Abstract
Plants are routinely exposed to biotic and abiotic stresses to which they have evolved by synthesizing constitutive and induced defense compounds. Induced defense compounds are usually made, initially, at low levels; however, following further stimulation by specific kinds of biotic and abiotic stresses, they can be synthesized in relatively large amounts to abate the particular stress. cDNA microarray hybridization was used to identify an array of genes that were differentially expressed in tomato plants 15 d after they were exposed to feeding by nonviruliferous whiteflies or by viruliferous whiteflies carrying Pepper golden mosaic virus (PepGMV) ( Begomovirus, Geminiviridae ). Tomato plants inoculated by viruliferous whiteflies developed symptoms characteristic of PepGMV, whereas plants exposed to nonviruliferous whitefly feeding or nonwounded (negative) control plants exhibited no disease symptoms. The microarray analysis yielded over 290 spotted probes, with significantly altered expression of 161 putative annotated gene targets, and 129 spotted probes of unknown identities. The majority of the differentially regulated “known” genes were associated with the plants exposed to viruliferous compared with nonviruliferous whitefly feeding. Overall, significant differences in gene expression were represented by major physiological functions including defense-, pathogen-, photosynthesis-, and signaling-related responses and were similar to genes identified for other insect–plant systems. Viruliferous whitefly-stimulated gene expression was validated by real-time quantitative polymerase chain reaction of selected, representative candidate genes (messenger RNA): arginase, dehydrin, pathogenesis-related proteins 1 and -4, polyphenol oxidase, and several protease inhibitors. This is the first comparative profiling of the expression of tomato plants portraying different responses to biotic stress induced by viruliferous whitefly feeding (with resultant virus infection) compared with whitefly feeding only and negative control nonwounded plants exposed to neither. These results may be applicable to many other plant–insect–pathogen system interactions.
Keywords: begomovirus, Bemisia tabaci, biotic stress, ethylene, Geminiviridae
During the 1970’s, whitefly-transmitted viruses in the genus Begomovirus (family, Geminiviridae ) were first recognized as new and emergent plant viral pathogens in subtropical and tropical locales worldwide. Begomoviral emergence and their whitefly vector, members of the Bemisia tabaci (Gennadius) (Aleyrodidae: Hempitera) cryptic, sibling species group ( Frohlich et al. 1999 , Brown et al. 2005 , Brown 2010 ), occurred due to the rapid expansion of monoculture production of crops grown under irrigation in tropical and mild climates. Initially, most begomoviral outbreaks were caused by endemic virus–vector complexes; however, beginning in about 1988–1989, a highly polyphagous exotic B. tabaci , referred to as the B biotype ( Costa and Brown 1991 , Brown et al. 1995 ), later, also known as Bemisia argentifolii Bellows & Perring ( Perring et al. 1993 , Bellows et al. 1994 ) was introduced into the United States and nearly worldwide, on plants through international trade routes ( Frohlich et al. 1999 , Gill and Brown 2010 ). It readily adapted to diverse environments, particularly in arid climates, where it displaced indigenous haplotypes in some locales ( Costa et al. 1993 ; Brown 1994 , 2007 ; Brown et al. 1995 ; Frohlich et al. 1999 ).
This event dramatically altered the epidemiology of certain begomoviral diseases, facilitating the emergence of previously unknown begomoviruses and the disappearance of once predominant viral species in cropping systems ( Brown 1990 , 1994 , 2001 , 2007 ; Brown and Bird 1992 ). One such newly discovered begomovirus was Pepper golden mosaic virus (PepGMV) ( Brown et al. 2005 ). It was first identified infecting Solanaceous species including tomato and pepper crops in the southern and southwestern United States, and also was discovered to be widespread in Mexico and Central America. PepGMV has a bipartite genome consisting of DNA-A and DNA-B components, both of which are required for infection of the plant host. Pepper plants that become infected with the distortion (Di) strain of PepGMV (PepGMV-Di) exhibit mild mottling of leaves within 8–10 d following inoculation, but the subsequently developing leaves are asymptomatic, referred to as “recovery,” and plants appear to be uninfected. In contrast, PepGMV-Di is highly virulent in inoculated tomato plants in which it causes downward curling of leaves, foliar chlorosis, and overall stunting ( Brown et al. 2005 ). The ability of PepGMV-Di to cause characteristic disease symptoms in tomato plants 10–12 d postinoculation (PI) makes this virus–vector–tomato host study system ideal for exploring plant host defenses that respond to different biotic stresses.
Members of the whitefly B. tabaci sibling species group ( Gill and Brown 2010 ) have evolved a specific relationship with the genus Begomovirus , which they transmit in a circulative, persistent manner ( Brown and Czosnek 2002 ). In addition, these whiteflies cause biotic stress to plants by stylet-mediated feeding in phloem cells. Whiteflies both elicit and abate the effects of plant innate defense responses when their stylets penetrate plant tissues, and they introduce salivary effectors into the phloem during feeding ( Walling 2000 , 2008 ; Van de Ven et al. 2000 ). In particular, the B biotype of B. tabaci induces the salicylic acid (SA)-mediated defense pathway, while at the same time, depressing the jasmonic acid (JA) pathway ( Zarate et al. 2007 ).
Plants have evolved specific defense responses that provide protection from insect herbivory and pathogen infection, some of which involve “cross-talk” through elicitors of the host defense pathways ( Koornneef and Pieterse 2008 ). The best-studied inducible defense pathways in plants are the systemic acquired resistance (SAR) and induced resistance (IR) pathways ( Karban and Baldwin 1997 ). The SAR pathway is usually associated with pathogen-induced responses and subtle damage caused by phloem feeders that use piercing/sucking mouthparts, as do whiteflies and aphids (Hemipterans) ( Walling 2000 ). In contrast, the IR pathway has been associated with large mechanical wounding and herbivory inflicted by chewing mouthparts, characteristic of caterpillars (Lepidopterans) and beetles (Coleopterans) ( Karban and Baldwin 1997 ).
The SAR response involves an oxidative burst in which reactive oxygen species are produced, typically H 2 O 2 and O 2− , that can function to 1) act directly as antimicrobial agents, 2) cross-link structural proteins in cells under attack to slow the pathogen infection process and systemic invasion of the plant, 3) upregulate genes in defense pathways, 4) signal initiation of the hypersensitive response (HR) causing local plant cell lesion formation in infected areas and halting pathogen progression, and 5) stimulate SA production that influences participating systemic responses ( Buchanan et al. 2001 ). Systemic resistance occurs when SA accumulates in distal plant parts resulting in production of defensive proteins, such as glucanases, chitinases, peroxidases, proteinases, and pathogenesis related (PR), in noninfected plant tissues ( Buchanan et al. 2001 ). The IR pathway is induced when a host plant experiences gross damage from chewing herbivores. This defense response is commonly due to systemin ( Ryan 2000 ) that is transported through the phloem ( Pearce et al. 1991 ). Systemin and systemin-like molecules trigger a cascade of events that result in the production of JA, ultimately providing antiherbivore defenses, through production of defensive proteins such as polyphenol oxidase, lipoxygenase, and wound-induced protease inhibitors.
The SAR and IR pathways can antagonize one another, a phenomenon known as “cross-talk.” SA, the signaling molecule involved in the induction of SAR plant defenses, has a negative effect on JA production and on plant responses to mechanical wounding ( Bostock et al. 2001 ). If the host plant responds to pathogen infection by synthesizing SA, this SA can interfere with the JA-mediated production of proteinase inhibitors, leaving the plant more vulnerable to attack from chewing insects ( Felton et al. 1999 ). Exogenous application of SA to artificially induce plant defenses against pathogens lowers JA production, a plant hormone involved in the herbivore defense pathway ( Stotz et al. 2002 ). In tomato, ethylene increases the ability of tomato to defend against wounding caused by insects, whereas for Arabidopsis, ethylene treatment increases plant vulnerability to chewing insects ( Stotz et al. 2000 ). Infection of plants by a begomovirus-associated betasatellite complex has been shown to result in the modulation of a subset of plant JA responses, resulting in attenuation of their expression. Additionally, the proteins expressed by begomoviruses during infection of the host plant can influence plant morphology and alter gene expression in the innate defense pathways ( Yang et al. 2008 , Lozano-Durán et al. 2011 ). Other pathways have been discovered that target plant viral transcripts and/or proteins interacting with antiviral signaling by transducing genes in plant defense pathways ( Santos et al. 2010 ).
Expression profiling has been implemented to identify genome-wide responses to plant–virus interactions ( Whitham et al. 2006 , Wise et al. 2007 , Alfenas-Zerbini et al. 2009 ) and plant–insect interactions ( Kazan et al. 2001 , Korth 2003 , Sinisterra et al. 2005 , Thompson and Goggin 2006 , Kempema et al. 2007 , Zarate et al. 2007 , Li et al. 2011 , Wan et al. 2002 ). Walling ( 2000 , 2008 ) demonstrated that stylet feeding by whitefly B. tabaci (Genn.) suppressed plant host responses ( Kempema et al. 2007 , Walling 2008 ), whereas earlier work identified the SLW3 gene from squash plants that was unresponsive to wounding or defense signaling ( Van de Ven et al. 2000 ), collectively suggesting that some plant defense cascades remain to be discovered. Even so, the emphasis regarding plant-specific responses to phytophagous arthropods that transmit viral and other plant pathogens has been limited, and yet these tritrophic interactions are extremely common. In fact, 75% of the viruses that infect plants are transmitted by Hemipteran vectors that feed in the plant vasculature ( Nault 1997 ).
