Abstract
Ecdysteroids, known as molting hormones, play central roles in the onset of molting, metamorphosis, and reproduction in arthropods. The ecdysteroids stored in eggs also play an important role in embryogenesis. In insects, ecdysteroids are stored as phosphate esters, which are converted to an active form by ecdysteroid-phosphate phosphatase (EPPase). Although EPPase is believed to be widely conserved in the Ecdysozoa, little is known about its expression in clades other than Insecta. In this study, we cloned a putative EPPase gene from a small fresh water crustacean known as a water flea, Daphnia magna Straus (Cladocera: Daphniidae), and examined its expression during embryogenesis. The amino acid sequence of the putative crustacean EPPase cDNA showed high similarity to insect EPPase and human suppressor of T-cell receptor signaling-1. We also found that the D. magna EPPase was highly expressed during early embryogenesis; its expression rapidly decreased 6 h after oviposition. This timing corresponds to the onset of organogenesis in D. magna . The expression of EPPase could not be detected in diapaused eggs. This is the first report of an EPPase from crustaceans, and the results suggest that the function of EPPase is conserved between insects and crustaceans.
Keywords: ecdysone, development, molecular cloning, mRNA quantification
Ecdysteroids are known as molting hormones in insects. These steroid hormones play pivotal roles not only in molting, but also in metamorphosis, development, and reproduction. In insect larvae, the prothoracic gland produces ecdysteroids ( Iga and Kataoka 2012 ), and during the adult insect stage, the ecdysteroids are produced in the gonads, particularly the ovaries ( Galikova et al. 2011 ). The ecdysteroids produced in the ovaries are transferred to the eggs where they are inactive as ecdysteroids, but are stored in conjugated forms such as glucosides ( Warren and Gilbert 1986 ), fatty acyl esters, and phosphate esters ( Tsoupras et al. 1982 ). Among these inactive ecdysteroids, the major products in insects are phosphate esters ( Sonobe and Ito 2009 ). It was first observed that inactive ecdysteroids were converted to their active form by ecdysteroid-phosphate phosphatase (EPPase) during embryogenesis in the silkworm Bombyx mori ( Yamada and Sonobe 2003 ). The cellular functions and metabolism in B. mori have been well studied ( Sonobe and Yamada 2004 ), and the ecdysteroid-phosphate was found localized in yolk granules as a form of vitellin-bound complex that is later released from the yolk granules and processed in the cytosol ( Yamada et al. 2005 ). Active ecdysteroids converted from phosphate esters by EPPase, together with de novo synthesized ecdysteroids, are indispensable for embryonic development. The importance of active ecdysteroids in the developing embryo was also shown in the fruit fly Drosophila melanogaster , in which morphological disruptions were observed when the metabolic pathway for 20-hydroxyecdysone biosynthesis was deleted. In addition, active ecdysteroids are converted from ovary-derived ecdysteroid phosphates in other insects such as the desert locust Schistocerca gregaria ( Scalia et al. 1987 ), the oriental migratory locust Locusta migratoria ( Lagueux et al. 1977 ), the cockroach Blaberus craniifer ( Bullière et al. 1979 ), and the tobacco hornworm Manduca sexta ( Warren et al. 1986 ), suggesting that they play an important role in controlling early embryonic development. Thus, the importance of free ecdysteroids in the eggs and their conversion by EPPase is becoming widely recognized in insects.
Although the function of ecdysteroids has been extensively studied in insects, little is known about ecdysteroids in other Ecdysozoa phyla. In particular, information on the role and function of ecdysteroids during embryonic stages in other Ecdysozoan species is still limited.
In order to clarify the function of ecdysteroids in other Ecdysozoan species, the water flea Daphnia magna Straus (Cladocera: Daphniidae), a small crustacean that commonly inhabits fresh water ponds, was chosen as a model. Because daphniids inhabit a diverse array of aquatic environments, they have been used as models for adaptation to environmental changes, evolution, ecology, toxicology ( Watanabe et al. 2008 ), and environmental science for decades ( Watanabe et al. 2007 ). In addition, the advent of whole genome sequencing ( Colbourne et al. 2011 ) and the development of gene manipulation techniques ( Kato et al. 2010 ) have made D. magna a suitable organism for molecular investigations in crustaceans. In this study, the presence and expression of EPPase in daphniids were examined. The putative crustacean EPPase identified had a structure similar to that of the insect EPPase. Its expression was detected from the onset of embryogenesis, suggesting that ecdysteroids are required at early stages of embryogenesis in crustaceans.
