that residues Tyr13, Arg347, Arg382 and Tyr383 may block the binding pocket; therefore, the side chains of these residues were trimmed off for initial docking and then refined in our in silico modelling. In the yeast Ndi1 structure, binding of NADH or ubiquinone is not associated with conformational changes around two highly conserved glutamine residues (Gln317 and Gln321), which hydrogen-bond to the short linker separating the NADH-binding site and the Q-site (Feng et al., 2012 ▸). Therefore, we assumed the same for our in silico quinone-docking experiments, and these hydrogen-bond contacts were maintained.
A ubiquinone molecule was comfortably placed in the Q-site of NDH-2 without major steric hindrance (Figs. 4 ▸ a and 4 ▸ b). The model showed that a ubiquinone head group nested into the hydrophobic slot-shaped Q-site, but with its carbon tail exposed to the solvent (Figs. 4 ▸ a and 4 ▸ b). The hydrophobic sections of the side chains of Gln317 and Ile379 held the planar quinone head. This observation is consistent with a previous study that showed that Ile379 is critical for quinone binding, and that any mutations greatly reduce its binding affinity (Blaza et al., 2017 ▸). Only one hydrogen-bonding interaction was observed in the present model, with one of the carbonyl O atoms on the ubiquinone head hydrogen-bonding to the N5 atom of the cofactor FAD (Fig. 4 ▸ b), as seen in the Ndi1–ubiquinone structure. Interestingly, the modelling program set this hydrogen-bond distance at 2.9 Å, which is consistent with the hydrogen-bonding distance found between water 2 and the N3 atom of the isoalloxazine in the new crystal structure (Figs. 2 ▸ and 3 ▸). A molecule of water 2 observed in the Q-site potentially predicts the position of a quinone carbonyl O atom in the Q-site. No other specific interactions were observed between ubiquinone and NDH-2.
Figure 4.
Ubiquinone and menadione docking at the quinone-binding site (Q-site) of NDH-2. (a) The docked ubiquinone in the Q-site, viewed from the si face of an FAD isoalloxazine. (b) A view of (a) rotated by 90° clockwise. (c) The docked menadione in the Q-site. (d) A view of (c) rotated by 90° clockwise. Potential hydrogen-bonding interactions are shown by dashed lines in (b) and (d).
Similar to ubiquinone, MD was docked into the Q-site (Figs. 4 ▸ c and 4 ▸ d). Generally, MD adopts a very similar binding pose to that predicted for ubiquinone. The predicted pose contained two hydrogen bonds. As expected from the ubiquinone model, there was a hydrogen bond between MD and the FAD cofactor. The model showed that the other ketone O atom of MD hydrogen-bonded to the side chain of Arg382. However, the significance of this contact remains unclear because the C-terminal membrane-anchoring domain containing the Q-site is presumably embedded in the membrane (Heikal et al., 2014 ▸). This residue most likely preferably contacts a phosphate group of the phospholipid in the membrane-bound environment. It is worth mentioning that it appears that MD can bind deeper in the pocket than ubiquinone, presumably because of the lack of methyl ether groups on its quinone head. The aromatic ring of MD makes a distant edge-to-face π–π stacking with the FAD cofactor, which is not observed for ubiquinone. This additional chemical interaction might confer higher affinity than ubiquinone species, which is consistent with the fact that menaquinone is the sole quinone species found in C. thermarum (Kalamorz et al., 2011 ▸). However, further experiments are required to prove this hypothesis.
4. Conclusions
We established a new crystallization platform to consistently produce high-quality NDH-2 crystals. Using these crystals, a high-resolution (2.15 Å) structure was determined for NDH-2. This platform will facilitate further crystallization of NDH-2 in the presence of both hydrophilic and hydrophobic ligands, which may ultimately contribute to the determination of substrate- and inhibitor-complex structures in the future. At 2.15 Å resolution, the molecular architecture, including water molecules at the two substrate-binding sites, was defined with confidence. Therefore, this high-resolution structure will be suitable for in silico screening and docking of inhibitors. Finally, we performed in silico MD and ubiquinone docking. These compounds adopt similar binding poses at the Q-site, with the only interactions occurring between the quinone head group and residues Gln317 and Ile379, and the N3 atom of the FAD isoalloxazine. Both the new co-crystallization platform and the in silico template model could be applied to determination of NDH-2–inhibitor complex structures, which is a step towards the rational development of new inhibitors for drug development.
Supplementary Material
PDB reference: type II NADH dehydrogenase, 5wed
Acknowledgments
We thank Gabrielle David PhD for editing a draft of this manuscript. Author contributions are as follows. YN, JP and YS expressed, purified and crystallized the protein. YN and DA solved the crystal structure. WJ performed the in silico docking studies. YN, WJ, EJP and GMC analysed the data and wrote the manuscript, with help from the other authors. YN, WJ, EJP and GMC designed the research and directed the project. The authors declare no competing financial interests.
Funding Statement
This work was funded by Maurice Wilkins Centre for Molecular Biodiscovery grant . Health Research Council of New Zealand grant .