Many studies exploring global plant gene expression in response to insect herbivores have focused on the model plant, Arabidopsis thaliana . Although model plants can yield important information about plant responses to various stimuli, the results are not transferable to all plant systems. Tomato is a host for whiteflies and many whitefly-transmitted viruses. Here, plant–insect–virus tritrophic interactions were investigated using tomato plants as the whitefly and virus host, the Arizona-B biotype whitefly that efficiently vectors PepGMV-Di, and PepGMV-Di, which is a phloem-limited begomovirus that systemically infects and causes severe symptoms in tomato plants ( Brown et al. 2005 ).
Microarray hybridization analysis was used to characterize plant mRNA transcript expression profiles resulting from biotic stress imposed by whitefly-mediated begomovirus (PepGMV-Di) transmission (involving wounding and the introduction of salivary constituents during feeding), virus-free tomato plants exposed to whitefly feeding alone, and nonwounded, virus-free plants, as the negative control. Although the responses of squash and tomato plants exposed to feeding by the whitefly B biotype ( Van de Ven et al. 2000 , McKenzie et al. 2005 , Kempema et al. 2007 , Walling 2008 ) and/or begomovirus infection of tomato by Tomato yellow leaf curl and Tomato mottle viruses (TMV) ( McKenzie et al. 2002 ) have been previously explored, this is the first study to use cDNA microarray hybridization to examine the global plant host response to begomovirus infection following whitefly-mediated inoculation of plants with virus.
Leaves were harvested 15 d PI to ensure that tomato plants with viruliferous whitefly feeding developed symptoms typical of PepGMV-Di and thus were clearly virus-infected, which also involved successful whitefly feeding. This time point was also selected because previous studies ( McKenzie et al. 2002 , Sinisterra et al. 2005 ) demonstrated that whitefly feeding on tomato plants stimulated a variety of defense-related genes, including ethylene proteinase inhibitor, 1-aminocyclopropane carboxylate oxidase (affects ethylene production), and wound induced proteinase inhibitor during this period, even at 25 d post whitefly infection of tomato plants ( McKenzie et al. 2005 ). Ascencio-Ibáñez et al. (2008) also reported significant differences in the transcripts of 5,365 genes at 12 d PI of Arabidopsis plants with a begomovirus, Cabbage leaf curl virus . Gorovits et al. (2007) also determined that tomato plants inoculated with Tomato yellow leaf curl virus exhibited significant upregulation of a PR enzyme for at least 4 wk PI.
Materials and Methods
Test Plants, Begomovirus, and Whitefly Treatments
Solanum lycopersicum, tomato plant (TMV resistant) seeds were sown in 4” plastic pots containing sterilized potting soil (3.8 cu. ft. peat moss, 4 cu. ft. vermiculite, 28 gallons of perlite, and 21 gallons of washed mortar sand) in a greenhouse at the University of Arizona (Tucson, AZ). The plants were fertilized weekly with Peter’s Professional fertilizer (Scotts, Marysville, OH), N:P:K 20:20:20, and grown in a whitefly-free growth chamber maintained at 27°C d and 18°C night temperatures with a 14-h photoperiod. At the 5–6 leaf stage, 18 tomato plants were transplanted into 6” pots and placed (six plants each) into separate, plexiglass boxes ventilated with fine nylon mesh on the sides, maintained under strict insect-free conditions. Plexiglass cages containing test plants were grown in a growth room and under very high output bulbs with a 12-h photoperiod with temperatures ranging from 24°C to 28°C. Plants at this developmental stage were selected, so that they could withstand the whitefly feeding pressure while still being susceptible to virus infection.
Adult B. argentifolii whiteflies (AZ-B colony) ( Costa and Brown 1991 ) were randomly collected with a hand-held aspirator. The AZ-B colony was established in the laboratory in 1987 at the University of Arizona (Tucson, AZ) ( Costa and Brown 1991 ) and has been maintained continuously on cotton Gossypium hirsutum (L . ) plants since 1990.
To obtain viruliferous whiteflies, three tomato plants at the 3–4 leaf stage were inoculated with an infectious clone of PepGMV-Di strain ( Brown et al. 2005 ) using a Biolistic Particle Delivery System Model PDS-1000 gene gun (BioRad, Hercules, CA) with tungsten particles (M-17) and a 900 pounds per square inch (PSI) rupture disk, according to Idris et al. (2001) . The tomato plants developed characteristic PepGMV-Di symptoms at 12–15 d PI. Three control plants biolistically mock inoculated with water did not display virus symptoms.
At 12 d PI, ∼500 colony-reared, virus-free adult whiteflies were placed on the symptomatic PepGMV-Di-infected tomato plants and given a 48-h acquisition-access period (AAP). Simultaneously, ∼500 nonviruliferous adult whiteflies were allowed to feed on the virus-free mock-inoculated tomato plants for a 48-h inoculation access period (IAP). Approximately 50 whiteflies were transferred from the virus-infected source or virus-free plants to each tomato seedling (one/pot) and given a 15 d IAP.
The treatments were 1) tomato plants infested for 15 d with PepGMV-Di viruliferous whiteflies (WF&V), 2) tomato plants infested for 15 d with nonviruliferous whiteflies (WF), and 3) nonwounded tomato plants that were whitefly free and virus free (NW). Four biological replicates were included for each treatment. Plants infested with PepGMV-Di viruliferous whiteflies developed symptoms characteristic of PepGMV-Di infection including interveinal chorosis, epinasty, stunting of the newly developing leaves, and shortening internodes.
No virus symptoms were observed on negative control plants infested with nonviruliferous whiteflies, or whitefly-free, nonwounded plant treatments. On day 15, leaves were collected from the newest growth, cleaned by gentle brushing to remove whiteflies and whitefly eggs, frozen in liquid nitrogen, and stored at −80°C. Four biological replicates were conducted for the real-time quantitative polymerase chain reaction (qPCR) analysis.
Microarray Hybridization
Preparation of the RNA and cDNA, and hybridization reactions were conducted according to Rodriguez-Saona et al. (2010) . Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA). Enrichment and purification of messenger RNA (mRNA) was conducted with Dynabeads Oligo (dT) 25 (Dynal Biotech, Lake Success, NY). The mRNA was reverse transcribed and labeled with Cy3- and Cy5-deoxyuridine triphosphate (Amersham Biosciences, Piscataway, NJ). The cDNA was purified using a Microcon YM30 column (Millipore, Billerica, MA). Tomato microarrays (TOM1, The Boyce Thompson Institute at Cornell University, Ithaca, NY) were hybridized at 65°C for 8 h. All treatments were dye reversed with Cy3 and Cy5 for a total of six microarray chips using a loop design for four biological replications for each treatment. The complete meta-grid size was 13,440 spots per array. Scanning was conducted with the Packard BioScience Scan Array 3000 equipped with red and green lasers at 10 µm resolution. Spot intensity was quantified using Imagene 5.1 (BioDiscovery, Inc., Marina Del Rey, CA). Gene identifications are available from the Sol Genomics Network ( http://solgenomics.net/ ).
Statistical Analysis of Microarray Results
Each microarray slide feature was log-2 transformed and each microarray chip was scaled on a median genomic level with the Genesifter Analysis Edition software (Geospiza, Inc., Seattle, WA). A one-way analysis of variance (ANOVA) for multiple groups was performed to determine the false discovery rate with a Benjamini and Hochberg statistical correction to determine the family-wise error rate in multiple hypotheses testing that take place in microarray studies ( Benjamini and Hochberg 1995 , Alba et al. 2004 ). For a post hoc multiple comparison analysis between treatments, a conservative Tukey's honestly significant differences test was used for genes of interest at an adjusted P value of <0.05. In addition, a hierarchal cluster was performed using the Manhattan method (GeneSifter) for the expressed sequence tags (ESTs) that were determined to be significantly different.
Real-Time qPCR
Although microarray data alone are widely accepted, to validate the microarray results, qPCR was conducted on several candidate plant defence-related genes: polyphenol oxidase F (PPO F), wound-induced proteinase inhibitor I (WIN1), wound-induced proteinase inhibitor II (WIN2), and PR protein 4 (PR-4) for four biological replicates. Total RNA was used as the template for cDNA synthesis using a 3:1 mix of random hexamer and Oligo (dT 20 ) primers following the Verso SYBR Green 2-Step qRT-PCR Low Rox Kit protocol (ThermoScientific, Waltham, MA). Gene-specific primers were designed using Primer3 software ( http://frodo.wi.mit.edu/primer3 ) based on sequences of the cDNA targets on the microarray. The amplicon for actin (Sol Genomics Network Unigene ID SGN-U579208) was used as the endogenous control. Primer sequences are provided in Musser et al. (2012) . The qPCR reactions were carried out in optical 96-well plates on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA) at Western Illinois University. The ROX dye was used as a (passive) reference dye included in the PCR master mix. A melt curve analysis was performed for all primer pairs, and all experimental samples for each amplicon had a single sharp peak at the amplicon’s melting temperature. Four genes (β-tubulin, eukaryotic initiation factor 4A, ribosomal protein S18, and actin) were examined as endogenous controls to correct for sample variation in the qPCR. Results indicated that actin was the best endogenous control transcript. The dynamic range of each primer set was examined by running three replicate reactions at five dilutions of the RNA. Because the target and the “normalizer” had similar dynamic ranges, the comparative quantification method delta-delta Ct (ΔΔCt) of Pfaffl (2001) was used, and data were transformed to absolute values with 2 -ΔΔCt to obtain fold changes between treatments. Each treatment consisted of four biological replicates.