Materials and Methods
D. magna Strain and Culture Conditions
The D. magna strain (NIES clone) was obtained from the National Institute for Environmental Studies (NIES; Tsukuba, Japan) and maintained as previously described ( Kato et al. 2010 ), except that ADaM ( Klüttgen et al. 1994 ) was used as the culture medium.
cDNA Synthesis from D. magna
Embryos at the appropriate developmental stage were dissected from the brood chamber and homogenized in the presence of Sepasol-RNAI reagent (Nacalai Tesque, Kyoto, Japan, http://www.nacalai.co.jp/ ). Total RNA was purified per the manufacturer instructions. The first-strand cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, http://www.lifetechnologies.com/ ) utilizing oligo (dT) primers (Toyobo, Osaka, Japan, http://www.toyobo.co.jp/ ).
Cloning of a Putative EPPase Gene
The nucleic acid sequence of the EPPase gene (AB609071) from B.mori was obtained from the NCBI database ( http://www.ncbi.nlm.nih.gov/ ). A BLAST search for related sequences was conducted on the D. magna Expression Sequence Tag (EST) database ( Watanabe et al. 2005 ). An EST sequence with high similarity to the B. mori EPPase was obtained from the database. Based on the sequence, Polymerase Chain Reaction (PCR) primers were designed and the first strand of cDNA was amplified by PCR using TaKaRa ExTaq (Takara, Ohtsu, Shiga, Japan, http://www.takara-bio.co.jp/ ). The primer sequences for the amplification of the EPPase gene fragment were as follows: forward, 5′-GAACTAC CAC CCAGGAAGA-3′; and reverse, 5′-GCGCATTGGGTGACGTAAT-3′. The amplified PCR product was subcloned into a pCRII vector (Invitrogen) and sequenced. The Gene Racer Kit (Invitrogen) and the SMART RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, http://www.clon tech.com/ ) were used for 5′ and 3′ rapid amplification of the cDNA ends (RACE), respectively, with the following primers: 5′-RACE primer, 5′-CTGGCCCAAAATTCCTGCAGCTGACT-3′; and 3′-RACE primer, 5′-CGTGAGTCAGCCGAGCAGTACTAT-3′.
Sequence Alignment and Phylogenetic Analysis of the Putative EPPase
A multiple sequence alignment was constructed using Clustal-W ( Thompson et al. 1994 ) with the following settings for the pairwise alignment parameters: gap opening penalty, 17.00; gap extension penalty, 0.1; and the identity protein weight matrix. The multiple alignment parameters were as follows: gap opening penalty, 17.00; gap extension penalty, 0.2; delay divergent cutoff, 30%; and gap separation distance, 4. The phylogenetic reconstruction was performed using the p-distance algorithm and the neighbor-joining method implemented in MEGA software version 5.05 ( Tamura et al. 2007 ). The known orthologs of EPPase and the sequences that showed high similarity were used to construct a phylogenetic tree. A BLAST search was conducted on the D. pulex EST-genome database for sequences related to EPPase in the D. pulex ( Colbourne et al. 2011 ). The amino acid sequence corresponding to the EPPase histidine phosphatase domain was used for the analysis.
Quantitative Real Time-PCR
Embryos were collected at 0, 6, 9, 12, 15, 18, and 24 h after oviposition, and cDNA was synthesized as described above. Quantitative PCR was performed using an Mx3005P real time (RT)-PCR system (Agilent Technologies, Santa Clara, CA, http://www.chem-agilent.com/ ) with SYBR GreenER qPCR SuperMix Universal (Invitrogen). The following EPPase primers were used: forward, 5′-GC C G TCTCTTCGCTCCTTAC-3′; and reverse, 5′-GAATA ATCC CG G TT CCAAACAA-3′. The PCR amplifications were performed in triplicate using the following conditions: 2 min at 50°C and 10 min at 95°C, followed by 40 two-temperature cycles of 15 s at 95°C and 1 min at 60°C. Gel electrophoresis and melting curve analyses were performed to confirm the correct amplicon size and the absence of nonspecific bands. The embryonic expression levels were normalized to the expression level of ribosomal protein L32.