References
- Adams, P. D., Mustyakimov, M., Afonine, P. V. & Langan, P. (2009). Acta Cryst. D65, 567–573. [DOI] [PMC free article] [PubMed]
- Biagini, G. A., Viriyavejakul, P., O’Neill, P. M., Bray, P. G. & Ward, S. A. (2006). Antimicrob. Agents Chemother. 50, 1841–1851. [DOI] [PMC free article] [PubMed]
- Blaza, J. N., Bridges, H. R., Aragão, D., Dunn, E. A., Heikal, A., Cook, G. M., Nakatani, Y. & Hirst, J. (2017). Sci. Rep. 7, 40165. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Farid, R., Day, T., Friesner, R. A. & Pearlstein, R. A. (2006). Bioorg. Med. Chem. 14, 3160–3173. [DOI] [PubMed]
- Feng, Y., Li, W., Li, J., Wang, J., Ge, J., Xu, D., Liu, Y., Wu, K., Zeng, Q., Wu, J.-W., Tian, C., Zhou, B. & Yang, M. (2012). Nature (London), 491, 478–482. [DOI] [PubMed]
- Gorrec, F. (2009). J. Appl. Cryst. 42, 1035–1042. [DOI] [PMC free article] [PubMed]
- Griffin, J. E., Gawronski, J. D., Dejesus, M. A., Ioerger, T. R., Akerley, B. J. & Sassetti, C. M. (2011). PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed]
- Heikal, A., Nakatani, Y., Dunn, E., Weimar, M. R., Day, C. L., Baker, E. N., Lott, J. S., Sazanov, L. A. & Cook, G. M. (2014). Mol. Microbiol. 91, 950–964. [DOI] [PubMed]
- Hirst, J. (2013). Annu. Rev. Biochem. 82, 551–575. [DOI] [PubMed]
- Iwata, M., Lee, Y., Yamashita, T., Yagi, T., Iwata, S., Cameron, A. D. & Maher, M. J. (2012). Proc. Natl. Acad. Sci. USA, 109, 15247–15252. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kalamorz, F. et al. (2011). J. Bacteriol. 193, 4290–4291. [DOI] [PMC free article] [PubMed]
- Kerscher, S., Dröse, S., Zickermann, V. & Brandt, U. (2008). Results Probl. Cell Differ. 45, 185–222. [DOI] [PubMed]
- Lagarde, N., Zagury, J.-F. & Montes, M. (2015). J. Chem. Inf. Model. 55, 1297–1307. [DOI] [PubMed]
- Lee, K. H., Humbarger, S., Bahnvadia, R., Sazinsky, M. H. & Crane, E. J. III (2014). Biochim. Biophys. Acta, 1844, 1708–1717. [DOI] [PubMed]
- Luft, J. R. & DeTitta, G. T. (1999). Acta Cryst. D55, 988–993. [DOI] [PubMed]
- Marreiros, B. C., Sena, F. V., Sousa, F. M., Batista, A. P. & Pereira, M. M. (2016). Environ. Microbiol. 18, 4697–4709. [DOI] [PubMed]
- Marreiros, B. C., Sena, F. V., Sousa, F. M., Oliveira, A. S. F., Soares, C. M., Batista, A. P. & Pereira, M. M. (2017). Sci. Rep. 7, 42303. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Melo, A. M. P., Bandeiras, T. M. & Teixeira, M. (2004). Microbiol. Mol. Biol. Rev. 68, 603–616. [DOI] [PMC free article] [PubMed]
- Saleh, A., Friesen, J., Baumeister, S., Gross, U. & Bohne, W. (2007). Antimicrob. Agents Chemother. 51, 1217–1222. [DOI] [PMC free article] [PubMed]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. (2013). J. Comput. Aided Mol. Des. 27, 221–234. [DOI] [PubMed]
- Sena, F. V., Batista, A. P., Catarino, T., Brito, J. A., Archer, M., Viertler, M., Madl, T., Cabrita, E. J. & Pereira, M. M. (2015). Mol. Microbiol. 98, 272–288. [DOI] [PubMed]
- Sherman, W., Beard, H. S. & Farid, R. (2006). Chem. Biol. Drug Des. 67, 83–84. [DOI] [PubMed]
- Sherman, W., Day, T., Jacobson, M. P., Friesner, R. A. & Farid, R. (2006). J. Med. Chem. 49, 534–553. [DOI] [PubMed]
- Sousa, F. M., Sena, F. V., Batista, A. P., Athayde, D., Brito, J. A., Archer, M., Oliveira, S. F., Soares, C. M., Catarino, T. & Pereira, M. M. (2017). Biochim. Biophys. Acta, 1858, 823–832. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Warren, G. L., Do, T. D., Kelley, B. P., Nicholls, A. & Warren, S. D. (2012). Drug Discov. Today, 17, 1270–1281. [DOI] [PubMed]
- Weinstein, E. A., Yano, T., Li, L.-S., Avarbock, D., Avarbock, A., Helm, D., McColm, A. A., Duncan, K., Lonsdale, J. T. & Rubin, H. (2005). Proc. Natl. Acad. Sci. USA, 102, 4548–4553. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, Y., Yu, Y., Li, X., Li, J., Wu, Y., Yu, J., Ge, J., Huang, Z., Jiang, L., Rao, Y. & Yang, M. (2017). J. Med. Chem. 60, 1994–2005. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: type II NADH dehydrogenase, 5wed