In a follow-up experiment, PepGMV-Di-viruliferous whiteflies were given a 48-h AAP on tomato plants and compared with nonwounded tomato plants; qPCR analysis was carried out for the defense genes: arginase, dehydrin, glutathione S-transferase (GST), ethylene-responsive proteinase inhibitor 1 (PI2Cev1), ethylene-responsive proteinase inhibitor 2 (PI2Cev2), PR protein-1A1, WIN1, WIN2, PPO F, and PR-4. qPCR analysis was also conducted on the gene for Ribulose 1,5-bisphosphase carboxylase (RuBisCO) small chain 3B (SGN-U578689: forward primer: 5’-TGAGACTGAGCACGGATTTG-3; reverse primer 5’-GCACCCAAACATAGGCAACT-3’). The relative fold changes for each gene were compared with the nonwounded treatment, which was set to one. Relative fold change was analyzed, and standard errors were determined to confirm the statistical significance of the fold differences obtained by the microarray analysis.
Results
Whitefly-Mediated PepGMV-Di Inoculation of Test Plants
Tomato plants inoculated with PepGMV-Di using viruliferous whiteflies developed typical PepGMV-Di symptoms (foliar chlorosis, shortened internodes, and reduced leaf size) 10–12 d PI ( Brown et al. 2005 ), confirming that whitefly-mediated virus transmission was successful. Symptoms of begomovirus infection were not observed on nonviruliferous whitefly-treated or on control plants.
Microarray Results
Of the 12,860 EST clones representing ∼8,500 independent tomato loci on the TOM1, a total of 290 tomato genes represented by the spotted probes were significantly differentially expressed at ANOVA ( P < 0.05) between the control NW tomato plants compared with plants exposed to whitefly feeding alone (WF) or to plants exposed to viruliferous whiteflies and PepGMV-Di infection (WF&V) 15 d after treatments were established (Supp. File [online only]). In nearly all instances, PepGMV-Di-viruliferous whitefly infested tomato plants dramatically stimulated plant gene expression more so than plants fed upon by nonviruliferous whiteflies or nonwounded plants ( Fig. 1 ). The 290 ESTs were annotated as 161 putative gene targets, and the remaining 129 ESTs have “unknown” functions or as having no available hit. Considering only genes with known functions, a conservative Tukey–Kramer analysis revealed 113 genes whose expression was significantly different among the three treatments. The expression profiles of 119 genes differed when comparing plants with viruliferous whitefly feeding and subsequent viral infection compared with the control treatment, and the expression of 44 genes was altered in tomato plants fed on by nonviruliferous whiteflies compared with nonwounded control plants.
Fig. 1.
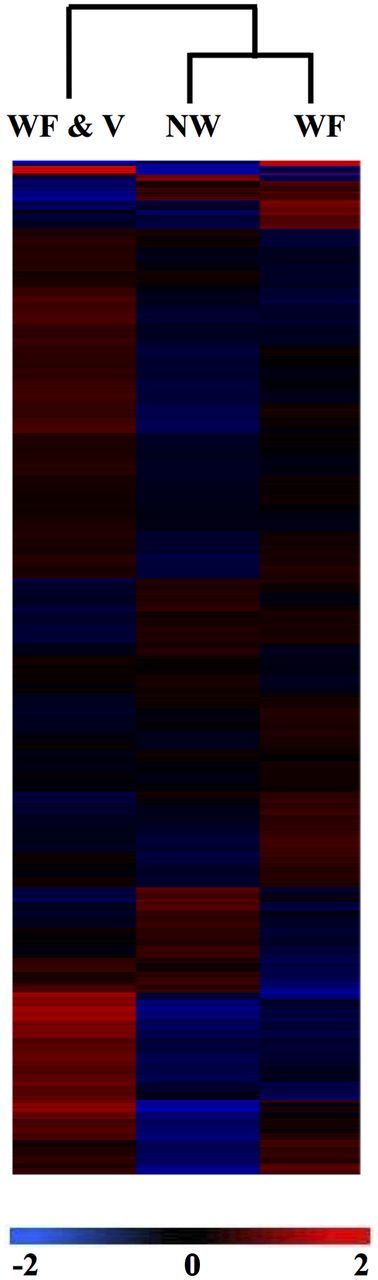
Clustering of the significantly different tomato gene expression levels in response to feeding by viruliferous whiteflies carrying PepGMV (WF & V), nonviruliferous whiteflies only (WF), or nonwounded plants (NW). Ratios (calculated between WF & V, W, and NW) are shown in colors as follows: black corresponds to a ratio of 1, red corresponds to a ratio ≥ 2 (upregulated genes), and blue corresponds to a ratio ≤ 2 (downregulated genes). Each row in the column corresponds to a single gene, and a color scale is presented below the heat map.
Selected putative tomato gene targets belonging to six different physiological functional groups were identified, and included defense-, pathogen-, photosynthesis-, signaling-, cell wall and growth-, and abiotic-related responses ( Table 1 ). Twelve putative genes grouped as defense-related genes with nine genes having been induced by viruliferous whiteflies at twofold to more than sevenfold expression levels, whereas only three defense genes were stimulated by the whitefly feeding alone. Both viruliferous and nonviruliferous whitefly feeding stimulated the expression of 1-aminocyclopropane 1-carboxylate oxidase (ACO) and Indole-3-acetic acid (IAA)-Ala hydrolase. The viruliferous whiteflies stimulated upregulation of arginase, several protease inhibitors, and polyphenol oxidase. Whitefly feeding alone resulted in increased expression of arginase, endochitinase, and syringolide-induced protein. Unexpectedly, the syringolide-induced protein putative gene was stimulated by nonviruliferous whitefly feeding, but not by the viruliferous whiteflies. Twelve of the 13 putative PR genes identified were stimulated by viruliferous whiteflies compared with 4 that were stimulated by whitefly feeding alone. In general, the four PR genes stimulated by whitefly feeding were upregulated to a greater extent by viruliferous whiteflies than by nonviruliferous whitefly feeding and included disease resistance protein, pentatricopeptide repeat containing protein, Pto-responsive protein, and a glycine-rich protein. Viruliferous whiteflies stimulated the expression of several PR proteins including PR-1a, PR-4, PR-6, and P23. Photosynthesis genes were suppressed by viruliferous whitefly and whitefly feeding, but more so by viruliferous whiteflies, and this result was likely due to virus infection. Both viruliferous whitefly feeding and nonviruliferous whitefly feeding significantly affected the expression of signaling-related genes.
Table 1.