RNAi Knockdown of EPPase
In order to downregulate EPPase expression, an siRNA (5′-CAUGGCGAAAGG AUCGACUUCA CA UTT-3′) corresponding to the EPPase gene was synthesized and injected as described previously ( Kato et al. 2011 ). As a control siRNA, a random sequence (5′-GGUUUAAGCCGCCUCACAUTT-3′) was selected and used. The random sequence was confirmed not to have similar sequence in D. magna EST database ( Watanabe et al. 2005 ) and others.
Results and Discussion
Molecular Cloning of the Putative Daphniid EPPase Gene
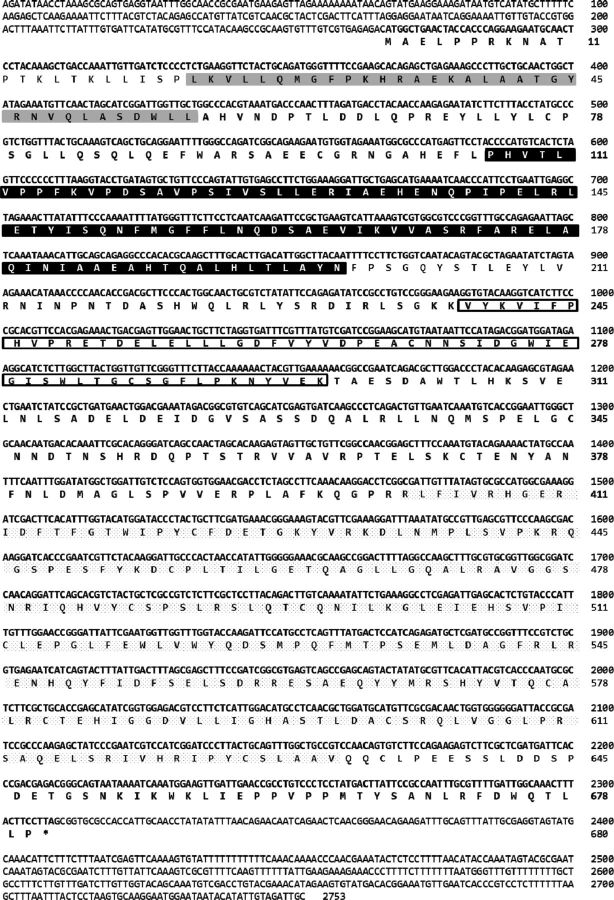
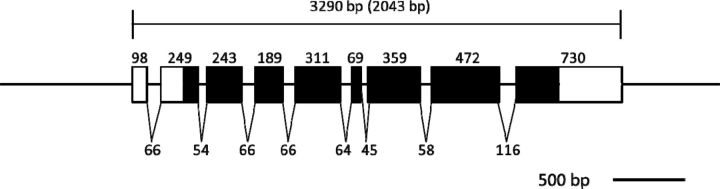
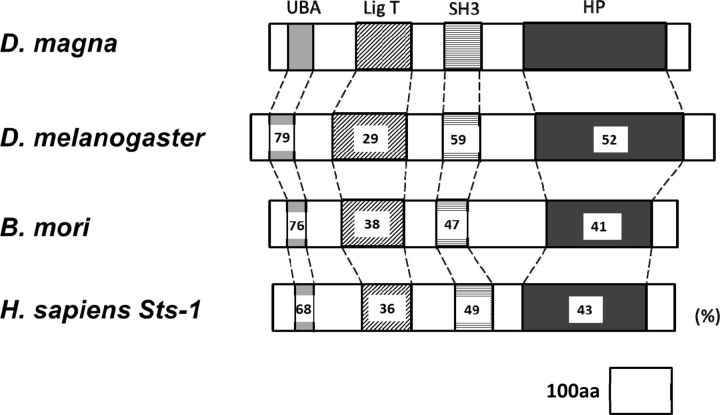
A gene encoding a putative EPPase in crustaceans was found by conducting a BLAST search of the D. magna EST database ( Watanabe et al. 2005 ) with the sequence for EPPase from B. mori . Based on the sequence, PCR primers were designed, and a partial DNA fragment of the putative EPPase gene was recovered from cDNA obtained from embryonic D. magna . To determine the sequences at both ends of the gene, 5′ and 3′ RACE PCR was performed. The sequences were assembled into a transcript encoding a protein of 680 amino acids ( Fig. 1 ). Mapping the putative EPPase transcript to the genomic sequence indicated that the gene had nine exons spread over 3,290 bp of genomic DNA ( Fig. 2 ). The sequence of the putative D. magna EPPase showed high similarity with that of the insect EPPases and the human suppressor of T-cell receptor signaling 1 ( Sts-1 ) gene, each of which has a ubiquitin-associated domain, a LigT-like phosphoesterase domain ( Mazumder et al. 2002 ) ( Fig. 3 ), an Src homology 3 domain, and a histidine phosphatase domain ( Rigden 2008 ). These motifs are highly conserved between the insect EPPase and the human Sts-1 gene ( Davies et al. 2007 ). The histidine phosphatase domain is found in a large, functionally diverse group of proteins, but they share a conserved catalytic core in which histidine is phosphorylated during the reaction. Because these histidine residues are conserved in the Daphnia EPPase ( Fig. 4 ), the gene cloned in this study may function in the same way as the other conserved EPPases. Hereafter, we designate this gene D. magna EPPase , or simply EPPase .