Relative fold change of the expression of selected tomato plant genes induced by viruliferous whiteflies carrying PepGMV (WF & V) or nonviruliferous whiteflies (WF), compared with nonwounded (NW) plants
| Gene classification | Gene name | NW | WF & V | WF |
|---|---|---|---|---|
| Defense-related annotation | ||||
| Acidic 26 kDa endochitinase precursor | SGN-U144297 | 1.00(a) | 2.90(b) | 1.10(a) |
| 1-Aminocyclopropane-1-carboxylate oxidase 1 (ACC oxidase 1) (Ethylene-forming) | SGN-U143274 | 1.00(a) | 2.75(b) | 2.74(b) |
| Arginase ( A. thaliana ) | SGN-U145219 | 1.00(a) | 6.01(b) | 1.69(b) |
| Aspartic protease inhibitor 1 precursor (pA1) (gCDI-A1) (STPIA) (STPID) | SGN-U143342 | 1.00(a) | 3.82(a) | −1.27(b) |
| Endochitinase 3 precursor | SGN-U143337 | 1.00(a) | 3.11(b) | 2.11(b) |
| PI2Cev1 precursor | SGN-U144127 | 1.00(a) | 6.91(b) | 1.60(a) |
| IAA-Ala hydrolase (IAR3) ( A. thaliana ) | SGN-U144992 | 1.00(a) | 3.07(b) | 3.89(b) |
| PPO F, chloroplast precursor (PPO) (Catechol oxidase) | SGN-U143365 | 1.00(a) | 3.51(b) | 1.14(a) |
| Subtilisin-like proteinase (EC 3.4.21.-) 4—tomato | SGN-U151318 | 1.00(a) | 3.06(b) | 2.01(a,b) |
| Syringolide-induced protein 19-1-5 ( Glycine max ) | SGN-U143593 | 1.00(a) | −1.14(a) | 3.26(b) |
| WIN1 precursor | SGN-U143552 | 1.00(a) | 7.56(b) | 1.45(a) |
| WIN2 precursor | SGN-U143329 | 1.00(a) | 7.24(b) | 1.04(a) |
| Wound-inducible carboxypeptidase ( Lycopersicon esculentum ) | SGN-U148185 | 1.00(a) | 2.38(b) | 1.31(a) |
| Pathogen-related annotation | ||||
| Avr9/Cf-9 rapidly elicited protein 216 ( Nicotiana tabacum ) | SGN-U152421 | 1.00(a) | 2.66(b) | −1.64(a) |
| Disease resistance protein BS2 ( Capsicum chacoense ) | SGN-U146088 | 1.00(a) | 3.22(b) | 4.27(b) |
| Glycine-rich protein ( Nicotiana glauca ) | SGN-U147446 | 1.00(a) | 2.99(b) | 1.55(c) |
| NP24 protein precursor (PR P23) (salt-induced protein) | SGN-U143414 | 1.00(a) | −1.55(b) | −1.10(a) |
| Pathogenesis-related leaf protein 4 precursor (P4) | SGN-U143238 | 1.00(a) | 3.59(b) | 1.08(a) |
| Pathogenesis-related leaf protein 6 precursor (P6) (ethylene-induced protein P1) | SGN-U143242 | 1.00(a) | 3.21(b) | 1.30(a,b) |
| PR-1 precursor ( Capsicum annuum ) | SGN-U143838 | 1.00(a) | 3.24(b) | 1.05(a) |
| Pentatricopeptide (PPR) repeat-containing protein ( A. thaliana ) | SGN-U154133 | 1.00(a) | 2.03(b) | 1.58(c) |
| Probable glucosyltransferase twi1 (EC 2.4.1.-)—tomato (fragment) | SGN-U144770 | 1.00(a) | 2.34(b) | 1.60(a) |
| Probable GST (PR-1) | SGN-U143280 | 1.00(a) | 2.32(b) | 1.46(a) |
| Probable GST (PR-1) | SGN-U143286 | 1.00(a) | 1.86(b) | 1.02(a) |
| Pto-responsive gene 1 protein ( L. esculentum ) | SGN-U144888 | 1.00(a) | 3.54(b) | 2.02(b) |
| Putative leucine rich repeat containing protein kinase | SGN-U155709 | 1.00(a) | 1.30(a) | −2.40(b) |
| Photosynthesis-related annotation | ||||
| Chlorophyll a/b-binding protein type I precursor—tomato | SGN-U143178 | 1.00(a) | −1.76(b) | −1.20(a) |
| RuBisCO small chain 3, chloroplast precursor | SGN-U155531 | 1.00(a) | −1.54(b) | −1.17(a) |
| RuBisCO small chain 3A/3C, chloroplast precursor | SGN-U155534 | 1.00(a) | −1.48(b) | −1.02(a) |
| RuBisCO 3A/3C, chloroplast precursor | SGN-U155538 | 1.00(a) | −1.57(b) | −1.06(a) |
| Photosystem II protein I ( Physcomitrella patens subsp. patens ) | SGN-U152054 | 1.00(a) | −1.75(b) | 1.00(a) |
| Photosystem II 22 kDa protein, chloroplast precursor (CP22) | SGN-U143807 | 1.00(a) | −3.13(b) | −1.89(b) |
| Signaling-related annotation | ||||
| ARG1 protein ( A. thaliana) | SGN-U150408 | 1.00(a) | 8.24(b) | 1.31(a) |
| Calcium-dependent protein kinase ( Solanum tuberosum ) | SGN-U148281 | 1.00(a) | −1.18(a) | −2.03(b) |
| Calcium-dependent protein kinase ( S. tuberosum ) | SGN-U145308 | 1.00(a) | 4.62(b) | 2.65(b) |
| Casein kinase 2 catalytic subunit ( N. tabacum ) | SGN-U144481 | 1.00(a) | 1.04(a) | 1.74(b) |
| Casein kinase I, putative ( A. thaliana ) | SGN-U147401 | 1.00(a) | 3.51(b) | 1.77(c) |
| Probable protein kinase—tomato | SGN-U147032 | 1.00(a) | −1.77(b) | −1.05(a) |
| Cell wall and growth annotation | ||||
| Elongation factor-1 alpha ( N. tabacum ) | SGN-U143402 | 1.00(a) | 1.17(a,b) | 1.97(b) |
| Expansin-like protein ( Quercus robur ) | SGN-U146351 | 1.00(a) | 5.72(b) | 2.92(a,b) |
| Extensin class I (clone w1-8 L)—tomato (fragment) | SGN-U146403 | 1.00(a) | 1.85(b) | 1.17(a,b) |
| F-box protein FKF1/ADO3, AtFBX2a ( A. thaliana ) | SGN-U153437 | 1.00(a) | 2.59(b) | 1.77(b) |
| Inorganic pyrophosphatase ( N. tabacum ) | SGN-U144182 | 1.00(a) | 1.83(b) | 1.57(ab) |
| Putative auxin-repressed protein ( Prunus armeniaca ) | SGN-U144576 | 1.00(a) | 4.17(b) | 1.56(a,b) |
| XET (XTR4), putative ( A. thaliana ) | SGN-U143928 | 1.00(a) | 2.16(b) | 1.93(b) |
| Abiotic-related annotation | ||||
| Cold-induced glucosyl transferase ( Solanum sogarandinum ) | SGN-U143772 | 1.00(a) | 2.72(b) | 2.27(ab) |
| Dehydration-induced protein ERD15 ( L. esculentum ) | SGN-U144231 | 1.00(a) | 2.62(b) | 1.02(a) |
| Shock protein SRC2 family ( A. thaliana ) | SGN-U147611 | 1.00(a) | 3.87(b) | 1.56(a) |
For each gene, ratios with different letters indicate significant differences among treatments ( P ≤ 0.05). Gene ID is the Sol Genomics Network identifier.
Validation of Microarray Data by qPCR
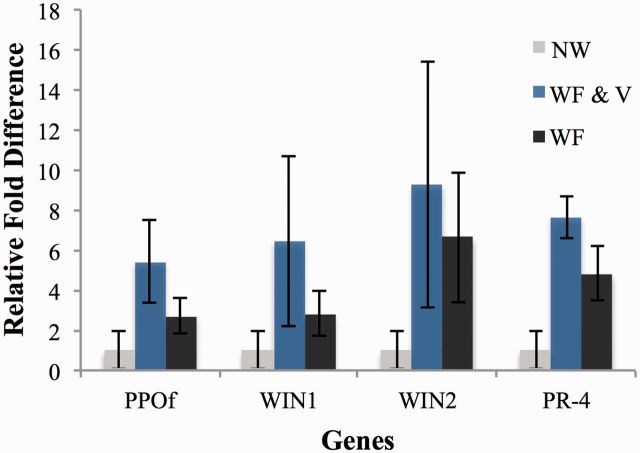
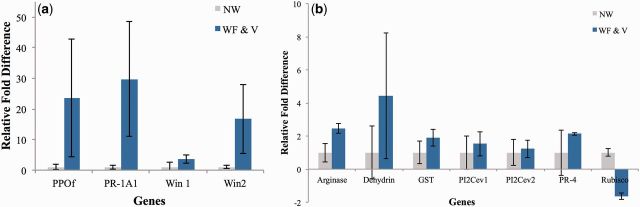
qPCR analysis was used for confirmation of the microarray results for tomato plant defense genes: PPO F, WIN1, WIN2, and PR-4 ( Fig. 2 ). Microarray results demonstrated higher levels of these plant defense- and pathogen-related genes stimulated by viruliferous whitefly feeding, followed by whitefly feeding alone in comparison to the unwounded control. The qPCR results tightly correlated with the microarray results. Other defense genes of interest including arginase, dehydrin, glutathione-S-transferase, ethylene-responsive proteinase inhibitors, and PR protein 1A1 were also stimulated to a greater degree by viruliferous whiteflies, compared with nonwounded treatment ( Fig. 3 ). Reverification of the expression levels for tomato defense genes in a separate set of biological replicates, including PPO F, WIN1, WIN2, and PR-4, was carried out and results corroborated the microarray and qPCR results. Also, expression of RuBisCO was downregulated in tomato plants subjected to viruliferous whiteflies, compared with the nonwounded treatment. In general, all of the overexpressed defense genes identified by microarray analysis and corroborated by qPCR were induced to a greater extent by the viruliferous whitefly treatment, followed by whitefly feeding alone, when compared with the nonwounded plant control.
Fig. 2.
Relative fold differences in the expression of selected tomato plant defense genes verified through qPCR analysis. Gene expression verified: PPO F, WIN1, WIN2, and PR-4. Tomato plants were nonwounded tomato plants (NW) or treated with viruliferous whiteflies carrying PepGMV (WF & V) or nonviruliferous whiteflies (WF). Leaf tissue was collected 15 d after treatment initiation for qPCR analysis. Bars indicate means ± SE. The relative fold differences correlated with the microarrays results as specified in Table 1 .
Fig. 3.
Relative fold differences in the expression of selected tomato plant defense genes verified through qPCR analysis. Gene expression verified: (a) PPO F, PR-1A1, WIN1, WIN2; (b), arginase, dehydrin, GST, PI2Cev1, PI2Cev2, PR-4, and RuBisCO. Treatments are nonwounded (NW) and viruliferous whiteflies carrying PepGMV (WF & V). Leaf tissue was collected 15 d after treatment initiation for qPCR analysis. Bars indicate means ± SE. The relative fold differences matched the microarrays results as specified in Table 1 .
Discussion
In this study, the global gene expression profiling of tomato, an important crop plant, by microarray analysis was carried out for plants exposed to feeding by viruliferous whiteflies harboring PepGMV-Di (and therefore virus infection), viruliferous whiteflies, and nonwounded plants. We found 290 tomato genes represented by the spotted probes were significantly differentially expressed at 12 d PI. This is similar to the results of McKenzie et al. (2005) who showed numerous genes upregulated at 25 d PI. Of these 290 probes, 161 of the ESTs could be annotated (putative) and 74% of these annotated genes were stimulated by viruliferous whiteflies (which resulted in virus infection in the host plant), whereas only 27% were stimulated by nonviruliferous whitefly feeding. Within 1 wk of the initial whitefly infestation, whitefly eggs had hatched, and nymphs were feeding on tomato plants infested with viruliferous and nonviruliferous treatments. Therefore, gene expression changes in both whitefly treatments would be expected to take into account feeding by the juvenile and adult whitefly instars.
Important differences in the tomato plant gene expression profiles resulted from both whitefly only feeding and from viruliferous whitefly feeding that resulted in virus infection of the tomato plants. However, viruliferous whitefly feeding resulted in greater significantly different expression of plant genes, compared with treatments involving whitefly only feeding, and the noninfested plants. Although there were certainly some differences, gene expression profiles for the nonviruliferous whitefly damaged plants and negative control plants were more similar to one another when compared with the viruliferous whitefly treatment, suggesting that important differences in gene expression were due to the presence of PepGMV-Di. Among Homopterans, whiteflies are considered “gentle” feeders even though their stylets penetrate intracellularly ( Johnson and Walker 1999 ), even considering that whiteflies feeding in great numbers on a susceptible plant can cause significant damage even in the absence of virus. This study demonstrated that begomovirus infection brought about by whitefly-mediated transmission that involves subtle interactions between whitefly stylets and plant tissues was correlated with the significant expression of host plant genes, compared with that associated with whitefly feeding alone. This suggests cross-talk between innate defense pathways owing to interactions between whitefly feeding together with viral infection. A sample of the gene expression results are discussed below according to assigned biological function.
Defense-Related Responses
In the defense category, arginase expression was significantly induced in both the viruliferous and nonviruliferous whitefly treatments compared with the nonwounded control. However, aphids, another piercing-sucking insect, did not stimulate arginase in aphid-infested tomato plants after 5 d of feeding ( Rodriguez-Saona et al. 2010 ). Chen et al. (2004) found that arginase expression is due to a JA signal from application of JA and also in tomato plants infected with Pseudomonas syringae pv . tomato . They suggested that similarities in the composition and expression profile of the mammalian arginase in wounding and pathogen infection imply that plant synthesized arginase might have a similar wound healing function. Arginase is considered to be antinutritive herbivore defense ( Zhu-Salzman et al. 2008 ). Arginase breaks down arginine, a nitrogen-rich storage amino acid, predominantly found in seeds and storage organs of numerous plant species, to urea and ornithine ( Van Etten et al. 1967 , Polacco and Holland 1993 ). Arginase activity is often increased during germination in many plants including Arabidopsis ( Zonia et al. 1995 ) and soybean ( Matsubara and Suzuki 1984 , Kang and Cho 1990 ). However, environmental stress induces accumulation of some nitrogen-containing compounds including arginine, proline, glutamine, asparagine, ammonium, and three polyamines ( Kao 1997 ). The increase in transcripts of proteins related to nitrogen accumulation is possibly due to the premature tissue senescence resulting from whitefly feeding and virus infection as the plant attempts to defend itself against herbivory damage and to control spread of the infection. Also, nitrogen reassimilation could be due to resource redistribution in response to wounding and virus infection as chlorosis of tissue progresses.
Several IR-related products, including proteinase inhibitors such as PI2Cev1 precursor, WIN1 and WIN2 precursors, and wound-inducible carboxy peptidase were significantly upregulated in the viruliferous whitefly-treated plants, compared with nonviruliferous whitefly treated or nonwounded plants. However, the aspartic proteinase inhibitor 1 precursor gene was significantly downregulated in the nonviruliferous whitefly treatment compared with the viruliferous whitefly and NW treatment. This result suggests that the combined effect of virus infection and whitefly feeding induced the expression of JA- and ethylene-response defense-related genes similar to the response of Cauliflower Mosaic Virus infection in A. thaliana ( Geri et al. 2004 ). Pathogen infection and feeding from piercing/sucking insects tends to induce defense genes related to the SA pathway; however, piercing sucking insects such as aphids also stimulate proteinase inhibitors but to a lesser degree than chewing insects (e.g., caterpillars) ( Rodriguez-Saona et al. 2010 ). Because of cross-talk between the SA, JA, and JA/ethylene pathways, these stresses can also elicit expression of genes related to the JA and JA-ethylene pathways as determined here and by other studies (reviewed in Walling 2000 ). McKenzie et al. (2005) also found upregulation of several JA/ethylene-related, e.g., proteinase inhibitors and ethylene-responsive proteinase inhibitors, 1-aminocyclopropane carboxylate oxidase 25 d after whitefly infestation of tomato plants. Thus, the ethylene produced in the viruliferous whitefly treatment could be due to induction of the PI2Cev1 precursor. We also found that ACO was significantly upregulated in the wounding treatments indicating that the ethylene pathway was affected by whiteflies, as seen with feeding from other phytophagous insects that utilize slender stylets for feeding. ACO is a regulated enzyme in the ethylene biosynthesis pathway. Ethylene is a critical plant hormone and signal involved in many areas of plant physiology including growth, development, senescence, fruit ripening, and defense ( Adams and Yang 1979 , Yang and Hoffman 1984 ).
The expression of PPO F, an antinutritive defense ( Zhu-Salzman et al. 2008 ), was significantly higher in the combined whitefly and virus treatment compared with the nonwounded and whitefly only treatments. Whitefly feeding alone did not substantially stimulate PPO F compared with nonwounded plants. A similar result was observed when aphids fed on tomato plants ( Rodriguez-Saona et al. 2010 ). PPO F can help plants detoxify during a variety of stresses and can serve as an herbivore deterrent ( Felton et al. 1989 ) by binding proteins causing agglutination and making them indigestible and are typically related to induction of the JA- and JA/ethylene pathways. The tomato plants experiencing the combined effect of feeding from whiteflies and virus infection appear to have possibly responded as though attacked by larger chewing insects, as also indicated in the upregulation of proteinase inhibitor precursors.
The expression of a syringolide-induced protein gene was significantly upregulated in the whitefly only treatment compared with the nonwounded control and viruliferous whitefly treatment. Syringolide is an elicitor from the bacterial plant pathogen P. syringae pv . glycinea that results in an HR in soybean carrying bacterial resistance genes, typical of the SA pathway. However, the viruliferous whitefly treatment did not result in upregulation of this gene. Although induction of the SA pathway by pathogens and piercing/sucking insects is more or less anticipated, this effect was not observed here, perhaps owing to the complexity of plant defense pathways, type of pathogen, and combined effects on insect–pathogen–host plant, as is demonstrated here.
The IAA-Ala hydrolase gene was significantly upregulated by both whitefly wounding treatments compared with the nonwounded control. IAA-Ala hydrolase regulates the levels of free IAA, which is a plant hormone that affects the regulation of many plant physiological processes such as growth and development ( Pengelly and Bandurski 1983 ). High levels of IAA can stimulate ethylene biosynthesis ( Lau and Yang 1973 ). The upregulation of IAA-Ala hydrolase is consistent with increased expression levels of the ethylene-forming ACO and the ethylene responsive proteinase inhibitor precursor proteins, reported above, and appears to be comparable to the result from a study showing ethylene involvement in the global plant response to whitefly herbivory, which resulted in increased production of plant defense proteins ( McKenzie et al. 2005 ).
Pathogen-Related Responses
With respect to pathogen-related responses, three PR protein precursor genes (PR-1, P4, and P6) were upregulated in the viruliferous whitefly treatment compared with whitefly feeding only or to the nonwounded plant treatment. No statistically significant differences were observed in the expression of these genes between the whitefly feeding only and nonwounded treatments. These results are consistent with McKenzie et al. (2002) that examined the effects of whitefly and viruliferous whiteflies on tomato PR protein expression and detected differences in PR protein levels at 14 d postwhitefly infestation. In this study, tomato plants developed obvious symptoms of virus infection by 10–12 d PI, and the leaves were harvested on d 15. Although it is not well understood how the PR proteins affect the response of plants to viral infection, PR protein expression is mediated by the SA pathway and induced by pathogens and damage from piercing and sucking insects. These results are consistent with those of other similar studies.
Two GST genes were significantly upregulated in the viruliferous whitefly treatment, compared with nonwounded or nonviruliferous whitefly treatments. GSTs catalyze the addition of glutathione to potentially harmful compounds, which detoxify them and prevent/reduce plant damage. Lipid hydroperoxides are harmful compounds produced in plants exposed to pathogen infection and salt and drought stress; GSTs are known to detoxify these compounds ( Plaisance and Gronwald 1999 , Csiszár et al. 2002 ) and are typically induced following biotic (e.g., pathogen infection) and abiotic (e.g., oxidative stress) stresses (reviewed in Marrs 1996 ). The expression of two GST genes was upregulated nearly threefold and fivefold, respectively, in A. thaliana plants following aphid feeding for 96 h ( Moran et al. 2002 ). Infection by the potyvirus Pepper yellow mosaic virus resulted in increased expression of GST in tomato ( Alfenas-Zerbini et al. 2009 ). However, Schenk et al. (2000) showed that GST was downregulated 2.9-fold in A. thaliana infected with the fungal pathogen Alternaria brassicicola. In this study, begomovirus PepGMV-Di infection of tomato plants was associated with upregulation of GST, which may assist in combatting the infection.