Fig. 1.
Nucleotide and predicted amino acid sequences of the putative EPPase in D. magna . The nucleotide sequence is shown from 266 bp before the start codon to 444 bp after the stop codon. The predicted amino acid sequence is indicated below the nucleotide sequence of the EPPase cDNA. The ubiquitin-associated domain is shaded in gray; the LigT-like phosphoesterase domain is shaded in black; the Src homology domain 3 is indicated by a box; and the histidine phosphatase domain is underlined.
Fig. 2.
Schematic diagram of the EPPase gene structure. Each exon is represented by a box, with the length of each exon indicated above. Filled boxes indicate coding regions and open boxes indicate untranslated regions. The numbers below the boxes indicate the lengths of introns.
Fig. 3.
Domain structures of the EPPase gene in D. magna and insect and human orthologs. The characteristic conserved domains of the EPPase gene are presented schematically for D. magna and the orthologs in D. melanogaster (fruit fly), B. mori (silkworm), and Homo sapiens (human). The percentage of amino acid identity in each conserved domain between the D. magna EPPase and that in the ortholog is indicated. UBA: ubiquitin-associated domain. LigT: LigT-like phosphoesterase domain. SH3: Src homology domain 3. HP: histidine phosphatase domain.
Fig. 4.
Amino acid sequence alignment of the histidine phosphatase domain of EPPase in D. magna and insect and human orthologs. The amino acid sequences for the EPPase histidine phosphatase domain are shown for D. magna and orthologs in D. melanogaster (fruit fly), B. mori (silkworm), and H. sapiens (human). The amino acids shaded in black are conserved.
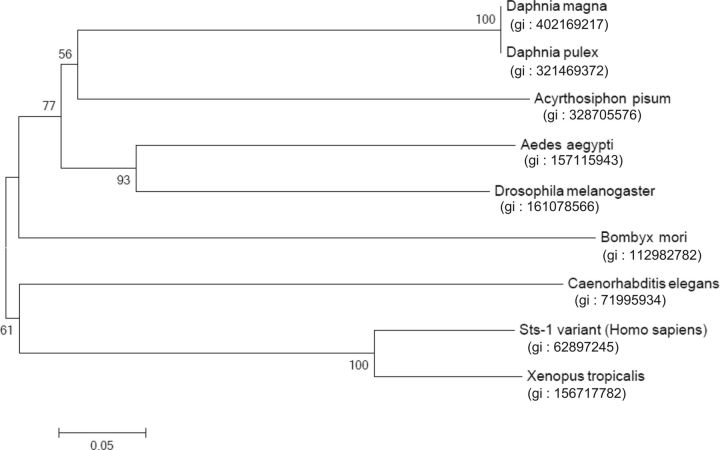
Phylogenetic Analysis of EPPase
A phylogenetic tree was constructed from the amino acid sequences of known EPPase proteins ( Fig. 5 ). The topology of the phylogenetic relationship between the EPPase orthologs was in good agreement with the taxonomic relationships of insects, and it indicated that the crustacean EPPase is closely related to the insect EPPase. The recent progress in phylogenetic analysis and other approaches suggest that insects originated from freshwater branchiopod crustaceans. The phylogenetic tree for EPPase is consistent with this hypothesis.
Fig. 5.
Phylogenetic tree of the amino acid sequences for the EPPase histidine phosphatase domains in D. magna and other species. Bootstrap values for 1,000 replicate analyses are shown at the branching points. The bar indicates branch length and corresponds to the mean number of differences ( P < 0.05) per residue along each branch.