Photosynthesis-Related Responses
In photosynthesis-related responses, several photosynthesis-related protein genes, three RuBisCO chloroplast precursors, two photosystem II proteins, and a chlorophyll a/b binding protein were significantly downregulated in the combined whitefly and virus treatment compared with the nonwounded control. Nonviruliferous whitefly feeding also resulted in downregulation of these genes, but the effect was not significantly different compared with the control plants. This result is consistent with the chlorotic appearance of the leaves due to PepGMV-Di infection. Often, photosynthetic protein translation is turned off following wounding ( Haldrup et al. 2000 ) because maintenance of the photosynthetic machinery represents a major expenditure of cellular energy. Repressing de novo synthesis of these proteins would save the plant energy following tissue damage ( Zhou and Thornburg 1999 ). Hui et al. (2003) found a twofold downregulation of RuBisCO after 24 hr of Manduca sexta larvae feeding on Nicotiana attenuata . However, in a microarray experiment by Schenk et al. (2000) involving A. thaliana plants treated with SA, a chlorophyll A/B-binding protein was upregulated by sevenfold. Our results show that gene expression involved in the photosynthetic machinery was reduced following the viruliferous whitefly and nonviruliferous whitefly treatments and could possibly be due to reallocation of resources for defense protein synthesis.
Signaling-Related Responses
Genes encoding two calcium-dependent protein casein kinases and a protein kinase were significantly affected by the wounding treatments. Protein kinases regulated by calcium (Ca 2+ ) play important roles in eukaryotic signal transduction ( Rutschmann et al. 2002 ). Calcium levels are modulated in response to various signals including light, mechanical manipulation, pathogens, abiotic stress, and hormones ( Sanders et al. 1999 , Evans et al. 2001 , Rudd and Franklin-Tong 2001 ), and calcium-dependent protein kinases perform physiological functions in plants from modulating hormone responses, regulating guard cells and stomatal movements, carbon and nitrogen metabolism, mediating abiotic stresses, and pathogen defense (reviewed in Cheng et al. 2002 ). Influx of calcium ions and the activity of a protein kinase both are required for the JA-pathway induction, systemin-triggered depolarization of the plasma membrane and alkalization of the extracellular space ( Felix and Boller 1995 , Moyen and Johannes 1996 , Moyen et al. 1998 , Schaller and Oecking 1999 , Schaller and Frasson 2001 ). Schenk et al. (2000) demonstrated that A. thaliana treated with ethylene upregulated a protein kinase nearly fivefold and expression of casein kinase I in regulation of membrane binding ( Yu and Roth 2002 ). Here, both viruliferous and nonviruliferous whitefly treatments significantly increased signal transduction associated with mediating the host plant response to disease and wounding, suggesting that tomato plants detected the subtle signals from whitefly feeding, and responded to viral pathogen attack.
An altered response to gravity (ARG1) protein gene ( Chen et al. 1998 ) was significantly upregulated in the viruliferous whitefly treatment, compared with the nonviruliferous or the nonwounded treatment. No significant differences were observed between nonviruliferous whitefly feeding and nonwounded plant treatments. AGR1 encodes a putative transmembrane protein, whose amino acid sequence shares some homology with bacterial transporters ( Chen et al. 1998 ). The high, increased expression (8.24-fold) of this protein in the virus-infected treatment, although speculative, suggests that this type of transporter may be important in transmembrane interactions with virus particles or viral–protein complexes that aid in aspects of viral infection and/or spread in the plant. It would be interesting to test for direct interaction between ARG1 and the PepGMV-Di proteins.
Cell Wall and Growth-Related Responses
In the cell wall and growth-related response category, a putative xyloglucan endotransglycosylase (XET) gene was significantly upregulated in both the whitefly treatments compared with the nonwounded treatment. XETs cleave and link xyloglucan chains resulting in increasing plasticity and elasticity of cell walls for cell growth ( Catalá et al. 2001 , Thompson and Fry 2001 ). They are also induced in response to various developmental and environmental factors such as mechanical stimuli, temperature changes, light, ( Malinowski and Filipecki 2002 ) wounding, and pathogen infection ( Maleck et al. 2000 , Schenk et al. 2000 ). Whitefly feeding activity and stylet penetration of the spaces between the cell wall and plasma membrane may be aided by induction of this gene, affecting the plasticity of the cell wall, and allowing for easier penetration of the plant tissue by the insect stylets ( Moran et al. 2002 ). Hui et al. (2003) determined that two XET genes were upregulated over twofold and threefold in N. attenuata after M. sexta larvae feeding for 24 h. This suggests that increasing the elasticity of cell walls is not unique to whitefly feeding but rather that it may be a feature associated with insect herbivore feeding in general.
Abiotic-Related Responses
A gene encoding glucosyltransferase was significantly upregulated in the virus treatment, compared with the nonwounded treatments, and increased expression of glucosyltransferase was observed in the nonviruliferous treatment, compared with the nonwounded plants. Glucosyl-transferases are involved in processes catalyzing transfer of glucose used for synthesis of oligosaccharides, polysaccharides, and other carbohydrates ( Malinowski and Filipecki 2002 ). This suggests that begomovirus infection was associated with the transfer of resources within the plant to aid in the production of compounds responding to begomoviral pathogen infection.
A dehydration-induced protein gene was significantly upregulated in the viruliferous whitefly treatment compared with both the nonviruliferous whitefly and nonwounded treatments. These proteins are expressed in response to low temperature stress. In addition, a heat shock protein gene was also significantly expressed in the virus treatment, compared with both the nonviruliferous whitefly and nonwounded treatments. These proteins act as molecular chaperons to aid organisms during stress by preventing denaturation of proteins critical to plant physiological processes. Their significant upregulation indicates that tomato plants detected and responded to whitefly feeding and virus infection, presumably, to protect themselves from damage.
Numerous genes were shown to be differentially expressed in tomato plants after 15 d of feeding by viruliferous (with PepGMV) or nonviruliferous whitefly feeding. Most of these were associated with the viruliferous whitefly and viral infection treatment, compared with nonviruliferous whitefly, and nonwounded plant treatments. Significantly different gene expression was observed for six major physiological function categories, including defense-, pathogen-, photosynthesis-, signaling-, cell wall and growth-, and abiotic-related responses. Although some of these transcripts or proteins encoded by them have been identified in studies involving whitefly–plant interactions, several were uniquely expressed in the begomovirus-infected tomato plants. This is the first report of the transcript profiling of tomato plants in which genes were differentially expressed owing to the combined effects of begomovirus–whitefly associated gene expression. The results provide new hypotheses regarding the selection of candidate genes for expression in transgenic plants to abate the assault on key defense responses to the invading insect vector–viral complex.
Acknowledgments
The National Science Foundation (NSF) funding provided R.O.M. and S.M.H.-M. Postdoctoral Fellowships under the Plant-Insect Interactions Training Grant to The University of Arizona. The NSF supported High School Teacher’s Training Program for M.G. This study was also supported by the National Science Foundation Plant Genome Research Initiative (0820367 to R.O.M. and S.M.H.-M.), the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (2004-01540 to R.O.M. and S.M.H.-M.) and the Western Illinois University research council. We thank Dr. Heiko Vogel of the Max Planck Institute for assistance with primer design.
References Cited
- Adams D. O., Yang S. F. . 1979. . Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene . Proc. Natl Acad. Sci. USA. 76 : 170 – 174 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba R., Fei Z., Payton P., Liu Y., Moore S. L., Debbie P., Cohn J., D'Ascenzo M., Gordon J. S., Rose J. K., et al. . 2004. . ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development . Plant J. 39 : 697 – 714 . [DOI] [PubMed] [Google Scholar]
- Alfenas-Zerbini P., Maia I. G., Fávaro R. D., Cascardo J.C.M., Brommonschenkel S. H., Zerbini F. M. . 2009. . Genome-wide analysis of differentially expressed genes during the early stages of tomato infection by a potyvirus . Mol. Plant Microbe Interact. 22 : 352 – 361 . [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J. T., Sozzani R., Lee T.-J., Chu T.-M., Wolfinger R. D., Cella R., Hanley-Bowdoin L. . 2008. . Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection . Plant Physiol. 148 : 436 – 454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellows T. S., Jr., Perring T. M., Gill R. J., Headrich D. H. . 1994. . Description of a species of Bemisia (Homoptera:Aleyrodidae) . Ann. Entomol. Soc. Am. 76 : 310 – 313 . [Google Scholar]
- Benjamini Y., Hochberg Y. . 1995. . Controlling the false discovery rate: a practical and powerful approach to multiple testing . J. R Stat. Soc. 57 : 289 – 300 . [Google Scholar]
- Bostock R. M., Karban R., Thaler J. S., Weyman P. D., Gilchrist D. . 2001. . Signal interactions in induced resistance to pathogens and insect herbivores . Eur. J. Plant Pathol. 107 : 103 – 111 . [Google Scholar]
- Brown J. K. 1990. . An update on the whitefly-transmitted geminiviruses in the Americas and the Caribbean Basin . Food Agric. Organ. United Nations Plant Prot. Bull. 39 : 5 – 23 . [Google Scholar]
- Brown J. K. 1994. . The status of Bemisia tabaci (Genn.) as a pest and vector in world agroecosystems . Food Agric. Organ. United Nations Plant Prot. Bull. 42 : 3 – 32 . [Google Scholar]
- Brown J. K. 2001. . The molecular epidemiology of begomoviruses , pp. 279 – 316 . InKhan J. A., Dykstra J. (eds.), Trends in plant virology . The Haworth Press, Inc. , Binghamton, NY: . [Google Scholar]
- Brown J. K. 2007. . The Bemisia tabaci complex: genetic and phenotypic variability drives begomovirus spread and virus diversification . pp. 25–56. In H. Czosnek (ed.), Tomato yellow leaf curl virus disease: management, molecular biology, breeding, breeding for resistance. Springer, Dordrecht . [Google Scholar]
- Brown J. K. 2010. . Phylogenetic biology of the Bemisia tabaci sibling species group , pp. 31 – 67 . InStansly P. A., Naranjo S. E. (eds.), Bemisia : bionomics and management of a global pest . Dordrecht-Springer; , Amsterdam, The Netherlands: . [Google Scholar]
- Brown J. K., Bird J. . 1992. . Whitefly-transmitted geminiviruses in the Americas and the Caribbean Basin: past and present . Plant Dis. 76 : 220 – 225. [Google Scholar]
- Brown J. K., Czosnek H. . 2002. . Whitefly transmission of plant diseases . Adv Biochem. Res. 36 : 65 – 100 . [Google Scholar]
- Brown J. K., Frohlich D. R., Rosell R. C. . 1995. . The sweetpotato or silverleaf whiteflies. Biotypes of Bemisia tabaci or a species complex . Annu. Rev. Entomol. 40 : 511 – 534 . [Google Scholar]
- Brown J. K., Idris A. M., Ostrow K. M., French R., Stenger D. C. . 2005. . Genetic and phenotypic variation of three strains of the Pepper golden mosaic virus complex . Phytopathology 95 : 1217 – 1224 . [DOI] [PubMed] [Google Scholar]
- Buchanan B. B., Gruissem W., Jones R. L. . 2001. . Biochemistry and molecular biology of plants . American Society of Plant Physiology; , Rockville, MD: . [Google Scholar]
- Catalá C., Rose J.K.C., York W. S., Albersheim P., Darvill A. G., Bennett A. B. . 2001Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls . Plant Physiol. 127 : 1180 – 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., Masson P. H. . 1998. . The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier . Proc. Natl Acad. Sci. USA. 95 : 15112 – 15117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaig B. C., Melotto M., He S. Y., Howe G. A. . 2004. . Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine . J. Biol. Chem. 279 : 45998 – 46007 . [DOI] [PubMed] [Google Scholar]
- Cheng S.-H., Willmann M. R., Chen H.-C., Sheen J. . 2002. . Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family . Plant Physiol. 129 : 469 – 485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa H. S., Brown J. K. . 1991. . Variation in biological characteristics and in esterase patterns among populations of Bemisia tabaci (Genn.) and the association of one population with silverleaf symptom development . Entomologia Experimantalis et Applicata 61 : 211 – 219 . [Google Scholar]
- Costa H. S., Brown J. K., Sivasupramaniam S., Bird J. . 1993. . Regional distribution, insecticide resistance, and reciprocal crosses between the ‘A’ and ‘B’ biotypes of Bemisia tabaci . Entomologia Experimantalis et Applicata 14 : 127 – 138. [Google Scholar]
- Csiszár J., Czabó M., Illés E., Kurucz K. . 2002. . Investigations of glutathione S-transferase and peroxidase activities in auxin heterotrophic and autotrophic tobacco calli under salt stress conditions . Acta Biologica Szegediensis 46 : 79 – 80 . [Google Scholar]
- Evans N. H., McAinsh M. R., Hetherington A. M. . 2001. . Calcium oscillations in higher plants . Curr. Opin. Plant Biol. 4 : 415 – 420 . [DOI] [PubMed] [Google Scholar]
- Felix G., Boller T. . 1995. . Systemin induces rapid ion fluxes and ethylene biosynthesis in Lycopersicon peruvianum cells . Plant J. 7 : 381 – 389 . [Google Scholar]
- Felton G. W., Donato K. K., Del Vecchio R. J., Duffey S. S. . 1989. . Activation of foliar oxidases by insect feeding reduces nutritive quality of dietary protein for foliage for noctuid herbivores . J. Chem. Ecol. 15 : 2667 – 2693 . [DOI] [PubMed] [Google Scholar]
- Felton G. W., Korth K. L., Bi J. L., Wesley S. V., Huhman D. V., Matthews M. C., Murphy J. B., Lamb C., Dixon R. A. . 1999. . Inverse relationship between systemic resistance of plants to microorganisms and to insect herbivory . Curr. Biol. 9 : 317 – 320 . [DOI] [PubMed] [Google Scholar]
- Frohlich D., Torres-Jerez I., Bedford I. D., Markham P. G., Brown J. K. . 1999. . A phylogeographic analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers . Mol. Ecol. 8 : 1593 – 1602 . [DOI] [PubMed] [Google Scholar]
- Geri C., Love A. J., Cecchini E., Barrett S. J., Laird J., Covey S. N., Milner J. J. . 2004. . Arabidopsis mutants that suppress the phenotype induced by transgene-mediated expression of cauliflower mosaic virus (CaMV) gene VI are less susceptible to CaMV-infection and show reduced ethylene sensitivity . Plant Mol. Biol. 56 : 111 – 24 . [DOI] [PubMed] [Google Scholar]
- Gill R., Brown J. K. . 2010. . Systematics of Bemisia and Bemisia relatives: can molecular techniques solve the Bemisia tabaci complex conundrum—a Taxonomist’s viewpoint , pp. 5 – 29 . InStansly P. A., Naranjo S. E. (eds.), Bemisia : bionomics and management of a global pest . Dordrecht-Springer; , Amsterdam, The Netherlands: . [Google Scholar]
- Gorovits R., Akad F., Beery H., Vidavsky F., Mahadav A., Czosnek H. . 2007. . Expression of stress-response proteins upon whitefly-mediated inoculation of Tomato yellow leaf curl virus in susceptible and resistant tomato plants . Mol. Plant Microbe Interact. 20 : 1376 – 83 . [DOI] [PubMed] [Google Scholar]
- Haldrup A., Simpson D. J., Scheller H. V. . 2000. . Down regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana . J. Biol. Chem. 275 : 31211 – 31218 . [DOI] [PubMed] [Google Scholar]
- Hui D., Iqbal J., Lehmann K., Gase K., Saluz H. P., Baldwin I. T. . 2003. . Molecular interactions between the specialist herbivore M. sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. V: microarray analysis and further characterization of large-scale changes in herbvore-induced mRNAs . Plant Physiol. 131 : 1877 – 1893 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris A. M., Smith S. E., Brown J. K. . 2001. . Ingestion, transmission, and persistence of Chino del tomate virus (CdTV), a New World begomovirus , by Old and New World biotypes of the whitefly vector Bemisia tabaci . Ann. Appl. Biol. 139 : 45 – 154 . [Google Scholar]
- Johnson D. D., Walker G. P. . 1999. . Intracellular punctures by the adult whitefly Bemisia argentifolii on DC and AC electronic feeding monitors . Entomologia Experimentalis et Applicata 92 : 257 – 270 . [Google Scholar]
- Kang J. H., Cho Y. D. . 1990. . Purification and properties of arginase from soybean ( Glycine max ) axes . Plant Physiol. 93 : 1230 – 1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C. H. 1997. . Physiological significance of stress-induced changes in polyamines in plants . Botanical Bulletin of Academia Sinica 38 : 141 – 144 . [Google Scholar]
- Karban R., Baldwin T. . 1997. . Induced responses to herbivory . The University of Chicago Press; , Chicago, IL: . [Google Scholar]
- Kazan K., Schenk P. M., Wilson I., Manners J. M. . 2001. . DNA microarrays: new tools in the analysis of plant defence responses . Mol. Plant Pathol. 2 : 177 – 185 . [DOI] [PubMed] [Google Scholar]
- Kempema L. A., Cui X., Holzer F. M., Walling L. L. . 2007. . Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids . Plant Physiol. 143 : 849 – 865 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth K. L. 2003. . Profiling plant responses to herbivorous insects . Genome Biol. 4 : 221 .1-221.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C.M.J. . 2008. . Cross talk in defense signaling . Plant Physiol. 146 : 839 – 844 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. L., Yang S. F. . 1973. . Mechanisms of a synergistic effect of kinetin on auxin induced ethylene production: suppression of auxin conjugation . Plant Physiol. 51 : 1011 – 1014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-M., Ruan Y.-M., Li F.-F., Liu S.-S., Wang X.-W. . 2011. . Gene expression profiling of the whitefly ( Bemisia tabaci ) Middle East–Asia Minor 1 feeding on healthy and Tomato yellow leaf curl China virus-infected tobacco . Insect Sci. 18 : 11 – 22 . [Google Scholar]