Expression Analysis of EPPase
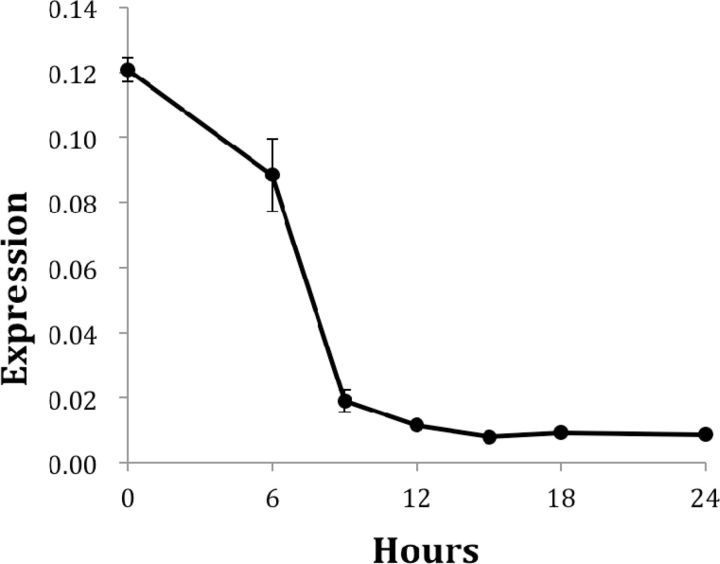
In insects, EPPase converts inactive ecdysteroid-phosphate stored in the eggs to active ecdysteroid. Thus, an analysis of the temporal changes in EPPase expression is important for understanding the function of ecdysteroids during the embryonic stages of crustaceans. Using quantitative PCR, EPPase expression was observed at early stages of embryonic development. Polyadenylated EPPase mRNA was detected just after oviposition (0 h), and higher levels of EPPase expression were sustained for ∼6 h. The expression level rapidly decreased after 6 h, which corresponds to the period before organogenesis ( Fig. 6 ).
Fig. 6.
Temporal expression profile of the putative EPPase in D. magna during the embryonic stage. Embryos were obtained at each time point, and the expression of EPPase and ribosomal protein 32 (L32) was evaluated by quantitative PCR. The expression of the EPPase was normalized to the expression of L32. Oviposition was designated as 0 h.
The expression level of EPPase in B. mori is very low at oviposition and increases gradually to its maximum level at 3–4 d after oviposition, corresponding to the late gastrulation phase before organogenesis ( Yamada and Sonobe 2003 ). The EPPase mRNA level in B. mori correlates well with EPPase activity and the presence of active ecdysteroids, suggesting that ecdysone stored in an inactive form is converted to its active form at the beginning of organogenesis. Because the embryonic development of D. magna is much faster than that of B. mori , it may be necessary to rapidly convert ecdysteroids from their inactive to active forms. The higher expression level of the EPPase gene in Daphnia may reflect a need for more rapid conversion of the ecdysteroid phosphates.