- Lozano-Durán R., Rosas-Díaz T., Gusmaroli G., Luna A. P., Taconnat L., Deng X. W., Bejarano E. R. . 2011. . Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana . Plant Cell 23 : 1014 – 1032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K. A., Dang J. L., Dietrich R. A. . 2000. . The transcriptome of Arabidopsis thaliana during systemic acquired resistance . Nat. Genet. 26 : 403 – 409 . [DOI] [PubMed] [Google Scholar]
- Malinowski R., Filipecki M. . 2002. . The role of cell wall in plant embryogenesis . Cell Mol. Biol. Lett. 7 : 1137 – 1151 . [PubMed] [Google Scholar]
- Marrs K. A. 1996. . The functions and regulation of glutathione S-transferases in plants . Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 : 127 – 158 . [DOI] [PubMed] [Google Scholar]
- Matsubara S., Suzuki Y. . 1984. . Arginase activity in the cotyledons of soybean seedlings . Physiologia Plantarum 62 : 309 – 314 . [Google Scholar]
- McKenzie C. L., Shatters R. G., Doostdar H., Lee S. D., Inbar M., Mayer R. . 2002. . Effect of geminivirus infection and Bemisia infestation on accumulation of PR proteins . Arch. Insect Biochem. Physiol. 49 : 203 – 214 . [DOI] [PubMed] [Google Scholar]
- McKenzie C. L., Sinisterra X. H., Powell C. A., Albano J. P., Bausher M. G., Shatters R. G. . 2005. . Deciphering changes in plant physiological response to whitefly feeding using microarray technology . Acta Horticulturae (ISHS) 695 : 347 – 352 . [Google Scholar]
- Moran M. J., Cheng Y., Cassell J. L., Thompson G. A. . 2002. . Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid Interactions . Arch. Insect Biochem. Physiol. 51 : 182 – 203 . [DOI] [PubMed] [Google Scholar]
- Moyen C., Johannes E. . 1996. . Systemin transiently depolarizes the tomato mesophyll cell membrane and antagonizes fusicoccin-induced extracellular acidification of mesophyll tissue . Plant Cell Environ. 19 : 464 – 470 . [Google Scholar]
- Moyen C., Hammond-Kosack K. E., Jones J., Knight M. R., Johanes E. . 1998. . Systemin triggers and increase of cytoplasmic calcium in tomato mesophyll cells: Ca2+ mobilization from intra- and extracellular compartments . Plant Cell Environ. 21 : 1101 – 1111 . [Google Scholar]
- Musser R. O., Hum-Musser S. M., Lee H. K., DesRochers B. L., Williams S. A, Vogel H. . 2012. . Caterpillar labial saliva alters tomato plant gene expression . J. Chem. Ecol. 38 : 1387 – 1401 . [DOI] [PubMed] [Google Scholar]
- Nault L. R. 1997. . Arthropod transmission of plant viruses: a new synthesis . Ann. Entomol. Soc. Am. 90 : 521 – 541 . [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C. A. . 1991. . A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins . Science 253 : 895 – 898 . [DOI] [PubMed] [Google Scholar]
- Pengelly W. L., Bandurski R. S. . 1983. . Analysis of indole-3-acetic acid metabolism in Zea mays using deuterium oxide as a tracer . Plant Physiol. 73 : 445 – 449 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perring T. M., Cooper A. D., Russell R. J., Farrar C. A., Bellows T. S., Jr . 1993. . Identification of a whitefly species by genomic and behavioral studies . Science 259 : 74 – 77 . [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. 2001. . A new mathematical model for relative quantification in real-time RT-PCR Nucleic Acids Res. 29 : 2002-2007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance K. L., Gronwald J. W. . 1999. . Enhanced catalytic constant for glutathione S-transferase (atrazine) activity in an atrazine resistant Abutilon theophrasti biotype . Pesticide Biochem. Physiol. 63 : 34 – 49 . [Google Scholar]
- Polacco J. C., Holland M. A. . 1993. . Roles of urease in plant cells . Int. Rev. Cytol. 145 : 65 – 103 . [Google Scholar]
- Rodriguez-Saona C. R., Musser R. O., Vogel H., Hum-Musser S. M., Thaler J. S. . 2010. . Molecular, biochemical, and organismal analyses of tomato plants simultaneously attacked by herbivores from two feeding guilds . J. Chem. Ecol. 36 : 1043 – 1057 . [DOI] [PubMed] [Google Scholar]
- Rudd J. J., Franklin-Tong V. E. . 2001. . Unravelling response-specificity in Ca2+ signalling pathways in plant cells . New Phytol. 151 : 7 – 33 . [DOI] [PubMed] [Google Scholar]
- Rutschmann F., Stalder U., Piotrowski M., Oecking C., Schaller A. . 2002. . LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization . Plant Physiol. 129 : 156 – 168 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C. A. 2000. . The systemin signaling pathway: differential activation of plant defensive genes . Biochimica et Biophysica Acta 1477 : 112 – 121 . [DOI] [PubMed] [Google Scholar]
- Sanders D., Brownlee C., Harper J. F. . 1999. . Communicating with calcium . Plant Cell 11 : 691 – 706 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. A., Lopes K.V.G., Apfata J.A.C., Fontes E.P.B. . 2010. . NSP-interacting kinase, NIK: a transducer of plant defence signaling . J. Exp. Bot. 61 : 3839 – 3845 . [DOI] [PubMed] [Google Scholar]
- Schaller A., Frasson D. . 2001. . Induction of wound response gene expression in tomato leaves by ionophores . Planta 212 : 431 – 435 . [DOI] [PubMed] [Google Scholar]
- Schaller A., Oecking C. . 1999. . Modulation of the plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants . Plant Cell 11 : 263 – 272 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P. M., Kazan K., Wilson I., Anderson J. P., Richmond T., Somerville S. C., Manners J. M. . 2000. . Coordinated plant defense responses in Arabidopsis revealed by macroarray analysis . Proc. Natl Acad. Sci. USA. 97 : 11655 – 11660 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinisterra X. H., McKenzie C. L., Hunter W. B., Shatters R. G., Jr . 2005. . Transcript expression of Begomovirus in the Whitefly Vector ( Bemisia tabaci , Gennadius: Hemiptera: Aleyrodidae) . J. Gen. Virol. 86 : 1525 – 32 . [DOI] [PubMed] [Google Scholar]
- Stotz H. U., Pittendrigh B. R., Kroymann J., Weniger K., Fritsche J., Bauke A., Mitchell-Olds T. . 2000. . Ethylene Signaling Reduces Resistance of Arabidopsis against Egyptian Cotton Worm But Not Diamondback Moth . Plant Physiol. 124 : 1007 – 1017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H. U., Koch T., Biedermann A., Weniger K., Boland W., Mitchell-Olds T. . 2002. . Evidence for regulation of resistance in Arabidopsis to Egyptian cotton worm by salicylic and jasmonic acid signaling pathways . Planta 214 : 648 – 652 . [DOI] [PubMed] [Google Scholar]
- Thompson J. E., Fry S. C. . 2001. . Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells . Plant J. 26 : 23 – 34 . [DOI] [PubMed] [Google Scholar]
- Thompson G. A., Goggin F. L. . 2006. . Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects . J. Exp. Bot. 5 : 755 – 766 . [DOI] [PubMed] [Google Scholar]
- Van de Ven W.T.G., LeVesque C. S., Perring T. M., Walling L. L. . 2000. . Local and systemic changes in squash gene expression in response to silverleaf whitefly feeding . Plant Cell 12 : 1409 – 1423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten C. H., Kwolek W. F., Peters J. E., Barclay A. S. . 1967. . Plant seeds as protein sources for food or feed . J. Agric. Food Chem. 15 : 1077 – 1085 . [Google Scholar]
- Walling L. L. 2000. . The myriad plant responses to herbivores . J. Plant Growth Regul. 19 : 195 – 216 . [DOI] [PubMed] [Google Scholar]
- Walling L. L. 2008. . Avoiding effective defenses: strategies employed by phloem-feeding insects . Plant Physiol. 146 : 859 – 866 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Dunning F. M., Bent A. F. . 2002. . Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays . Funct. Integr. Genomics 2 : 259 – 273 . [DOI] [PubMed] [Google Scholar]
- Whitham S. A., Yang C., Goodin M. M. . 2006. . Global impact: elucidating plant responses to viral infection . Mol. Plant Microbe Interact. 19 : 1207 – 1215 . [DOI] [PubMed] [Google Scholar]
- Wise R. P., Moscou M. M., Whitham S. A., Bogdanove A. J. . 2007. . Transcript Profiling in Host-pathogen Interactions . Annu. Rev. Phytopathol. 45 : 329 – 369 . [DOI] [PubMed] [Google Scholar]
- Yang S. F., Hoffman N. E. . 1984. . Ethylene biosynthesis and its regulation in higher plants . Annu. Rev. Plant Physiol. 35 : 155 – 189 . [Google Scholar]
- Yang J.-Y., Iwasaki M., Machida C., Machinda Y., Zhou X., Chua N. H. . 2008. . βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses . Genes Dev. 22 : 2564 – 2577 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Roth M. G. . 2002. . Casein kinase I regulates membrane binding by ARF GAP1 . Mol. Biol. Cell 13 : 2559 – 2570 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate S. I., Kempema L. A., Walling L. L. . 2007. . Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses . Plant Physiol. 143 : 866 – 875 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Thornburg R. W. . 1999. . Wound-inducible genes in plants , pp. 127 – 167 . InReynolds P. (ed.), Inducible gene expression in plants . CAB International; , Wallingford, United Kingdom: . [Google Scholar]
- Zhu-Salzman K., Luthe D. S., Felton G. W. . 2008. . Arthropod-inducible proteins: broad spectrum defenses against multiple herbivores . Plant Physiol. 146 : 852 – 858 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L. E., Stebbins N. E., Polacco J. C. . 1995. . Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds . Plant Physiol. 107 : 1097 – 1103 . [DOI] [PMC free article] [PubMed] [Google Scholar]