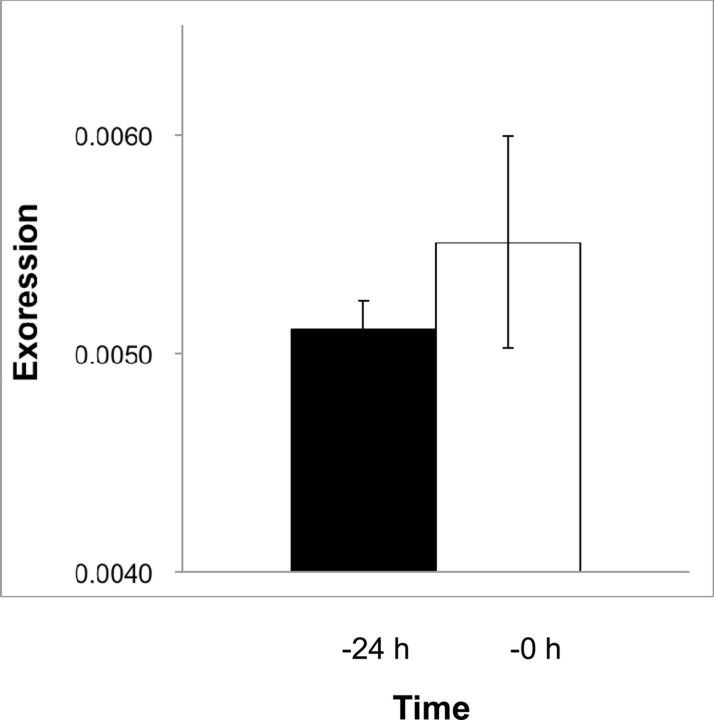
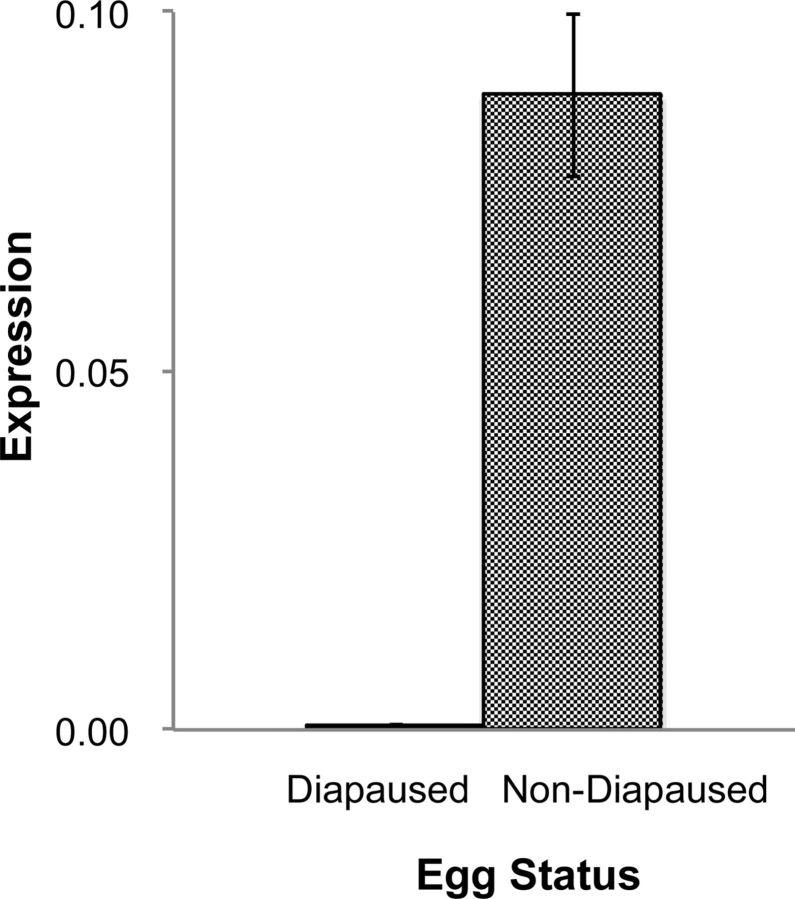
The detection of EPPase mRNA just after oviposition suggested that EPPase mRNA was synthesized during oogenesis. We, therefore, investigated the expression level of EPPase prior to oviposition. Although a direct comparison of mRNA expression levels between the ovary and eggs was not possible due to the inclusion of other nonovarian cells with the ovaries, EPPase mRNA was detected 24 h before oviposition in addition to just prior to oviposition ( Fig. 7 ). These results are consistent with the accumulation of EPPase mRNA that we observed shortly after oviposition. Given that the polyadenylation of mRNA is a critical function during embryogenesis, an analysis of posttranscriptional modifications and a direct estimation of protein expression may be necessary for the evaluation of actual EPPase activity. EPPase was not detected in diapaused eggs from B. mori . We, therefore, examined daphniid diapaused eggs for EPPase expression as a comparison, and we were unable to detect EPPase mRNA in the diapaused eggs ( Fig. 8 ). This result was consistent with that of B. mori , indicating that specific expression of EPPase only in nondiapaused eggs is conserved, irrespective of the evolutionary differences between D. magna and B. mori .
Fig. 7.
Expression of the putative EPPase in the ovaries of D. magna . Ovaries were isolated 24 h before or just prior to oviposition, and the total RNA was isolated. After reverse transcribing the RNA to cDNA, the EPPase was quantified by RT-PCR and normalized to the expression level of the ribosomal protein L32.
Fig. 8.
Expression of the putative EPPase in nondiapaused eggs. Diapaused eggs and nondiapaused eggs were collected, and total RNA was isolated. The RNA was isolated from nondiapaused eggs at the late blastula stage, which is when cell division stops. After reverse transcription of the RNA to cDNA, the EPPase was quantified by RT-PCR and normalized to the expression level of the ribosomal protein L32.
EPPase is Essential for Embryonic Development in Daphnia
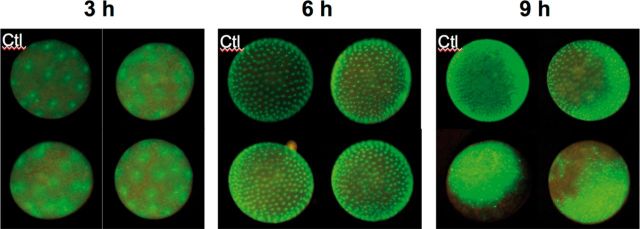
In order to examine if EPPase plays a critical role in the embryonic development of D. magna , we used a recently developed RNAi technique for Daphnia ( Kato et al. 2011 ) to downregulate EPPase expression in a transgenic Daphnia carrying the GFP protein fused to the histone H2B gene, which facilitated visualization of the developmental stages. When the EPPase siRNA was injected into the eggs, embryonic development stopped by 9 h after oviposition, while the developmental status 6 h after the injection was similar to that of mock-injected eggs ( Fig. 9 ). This result suggested that EPPase was essential for early development. As EPPase expression rapidly declined between 6 and 9 h ( Fig. 6 ), the ecdysone supplied by 6 h from EPPase conversion of the inactive form may be essential for the early development of Daphnia .
Fig. 9.
RNAi knockdown of EPPase in Daphnia . An EPPase siRNA was injected into a transgenic Daphnia constitutively expressing GFP just after oviposition, and an unrelated siRNA was injected as a control. The images were obtained with a fluorescent stereomicroscope at 3, 6, and 9 h after oviposition. Ctl: control. Note that GFP signals are diffused and number of spots is limited at 9 h.
EPPase is known to specifically catalyze the conversion of ecdysteroid-phosphates to free ecdysteroids, which are required for embryonic development. However, EPPase also has an ubiquitin-associated domain, a LigT-like phosphoesterase domain ( Mazumder et al. 2002 ), an Src homology domain 3, and a histidine phosphatase domain ( Rigden 2008 ), which are conserved in the human Sts-1 gene. Thus, it is possible that additional unknown functions of EPPase activity are important during early embryogenesis.
EPPase has been well studied in B. mori , and our study has shown the similarities and differences between the insect and daphniid EPPases. To the best of our knowledge, this is the first study of EPPase in crustaceans, which will contribute to understanding the functions of EPPase.
Acknowledgments
This work was partially supported by the Asahi Glass Foundation.
References Cited
- Bullière D., Bullière F., de Reggi M. . 1979. . Ecdysteroid titres during ovarian and embryonic development in Blaberus craniifer . Roux’s Arch. Develop. Biol. 186 : 103 – 114 . [DOI] [PubMed] [Google Scholar]
- Colbourne J. K., Pfrender M. E., Gilbert D., Thomas W. K., Tucker A., Oakley T. H., Tokishita S., Aerts A., Arnold G. J., Basu M. K., et al. . 2011. . The ecoresponsive genome of Daphnia pulex . Science 331 : 555 – 561 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L., Anderson I. P., Turner P. C., Shirras A. D., Rees H. H., Rigden D. J. . 2007. . An unsuspected ecdysteroid/steroid phosphatase activity in the key T-cell regulator, Sts-1 : surprising relationship to insect ecdysteroid phosphate phosphatase . Proteins 67 : 720 – 731 . [DOI] [PubMed] [Google Scholar]
- Galikova M., Klepsatel P., Senti G., Flatt T. . 2011. . Steroid hormone regulation of C. elegans and Drosophila aging and life history . Exp. Gerontol. 46 : 141 – 147 . [DOI] [PubMed] [Google Scholar]
- Iga M., Kataoka H. . 2012. . Recent Studies on insect hormone metabolic pathways mediated by cytochrome P450 enzymes . Biol. Pharm. Bull. 35 : 838 – 843 . [DOI] [PubMed] [Google Scholar]
- Kato Y., Kobayashi K., Watanabe H., Iguchi T. . 2010. . Introduction of foreign DNA into the water flea, Daphnia magna , by electroporation . Ecotoxicology 19 : 589 – 592 . [DOI] [PubMed] [Google Scholar]
- Kato Y., Shiga Y., Kobayashi K., Tokishita S.-I., Yamagata H., Iguchi T., Watanabe H. . 2011. . Development of an RNA interference method in the cladoceran crustacean Daphnia magna . Develop. Genes Evol. 220 : 337 – 345 . [DOI] [PubMed] [Google Scholar]
- Klüttgen B., Dülmer U., Engels M., Ratte H. T. . 1994. . ADaM, An artificial freshwater for the culture of zooplankton . Water Res. 28 : 743 – 746 . [Google Scholar]
- Lagueux M., Hirn M., Hoffmann J. A. . 1977. . Ecdysone during ovarian development in Locusta migratoria . J. Insect Physiol. 23 : 109 – 119 . [DOI] [PubMed] [Google Scholar]
- Mazumder R., Iyer L. M., Vasudevan S., Aravind L. . 2002. . Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily . Nucl. Acids Res. 30 : 5229 – 5243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden D. J. 2008. . The histidine phosphatase superfamily: structure and function . Biochem. J. 409 : 333 – 348 . [DOI] [PubMed] [Google Scholar]
- Scalia S., Sbrenna-Micciarelli A., Sbrenna G., Morgan E. . 1987. . Ecdysteroid titres and location in developing eggs of Schistocerca gregaria . Insect Biochem. 17 : 227 – 236 . [Google Scholar]
- Sonobe H., Yamada R. . 2004. . Ecdysteroids during early embryonic development in silkworm Bombyx mori : metabolism and functions . Zool. Sci. 21 : 503 – 516 . [DOI] [PubMed] [Google Scholar]
- Sonobe H., Ito Y. . 2009. . Phosphoconjugation and dephosphorylation reactions of steroid hormone in insects . Mol. Cell. Endocrinol. 307 : 25 – 35 . [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. . 2007. . MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0 . Mol. Biol. Evol. 24 : 1596 – 1599 . [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. . 1994. . Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice . Nucl. Acids Res. 22 : 4673 – 4680 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoupras G., Luu B., Hoffmann J. A. . 1982. . Isolation and identification of three ecdysteroid conjugates with a C-20 hydroxy group in eggs of Locusta migratoria . Steroids 40 : 551 – 560 . [DOI] [PubMed] [Google Scholar]
- Warren J. T., Gilbert L. I. . 1986. . Ecdysone metabolism and distribution during the pupal adult development of Manduca sexta . Insect Biochem. 16 : 65 – 82 . [Google Scholar]
- Warren J. T., Steiner B., Dorn A., Pak M., Gilber L. . 1986. . Metabolism of ecdysteroids during the embryogenesis of Manduca sexta . J. Liquid Chromatogr. 9 : 1759 – 1782 . [Google Scholar]
- Watanabe H., Tatarazako N., Oda S., Nishide H., Uchiyama I., Morita M., Iguchi T. . 2005. . Analysis of expressed sequence tags of the water flea Daphnia magna . Genome 48 : 606 – 609 . [DOI] [PubMed] [Google Scholar]
- Watanabe H., Takahashi E., Nakamura Y., Oda S., Tatarazako N., Iguchi T. . 2007. . Development of a Daphnia magna DNA microarray for evaluating the toxicity of environmental chemicals . Environ. Toxicol. Chem. 26 : 669 – 676 . [DOI] [PubMed] [Google Scholar]
- Watanabe H., Kobayashi K., Kato Y., Oda S., Abe R., Tatarazako N., Iguchi T. . 2008. . Transcriptome profiling in crustaceans as a tool for ecotoxicogenomics . Cell Biol. Toxicol. 24 : 641 – 647 . [DOI] [PubMed] [Google Scholar]
- Yamada R., Sonobe H. . 2003. . Purification, kinetic characterization, and molecular cloning of a novel enzyme ecdysteroid-phosphate phosphatase . J. Biol. Chem. 278 : 26365 – 26373 . [DOI] [PubMed] [Google Scholar]
- Yamada R., Yamahama Y., Sonobe H. . 2005. . Release of ecdysteroid-phosphates from egg yolk granules and their dephosphorylation during early embryonic development in silkworm, Bombyx mori . Zool. Sci. 22 : 187 – 198 . [DOI] [PubMed] [Google Scholar]